Abstract
Cronobacter sakazakii is an opportunistic pathogen associated with outbreaks of life-threatening necrotizing enterocolitis, meningitis and sepsis in neonates and infants. The pathogen possesses an array of virulence factors which aid in tissue adhesion, invasion and host cell injury. Although the identification and validation of C. sakazakii virulence factors has been hindered by availability of suitable neonatal animal model, various studies has reported outer membrane protein A (ompA) as a potential virulence marker. Various other plasmid associated genes such as filamentous hemagglutinin (fhaBC), Cronobacter plasminogen activator (cpa) and genes responsible for iron acquisition (eitCBAD and iucABD/iutA) have been reported in different strains of C. sakazakii. Besides these proposed virulence factors, several biophysical growth factors such as formation of biofilms and resistance to various environmental stresses also contributes to the pathogenic potential of this pathogen. This review provides an update on virulence determinants associated with the pathogenesis of C. sakazakii. The potential reservoirs of the pathogen, mode of transmission and epidemiology are also discussed.
Keywords: Cronobacter sakazakii, powdered infant formula, pathogenesis, virulence
Introduction
The genus Cronobacter (previously known as Enterobacter sakazakii) is recognized as an emerging opportunistic pathogen causing life-threatening infections in neonates and immune-compromised infants.1–5 Urmenyi and Franklin6 firstly, reported severe C. sakazakii induced systemic infection in neonates in England. Since then, there have been around 150 reported cases of this contagion with 26 deaths worldwide.7-9 The pathogen received world-wide attention after an outbreak of meningitis in Tennessee in 2001.10 It was Farmer et al.11 who foremost established the taxonomic position of novel species named C. sakazakii which was initially referred to as Gram negative, facultative anaerobe yellow-pigmented Enterobacter cloacae. Till the date, the Cronobacter consists of 11 species that include C. sakazakii, C. malonaticus, C. dublinensis, C. turicensis, C. muytjensii, C. condimenti, C. universalis, C. helveticus, C. zurichenesis, C. pulveris and C. colletis.12-15 The species C. sakazakii, C. turicensis and C. malonaticus are reported to cause neonatal infections.12 Although the incidence of disease is very low, the fatality rates range between 40 to 80% and survivors often had severe neurological and developmental disorders.8,16,17,18 It is worth mentioning that premature birth and/or low birth weight are often cited as highest risk individuals for Cronobacter infection because they lack normal gut microflora and established gut epithelial lining which makes them more susceptible to increased mucosal permeability.19 Reconstituted powdered infant formula (PIF) is reported to be the most associated vehicle for transmission of pathogen. Voluntary recalls of PIF contaminated with Cronobacter in the United States, Europe and Asia-Pacific region suggested the need of a collective effort among PIF manufacturers, health-care facilities and governing bodies to develop hygienic practices and maintain higher microbiological standards.20
The application of genome sequencing data and multilocus sequence typing (MLST) validated the revision of the taxonomic position of the 11 identified Cronobacter species.21-31 The MLST utilizing 7 (atpD, fusA, glnS, gltB, gyrB, infB, ppsA) is a more successful typing method for the Cronobacter genus and has exhibited a high caliber of discrimination between the isolates.32-36 The Cronobacter PubMLST database curated by Stephen Forsythe comprised the enteries for 1193 Cronobacter isolates reported worldwide. The MLST database of 739 C. sakazakii isolates indicates clonal complex 4 (CC4) as stable and predominantly coupled with neonatal meningitis. The ribosomal-MLST (53-loci) and Clusters of Orthologous Groups–core genome (COG-cg) MLST (1865 loci) has also confirmed CC4 as dominant lineage.37 However, due to limited information on the virulence characteristics of CC4, its association with neonatal meningitis is unclear.34 Therefore, unveiling the virulence characteristics of this pathogen would contribute toward underpinning the association of the pathogen to infant foods and to develop mitigation strategies.
The little information on the ecology, pathogenesis and virulence of C. sakazakii warrants an update on this enteric pathogen with special emphasis on virulence factors associated with the pathogenesis of C. sakazakii.
Reservoir and mode of transmission
The bacterium is ubiquitous and has been isolated from a wide variety of foods, including cheese products, infant cereal, dried foods, fruits, vegetables, meats, water, medicinal plants, herbs and spices, bread, rice and PIF.38-50 Moreover, the Cronobacter spp. has also been reported from clinical sources, including cerebrospinal fluid, blood, intestinal and respiratory tracts, bone marrow and skin wounds.51 The pathogen has also been detected from domestic vacuum cleaner bags, river water, the gut of a Mexican fruit fly and stable fly and faecal sample of animals.19,52-54 Reconstituted PIF is the most associated vehicle for transmission of the pathogen, being an intrinsic or extrinsic contaminant during manufacturing under poor good manufacturing practices (GMP) or reconstitution of PIF (Table 1).2,16,38,55-64 The presence of the pathogen as vaginal microflora has been neglected by several studies however, babies delivered through birth canal or Caesarean section (C-section) have been contracted with the pathogen few days after birth.4,22,65
Table 1.
Powdered Infant Formula (PIF) implicated in worldwide outbreaks of Cronobacter infection
| Country | Year | No. of cases / No. of deaths | Reference(s) |
|---|---|---|---|
| New Mexico | 2008 | 2/2 | 106 |
| France | 2004 | 4/2 | 107 |
| USA | 2004 | 1/0 | 108 |
| New Zealand | 2004 | 1/1 | 109 |
| Tennessee | 2001 | 10/1 | 55 |
| Israel | 1999–2000 | 2/0 | 2 |
| Belgium | 1998 | 12/2 | 16 |
| India | 1992 | 1/1 | 110 |
| Maryland | 1990 | 1/0 | 111 |
| Tennessee | 1988 | 4/0 | 112,113 |
| Iceland | 1986–1987 | 3/1 | 36 |
| Denmark | 1983 | 1/1 | 114 |
Epidemiology
The epidemiology of Cronobacter species is incomplete and poorly described because of its rare infections and often underreported cases due to missing or different reporting criteria in developed and some developing countries.66 Feeding with reconstituted PIF has been epidemiologically implicated in numerous clinical cases (Table 2). Cases are somewhat sporadic, but epidemics are not unusual; the utmost-risk group is neonates (<28 days old) that have low birth weights (<2,000 to 2,500 g) or the premature (<37 weeks of gestation stage). Friedemann8 reported the lethality of Cronobacter meningitis, bacteraemia and NEC to be 41.9% (P < 0.0001), <10% and 19.0% (P < 0.05), respectively, for 120–150 microbiologically Cronobacter confirmed neonatal infections occurred between 2000 and 2008. The annual occurrence rate among the premature and underweight infant is reported to a figure of 8.7 per 100,000 low-birth weight neonates in the USA, and one Cronobacter infection per 10,660 every low-birth neonates.59,67 Hunter and Bean65 demonstrated the worldwide distribution of reported cases; the majority of them are within the developed countries (approx. 45%). This distribution may be underestimated since not all clinical analysis laboratories carry out research on the pathogen and not all countries have a system for reporting diseases. The Food and Drug Administration (FDA) has accounted a series of neonatal disease outbreaks in Florida, Missouri Illinois and Oklahoma in December 2011.68 The limited information on its epidemiology necessitates that the researchers should record consistent and sufficiently informative data of invasive neonatal Cronobacter infections as developed under PubMLST database.
Table 2.
Characteristics of major known virulence factors of Cronobacter sakazakii
| Factors | Genes | Potential role | Reference(s) |
|---|---|---|---|
| Outer membrane proteins (OMPs) | ompXompA | Involved in the basolateral invasion of enterocyte-like human epithelial cells | 59,60–62 |
| Enterotoxin | Not known yet | Heat stable toxin elaborated by the pathogen | 63,64 |
| Outer membrane protease | cpa | Provides resistance against bactericidal activity of serum; activates plasminogen and inactivates α2-AP | 65,66 |
| Sialic acid utilization | nanAKT | Confers in pathogenesis | 71 |
| Iron acquisition system | iuc | Encodes an iron-uptake system mediated by the active siderophorethat plays a role in iron transport and regulation | 67,73 |
| Efflux system | ibeB | Encodes copper and silver resistance cation efflux system facilitating invasion of brain microvascular endothelial cells (BMEC) | 23 |
| Proteolytic enzymes | zpx | Cause cell deformation and rounding of cells | 83 |
| Lipopoysaccarides | Chromosomal encoded genes | Disrupt epithelial tight junctions | 91,102 |
| Type III hemolysin | hly | Hemolytic activity | 66,90 |
Pathogenicity and virulence factors
The high mortality and fatality rate caused by C. sakazakii is still poorly understood, and the list of virulence factors (Table 2) is probably far from complete. The specific virulence factors associated with the pathogenesis are discussed in this section.
Outer membrane proteins (OMPs)
Outer membrane proteins (OMPs) are of peculiar interest, owing to their cell-surface exposure and contribution in export of extracellular virulence factors, and in anchoring the structures that mediate adhesion and motility.
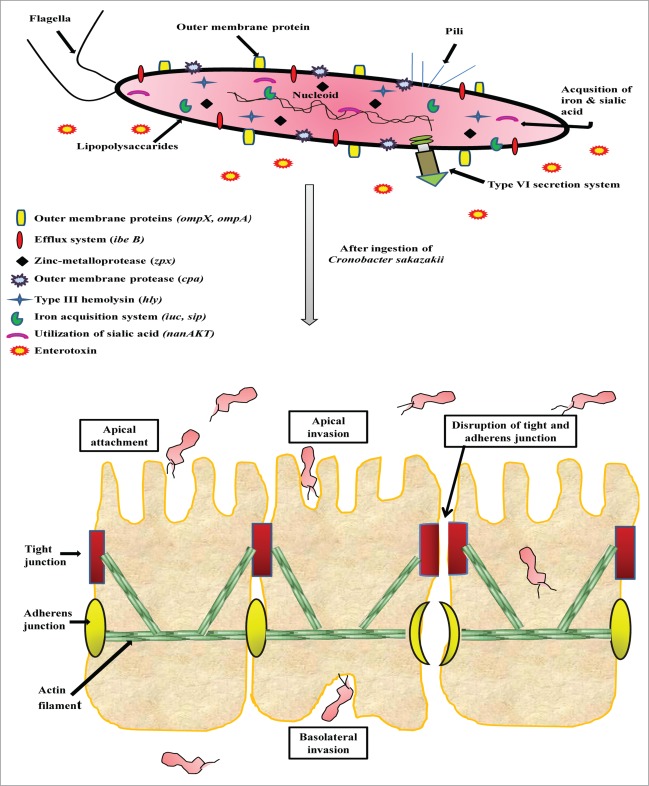
Several studies put forwarded that outer membrane protein A (ompA), contributes significantly to the virulence potential of Cronobacter spp by invading various epithelial and endothelial cells of human and animal origin. The invasion studies with human intestinal (INT407) cells showed the contribution of both microfilaments and microtubules from host and bacterial ompA.69–71 Mittal and co-workers72 reported that OmpA-positive isolates breach blood-brain barrier and invade central nervous system (CNS) causing clinical manifestations. In addition to ompA, Kim et al.73 reported that ompX also played vital roles in the invasion not only the apical side, but also the basolateral side of the host cells and can translocate into the deeper organs (spleen and liver) of rats (Fig. 1).
Figure 1.
Proposed model for Cronobacter sakazakii infection and pathogenesis. The pathogen encodes several illustrated pathogenicity-associated factors engaged in imperative processes including adhere to host surfaces, transmigration across, invasion into and disrupt the intestinal barrier within intestinal epithelial cells.
Enterotoxin
Pagotto et al.74 were the first to study the dose-response of C. sakazakii in suckling mouse and reported a minimum lethal dose of 108 colony-forming units (cfu) in neonatal mouse suggesting the possibility of enterotoxin analog in infections. The function of this toxin may act in a parallel fashion to lipopolysaccaride (LPS), mediating toll-like receptor 4 (TLR4) activation and stimulating a host inflammatory response.74 However, later it was Raghav and Aggarwal75 who identified a thermostable putative toxin with molecular mass of 66 kDa. The potent activity of the toxin (LD50 = 56 pg) emphasizes the emerging risk to neonates fed reconstituted PIF contaminated with C. sakazakii. The implication of the enterotoxin is still blurred as the genes encoding the putative toxin and the protein itself remain unidentified. Further studies using functional genomics and system biology might help in characterization of toxin-related genes.
Cronobacter plasminogen activator (cpa)
Recently, the study on C. sakazakii BAA-894 reported presence of a plasmid (pESA3) encoding an outer membrane proteases (cpa) that has significant identity to proteins that belong to Pla subfamily of omptins. This protease has an ability to render serum resistance by cleaving complement components, activating plasminogen and inactivating the plasmin inhibitor α2-AP.76 Franco et al. and Cruz et al. also portray the prevalence and distribution of plasmid-encoded virulence genes cpa, a type 6 secretion system (T6SS, also encoded on pESA3) and a filamentous haemaggltunin/adhesion (FHA) gene locus (located on pCTU1) among 231 Cronobacter strains.77,78
Sialic acid utilization
Sialic acid is found in human milk and in infant formulae in the form of sialyloligosaccharides.79 These oligosaccharides remain undigested in neonates and infants, therefore the intestinal microvilli of neonates have increased sialic acid and N-acetylglucosamine residues leading to proliferation of gut microbiota.80,81 Recently, Joseph et al.82 explained a plausible linkage between sialic acid metabolism and the pathogenicity of C. sakazakii as it is the only Cronobacter species possessing the nanAKT gene cluster encoding for sialic acid utilization.
Iron acquisition gene system
Iron is an essential micro element for bacterial growth and metabolism and a vital factor for bacterial pathogenesis.83 In Cronobacter, Franco et al.78 reported that plasmid pESA3 contain 2 clusters of genes, a homolog of an ABC transport-mediated iron uptake siderophore system (eitCBAD operon) and a siderophore-mediated iron acquisition system (iucABCD/iutA operon). This characteristic may contribute to the systemic survival of C. sakazakii and subsequent invasion of the CNS to cause diseases. It was later, Grim et al.84 who identified both the feo and efe systems for acquisition of ferrous iron. They confirmed that 98% of the plasmid-harboring Cronobacter strains have the aerobactin-like siderophore, cronobactin, for transport of ferric iron in Cronobacter. Cruz et al.77 have also revealed that C. sakazakii isolates harbour siderophore-interacting protein (sip) gene. The sip gene has a ferrodoxin-reductase domain with binding sites to FAD and NAD(P), capable of transfer an electron from reduced ferrodoxin to FAD and then convert NADP+ to NADPH.85
Efflux system
Active efflux system is a recognized virulence mechanism contributing to survival of members of Enterobacteriaceae in the host's gastrointestinal tract.86 Interestingly, ibeB (a gene synonymous with cusC) in C. sakazakii has been reported, belonging to constellate of genes encoding a copper and silver resistance cation efflux system, ultimately allowing the invasion to brain microvascular endothelial cells (BMEC) cells.23,87 When assessed by Kucerova et al.23 it was discovered that the entire cation efflux operon (cusA, cusB and cusC) and its regulatory gene cusR were present in isolates colligated with neonatal infections (including C. sakazakii ATCC 29544T, 696, 701, 767, C. malonaticus and C. turicensis) and absent in the other strains evaluated (C. sakazakii B894, ATCC 12868, 20, C. dublinensis and C. muytjensii).
Biofilm formation
Biofilms are interface-associated consortia of microorganisms embedded in an endogenous slimy matrix referred to as extracellular polysaccharides (EPS) and are well-known to contribute to survival and increased resistance to antimicrobial treatments.88–90 Two hypothetical proteins have been newly described as possible adhesins engaged in biofilm formation in Cronobacter (ESA_00281 and ESA_00282).91 Iverson et al.92 reported that Cronobacter was able to adhere to silicon, stainless steel, polycarbonate and latex with apparently greater attachment occurring with EPS producing bacteria. Colanic acid (CA) was identified as an EPS component in Cronobacter spp. contributing to adherence to various surfaces and increased resistance to environmental stresses thermal, desiccation and pH.93
Other potential factors
Among the minor but important virulent factors, the proteolytic enzymes of Cronobacter strains have been found to cause deformation of the tissue cells in mice.74 Kothary et al.94 isolated and characterized a cell-bound zinc-containing metalloprotease encoded by a nucleotide sequence (zpx), unique among all the 135 Cronobacter strains tested. The protease was active in against azocasein, caused rounding of Chinese's hamster ovarian cells. It is hypothesized that proteolytic enzymes may permit the organism to cross the blood–brain barrier or cause extensive cellular destruction in neonates with NEC.
Recently, Hamby et al.95 investigated the genomes of C.sakazakii and C.turicensis and reported that the gene for inositol monophosphatase is also associated with virulence of this pathogen.
Current studies revealed that the plasmid-encoded methyl-accepting chemotaxis protein (MCP) sequences present in C. sakazakii sequence type 8 (ST8) lineage are involved in virulence, invasion/adhesion, motility and biofilm formation.96 It was also observed that this sequence was not found in any other lineages, implying that the MCP association with virulence is probably specific to the ST8 lineage.
LysR-type transcriptional regulator (LTTRs) are known to regulate a range of regulons involved in quorum sensing and virulence of bacteria.97,98 Recently, Choi et al. (2012) characterized LysR-type transcriptional regulator (LTTR) gene (ESA_01081 homolog) as a potential regulator for C. sakazakii ATCC 29544 pathogenesis. They reported that the putative LysR-type protein plays a role in regulating genes involved in a host cell invasion, but not in adhesion.99
In another study, Cruz et al.77 in addition to sip and cpa identified putative virulence genes, including type III haemolysin (hly) in Cronobacter isolated from human and non-human sources. The type III hemolysin, a virulence factor in numerous pathogenesis, is an integral outer membrane protein with hemolytic activity.100,101
Lipopolysaccarides (LPS) is an outer membrane virulence factor of C. sakazakii, which interacts with enterocytes through LPS mediated binding to TLR4 inducing NEC in animals.102-105 In the NEC patients, the elevated level of LPS in serum and stools has been reported.106-111 Altogether, these findings raise the intriguing possibility that LPS may engage in the pathogenesis of NEC and the role of TLR4 within the intestinal epithelium seeks detailed consideration. It has been also reported that PIF is frequently contaminated with elevated levels of LPS, which disrupts tight junctions thereby increasing the permeability of the host cell membrane.112,113
The genome study of Cronobacter has revealed the presence of gene for type IV pili in addition to a P pilus homologous to other pathogens such as E.coli.28 Additionally the role of fibronectin, a glycoprotein in an extracellular matrix of Cronobacter has been postulated in the adherence to intestinal epithelial or endothelial cells.69,72,114 However, the implications from these findings in pathogenesis and virulence have not been fully understood.
Recently, the role of hfq in pathogenesis of C. sakazakii ATCC 29544 has been demonstrated by generating the mutants using lambda red recombination where the mutants indicated defects in survival and invasion within host cells and exhibited low resistance to hydrogen peroxide.115 Hfq, identified as RNA chaperone, is considered as a post-transcriptional regulator engaged in the biogenesis of quorum sensing, OMPs and various stress responses.116–118 The studies in other Gram negative pathogens i.e. Escherichia coli, Listeria monocytogenes, Salmonella typhimurium, Yersinia pseudotuberculosis, and Francisella tularensis have also expressed the importance of Hfq in the pathogenesis.119-123
Limited studies regarding the effect of Cronobacter invasion on immune response have been done. The pathogen is reported to persist within human macrophages indicating that the Cronobacter possessed virulence properties that make it allows to tolerate the intracellular environment of macrophages.124,125
Conclusions and future perspectives
Cronobacter spp is a newly classified genus and more research is yet to be completed for better understanding this unique group of organism. As a virulent species, it causes high mortalities in the neonates, therefore, it is important to understand which gene products are responsible for the pathogenicity of the bacteria and how the expression of these virulence factors is regulated. Work is therefore required to address better understanding of the progression and pathogenesis of Cronobacter spp. related diseases, particularly using in vitro cell-based assays combined with animal models.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The authors are thankful to Department of Science and Technology (DST) and Department of Biotechnology (DBT), Government of India, for providing financial support.
References
- 1.Anonymous, Milk and Milk Products – Detection of Enterobacter sakazakii Technical Specification ISO/ TS 22964 ISO/ TS 22964, 2006(E) and IDF / RM 210, 2006(E), 2006; 1st edn Geneva, International Organization for Standardization [Google Scholar]
- 2.Bar-Oz B, Preminger A, Peleg O, Block C, Arad I. Enterobacter sakazakii infection in the newborn. Acta Paediatr 2001; 90:356-358; PMID:11332182; http://dx.doi.org/ 10.1080/080352501300067857 [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) . Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee. MMWR Morb Mortal Wkly Rep 2002; 51:297-300; PMID:12002167 [PubMed] [Google Scholar]
- 4.Block C, Peleg O, Minster N, Bar-Oz B, Simhon A, Arad I, Shapiro M. Cluster of neonatal infections in Jerusalem due to unusual biochemical variant of Enterobacter sakazakii. Eur J Clin Microbiol Infect Dis 2002; 21:613-616; PMID:12226694; http://dx.doi.org/ 10.1007/s10096-002-0774-5 [DOI] [PubMed] [Google Scholar]
- 5.Mullane NR, Iversen C, Healy B, Walsh C, Whyte P, Wall PG, Quinn T, Fanning S. Enterobacter sakazakii an emerging bacterial pathogen with implications for infant health. Minerva Pediatr 2007; 59:137-148; PMID:17404564 [PubMed] [Google Scholar]
- 6.Urmenyi AMC, Franklin AW. Neonatal death from pigmented coliform infection. Lancet 1961; 1:313-315; PMID:13779326 [DOI] [PubMed] [Google Scholar]
- 7.Gurtler JB, Kornacki JL, Beuchat LR. Enterobacter sakazakii: a coliform of increased concern to infant health. Int J Food Microbiol 2005; 104:1-34; PMID:16039742; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2005.02.013 [DOI] [PubMed] [Google Scholar]
- 8.Friedemann M. Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur J Clin Microbiol Infect Dis 2009; 28:1297-1304; PMID:19662446; http://dx.doi.org/ 10.1007/s10096-009-0779-4 [DOI] [PubMed] [Google Scholar]
- 9.Yan Q, Power KA, Cooney S, Fox E, Gopinath GR, Grim CJ, Tall BD, McCusker MP, Fanning S. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Front Microbiol 2013; 4:256; PMID:24032028; http://dx.doi.org/ 10.3389/fmicb.2013.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iversen C, Forsythe SJ. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci Technol 2003; 14:443-454; http://dx.doi.org/ 10.1016/S0924-2244(03)00155-9 [DOI] [Google Scholar]
- 11.Farmer JJ III, Asbury MA, Hickman FW, Brenner DJ. The Enterobacteriaceae Study Group (USA) Enterobacter sakazakii, a new species of “Enterobacteriaceae” isolated from clinical specimens. Int J Syst Bacteriol 1980; 30: 569-584; http://dx.doi.org/ 10.1099/00207713-30-3-569 [DOI] [Google Scholar]
- 12.Joseph S, Cetinkaya E, Drahovska H, Levican A, Figueras MJ, Forsythe SJ. Cronobacter condimenti sp nov, isolated from spiced meat and Cronobacter universalis sp nov, a novel species designation for Cronobacter sp genomospecies 1, recovered from a leg infection, water, and food ingredients. Int J Syst Evol Microbiol 2012a; 62:1277-1283; PMID:22661070; http://dx.doi.org/ 10.1099/ijs.0.032292-0 [DOI] [PubMed] [Google Scholar]
- 13.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 2013; 36:309-319; PMID:23632228; http://dx.doi.org/ 10.1016/j.syapm.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 14.Oren A, Garrity GM. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2013; 63:3931-3934; PMID:25795763; http://dx.doi.org/ 10.1099/ijs.0.058222-0 [DOI] [PubMed] [Google Scholar]
- 15.Masood N, Jackson E, Moore K, Farbos A, Paszkiewicz K, Dickins B, McNally A, Forsythe S. Draft genome sequence of “Candidatus Cronobacter colletis” NCTC 14934T, a new species in the genus Cronobacter. Gen Announ 2014; 2:e00585-14; PMID:24948763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Acker J, de Smet F, Muyldermans G, Bougatef A, Naessens A, Lauwers S. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J Clin Microbiol 2001; 39:293-297; PMID:11136786; http://dx.doi.org/ 10.1128/JCM.39.1.293-297.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen AB, Braden CR. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis 2006; 12:1185-1189; PMID:16965695; http://dx.doi.org/ 10.3201/eid1208.051509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsythe SJ. Enterobacter sakazakii and other bacteria in powdered infant milk formula. J Matern Child Nutr 2005; 1:44-50; PMID:16881878; http://dx.doi.org/ 10.1111/j.1740-8709.2004.00008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M. Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 2004; 363:39-40; PMID:14723994; http://dx.doi.org/ 10.1016/S0140-6736(03)15169-0 [DOI] [PubMed] [Google Scholar]
- 20.Food and Drug Administration FDA 2002. Available from, http,//wwwfdagov/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm154581htm [Google Scholar]
- 21.Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, Stephan R, Joosten H. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 2008; 58:1442-1447; PMID:18523192; http://dx.doi.org/ 10.1099/ijs.0.65577-0 [DOI] [PubMed] [Google Scholar]
- 22.Yan QQ, Condell O, Power K, Butler F, Tall BD, Fanning S. Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium. J Appl Microbiol 2012; 113:1-15; PMID:22420458; http://dx.doi.org/ 10.1111/j.1365-2672.2012.05281.x [DOI] [PubMed] [Google Scholar]
- 23.Kucerova E, Clifton SW, Xia X-Q, Long F, Porwollik S, Fulton L, Fronick C, Minx P, Kyung K, Warren W, et al.. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 2010; 5:e9556; PMID:20221447; http://dx.doi.org/ 10.1371/journal.pone.0009556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Strain EA, Allard M, Brown EW. Genome sequence of Cronobacter sakazakii E899, a strain associated with human illness. J Bacteriol 2011; 193(20):5861; PMID:21952538; http://dx.doi.org/ 10.1128/JB.05913-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, Forsythe S. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol 2009; 9:223; PMID:19852808; http://dx.doi.org/ 10.1186/1471-2180-9-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin H, Lee JH, Choi Y, Ryu S. Complete genome sequence of the opportunistic food-borne pathogen Cronobacter sakazakii ES15. J Bacteriol 2012; 194(16):44438-9; PMID:22843579; http://dx.doi.org/ 10.1128/JB.00841-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Q, Power KA, Cooney S, Fox E, Gopinath GR, Grim CJ, Tall BD, McCusker MP, Fanning S. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Front Microbiol 2013; 2(4):256; PMID:24032028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grim CJ, Kotewicz ML, Power KA, Gopinath G, Franco AA, Jarvis KG, Yan QQ, Jackson SA, Sathyamoorthy V, Hu L, et al.. Pan-genome analysis of the emerging foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence driven by niche adaptation. BMC Genomics 2013; 14:366; PMID:23724777; http://dx.doi.org/ 10.1186/1471-2164-14-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masood N, Moore K, Farbos A, Hariri S, Block C, Paszkiewicz K, Dickins B, McNally A, Forsythe S. Draft Genome Sequence of a Meningitic Isolate of Cronobacter sakazakii Clonal Complex 4, Strain 8399. Genome Announc 2013; 1(5); PMID:24115548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z, Wang L, Wang B, Liang H, Ye Q, Zeng M. Complete Genome Sequence of Cronobacter sakazakii Strain CMCC 45402. Genome Announc 2014; 2(1); PMID:24435860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pightling AW, Pagotto F. Draft Genome Sequence of Cronobacter sakazakii Clonal Complex 45 Strain HPB5174, Isolated from a Powdered Infant Formula Facility in Ireland. Genome Announc 2014; 2(4); PMID:25103765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph S, Forsythe SJ. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front Microbiol 2012; 3:397; PMID:23189075; http://dx.doi.org/ 10.3389/fmicb.2012.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, Rico A, Brzoska P, Hamby SE, Masood N, et al.. Comparative analysis of genome sequences covering the seven cronobacter species. PLoS One 2012; 7:e49455; PMID:23166675; http://dx.doi.org/ 10.1371/journal.pone.0049455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, Forsythe SJ. Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J Clin Microbiol 2012; 50:3031-3039; PMID:22785185; http://dx.doi.org/ 10.1128/JCM.00905-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseph S, Hariri S, Forsythe SJ. Lack of continuity between Cronobacter biotypes and species as determined using multilocus sequence typing. Mol Cell Probes 2013; 27:137-139; PMID:23474194; http://dx.doi.org/ 10.1016/j.mcp.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 36.Sonbol H, Joseph S, McAuley C, Craven H, Forsythe SJ. Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int Dairy J 2013; 30:1-7; http://dx.doi.org/ 10.1016/j.idairyj.2012.11.004 [DOI] [Google Scholar]
- 37.Forsythe SJ, Dickins B, Jolley KA. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 2014; 15:1121; PMID:25515150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biering G, Karlsson S, Clark NC, Jonsdottir KE, Ludvigsson P, Steingrimsson O. Three cases of neonatal meningitis caused by Enterobacter sakazakii in powdered milk. J Clin Microbiol 1989; 27:2054-2056; PMID:2778070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H, Beuchat LR. Survival and growth of Enterobacter sakazakii on fresh-cut fruits and vegetables and in unpasteurized juice as affected by storage temperature. J Food Prot 2005; 68:2541-2552; PMID:16355824 [DOI] [PubMed] [Google Scholar]
- 40.Estuningsih S, Kress C, Hassan AA, Akineden O, Schneider E, Usleber E. Enterobacteriaceae in dehydrated powdered infant formula manufactured in Indonesia and Malaysia. J Food Prot 2006; 69:3013-3017; PMID:17186672 [DOI] [PubMed] [Google Scholar]
- 41.Friedemann M. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 2007; 116:1-10; PMID:17331606; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2006.12.018 [DOI] [PubMed] [Google Scholar]
- 42.Lin LC, Beuchat LR. Survival and growth of Enterobacter sakazakii in infant cereal as affected by composition, reconstitution liquid, and storage temperature. J Food Prot 2007; 70:1410-1422; PMID:17612071 [DOI] [PubMed] [Google Scholar]
- 43.Baumgartner A, Grand M, Liniger M, Iversen C. Detection and frequency of Cronobacter spp (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula. Int J Food Microbiol 2009; 136:189-192; PMID:19419789; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 44.Jaradat ZW, Ababneh QO, Saadoun IM, Samara NA, Rashdan AM. Isolation of Cronobacter spp (formerly Enterobacter sakazakii) from infant food, herbs and environmental samples and the subsequent identification and confirmation of the isolates using biochemical, chromogenic and molecular methods. BMC Microbiol 2009; 9:225-235; PMID:19860874; http://dx.doi.org/ 10.1186/1471-2180-9-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belal M, Al-Mariri A, Hallab L, Hamad I. Detection of Cronobacter spp (formerly Enterobacter sakazakii) from medicinal plants and spices in Syria. J Infect Dev Ctries 2013; 17:82-89; PMID:23416653 [DOI] [PubMed] [Google Scholar]
- 46.Dong X, Li C, Wu Q, Zhang J, Mo S, Guo W, Yang X, Xu X. Isolation and identification of Cronobacter (Enterobacter sakazakii) strains from food. Wei Sheng Wu Xue Bao 2013; 53:429-436; PMID:23957146 [PubMed] [Google Scholar]
- 47.Mozrová V, Břeňová N, Mrázek J, Lukešová D, Marounek M. Surveillance and characterisation of Cronobacter spp. in Czech retail food and environmental samples. Folia Microbiol (Praha) 2013; 5:2077-2091; PMID:23873391 [DOI] [PubMed] [Google Scholar]
- 48.Muller A, Stephan R, Fricker-Feer C, Lehner A. Genetic diversity of Cronobacter sakazakii isolates collected from a Swiss infant formula production facility. J Food Prot 2013; 76:883-887; PMID:23643134 [DOI] [PubMed] [Google Scholar]
- 49.Singh N, Goel G, Raghav M, Rajani CS, Puniya AK. Indian spices as modulators of quorum sensing in Cronobacter sakazakii, a food borne pathogen. EUROBIOFILMS Third European Congress on Microbial Biofilms Ghent; 2013 [Google Scholar]
- 50.Siqueira Santos RF, da Silva N, Amstalden Junqueira VC, Kajsik M, Forsythe S, Pereira JL. Screening for Cronobacter Species in Powdered and Reconstituted Infant Formulas and from Equipment Used in Formula Preparation in Maternity Hospitals. Ann Nutr Metab 2013; 63:62-68; PMID:23941974; http://dx.doi.org/ 10.1159/000353137 [DOI] [PubMed] [Google Scholar]
- 51.Gallaher PG, Ball WS. Cerebral infarcations due to CNS infection with Enterobacter sakazakii. Pediatr Radiol 1991; 21:135-136; PMID:2027718; http://dx.doi.org/ 10.1007/BF02015629 [DOI] [PubMed] [Google Scholar]
- 52.Hamilton JV, Lehane MJ, Braig HR. Isolation of Enterobacter sakazakii from midgut of Stomoxys calcitrans. Emerg Infect Dis 2003; 9:1355-1356; PMID:14626227; http://dx.doi.org/ 10.3201/eid0910.030218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raghav M, Aggarwal PK. Isolation and characterization of Enterobacter sakazakii from milk foods and environment. Milchwissenchaft 2007a; 62:266-269 [Google Scholar]
- 54.Goel G, Raghav M, Kaushik S, Puniya AK, Singh K. Tannic acid degradation by Cronobacter sakazakii isolated from Goat. 4th International Conference on Environmental, Industrial and Applied Microbiology Spain 2011 [Google Scholar]
- 55.CDC . Cronobacter species isolation in two infants – New Mexico, 2008. MMWR Morb Mortal Wkly Rep 2009; 58:1179-1183; PMID:19875980 [PubMed] [Google Scholar]
- 56.Food Safety Authority of Ireland Pregestimil infant formula recall. Available at: http://www.fsai.ie/alerts/archive/fa20050120.asp. Accessed 13February2006 [Google Scholar]
- 57.FAO/WHO Enterobacter sakazakii (Cronobacter spp.) in Powdered Follow-Up Formulae Microbiological Risk Assessment Series 15. Rome: Food and Agriculture Organization of the United Nations/World Health Organization; 2008 [Google Scholar]
- 58.Jarvis C. Fatal Enterobacter sakazakii infection associated with powdered infant formula in a neonatal intensive care unit in New Zealand. Am J Infect Control 2005; 33:e19; http://dx.doi.org/ 10.1016/j.ajic.2005.04.012 [DOI] [Google Scholar]
- 59.Himelright I, Harris E, Lorch V, Anderson M. Enterobacter sakazakii infections associated with the use of powdered infant formula – Tennessee, 2001. J Am Med Assoc 2002; 7:204-2205 [Google Scholar]
- 60.Ray P, Das A, Gautam V, Jain N, Narang A, Sharma M. Enterobacter sakazakii in infants: novel phenomenon in India. A case report in. Ind J Medical Microbio 2007; 25:408-410; PMID:18087097; http://dx.doi.org/ 10.4103/0255-0857.37351 [DOI] [PubMed] [Google Scholar]
- 61.Noriega FR, Kotloff KL, Martin MA, Schwalbe RS. Nosocomial bacteremia caused by Enterobacter sakazakii and Leuconostoc mesenteroides resulting from extrinsic contamination of infant formula. Pediatr Infect Dis J 1990; 9:447-449; PMID:2114609; http://dx.doi.org/ 10.1097/00006454-199006000-00018 [DOI] [PubMed] [Google Scholar]
- 62.Simmons BP, Gelfand MS, Haas M, Metts L, Ferguson J. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect Control Hosp Epidemiol 1989; 10:398-401; PMID:2794464; http://dx.doi.org/ 10.2307/30144207 [DOI] [PubMed] [Google Scholar]
- 63.Nazarowec-White M, Farber JM. Enterobacter sakazakii: a review. Int J Food Microbiol 1997; 34:103-111; PMID:9039558; http://dx.doi.org/ 10.1016/S0168-1605(96)01172-5 [DOI] [PubMed] [Google Scholar]
- 64.Muytjens HL, Zanen HC, Sonderkamp HJ, Kollee LA, Wachsmuth IK, Farmer JJ III. Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J Clin Microbiol 1983; 18:115-20; PMID:6885983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter CJ, Bean JF. Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J Perinatol 2013; 33:581-585; PMID:23538645; http://dx.doi.org/ 10.1038/jp.2013.26 [DOI] [PubMed] [Google Scholar]
- 66.Communicable Disease Centre CDC update: Investigation of Cronobacter infections among infants in the United States Atlanta: CDC; 2011 [Google Scholar]
- 67.Stoll BJ, Hansen N, Fanaroff AA, Lemons JA. Enterobacter sakazakii is a rare cause of neonatal septicemia or meningitis in VLBW infants. J Pediatr 2004; 144:821-823; PMID:15192634 [DOI] [PubMed] [Google Scholar]
- 68.Food and Drug Administration FDA Investigation of Cronobacter Bacteria Illness in Infants. 2012; Available from, http,//wwwfdagov/NewsEvents/PublicHealthFocus/ucm285401htm [Google Scholar]
- 69.Nair MK, Venkitanarayanan K. Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr Res 2007; 62:664-669; PMID:17957161; http://dx.doi.org/ 10.1203/PDR.0b013e3181587864 [DOI] [PubMed] [Google Scholar]
- 70.Singamsetty VK, Wang Y, Shimada H, Prasadarao NV. Outer membrane protein A expression in Enterobacter sakazakii is required to induce microtubule condensation in human brain microvascular endothelial cells for invasion. Microb Pathog 2008; 45:181-191; PMID:18606523; http://dx.doi.org/ 10.1016/j.micpath.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nair MK, Venkitanarayanan K, Silbart LK, Kim KS. Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog Dis 2009; 6:495-501; PMID:19415974; http://dx.doi.org/ 10.1089/fpd.2008.0228 [DOI] [PubMed] [Google Scholar]
- 72.Mittal R, Wang Y, Hunter CJ, Gonzalez-Gomez I, Prasadarao NV. Brain damage in newborn rat model of meningitis by Enterobacter sakazakii, a role for outer membrane protein. Lab Invest 2009; 89:263-277; PMID:19139724; http://dx.doi.org/ 10.1038/labinvest.2008.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim K, Kim KP, Choi J, Lim JA, Lee J, Hwang S, Ryu S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl Environ Microbiol 2010; 76:5188-5198; PMID:20543055; http://dx.doi.org/ 10.1128/AEM.02498-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pagotto FJ, Nazarowec-White M, Bidawid S, Farber JM. Enterobacter sakazakii, infectivity and enterotoxin production in vitro and in vivo. J Food Prot 2003; 66:370-375; PMID:12636287 [DOI] [PubMed] [Google Scholar]
- 75.Raghav M, Aggarwal PK. Purification and characterisation of Enterobacter sakazakii Enterotoxin. Can J Microbiol 2007b; 5:750-755; PMID:17668035 [DOI] [PubMed] [Google Scholar]
- 76.Franco AA, Hu L, Grim CJ, Gopinath G, Sathyamoorthy V, Jarvis KG, Lee C, Sadowski J, Kim J, Kothary MH, et al.. Characterization of putative virulence genes encoded on the related RepFIB plasmids harboured by Cronobacter spp. Appl Environ Microbiol 2011a; 77:3255-3267; PMID:21421789; http://dx.doi.org/ 10.1128/AEM.03023-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cruz A, Xicohtencatl-Cortes J, González-Pedrajo B, Bobadilla M, Eslava C, Rosas I. Virulence traits in Cronobacter species isolated from different sources. Can J Microbiol 2011; 57:735-744; PMID:21859256; http://dx.doi.org/ 10.1139/w11-063 [DOI] [PubMed] [Google Scholar]
- 78.Franco AA, Kothary MH, Gopinath G, Jarvis KG, Grim CJ, Hu L, Datta AR, McCardell BA, Tall BD. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect Immun 2011b; 79:1578-1587; PMID:21245266; http://dx.doi.org/ 10.1128/IAI.01165-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang B. Sialic acid is an essential nutrient for brain development and cognition. Ann Rev Nutr 2009; 29: 177-222; PMID:19575597; http://dx.doi.org/ 10.1146/annurev.nutr.28.061807.155515 [DOI] [PubMed] [Google Scholar]
- 80.Lewis AL, Lewis WG. Host sialoglycans and bacterial sialidases, a mucosal perspective. Cell Microbiol 2012; 14:1174-1182; PMID:22519819; http://dx.doi.org/ 10.1111/j.1462-5822.2012.01807.x [DOI] [PubMed] [Google Scholar]
- 81.Sprenger N, Duncan PI. Sialic acid utilization. Adv Nutr 2012; 3:92S-97S; PMID:22585917; http://dx.doi.org/ 10.3945/an.111.001479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joseph S, Hariri S, Masood N, Forsythe S. Sialic acid utilization by Cronobacter sakazakii. Microb Inform Exp 2013; 3(1):3; PMID:23706082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Negre VL, Bonacorsi S, Schubert S, Bidet P, Nassif X, Bingen E. The siderophore receptor iron, but not the high- pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect Immun 2004; 72:1216-1220; PMID:14742579; http://dx.doi.org/ 10.1128/IAI.72.2.1216-1220.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grim CJ, Kothary MH, Gopinath G, Jarvis KG, Beaubrun JJ, McClelland M, Tall BD, Franco AA. Identification and characterization of Cronobacter iron acquisition systems. Appl Environ Microbiol 2012; 78:6035-6050; PMID:22706064; http://dx.doi.org/ 10.1128/AEM.01457-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miethke M, Marahiel MA. Siderophores-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 2007; 71:413-451; PMID:17804665; http://dx.doi.org/ 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Touze T, Eswaran J, Bokma E, Koronakis E, Hughes C, Koronakis V. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol 2004; 53:697-706; PMID:15228545; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04158.x [DOI] [PubMed] [Google Scholar]
- 87.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system Cus CFBA of Escherichia coli. J Bacteriol 2003; 185:3804-3812; PMID:12813074; http://dx.doi.org/ 10.1128/JB.185.13.3804-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donlan RM, Costerton JW. Biofilms, survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 2002; 15:167-193; PMID:11932229; http://dx.doi.org/ 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim H, Ryu JH, Beuchat LR. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl Environ Microbiol 2006; 72:5846-5856; PMID:16957203; http://dx.doi.org/ 10.1128/AEM.00654-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beuchat LR, Kim H, Gurtler JB, Lin LC, Ryu JH, Richards GM. Cronobacter sakazakii in foods and factors affecting its survival, growth, and inactivation. Int J Food Microbiol 2009; 136: 204-213; PMID:19346021; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2009.02.029 [DOI] [PubMed] [Google Scholar]
- 91.Hartmann I, Carrranza P, Lehner A, Stephan R, Eberl L, Riedel K. Genes involved in Cronobacter sakazakii biofilm formation. Appl Environ Microbiol 2010; 76:2251-2261; PMID:20118366; http://dx.doi.org/ 10.1128/AEM.00930-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iversen C, Lane M, Forsythe SJ. The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula. Lett Appl Microbiol 2004; 38:378-382; PMID:15059207; http://dx.doi.org/ 10.1111/j.1472-765X.2004.01507.x [DOI] [PubMed] [Google Scholar]
- 93.Scheepe-Leberkuhne Wagner F. Optimization and preliminary characterization of an exopolysaccharide synthesized by Enterobacter sakazakii. Biotechnol Lett 1986; 8:695-700; http://dx.doi.org/ 10.1007/BF01032564 [DOI] [Google Scholar]
- 94.Kothary MH, McCardell BA, Frazar CD, Deer D, Tali BD. Characterization of the zinc-containing metalloprotaese encoded by zpx and development of a species-specific detection method for Enterobacter sakazakii. Appl Environ Microbiol 2007; 73:4142-4151; PMID:17483271; http://dx.doi.org/ 10.1128/AEM.02729-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamby SE, Joseph S, Forsythe SJ, Chuzhanova N. In Silico identification of pathogenic strains of Cronobacter from biochemical reveals association of inositol fermentation with pathogenicity. BMC Microbiol 2011; 11:204; PMID:21933417; http://dx.doi.org/ 10.1186/1471-2180-11-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi Y, Kim S, Hwang H, Kim KP, Kang DH, Ryu S. Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect Immun 2015; 83:197-204; PMID:25332122; http://dx.doi.org/ 10.1128/IAI.02633-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology (SGM) 2008; 154: 3609-3623; PMID:19047729; http://dx.doi.org/ 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- 98.MacLean AM, Anstey MI, Finan TM. Binding site determinants for the LysR-type transcriptional regulator PcaQ in the legume endosymbiont Sinorhizobium meliloti. J Bacteriol 2008; 190:1237-1246; PMID:18055594; http://dx.doi.org/ 10.1128/JB.01456-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi Y, Kim KP, Kim K, Choi J, Shin H, Kang DH, Ryu S. Possible roles of LysR-type transcriptional regulator (LTTR) homolog as a global regulator in Cronobacter sakazakii ATCC 29544. Int J Med Microbiol 2012; 302:270-275; PMID:22770741; http://dx.doi.org/ 10.1016/j.ijmm.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 100.Baida GE, Kuzmin NP. Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophys Acta 1996; 1284:122-124; PMID:8962879; http://dx.doi.org/ 10.1016/S0005-2736(96)00168-X [DOI] [PubMed] [Google Scholar]
- 101.Chen YC, Chang MC, Chuang YC, Jeang CL. Characterization and virulence of hemolysin III from Vibrio vulnifucus. Curr Microbiol 2004; 49:175-179; PMID:15386100; http://dx.doi.org/ 10.1007/s00284-004-4288-5 [DOI] [PubMed] [Google Scholar]
- 102.Hunter CJ, Singasmetty VK, Chokshi NK, Boyle P, Camerini V, Grishin AV, Upperman JS, Ford HR, Prasadrao VN. Enterobacter sakazakii enhances epithelial cell injury by inducing apoptosis in a rat model of necrotizing enterocolitis. J Infect Dis 2008; 198:586-593; PMID:18588483; http://dx.doi.org/ 10.1086/590186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hotta T, Yoshida N, Yoshikawa T, Sugino S, Kondo M. Lipopolysaccharide induced colitis in rabbits. Res Exp Med (Berl) 1986; 186:61-69; PMID:3961278; http://dx.doi.org/ 10.1007/BF01851834 [DOI] [PubMed] [Google Scholar]
- 104.Feng J, El-Assal ON, Besner GE. Heparin-binding EGF-like growth factor (HB-EGF) and necrotizing enterocolitis. Semin Pediatr Surg 2005; 14:167-174; PMID:16084404; http://dx.doi.org/ 10.1053/j.sempedsurg.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 105.Feng J, Besner GE. Heparin-binding epidermal growth factor-like growth factor promotes enterocyte migration and proliferation in neonatal rats with necrotizing enterocolitis. J Pediatr Surg 2007; 42:214-220; PMID:17208569; http://dx.doi.org/ 10.1016/j.jpedsurg.2006.09.055 [DOI] [PubMed] [Google Scholar]
- 106.Kruis W, Schussler P, Weinzierl M, Galanos C, Eisenburg J. Circulating lipid A antibodies despite absence of systemic endotoxemia in patients with Crohn's disease. Dig Dis Sci 1984; 29:502-507; PMID:6144475; http://dx.doi.org/ 10.1007/BF01296269 [DOI] [PubMed] [Google Scholar]
- 107.Caradonna L, Amati L, Lella P, Jirillo E, Caccavo D. Phagocytosis, killing, lymphocyte-mediated antibacterial activity, serum autoantibodies, and plasma endotoxins in inflammatory bowel disease. Am J Gastroenterol 2000; 95:1495-1502; PMID:10894586; http://dx.doi.org/ 10.1111/j.1572-0241.2000.02085.x [DOI] [PubMed] [Google Scholar]
- 108.Noerr B. Current controversies in the understanding of necrotizing enterocolitis. Adv Neonatal Care 2003; 3:107-120; PMID:12891835; http://dx.doi.org/ 10.1016/S1536-0903(03)00072-9 [DOI] [PubMed] [Google Scholar]
- 109.Sharma R, Tepas JJ III, Hudak ML, Mollitt DL, Wludyka PS, Teng RJ, Premachandra BR. Neonatal gut barrier and multiple organ failure: role of endotoxin and proinflammatory cytokines in sepsis and necrotizing enterocolitis. J Pediatr Surg 2007; 42:454-461; PMID:17336180; http://dx.doi.org/ 10.1016/j.jpedsurg.2006.10.038 [DOI] [PubMed] [Google Scholar]
- 110.Duffy LC, Zielezny MA, Carrion V, Griffiths E, Dryja D, Hilty M, Rook C, Morin F III. Concordance of bacterial cultures with endotoxin and interleukin-6 in necrotizing enterocolitis. Dig Dis Sci 1997; 42:359-365; PMID:9052520; http://dx.doi.org/ 10.1023/A:1018826204819 [DOI] [PubMed] [Google Scholar]
- 111.Leaphart CL, Cavallo JC, Gribar SC, Cetin S, Li J, Branca MF, Dubowski TD, Sodhi CP, Hackam DJ. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J Immunol 2007; 179:4808-4820; PMID:17878380; http://dx.doi.org/ 10.4049/jimmunol.179.7.4808 [DOI] [PubMed] [Google Scholar]
- 112.Townsend S, Barron JC, Loc-Carrillo C, Forsythe S. The presence of endotoxin in powdered infant formula milk and the influence of endotoxin and Enterobacter sakazakii on bacterial translocation in the infant rat. Food Microbiol 2007; 24:67-74; PMID:16943096; http://dx.doi.org/ 10.1016/j.fm.2006.03.009 [DOI] [PubMed] [Google Scholar]
- 113.Moriez R, Salvador-Cartier C, Theodorou V, Fioramonti J, Eutamene H, Bueno L. Myosin light chain kinase is involved in lipopolysaccharide-induced disruption of colonic epithelial barrier and bacterial translocation in rats. Am J Pathol 2005; 267:1071-1079; PMID:16192642; http://dx.doi.org/ 10.1016/S0002-9440(10)61196-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mange JP, Stephan R, Borel N, Wild P, Kim KS, Pospischil A, Lehner A. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol 2006; 6:58; PMID:16800879; http://dx.doi.org/ 10.1186/1471-2180-6-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim S, Hwang H, Kim KP, Yoon H, Kang DH, Ryu S. The hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect Immun 2015; 8:2089-98; PMID:25754196; http://dx.doi.org/ 10.1128/IAI.03161-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev 2006; 20:2338-2348; PMID:16951250; http://dx.doi.org/ 10.1101/gad.1457506 [DOI] [PubMed] [Google Scholar]
- 117.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 2004; 118:69-82; PMID:15242645; http://dx.doi.org/ 10.1016/j.cell.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 118.Repoila F, Majdalani N, Gottesman S. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 2003; 48:855-861; PMID:12753181; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03454.x [DOI] [PubMed] [Google Scholar]
- 119.Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol 2004; 186:3355-3362; PMID:15150220; http://dx.doi.org/ 10.1128/JB.186.11.3355-3362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schiano CA, Bellows LE, Lathem WW. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect Immun 2010; 78:2034-2044; PMID:20231416; http://dx.doi.org/ 10.1128/IAI.01046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, Forsberg A, Charbit A. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun 2009; 77:1866-1880; PMID:19223477; http://dx.doi.org/ 10.1128/IAI.01496-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 2007; 63:193-217; PMID:17163975; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect Immun 2008; 76:3019-3026; PMID:18458066; http://dx.doi.org/ 10.1128/IAI.00022-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Townsend SM, Hurrell E, Gonzalez-Gomez I, Lowe J, Frye JG, Forsythe S, Badger JL. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 2007b; 153:3538-3547; PMID:17906151; http://dx.doi.org/ 10.1099/mic.0.2007/009316-0 [DOI] [PubMed] [Google Scholar]
- 125.Emami CN, Mittal R, Wang L, Ford HR, Prasadarao NV. Role of neutrophils and macrophages in the pathogenesis of necrotizing enterocolitis caused by Cronobacter sakazakii. J Surg Res 2012; 172:18-28; PMID:21601887; http://dx.doi.org/ 10.1016/j.jss.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]