Abstract
D-Amino acid oxidase (DAO) is a flavin enzyme that catalyzes the oxidative deamination of d-amino acids. This enzyme has been studied extensively both biochemically and structurally as a model for the oxidase-dehydrogenase class of flavoproteins. This enzyme also has various applications, such as the determination of d-amino acids and production of building blocks for a number of pharmaceuticals. DAO has been found mainly in eukaryotic organisms and has been suggested to play a significant role in various cellular processes, one of which includes neurotransmission in the human brain. In contrast, this enzyme has not been identified in prokaryotic organisms. Some studies have recently identified and characterized DAO enzyme in some actinobacteria. In addition, a genome database search reveals a wide distribution of DAO homologous genes in this bacterial group. The bacterial DAOs characterized so far have certain distinct properties in comparison to eukaryotic DAOs. These enzymes also exhibit some important applicable properties, suggesting that bacteria could be used as a source for obtaining novel and useful DAOs. The physiological function of bacterial DAO have been proposed to include the degradation of non-canonical d-amino acids released from cell wall, but is still largely unknown and need to be studied in depth.
Keywords: bacteria, d-amino acid oxidase, enzymatic properties, flavin enzyme, physiological role, potential applications, structure
Introduction
DAO (EC 1.4.3.3) catalyzes the oxidative deamination of neutral and basic d-amino acids and contains a non-covalently bound flavin adenine dinucleotide (FAD) moiety as a cofactor.1 This enzyme reaction produces imino acids from d-amino acids concomitant with the reduction of FAD (Fig. 1). The imino acids are then non-enzymatically hydrolyzed to give corresponding α-keto acids and ammonium. The reduced FAD is reoxidized by molecular oxygen to produce hydrogen peroxide. The substrate stereoselectivity of DAO is specific, and thus l-amino acids are not accepted as substrates. In addition, this enzyme exhibits a negligible or no activity toward acidic d-amino acids, which are substrates for another flavin oxidase d-aspartate oxidase (DDO, EC 1.4.3.1).2 DAO has been extensively studied for its biochemical and structural properties as a model enzyme of the oxidase-dehydrogenase class of flavoproteins.3,4 Further, the enzyme can be utilized for a broad range of applications, such as the determination of d-amino acids, production of building blocks for pharmaceuticals, and diagnosis and treatment of certain diseases, now desiring a highly stable DAO.3,5
Figure 1.
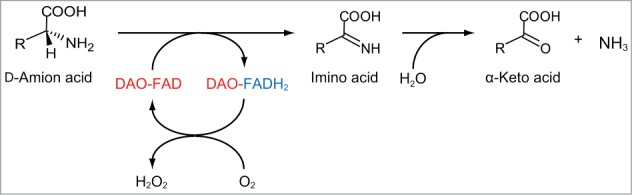
Reaction of oxidation of d-amino acids catalyzed by DAO.
DAO was first identified by Dr. Krebs in pig kidney, and since then this enzyme has also been found in various eukaryotic organisms, such as fungi, nematodes, fish, plants, animals, and human.1,4,6-8 The homologs of this enzyme show amino acid sequence variations; however, some of the residues responsible for substrate binding and interactions with FAD are highly conserved.9 The eukaryotic DAOs possess a peroxisome-targeting signal peptide at their carboxy terminus to localize in the peroxisome for removing the hydrogen peroxide produced by the enzyme reaction. Moreover, DAO plays an important role in various cellular processes in the eukaryotic organisms.4 In fungi, the enzyme degrades d-amino acids to utilize them for cell growth and to protect them from the toxic effect of d-amino acids. In animals, this enzyme plays a role in the degradation of endogenous and exogenous d-amino acids. It has been recently found to be involved in the regulation of neurotransmission in the human brain through the degradation of d-serine, which plays as a co-agonist of N-methyl-d-aspartate receptor. In contrast, DAO has been for a long time considered to be absent in prokaryotic organisms. However, this enzyme has been recently identified, characterized, and cloned from some bacterial species.10-12 In addition, a database homology search reveals DAO homologous genes in various bacterial species, suggesting the possibility that DAO might be widely distributed in bacteria also.10,13
In this addendum, we have described the occurrence, structure, and enzymatic properties of bacterial DAOs. We also discuss the putative physiological role and the potential biotechnological applications of the bacterial DAOs.
Occurrence of Bacterial DAOs and their Homologous Genes
The first bacterial DAO was reported by Gueueke et al.11 They isolated some bacterial species from soil samples that exhibited DAO activity. Among these species, the bacterium that exhibited the highest DAO activity was identified as Arthrobacter protophormiae based on its 16S rRNA gene sequence. The crude extract from the bacterial cells however exhibits only a small activity (3.0 mU/mg-protein) even against the most preferred substrate i.e. d-methionine. In contrast, DAO activities in yeast are detected at approximately 1.0 U/mg-protein.14,15 A homology search in a prokaryotic genome database reveals that DAO homologous genes are widely distributed in various bacteria, except for Archaea that appear to have no homologous genes. The bacterial species having DAO homologous genes are predominantly members of the phylum Actinobacteria, which include A. protophormiae and some pathogenic bacteria, such as Mycobacterium spp. and Nocardia spp.10 Exceptionally, the gram-negative bacteria Stigmatella aurantiaca and Myxococcus stipitatus possess the homologous genes. We have demonstrated that the homologous genes found in the actinobacteria Streptomyces coelicolor and Rubrobacter xylanophilus encode active DAO proteins (ScDAO and RxDAO, respectively) by recombinant expression in Escherichia coli.10,12 Additionally, the crude extract of S. coelicolor oxidizes d-valine, although the activity is quite low (1.5 mU/mg-protein), as observed in the case of A. protophormiae.10
Structure of Bacterial DAOs
The primary sequence of bacterial DAO was first identified in A. protophormiae DAO (ApDAO) by cloning the encoding gene.11 The primary structures of ScDAO and RxDAO were identified from a genome database, but the sequence of RxDAO is slightly different from that of the isolated gene.10 The primary sequence of these bacterial DAOs show high amino acid identities, approximately 38–46%. However, these percentage identity values are only slightly higher in comparison to those obtained for eukaryotic DAOs (approximately 30–36%). In the primary structure of the DAOs, the functional amino acid residues are conserved between eukaryotic and bacterial DAOs. These include the dinucleotide-binding motif (GXGXXG) at the N-terminal region and an arginine, a tyrosine, and a glycine residues binding to the α-carboxy and α-amino groups of the substrate.9 However, peroxisome-targeting signal peptides found in eukaryotes at the C-terminus is lacking in the bacterial enzyme, resulting in a shorter length of the amino acid sequence (approximately a difference of 20–40 amino acid residues).
ApDAO and RxDAO are dimeric and monomeric proteins, respectively.10,11 Almost all eukaryotic DAOs are dimers, except the enzymes found in rat and yeast, Candida boidinii, are monomers.15,16 Animal DAOs are arranged as head-to-head dimers and the yeast Rhodotorula gracilis DAO (RgDAO) is a head-to-tail dimer. The head-to-tail dimer formation in RgDAO results due to an extra stretch of amino acids not found in other eukaryotic DAOs.17 This additional stretch is also not found in ApDAO, suggesting its head-to-head dimer formation. It has been proposed that the surface potential of the dimer interface might be involved in the interactions between the monomers, i.e., the positively and negatively charged interfaces possibly cause a weak and a strong interaction, respectively.18 This argument is further strengthened by the fact that the surface of RxDAO model structure is abundant in positively charged residues.10
The crystal structures of some eukaryotic DAOs have been reported, whereas no crystal structure for bacterial DAOs have been determined. However, a 3-dimensional structure model for RxDAO has been built.10 In this model, the substrate binding at the active site is very similar to the one in eukaryotic DAOs. The α-carboxy and α-amino groups of the substrate interact with Arg272 and Tyr217 residues, and with the carbonyl oxygen of Gly299, respectively (Fig. 2). In addition, similar to eukaryotic DAOs, the side chain of the substrate is covered by the hydrophobic amino acid residues of the enzyme. However, the composition of these hydrophobic residues is different from eukaryotic ones, thus resulting in a different active site structure that might provide different substrate specificity. The RxDAO model also shows the presence of a loop covering the active site, which is called the active site lid that opens and closes when substrate enters and product exits. This lid in RxDAO model is relatively longer in comparison to that in RgDAO; however, it is shorter in comparison to the mammalian DAOs.
Figure 2.
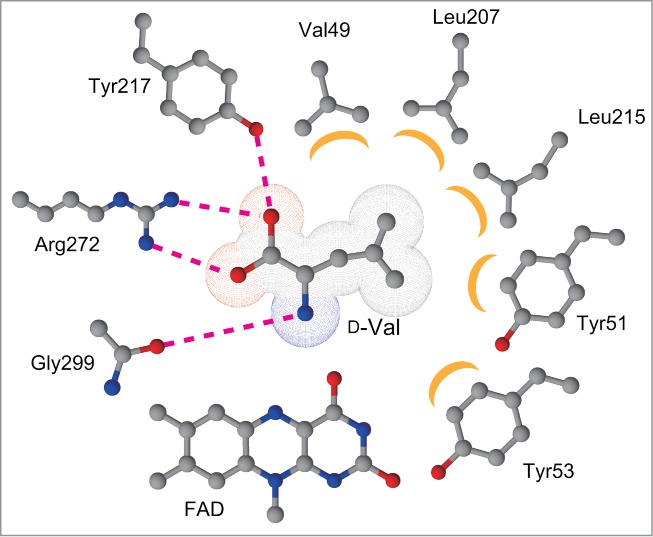
Putative substrate interactions in the active site of a model RxDAO. Hydrogen bonds and electrostatic interactions are indicated by dashed lines, while hydrophobic interactions are indicated by crescent-shaped symbols.
Enzymatic Properties of Bacterial DAOs
The enzymatic properties of bacterial DAOs are summarized in Table 1. Three bacterial DAOs analyzed so far are divided into 2 groups based on substrate specificity. ScDAO and RxDAO show higher activities toward branched chain d-amino acids and d-methionine.10,12 In contrast, ApDAO exhibits higher activities for basic d-amino acids as well as d-methionine.11 This higher activity toward basic d-amino acids is a unique feature, as other DAOs show poor or no activity for basic d-amino acids.
Table 1.
Comparison of bacterial d-amino acid oxidases
| A. protophormiae4 | R. xylanophilus26 | S. coelicolor23 | |
|---|---|---|---|
| No. of amino acids | 326 | 324 | 320 |
| Oligomeric state | Dimer | Monomer | — |
| Substrate specificity | d-Met ≈ d-Lys > d-Arg > d-Phe | d-Leu ≈ d-Ile > d-Val > d-Tyr | d-Val ≈ d-Ile > d-Met ≈ d-Leu |
| Specific activity | 180 U/mg (d-Met) | 21.1 U/mg (d-Val) | — |
| Optimum temp. | — | 65°C | — |
| Half-life at 50°C | 2 min | 5 days | — |
| Optimum pH | 6.5–8.5 | 7.5–10 | — |
| pH stability | 6.0–9.0 | 5.0–8.0 | — |
| Activity in original cells (mU/mg-protein) | 3.0 (d-Met) | — | 1.5 (d-Val) |
| Production in E. coli (U/g-wet cells) | 1,200 (d-Val) | 3.96 (d-Val) | 7.05 (d-Val) |
The highest activity shown by ApDAO is approximately 180 U/mg with d-methionine, which is comparable to the activity of yeast DAOs. In contrast, RxDAO exhibits an activity of approximately 20 U/mg with d-valine, which is similar to that of animal DAOs. The kinetic parameters of bacterial DAOs have been investigated in detail only for RxDAO. The highest calculated kcat value (53 s−1) for RxDAO is obtained for d-tyrosine as substrate, but the apparent kcat value may be much lower, as d-tyrosine exhibits substrate inhibition.10 Similar substrate inhibition was also found in human DAOs.19 The apparent kcat values of RxDAO for branched chain d-amino acids are 23–31 s−1, and the substrate affinity is the highest for d-leucine and d-isoleucine with a Km value of approximately 0.05 mM, resulting in d-leucine is the most preferred substrate for this enzyme. The Km value is one order magnitude lower in comparison to that for preferred substrates of other DAOs. Among the kinetic parameters for ApDAO, only Km value (1 mM for d-methionine) has been reported. This value is comparable to that of other eukaryotic DAOs.
Although, the optimum temperature for ApDAO has been not reported, the incubation of the enzyme at 50°C for 2 min decreases the activity by 50%.11 The optimum temperature of RxDAO is 65°C, and the enzyme is stable up to 60°C for a 1 h incubation period.10 The T50 of RxDAO, the temperature at which 50% of the initial activity is lost during incubation, is 64°C. This temperature is approximately 15°C higher in comparison to the yeast Trigonopsis variabilis DAO, which has greater stability among eukaryotic DAOs.20 In addition, approximately 25% of the residual activity was observed after incubation at 50°C for 8 d These results suggest that RxDAO has the highest thermal stability among the known DAOs. This stability has been suggested to be mainly due to the tight binding of FAD.10 The optimum pH for ApDAO is between 6.5 and 8.5 and is stable between pH 6.0 and 9.0, whereas RxDAO has optimum pH between 7.0 and 10.0. This activity-pH profile of RxDAO is similar to that of porcine kidney DAO but not like yeast DAOs, which exhibit higher activity at acidic pH values.9 RxDAO is stable around neutral values of pH i.e. between 5.0 and 8.0.10
Production of Recombinant Bacterial DAOs
The first report of the expression of bacterial DAO in E. coli is for the ApDAO.11 This enzyme is successfully expressed in E. coli with 1,200 U/g-wet cells and the specific activity of 15–30 U/mg-protein in the crude extract (Table 1). The total activity per culture for ApDAO is 1.53 × 105 U/L-culture medium. In contrast, ScDAO is produced mainly in the inclusion bodies, and the soluble enzyme occupies approximately 2% of the total soluble proteins with 7.05 U/g-wet cells and 33.7 U/L-culture medium under optimized conditions.12 The specific activity of the crude extract is 0.095 U/mg-protein. Further, RxDAO is also produced mainly in the inclusion bodies, and the soluble enzyme occupies approximately 2% of the total soluble protein.10 The decrease in the expression temperature increases the soluble enzyme content to approximately 6%; however, this results in a simultaneous decrease in the activity. The incubation of the crude extract at 70°C increases the activity, implying the presence of a folding intermediate. The specific activity of RxDAO in the crude extract (0.063 U/mg-protein) is comparable to that of ScDAO but is considerably lower in comparison to that of ApDAO. The activities for RxDAO per culture and per wet cell weight are 32.3 U/L and 3.96 U/g, respectively.
Possible roles of Bacterial DAOs
Bacteria produce d-amino acids, usually d-alanine and d-glutamate, to synthesis the peptidoglycans in the cell wall. Recently, the other d-amino acids, such as d-valine, d-leucine, d-tyrosine, d-methionine, and d-tryptophan are found to be produced by various bacterial species and are called non-canonical d-amino acids.21 These non-canonical d-amino acids have been shown to be incorporated into the bacterial cell wall and have been suggested to be involved in cell wall remodeling and biofilm disassembly.22,23
ScDAO preferentially catalyzes some of the non-canonical d-amino acids as substrates, giving an indication that this enzyme was possibly involved in cell wall remodeling or biofilm disassembly through the degradation of the d-amino acids.12 The addition of these d-amino acids in the culture medium of S. coelicolor retards the cell growth and alters the cell shape, suggesting the inhibition of cell wall synthesis. However, these non-canonical d-amino acids do not affect the biofilm formation (attachment to solid surfaces in the standing liquid cultures). In this bacterium, several putative peptidase genes possibly involved in the cell wall degradation are located around the DAO gene. These findings suggest the possibility that ScDAO may play an important role in the degradation of non-canonical d-amino acids released from the cell wall.12 Although the role of other bacterial DAOs has yet to be analyzed, the largely different activity and substrate specificity of ApDAO suggests that ApDAO might have a distinct physiological role in comparison to ScDAO.
Potential Applications of Bacterial DAOs
DAO is an effective catalyst that can be utilized in diverse applications.3,5,13 The potential applications of DAO are as follows: the determination of d-amino acids, the optical resolution of amino acid racemate, the analysis of optical purity of l-amino acids, the production of α-keto acids, the production of 7-aminocephalosporanic acid (7-ACA) from cephalosporin C, the conversion of d-amino acids to l-amino acids, the production of unnatural amino acids, the diagnosis of psychotic disorders, and the treatment of cancer. Of these, an important commercialized application of DAO is the production of 7-ACA, which is an important intermediate in the production of semisynthetic cephalosporins that are one of the best-selling antibiotics. However, ApDAO is unable to catalyze cephalosporin C, and it is unknown with clarity whether ScDAO or RxDAO could catalyze the compound.11 The determination of d-amino acids in various samples has recently gained much attention, because d-amino acids are found to play a significant role in various biological processes, and their presence in different foods and beverages can affect the taste and nutritional value of these items.24,25 Currently, d-amino acids are mainly determined using high-performance liquid chromatography; while, the enzymatic determination method using DAO has also been developed for the purpose of more rapid and convenient estimation.26 However, each d-amino acid cannot be determined individually using eukaryotic enzymes because of their broad substrate specificities.26,27 In contrast, the substrate specificity of bacterial DAOs is relatively narrow. If protein engineering deletes only the activity of ApDAO toward d-methionine, the engineered ApDAO enzyme could be useful for determining the basic d-amino acids. RxDAO has a higher binding affinity toward branched chain d-amino acids, which might be useful for determining lower concentrations of the d-amino acids in a sample. In addition, the higher stability of RxDAO may be useful as a scaffold for creating a novel DAO suitable for each application mentioned above.
Conclusions and Future Perspectives
DAO is a versatile enzyme that not only acts as a model enzyme for the flavin-dehydrogenase class of flavoproteins but also acts as a catalyst in various applications. This enzyme was found in animal tissues approximately 80 y ago, and it was until recently identified only in eukaryotic organisms. Therefore, the studies and the applications of DAO have been conducted in eukaryotes. However, recent studies reveal the presence of DAOs in bacteria and suggest the presence of this enzyme in a wide variety of bacterial species. Bacterial DAOs have been shown to possess unique enzymatic properties, such as, substrate specificity, substrate-binding affinity, and stability, in comparison to eukaryotic enzymes, indicating their potential use in various applications. On the other hand, the physiological role of bacterial DAOs is still largely unknown, as various studies on these enzymes have commenced only recently. Further studies could help to understand the physiological functions of not only these enzymes but also the non-canonical d-amino acids in bacteria. In addition, several other key topics might be considered in future research. One of them is to understand the mechanism responsible for the higher thermal stability of RxDAO. This study might help to improve the stability of valuable DAOs, especially used for the production of the antibiotic intermediate. It is also important to investigate the mechanism of the higher activity of ApDAO toward basic d-amino acids. This investigation might contribute to understand the substrate recognition mechanism of DAO and to the creation of biosensor for the enzymatic detection of basic d-amino acids. To elucidate these mechanisms, the determination of crystal structure of these bacterial DAOs would be a critical challenge. Furthermore, because it has been shown that d-amino acids or their derivatives are possibly used to prevent the infection by pathogenic bacteria, it is also important to investigate DAO as well as d-amino acids-metabolizing enzymes of pathogenic bacteria, such as Nocardia spp. and Mycobacterium spp. These future researches on bacterial DAOs would not only provide us new fundamental findings of the enzyme but also contribute to expand the use of this enzyme in a wide variety of applications.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest are disclosed.
Funding
A part of our works in this article was supported by a Grant-in-Aid for Scientific Research (C) (23580106), provided to ST from the Japan Society for the Promotion of Science.
References
- 1.Krebs HA. Metabolism of amino-acids: Deamination of amino-acids. Biochem J 1935; 29:1620-44; PMID:16745832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Negri A, Massey V, Williams CH Jr.. D-Aspartate oxidase from beef kidney. Purification and properties. J Biol Chem 1987; 262:10026-34; PMID:3611051. [PubMed] [Google Scholar]
- 3.Khoronenkova SV, Tishkov VI. D-Amino acid oxidase: Physiological role and applications. Biochemistry (Mosc) 2008; 73:1511-8; PMID:19216715; http://dx.doi.org/ 10.1134/S0006297908130105 [DOI] [PubMed] [Google Scholar]
- 4.Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. Physiological functions of D-amino acid oxidases: from yeast to humans. Cell Mol Life Sci 2007; 64:1373-94; PMID:17396222; http://dx.doi.org/ 10.1007/s00018-007-6558-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollegioni L, Molla G. New biotech applications from evolved D-amino acid oxidases. Trends Biotechnol 2011; 29:276-83; PMID:21397351; http://dx.doi.org/ 10.1016/j.tibtech.2011.01.010 [DOI] [PubMed] [Google Scholar]
- 6.Gholizadeh A, Kohnehrouz BB. Molecular cloning and expression in Escherichia coli of an active fused Zea mays L. D-amino acid oxidase. Biochemistry (Mosc) 2009; 74:137-44; PMID:19267668; http://dx.doi.org/ 10.1134/S0006297909020035 [DOI] [PubMed] [Google Scholar]
- 7.Katane M, Saitoh Y, Seida Y, Sekine M, Furuchi T, Homma H. Comparative characterization of three D-aspartate oxidases and one D-amino acid oxidase from Caenorhabditis elegans. Chem Biodivers 2010; 7:1424-34; PMID:20564561; http://dx.doi.org/ 10.1002/cbdv.200900294 [DOI] [PubMed] [Google Scholar]
- 8.Sarower MG, Okada S, Abe H. Molecular characterization of D-amino acid oxidase from common carp Cyprinus carpio and its induction with exogenous free D-alanine. Arch Biochem Biophys 2003; 420:121-9; PMID:14622982; http://dx.doi.org/ 10.1016/j.abb.2003.09.035 [DOI] [PubMed] [Google Scholar]
- 9.Pollegioni L, Sacchi S, Caldinelli L, Boselli A, Pilone MS, Piubelli L, Molla G. Engineering the properties of D-amino acid oxidases by a rational and a directed evolution approach. Curr Protein Pept Sci 2007; 8:600-18; PMID:18220846; http://dx.doi.org/ 10.2174/138920307783018677 [DOI] [PubMed] [Google Scholar]
- 10.Takahashi S, Furukawara M, Omae K, Tadokoro N, Saito Y, Abe K, Kera Y. A highly stable D-amino acid oxidase of the thermophilic bacterium Rubrobacter xylanophilus. Appl Environ Microbiol 2014; 80:7219-29; http://dx.doi.org/ 10.1128/AEM.02193-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geueke B, Weckbecker A, Hummel W. Overproduction and characterization of a recombinant D-amino acid oxidase from Arthrobacter protophormiae. Appl Microbiol Biotechnol 2007; 74:1240-7; PMID:17279391; http://dx.doi.org/ 10.1007/s00253-006-0776-9 [DOI] [PubMed] [Google Scholar]
- 12.Saito Y, Takahashi S, Kobayashi M, Abe K, Kera Y. D-Amino acid oxidase of Streptomyces coelicolor and the effect of D-amino acids on the bacterium. Ann Microbiol 2014; 64:1167-77 [Google Scholar]
- 13.Pollegioni L, Molla G, Sacchi S, Rosini E, Verga R, Pilone MS. Properties and applications of microbial d-amino acid oxidases: current state and perspectives. Appl Microbiol Biotechnol 2008; 78:1-16; PMID:18084756; http://dx.doi.org/ 10.1007/s00253-007-1282-4 [DOI] [PubMed] [Google Scholar]
- 14.Molla G, Motteran L, Piubelli L, Pilone MS, Pollegioni L. Regulation of D-amino acid oxidase expression in the yeast Rhodotorula gracilis. Yeast 2003; 20:1061-9; PMID:12961754; http://dx.doi.org/ 10.1002/yea.1023 [DOI] [PubMed] [Google Scholar]
- 15.Yurimoto H, Hasegawa T, Sakai Y, Kato N. Characterization and high-level production of D-amino acid oxidase in Candida boidinii. Biosci Biotechnol Biochem 2001; 65:627-33; PMID:11330678; http://dx.doi.org/ 10.1271/bbb.65.627 [DOI] [PubMed] [Google Scholar]
- 16.Sacchi S, Caldinelli L, Cappelletti P, Pollegioni L, Molla G. Structure-function relationships in human d-amino acid oxidase. Amino Acids 2012; 43:1833-50; PMID:22865246; http://dx.doi.org/ 10.1007/s00726-012-1345-4 [DOI] [PubMed] [Google Scholar]
- 17.Piubelli L, Molla G, Caldinelli L, Pilone MS, Pollegioni L. Dissection of the structural determinants involved in formation of the dimeric form of D-amino acid oxidase from Rhodotorula gracilis: role of the size of the βF5-βF6 loop. Protein Eng 2003; 16:1063-9; PMID:14983088; http://dx.doi.org/ 10.1093/protein/gzg125 [DOI] [PubMed] [Google Scholar]
- 18.Kawazoe T, Park HK, Iwana S, Tsuge H, Fukui K. Human D-amino acid oxidase: an update and review. Chem Rec 2007; 7:305-15; PMID:17924443; http://dx.doi.org/ 10.1002/tcr.20129 [DOI] [PubMed] [Google Scholar]
- 19.Kawazoe T, Tsuge H, Imagawa T, Aki K, Kuramitsu S, Fukui K. Structural basis of D-DOPA oxidation by d-amino acid oxidase: alternative pathway for dopamine biosynthesis. Biochem Biophys Res Commun 2007; 355:385-91; PMID:17303072; http://dx.doi.org/ 10.1016/j.bbrc.2007.01.181 [DOI] [PubMed] [Google Scholar]
- 20.Wang SJ, Yu CY, Lee CK, Chern MK, Kuan IC. Subunit fusion of two yeast D-amino acid oxidases enhances their thermostability and resistance to H2O2. Biotechnol Lett 2008; 30:1415-22; PMID:18330517; http://dx.doi.org/ 10.1007/s10529-008-9694-5 [DOI] [PubMed] [Google Scholar]
- 21.Alvarez L, Espaillat A, Hermoso JA, de Pedro MA, Cava F. Peptidoglycan remodeling by the coordinated action of multispecific enzymes. Microb Drug Resist 2014; 20:190-8; PMID:24799190; http://dx.doi.org/ 10.1089/mdr.2014.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-Amino acids govern stationary phase cell wall remodeling in bacteria. Science 2009; 325:1552-5; PMID:19762646; http://dx.doi.org/ 10.1126/science.1178123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-Amino acids trigger biofilm disassembly. Science 2010; 328:627-9; PMID:20431016; http://dx.doi.org/ 10.1126/science.1188628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solms J, Vuataz L, Egli RH. The taste of L- and D-amino acids. Experientia 1965; 21:692-4; PMID:5869699; http://dx.doi.org/ 10.1007/BF02138474 [DOI] [PubMed] [Google Scholar]
- 25.Friedman M. Origin, microbiology, nutrition, and pharmacology of D-amino acids. Chem Biodivers 2010; 7:1491-530; PMID:20564567; http://dx.doi.org/ 10.1002/cbdv.200900225 [DOI] [PubMed] [Google Scholar]
- 26.Molla G, Piubelli L, Volonte F, Pilone MS. Enzymatic detection of D-amino acids. Methods Mol Biol 2012; 794:273-89; PMID:21956570 [DOI] [PubMed] [Google Scholar]
- 27.Gabler M, Hensel M, Fischer L. Detection and substrate selectivity of new microbial D-amino acid oxidases. Enzyme Microb Technol 2000; 27:605-11; PMID:11024524; http://dx.doi.org/ 10.1016/S0141-0229(00)00262-3 [DOI] [PubMed] [Google Scholar]


