Abstract
Owing to our limited understanding of the relationship between sequence and function and the interaction between intracellular pathways and regulatory systems, the rational design of enzyme-coding genes and de novo assembly of a brand-new artificial genome for a desired functionality or phenotype are difficult to achieve. As an alternative approach, directed evolution has been widely used to engineer genomes and enzyme-coding genes. In particular, significant developments toward DNA synthesis, DNA assembly (in vitro or in vivo), recombination-mediated genetic engineering, and high-throughput screening techniques in the field of synthetic biology have been matured and widely adopted, enabling rapid semi-rational genome engineering to generate variants with desired properties. In this commentary, these novel tools and their corresponding applications in the directed evolution of genomes and enzymes are discussed. Moreover, the strategies for genome engineering and rapid in vitro enzyme evolution are also proposed.
Keywords: directed evolution, DNA assembly, enzyme, genome engineering, metabolic engineering, recombineering, synthetic biology
Abbreviations
- dsDNA
double-stranded DNA
- HTS
high-throughput screening
- LCR
Ligase Cycling Reaction
- MAGE
multiplex automated genome engineering
- ssDNA
single-stranded DNA
Traditional Approaches for the Directed Evolution of Genomes and Enzyme-coding Genes
Currently, more than 2000 classes of enzymes that catalyze various synthetic reactions have been recognized.1 However, in most cases, the naturally occurring enzymes often lack features that are necessary for commercial applications because of their natural, complicated regulation and harsh biocatalytic process conditions.2 Therefore, natural enzymes always need to be engineered to possess the desirable catalytic properties that are required for practical applications.3 In this regard, many directed evolution techniques such as error-prone PCR, site-directed saturation mutagenesis, iterative saturation mutagenesis, and DNA shuffling have been developed and widely used to optimize many catalytic parameters including thermostability, activity, substrate specificity, and enantioselectivity in artificial environments.4,5 In addition, the construction of robust cell factories for whole-cell biocatalysis or de novo synthesis of the target products with an optimized background is also attractive and indispensable. As a result, many studies to improve the titer of target products or cell resistance to environments by genome engineering with the above-mentioned directed evolution techniques have been reported.6 In particular, the construction of stable synthetic pathways to the desired end products with multiple genome modifications has been intensively studied recently.7-9 In contrast, rapid semi-rational engineering of enzyme-coding genes and genomes with novel directed evolution and synthetic biology strategies should be preferred and considered more promising because these strategies avoid the side effects introduced by random approaches.
Recombineering as a Powerful Tool for Rapid Engineering of Genomes and Enzyme-coding Genes
Recombination-mediated genetic engineering (recombineering) emerged as an in vivo technique and has been widely used in bacterial genome evolution because of its efficiency and simplicity.10 Especially since 2000, linear DNA-mediated integration with the help of phage recombinases (RecET and λ-Red) has been developed and routinely applied in the genome engineering of bacteria and Saccharomyces cerevisiae.11,12 Through synergistic actions with the three λ-Red proteins Exo, Bet, and Gam, the transformed double-stranded linear DNAs (dsDNA) with short (36–100 bp) homologous arms were digested into intermediates with a single-stranded sticky end, which promotes efficient homologous recombination. Subsequently, the integrated antibiotic resistance gene flanked by the FRT or loxP sites were recognized and removed by the site-specific recombinases FLP recombinase (for FRT) or Cre recombinase (for loxP), resulting in the desired mutant strains.12 Although this method is simple and effective, multiple fragment deletions or modifications in one strain are time-consuming. In addition, residual small loxP scars that were generated during the elimination of the helper plasmid tend to trigger unwanted recombinations and result in unexpected phenotypes. To solve this problem, Zhang et al. developed a modified sacB-based counterselection method with two steps of homologous recombination to perform scarless point mutation: gene knock-out, and gene integration on the chromosome.13 Applying these recombineering tools (the efficiency with dsDNA is about 0.1%),14 many recombinant strains with engineered genes or regulatory elements have been constructed for various applications.8,10 Even so, simple methods for the transformation and integration of multiple large segments remain undeveloped.
To simplify the operation and improve the recombination efficiency, short and synthesized single-stranded DNA (ssDNA) oligonucleotides for genome engineering (Fig. 1A) have been demonstrated to have high efficiencies in Saccharomyces cerevisiae 15 and many bacteria including Escherichia coli.16,17 Accordingly, several recombineering strategies depending on ssDNA (with only 35 bases of homology) including multiplex automated genome engineering (MAGE),18 conjugative assembly genome engineering and MAGE Oligo Design Tool9 have been established and applied for precise manipulation and rapid evolution of chromosomes (Fig. 1A). In light of high-throughput screening (HTS) strategies, variant cells with desirable phenotypes can be generated and isolated after several repeated rounds of recombination. As a proof of concept, Wang et al. successfully optimized the 1-deoxy-D-xylulose-5-phosphate pathway to improve lycopene accumulation, transforming and integrating 90-mer oligos that were designed to target the ribosome-binding sites of 20 genes and 4 genes.18 Similarly, Isaacs et al. replaced all 314 TAG stop codons with TAA codons in E. coli strains.9 To improve the insertion efficiency of short oligonucleotides (>10 bases), Wang et al. proposed a co-selection strategy and combinatorially inserted multiple T7 promoters simultaneously into 12 genomic operons, enabling the rapid optimization of the biosynthesis of aromatic amino acid derivatives.19 Moreover, by applying over 110 MAGE cycles, they simultaneously inserted hexa-histidine sequences into 38 essential genes that encode the complete translation machinery and realized its in vitro co-purification.20 Although MAGE-related techniques are efficient and easy to perform, their application is restricted by the development of high-throughput methodologies to screen mutants with desired phenotypes.
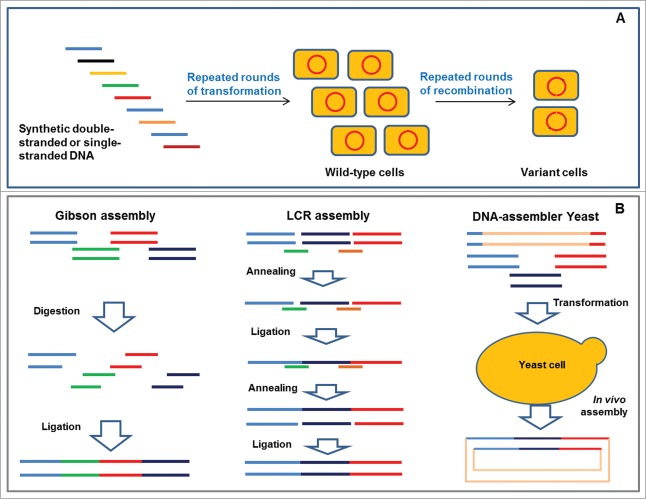
Figure 1.
Schematic overview of multiplex genome engineering and DNA-assembly methods. (A) Recombineering with synthetic double-stranded or single-stranded DNA fragments. Short segments containing different mutation sites were designed and synthesized. After multiple rounds of transformation and recombination, the variants with desired phenotypes were isolated by high-throughput screening methods. (B) Illustration of the Gibson, ligase cycling reaction, and yeast-dependent DNA-assembly methods.
More recently, the clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 (endonuclease)-mediated genome-editing technology, which depends on specific homologous recombination and nuclease-specific cleavage, has been developed and applied in genome engineering.21,22 To simplify the CRISPR process and broaden its applications, the small guide RNA, a hybrid of the trans-activating crRNA and the precursor CRISPR RNA, was constructed and employed in genome editing such as gene inactivation, precise mutations, and insertions.22,23 In addition, the CRISPR-Cas9 system was also engineered to down-regulate gene expression at the transcriptional level by inactivating the Cas9 nuclease,23 demonstrating its versatile applications in genome engineering. Consequently, constructing an optimized biosynthesis pathway at the genome level by applying these bioengineering tools is a promising and attractive option for the near future (Fig. 2).
Figure 2.
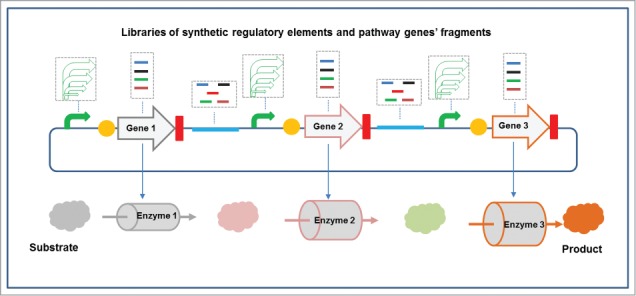
Combinatorial recombineering to optimize the biosynthesis pathway of interest. To construct a balanced synthetic pathway to the end product, different libraries of pathway functional genes and regulatory elements—including promoters, intergenic spacers, and ribosome-binding sites were designed and synthesized. Applying these recombineering tools, all of the above segments can be combinatorially optimized.
Rapid Assembly Tools Enable Rapid Evolution of Genes and Genomes
DNA assembly is one of the most important foundational technologies for rapid prototyping of metabolic pathways or genetic circuits of interest.24 Especially today, the rapid development of synthetic biology25 and metabolic engineering26 require efficient, flexible, and faithful DNA assembly approaches because of the low capacity of traditional restriction, digestion, and ligation methods. In fact, modular and combinatorial assemblies of various genetic segments, particularly the assembly of large DNA fragments without scars by restriction, digestion, and ligation methods are extremely difficult.24,27 As a result, several in vitro methods, such as circular polymerase extension cloning,28 sequence and ligation-independent cloning,29 Gibson assembly method,30 Ligase Cycling Reaction (LCR),31 and in vivo methods including DNA assembly with homologous recombination in Saccharomyces cerevisiae (DNA assembler-yeast)27,32 have been developed recently. The Gibson assembly method is the most well-known method, and involves the generation of single-stranded complementary overhangs by T5 exonuclease and covalent joining by fusion DNA polymerase and Taq DNA ligase.30 However, digestion with T5 exonuclease to generate sticky ends is difficult to precisely regulate. In addition, the method is comparatively complicated. More recently, experiments demonstrated that the LCR and DNA assembler-yeast methods (Fig. 1B) have a higher assembly capacity (up to 12 DNA parts) than the other in vitro methods mentioned above,31 indicating their great potential to assemble different regulatory and functional fragments.
Since the advent of these assembly methods, many successful applications have been reported. Impressively, a brand new functional bacterial genome and eukaryotic chromosome have been successfully assembled using rational design and chemical synthesis.33,34 Through random assembly of a set of constitutive promoters, a silent spectinabilin pathway from Streptomyces orinoci and a cryptic polycyclic tetramate macrolactams biosynthetic gene cluster from Streptomyces griseus were respectively discovered and characterized,35,36 which confirmed the powerful ability of DNA-assembly methods in the discovery of novel natural products. More recently, using both combinatorial transcriptional engineering and directed evolution strategies, a library of promoters with varying strengths were assembled with the xylose-utilizing pathway or the cellobiose-utilizing pathway functional structural genes. As expected, highly efficient heterologous xylose- and cellobiose-utilizing pathways were generated and isolated with a cell growth–based HTS strategy,37,38 confirming the practicality of DNA-assembly methods in metabolic engineering.
In the last two decades, although DNA shuffling has been used as an alternative technique to assemble mutations that were introduced by random or site-directed mutagenesis methods, the efficiency is relatively low, and the method requires multiple rounds, which is time-consuming. Consequently, more efficient directed evolution techniques are desired to rapidly engineer target enzymes. Here, we propose a rapid in vitro evolution method that depends on rapid and scarless in vitro DNA assembly tools. As shown in Figure 3, the target enzyme-encoding gene is separated into several parts, and all of the potential mutations are introduced during in vitro amplification with the designed oligonucleotides or degenerate primers. Subsequently, the cloned variant segments with short homologous arms are assembled and transformed into the expression host strains. By applying an appropriate HTS approach, enzyme variants with desirable phenotypes can be rapidly isolated. In particular, this rapid directed evolution approach will be applicable to semi-rational and multiple engineering of the desired enzyme with the help of crystal structure analysis.
Figure 3.
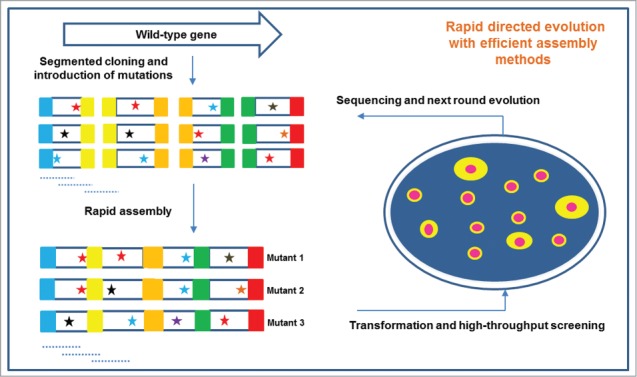
Illustration of a rapid in vitro directed evolution technique for enzyme engineering. The native gene of interest is divided into several segments. The designed potential mutation sites are introduced during amplification. Subsequently, the mutant fragments are assembled and overexpressed in the host expression strains. With high-throughput screening approaches, the variants with desirable phenotypes are quickly isolated.
Conclusion
With the development of directed evolution, synthetic biology, the associated powerful tools,39 and our knowledge on enzyme functions and genome regulatory mechanisms, the creation of novel enzymes and functional genomes with targeted properties and phenotypes of interest will be more efficient and convenient. For instance, by applying the available strategies, we can rapidly construct or optimize biosynthesis pathways (Fig. 2). In addition, the direct de novo synthesis of designed DNA fragments will be affordable because of decreasing costs, which will further accelerate the downstream directed evolution. In return, directed evolution with these novel tools enables us to gain more insight into the complex living systems composed of proteins, metabolic pathways, and regulatory circuits.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China (31200020), the Major State Basic Research Development Program of China (973 Program, 2014CB745103), the Natural Science Foundation of Jiangsu Province (BK20141107) and a grant from the Key Technologies R & D Program of Jiangsu Province, China (BE2014607).
References
- 1. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27-30; PMID:10592173; http://dx.doi.org/ 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hibbert EG, Baganz F, Hailes HC, Ward JM, Lye GJ, Woodley JM, Dalby PA. Directed evolution of biocatalytic processes. Biomol Eng 2005; 22:11-9; PMID:15857779; http://dx.doi.org/ 10.1016/j.bioeng.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 3. Turner NJ. Directed evolution drives the next generation of biocatalysts. Nat Chem Biol 2009; 5:567-73; PMID:19620998; http://dx.doi.org/ 10.1038/nchembio.203 [DOI] [PubMed] [Google Scholar]
- 4. Cobb RE, Si T, Zhao H. Directed evolution: an evolving and enabling synthetic biology tool. Curr Opin Chem Biol 2012; 16:285-91; PMID:22673064; http://dx.doi.org/ 10.1016/j.cbpa.2012.05.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dalby PA. Strategy and success for the directed evolution of enzymes. Curr Opin Struct Biol 2011; 21:473-80; PMID:21684150; http://dx.doi.org/ 10.1016/j.sbi.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 6. Biot-Pelletier D, Martin VJ. Evolutionary engineering by genome shuffling. Appl Microbiol Biotechnol 2014; 98:3877-87; PMID:24595425; http://dx.doi.org/ 10.1007/s00253-014-5616-8 [DOI] [PubMed] [Google Scholar]
- 7. Pal C, Papp B, Posfai G. The dawn of evolutionary genome engineering. Nat Rev Genet 2014; 15:504-12; PMID:24866756; http://dx.doi.org/ 10.1038/nrg3746 [DOI] [PubMed] [Google Scholar]
- 8. Boyle NR, Gill RT. Tools for genome-wide strain design and construction. Curr Opin Biotechnol 2012; 23:666-71; PMID:22357141; http://dx.doi.org/ 10.1016/j.copbio.2012.01.012 [DOI] [PubMed] [Google Scholar]
- 9. Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA,Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 2011; 333:348-53; PMID:21764749; http://dx.doi.org/ 10.1126/science.1205822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song CW, Lee J, Lee SY. Genome engineering and gene expression control for bacterial strain development. Biotechnol J 2014; PMID:25155412; http://dx.doi.org/ 10.1002/biot.201400057 [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 1998; 20:123-8; PMID:9771703; http://dx.doi.org/ 10.1038/2417 [DOI] [PubMed] [Google Scholar]
- 12. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 2000; 97:6640-5; PMID:10829079; http://dx.doi.org/ 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang X, Jantama K, Moore JC, Shanmugam KT, Ingram LO. Production of L-alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 2007; 77:355-66; PMID:17874321; http://dx.doi.org/ 10.1007/s00253-007-1170-y [DOI] [PubMed] [Google Scholar]
- 14. Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 2000; 97:5978-83; PMID:10811905; http://dx.doi.org/ 10.1073/pnas.100127597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DiCarlo JE, Conley AJ, Penttila M, Jantti J, Wang HH, Church GM. Yeast oligo-mediated genome engineering (YOGE). ACS Synth Biol 2013; 2:741-9; PMID:24160921; http://dx.doi.org/ 10.1021/sb400117c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA 2001; 98:6742-6; PMID:11381128; http://dx.doi.org/ 10.1073/pnas.121164898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu D, Sawitzke JA, Ellis H, Court DL. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc Natl Acad Sci USA 2003; 100:7207-12; PMID:12771385; http://dx.doi.org/ 10.1073/pnas.1232375100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009; 460:894-8; PMID:19633652; http://dx.doi.org/ 10.1038/nature08187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang HH, Kim H, Cong L, Jeong J, Bang D, Church GM. Genome-scale promoter engineering by coselection MAGE. Nat Methods 2012; 9:591-3; PMID:22484848; http://dx.doi.org/ 10.1038/nmeth.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang HH, Huang PY, Xu G, Haas W, Marblestone A, Li J, Gygi SP, Forster AC, Jewett MC, Church GM. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth Biol 2012; 1:43-52; PMID:22737598; http://dx.doi.org/ 10.1021/sb3000029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819-23; PMID:23287718; http://dx.doi.org/ 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013; 31:233-9; PMID:23360965; http://dx.doi.org/ 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816-21; PMID:22745249; http://dx.doi.org/ 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chao R, Yuan Y, Zhao H. Recent advances in DNA assembly technologies. FEMS Yeast Res 2014; PMID:24903193; http://dx.doi.org/ 10.1111/1567-1364.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 2009; 10:410-22; PMID:19461664; http://dx.doi.org/ 10.1038/nrm2698 [DOI] [PubMed] [Google Scholar]
- 26. Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 2013; 4:1409; PMID:23361000; http://dx.doi.org/ 10.1038/ncomms2425 [DOI] [PubMed] [Google Scholar]
- 27. Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA, 3rd. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A 2008; 105:20404-9; PMID:19073939; http://dx.doi.org/ 10.1073/pnas.0811011106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PloS one 2009; 4:e6441; PMID:19649325; http://dx.doi.org/ 10.1371/journal.pone.0006441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 2007; 4:251-6; PMID:17293868; http://dx.doi.org/ 10.1038/nmeth1010 [DOI] [PubMed] [Google Scholar]
- 30. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009; 6:343-5; PMID:19363495; http://dx.doi.org/ 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 31. de Kok S, Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol 2014; 3:97-106; PMID:24932563; http://dx.doi.org/ 10.1021/sb4001992 [DOI] [PubMed] [Google Scholar]
- 32. Shao Z, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res 2009; 37:e16; PMID:19074487; http://dx.doi.org/ 10.1093/nar/gkn991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, et al. Total synthesis of a functional designer eukaryotic chromosome. Science 2014; 344:55-8; PMID:24674868; http://dx.doi.org/ 10.1126/science.1249252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010; 329:52-6; PMID:20488990; http://dx.doi.org/ 10.1126/science.1190719 [DOI] [PubMed] [Google Scholar]
- 35. Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun 2013; 4:2894; PMID:24305602; http://dx.doi.org/ 10.1038/ncomms3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol 2013; 2:662-9; PMID:23968564; http://dx.doi.org/ 10.1021/sb400058n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du J, Yuan Y, Si T, Lian J, Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res 2012; 40:e142; PMID:22718979; http://dx.doi.org/ 10.1093/nar/gks549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan Y, Zhao H. Directed evolution of a highly efficient cellobiose utilizing pathway in an industrial Saccharomyces cerevisiae strain. Biotechnol Bioeng 2013; 110:2874-81; PMID:23616289; http://dx.doi.org/ 10.1002/bit.24946 [DOI] [PubMed] [Google Scholar]
- 39. Kang Z, Zhang C, Zhang J, Jin P, Du G, Chen J. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 2014; 98:3413-24; PMID:24519458; http://dx.doi.org/ 10.1007/s00253-014-5569-y [DOI] [PubMed] [Google Scholar]