Abstract
Yeasts are widely used for the production of heterologous proteins. Improving the expression of such proteins is a top priority for pharmaceutical and industrial applications. N-Glycosylation, a common form of protein modification in yeasts, facilitates proper protein folding and secretion. Accordingly, our previous study revealed that the attachment of additional N-glycans to recombinant elastase by introducing an N-glycosylation sequon at suitable locations could stimulate its expression. Interestingly, the sequon Asn-Xaa-Thr is N-glycosylated more efficiently than Asn-Xaa-Ser, so improving the N-glycosylation efficiency via the conversion of Ser to Thr in the sequon would enhance the efficiency of N-glycosylation and increase glycoprotein expression. Recently, the expression level of recombinant elastase was enhanced by this means in our lab. Actually, the modification of N-glycosylation sites can generally be achieved through site-directed mutagenesis; thus, the method described in this report represents a feasible means of improving heterologous protein expression in yeasts.
Keywords: heterologous protein, N-glycosylation site, protein folding, protein secretion, yeast
Introduction
Yeast species have been widely utilized as an ideal platform for the production of recombinant proteins on an industrial scale.1 Such systems are becoming increasingly popular due to the many advantages they offer, including simple molecular genetic manipulation, high production levels of foreign proteins, elimination of endotoxin and bacteriophage contamination, effective posttranslational modification, high cell densities, and simple purification of secreted target proteins.1,2 Although yeasts are excellent hosts for producing heterologous proteins, the levels of secretion for many recombinant secretory proteins are so low that the applicability of such systems to industrial applications is limited; thus, improving the expression of heterologous proteins remains a top priority.1-3
Recent attempts to enhance target protein expression in yeasts have largely focused on increasing the gene copy number and mRNA content, optimizing the fermentation process, or selecting stronger promoters and more effective secretion signals.3,4 On the other hand, studies have shown that the folding and processing of a nascent polypeptide chain in the endoplasmic reticulum (ER) greatly affect protein expression.5
N-Glycosylation is one of the most common forms of protein modification occurring across all kingdoms of life,6 in which, an oligosaccharide was attached to a suitable Asn residue in the sequon of Asn-Xaa-Thr/Ser (Xaa is any amino acid except proline, Thr is threonine, and Ser is serine). Furthermore, the N-glycosylation efficiency of the sequon Asn-Xaa-Thr exceeds that of Asn-Xaa-Ser. It was widely reported that N-glycosylation is involved in protein folding and secretion6; thus, it was suggested that modifying the N-glycosylation sites in target proteins would be an effective way to enhance their production in yeast cells.
The expression level of the recombinant elastase (rPAE) was enhanced by this means in our labs. In our recent work, the addition of an N-glycosylation site to the propeptide at N51 or N93 stimulated rPAE production by 104 or 57% in P. pastoris, respectively.7 On other hand, the substitution of Thr for Ser in the N-glycosylation sites at N212 and N280 enhanced the N-glycosylation of the protein, and also successfully increased its production by 43 and 25%, respectively.8
Here, we sum up the strategy of enhancing expression of heterologous proteins in yeast cells via the modification of N-glycosylation sites.
Quality control of protein folding
Generally, the folding of secretory proteins in the ER is one of the main rate-limiting steps in their expression.4,5 The folding of a nascent protein is subject to a strict quality control system in the ER. The accumulation of misfolded proteins in the ER is harmful to cells due to a self-defense mechanism known as the unfolded protein response (UPR). Proteins that are unlikely or unable to fold are distinguished from the far more abundant sea of newly synthesized proteins that are in the process of folding.5,9 In most cases, misfolded polypeptides are subjected to ER-associated degradation (ERAD) by ubiquitination via ER-associated ubiquitin-conjugating enzymes, and finally degraded in the cytoplasm by the proteasome (Fig. 1a). Generally, the proper folding of a secretory protein is a prerequisite for its export from the ER and subsequent transport to the Golgi apparatus for further processing.5,9
Figure 1.
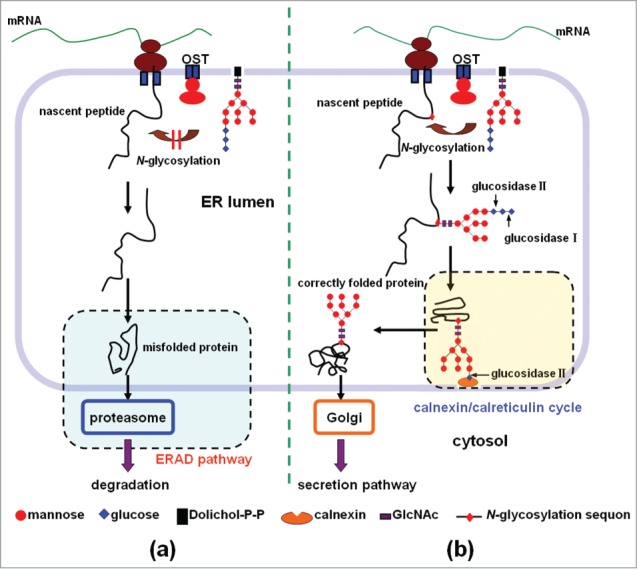
The role of N-glycosylation in the process of protein folding in the ER and secretion. (a) Lack of the N-glycosylation of the nascent peptide often leads to the protein misfolding. The misfolded protein will be subjected to ER-associated degradation (ERAD), and finally degraded in the cytoplasm by the proteasome. (b) N-glycosylation of the nascent peptide at the proper sites facilitates the protein folding and secretion. In the introduced sequon of Asn-Xaa-Thr/Ser (Xaa is any amino acid except proline, Thr is threonine, and Ser is serine), a lipid-linked oligosaccharide unit (Glc3Man9GlcNAc2) is transferred co-translationally to an Asn residue, and the oligosaccharide unit is then trimmed to Glc1Man9GlcNAc2 by glucosidase I and glucosidase II. The resulting glycoprotein enters the calnexin/calreticulin cycle to become properly folded, and exit the ER, and then go to the Golgi apparatus for secretion.
N-Glycosylation is critical for protein folding and secretion
Glycosylation is one of the most common forms of protein modification, occurring across all kingdoms of life.6,10 Both N- and O-linked glycosylation has been reported in yeasts, and the N-linked glycosylation pathway in the ER is conserved between yeasts and most higher eukaryotes.2–4,11 Most N-glycosylation sites are found in putative loop and turn domains of proteins, but a significant number of N-glycosylation sites are present in sites important for the secondary structure of proteins (e.g., β-sheets); approximately, 70–90% of the asparagine (Asn, N) residues in potential N-glycosylation sites are N-glycosylated.6
N-Glycosylation, which is involved in the process of protein folding in the ER, plays an important role in the production of heterologous secretory proteins requiring N-glycosylation for proper folding.11 The folding cycle for mono-glucosylated glycoproteins in eukaryotes is well established (Fig. 1b). Correctly folded glycoproteins can exit the ER and go to the Golgi apparatus for secretion (Fig. 1b); in comparison, misfolded proteins are mostly recognized by chaperones on the basis of their exposed hydrophobic patches, and if the proteins cannot be refolded to their native state, they are targeted to proteolytic pathways (Fig. 1a).10–12
Because N-linked oligosaccharides are large and hydrophilic (e.g., yeasts can produce high mannose-type glycan structures such as Man9GlcNAc2 and have a tendency to hypermannosylate saccharides), their attachment to glycoproteins has the potential to strongly influence the final structure and folding kinetics.13 Many studies have shown that N-linked glycans can affect or facilitate protein folding in the ER, while their removal from proteins do not affect the biological function, similar to protein chaperones.14 These observations suggest that N-linked glycans possess ‘chaperone-like’ activity.
Accordingly, in most cases, the deletion of N-glycosylation sites causes a significant decrease in glycoprotein production.
The role of 2 potential N-glycosylation sites (N134 and N229) in the secretion of Schwanniomyces occidentalis SWA2 α-amylase expressed in Saccharomyces cerevisiae was analyzed by site-directed mutagenesis by Yanez et al.15 in 1998. The Asn residues in the 2 sites were replaced by alanine (Ala, A) or glycine (G) individually or in combination. Any substitution at N229 caused a drastic decrease in expression, whereas the substitution of N134 had a trivial effect on expression.
In 2006, Turner et al.16 reported the deletion of the N-glycosylation sites in wild-type PLB1 and its glycosylphosphatidylinositol (GPI) anchorless version (PLB1GPI−) expressed in S. cerevisiae by the conversion of Asn to Ala by site-directed mutagenesis. They found that 2 mutations, N56A and N550A, inhibited protein secretion completely, while N430A reduced the level of secretion by 60%. These results demonstrate that the N-glycosylation sites at N56, N430, and N550 facilitated protein expression.
Ovalbumin (OVA), a major protein in hen egg white, contains 2 potential N-glycosylation sites at N292 and N311. The mutation of these N-glycosylation sites (Asn to glutamine [Q]: N292Q and N292/311Q) followed by expression in Pichia pastoris revealed that the secretion and expression of the proteins were greatly reduced compared with wild-type OVA.17 Furthermore, they found that the N-glycan at N292 of OVA was essential for its secretion and folding in P. pastoris cells.
The two N-glycosylation sites at N10 and N236 in ricin A chain were mutated by the substitution of Gln for Asn in the N-glycosylation sequons by Yan et al. in 2012.18 Their results indicate that N-glycosylation did not affect the catalytic activity of the protein but that it promoted the transport of the protein out of the ER in S. cerevisiae.
In 2014, Han et al.19 investigated the role of N-glycosylation sites in recombinant Pseudomonas aeruginosa elastase (rPAE) expressed in P. pastoris by converting the Asn residues to Gln at 3 N-glycosylation sites (N43, N212, and N280). The mutation of any site was detrimental to the expression of the protein. The observed decreases in expression were 23.9% for the N43Q mutant, 63.6% for the N212Q mutant, and 63.7% for the N280Q mutant compared with wild type. Furthermore, the combined mutation of these sites resulted in an additional decrease in rPAE expression.
Introducing N-glycosylation sites into proteins affects their expression
The deletion of N-glycosylation sites mostly inhibits protein expression in yeast cells, whereas the introduction of N-glycosylation sites to proteins could greatly affect, and possibly enhance, their expression. Although this could be an effective approach for improving the production of target proteins in yeasts, few related reports exist.
The insertion of a hydrophobic pentapeptide (Phe-Phe-Val-Ala-Pro) into the C-terminus of hen egg white lysozyme by genetic modification resulted in little secretion in a yeast expression system, even though this modification is useful to enhance bactericidal action against Gram-negative bacteria. In 1998, Arima et al.20 attempted to introduce N-glycosylation sites into lysozyme via a G49N mutation in order to enhance the secretion of the fused lysozyme in S. cerevisiae. According to their results, the enzyme containing the N-glycosylation sequon was expressed in the medium at 3.4 times the level of the unglycosylated version.
The effect of adding N-glycosylation sites to cutinase on its production in S. cerevisiae was studied by Sagt et al.21 in 2000. When an N-glycosylation site was introduced to the N-terminal region of cutinase, enzyme secretion increased 5 fold. If an N-glycosylation site was added to the C-terminal region, however, secretion increased 1.8 fold.
In 2014, Han et al.7 studied the effect of introducing N-glycosylation sites to the propeptide of rPAE on its expression level in P. pastoris. The addition of an N-glycosylation site to the propeptide at N51 or N93 enhanced rPAE production by 104 or 57%, respectively, while the same addition at N11 or N127 led to a 25 or 50% decrease, respectively.
Enhancing the role of N-glycosylation sites in protein expression by the conversion of Asn-Xaa-Ser to Asn-Xaa-Thr
Oligosaccharyltransferase, the central enzyme in the N-glycosylation pathway in the ER, catalyzes the transfer of an oligosaccharide to the amide group of select Asn residues, showing a preference for Asn-Xaa-Thr over Asn-Xaa-Ser. The N-glycosylation efficiency of the sequon Asn-Xaa-Thr exceeds that of Asn-Xaa-Ser; in fact, the former is reported to be glycosylated 2 to 3 times more often than the latter.10,22 The attachment of oligosaccharides to glycoproteins greatly affects their folding and secretion, suggesting that the enhanced N-glycosylation efficiency resulting from the substitution of Thr for Ser in the N-glycosylation sequon would significantly affect protein production.
An example of this was reported by Kasturi et al. in 1995.23 A variant of rabies virus glycoprotein containing a single Asn-Xaa-Ser sequon at N37 was changed by site-directed mutagenesis to the sequon Asn-Xaa-Thr. The substitution increased the core glycosylation efficiency of the protein at N37 and dramatically increased its expression.
Recently, Han et al.8 studied the effect of substituting Thr for Ser in the N-glycosylation sites (N36, N43, N212, N264, and N280) of rPAE on expression of the protein in P. pastoris. As expected, substitution at N36, N43, N212, and N280 enhanced the N-glycosylation of the protein. On the other hand, the influence of changing Asn-Xaa-Ser to Asn-Xaa-Thr on rPAE secretion was site-specific: conversion in the N-glycosylation sequon at N212 and N280 successfully increased rPAE production by 43 and 25%, respectively, while that at N36 and N43 led to a decrease in its production by 31 and 9%, respectively. In comparison, conversion in the sequon at N264 had only a trivial effect on protein production. The same study proposed that the increase in N-glycosylation efficiency caused by the substitution of Thr for Ser in the N-glycosylation sequon would intensify the stimulatory or inhibitory effects of N-glycosylation on glycoprotein expression.
Prospects and Summary
Yeast species have been used extensively to express many industrial and pharmaceutical recombinant proteins; however, in most cases increasing their production levels remains a major challenge. N-Glycosylation, a common form of protein modification in yeasts, is of key importance for proper protein folding and secretion. In this study, we found that introducing N-glycosylation sequons to proteins at appropriate sites could increase their expression in yeast cells. Also, enhancement of the N-glycosylation efficiency by the conversion of Asn-Xaa-Ser to Asn-Xaa-Thr intensified the stimulatory effects of N-glycosylation on glycoprotein expression.
It is important to note that not all N-glycosylation sequons in polypeptides are glycosylated in vivo; thus, the N-glycosylation efficiency should be evaluated before a new N-glycosylation sequon is introduced to a protein at the target site. Luckily, several web-based servers for predicting N-glycosylation sites are available.24-27 This will simplify the process of estimating the N-glycosylation efficiency for a given site in a protein. Furthermore, the effect of N-glycosylation on protein folding and secretion was site-specific: the attachment of N-glycan to a given protein at different sites exerted different effects on protein expression.7,8,15,16,19 Determining the correlation between the position of an individual N-glycosylation site and its effect on protein folding and secretion is of great value, though it is also a challenging task. Even so, modifying N-glycosylation sites to stimulate protein expression is a feasible strategy for increasing protein expression that can be conveniently implemented by site-directed mutagenesis with ease and efficiency in a laboratory setting.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Celik E, Calik P. Production of recombinant proteins by yeast cells. Biotechnol Adv 2012; 30:1108-18; PMID:21964262; http://dx.doi.org/ 10.1016/j.biotechadv.2011.09.011 [DOI] [PubMed] [Google Scholar]
- 2. Martinez JL, Liu L, Petranovic D, Nielsen J, Martinez JL. Pharmaceutical protein production by yeast: towards production of human blood proteins by microbial fermentation. Curr Opin Biotechnol 2012; 23:965-71; PMID:22503236; http://dx.doi.org/ 10.1016/j.copbio.2012.03.011 [DOI] [PubMed] [Google Scholar]
- 3. Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D. Recombinant protein production in yeasts. Methods Mol Biol 2012; 824:329-58; PMID:22160907; http://dx.doi.org/ 10.1007/978-1-61779-433-9_17 [DOI] [PubMed] [Google Scholar]
- 4. Damasceno LM, Huang CJ, Batt CA. Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol 2012; 93:31-9; PMID:22057543; http://dx.doi.org/ 10.1007/s00253-011-3654-z [DOI] [PubMed] [Google Scholar]
- 5. Roth J, Zuber C, Park S, Jang I, Lee Y, Kysela KG, Le Fourn V, Santimaria R, Guhl B, Cho JW. Protein N-glycosylation, protein folding, and protein quality control. Mol Cells 2010; 30:497-506; PMID:21340671; http://dx.doi.org/ 10.1007/s10059-010-0159-z [DOI] [PubMed] [Google Scholar]
- 6. Skropeta D. The effect of individual N-glycans on enzyme activity. Bioorg Med Chem 2009; 17:2645-53; PMID:19285412; http://dx.doi.org/ 10.1016/j.bmc.2009.02.037 [DOI] [PubMed] [Google Scholar]
- 7. Han M, Wang W, Jiang G, Wang X, Liu X, Cao H, Tan Y, Yu X. Enhanced expression of recombinant elastase in Pichia pastoris through addition of N-glycosylation sites to the propeptide. Biotechnol Lett 2014; 36:2467-71; PMID:25048243; http://dx.doi.org/ 10.1007/s10529-014-1620-4 [DOI] [PubMed] [Google Scholar]
- 8. Han M, Wang W, Wang X, Liu X, Cao H, Tao Y, et al. Enhanced expression of recombinant elastase in Pichia pastoris through the substitution of Thr for Ser in Asn-Xaa-Ser sequons. Appl Biochem Biotechnol 2015; 175:428-35; PMID:25308616; http://dx.doi.org/ 10.1007/s12010-014-1284-5 [DOI] [PubMed] [Google Scholar]
- 9. Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol 2010; 22:437-46; PMID:20570125; http://dx.doi.org/ 10.1016/j.ceb.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta 2013; 1833:2430-7; PMID:23583305; http://dx.doi.org/ 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 11. Yu P, Zhu Q, Chen K, Lv X. Improving the secretory production of the heterologous protein in Pichia pastoris by focusing on protein folding. Appl Biochem Biotechnol 2015; 175:535-48; PMID:25326186; http://dx.doi.org/ 10.1007/s12010-014-1292-5 [DOI] [PubMed] [Google Scholar]
- 12. Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta 2014; 1843:182-96; PMID:23850760; http://dx.doi.org/ 10.1016/j.bbamcr.2013.06.031 [DOI] [PubMed] [Google Scholar]
- 13. Imperiali B, O'Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol 1999; 3:643-49; PMID:10600722 [DOI] [PubMed] [Google Scholar]
- 14. Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci 2006; 31:156-63; PMID:16473013; http://dx.doi.org/ 10.1016/j.tibs.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 15. Yanez E, Carmona TA, Tiemblo M, Jimenez A, Fernandez-Lobato M. Expression of the Schwanniomyces occidentalis SWA2 amylase in Saccharomyces cerevisiae: role of N-glycosylation on activity, stability and secretion. Biochem J 1998; 329:65-71; PMID:9405276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turner KM, Wright LC, Sorrell TC, Djordjevic JT. N-linked glycosylation sites affect secretion of cryptococcal phospholipase B1, irrespective of glycosylphosphatidylinositol anchoring. Biochim Biophys Acta 2006; 1760:1569-79; PMID:16919392; http://dx.doi.org/ 10.1016/j.bbagen.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 17. Ito K, Ishimaru T, Kimura F, Matsudomi N. Importance of N-glycosylation positioning for secretion and folding of ovalbumin. Biochem Biophys Res Commun 2007; 361:725-31; PMID:17678626; http://dx.doi.org/ 10.1016/j.bbrc.2007.07.066 [DOI] [PubMed] [Google Scholar]
- 18. Yan Q, Li XP, Tumer NE. N-glycosylation does not affect the catalytic activity of ricin a chain but stimulates cytotoxicity by promoting its transport out of the endoplasmic reticulum. Traffic 2012; 13:1508-21; PMID:22882900; http://dx.doi.org/ 10.1111/j.1600-0854.2012.01404.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han M, Wang X, Ding H, Jin M, Yu L, Wang J, Yu X. The role of N-glycosylation sites in the activity, stability, and expression of the recombinant elastase expressed by Pichia pastoris. Enzyme Microb Technol 2014; 54:32-7; PMID:24267565; http://dx.doi.org/ 10.1016/j.enzmictec.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 20. Arima H, Kinoshita T, Ibrahim HR, Azakami H, Kato A. Enhanced secretion of hydrophobic peptide fused lysozyme by the introduction of N-glycosylation signal and the disruption of calnexin gene in Saccharomyces cerevisiae. FEBS Lett 1998; 440:89-92; PMID:9862432 [DOI] [PubMed] [Google Scholar]
- 21. Sagt CM, Kleizen B, Verwaal R, de Jong MD, Muller WH, Smits A, Visser C, Boonstra J, Verkleij AJ, Verrips CT. Introduction of an N-glycosylation site increases secretion of heterologous proteins in yeasts. Appl Environ Microbiol 2000; 66:4940-4; PMID:11055947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones J, Krag SS, Betenbaugh MJ. Controlling N-linked glycan site occupancy. Biochim Biophys Acta 2005; 1726:121-37; PMID:16126345; http://dx.doi.org/ 10.1016/j.bbagen.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 23. Kasturi L, Eshleman JR, Wunner WH, Shakin-Eshleman SH. The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J Biol Chem 1995; 270:14756-61; PMID:7782341 [DOI] [PubMed] [Google Scholar]
- 24. Lam PV, Goldman R, Karagiannis K, Narsule T, Simonyan V, Soika V, et al. Structure-based comparative analysis and prediction of N-linked glycosylation sites in evolutionarily distant eukaryotes. Genomics Proteomics Bioinformatics 2013; 11:96-104; PMID:23459159; http://dx.doi.org/ 10.1016/j.gpb.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamby SE, Hirst JD. Prediction of glycosylation sites using random forests. BMC Bioinformatics 2008; 9:500; PMID:19038042; http://dx.doi.org/ 10.1186/1471-2105-9-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chuang GY, Boyington JC, Joyce MG, Zhu J, Nabel GJ, Kwong PD, et al. Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics 2012; 28:2249-55; PMID:22782545; http://dx.doi.org/ 10.1093/bioinfor-matics/bts426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004; 4:1633-49; PMID:15174133; http://dx.doi.org/ 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]


