ABSTRACT
Yeast [PSI+] prion is one of the most suitable and well characterized system for the investigation of the prion phenomenon. However, until recently, the lack of data on the 3D arrangement of Sup35p prion fibrils hindered progress in this area. The recent arrival in this field of new experimental techniques led to the parallel and in-register superpleated β-structure as a consensus model for Sup35p fibrils. Here, we analyzed the effect of amino acid substitutions of the Sup35 protein through the prism of this structural model. Application of a newly developed computational approach, called ArchCandy, gives us a better understanding of the effect caused by mutations on the fibril forming potential of Sup35 protein. This bioinformatics tool can be used for the design of new mutations with desired modification of prion properties. Thus, we provide examples of how today, having progress toward elucidation of the structural arrangement of Sup35p fibrils, researchers can advance more efficiently to a better understanding of prion [PSI+] stability and propagation.
Keywords: amyloid, prion, protein misfolding, protein structure, Saccharomyces cerevisiae, superpleated β-structure, [PSI+]
Abbreviations
- Asu mutations
antisupressor mutations
- EM
electron microscopy
- NMR
nuclear magnetic resonance
- PNM
[PSI+] no more
- STEM
scanning transmission electron microscopy
1. Structural arrangement of Sup35p FIBRILS
The [PSI+] prion is a self-propagating amyloid of the release factor, Sup35p, of Saccharomyces cerevisiae. Sup35p aggregated into amyloid fibrils poorly performs its native function that leads to a decrease of translation termination efficiency and consequently to a nonsense suppression phenotype of [PSI+] yeast cells (for a review see ref. 1). Several [PSI+] variants of the same Sup35 protein with different strength of phenotype, mitotic stability, size of aggregates and morphology of the fibrils are described.2–4 The structure of the Sup35p fibrils affects the properties of [PSI+] variants.4,5
The Sup35p consists of 3 distinct domains (Fig. 1A). The amino-terminal domain (N-domain) is nonessential for viability or translation termination 6,7 but critical for prion propagation.8 This region is uniformly rich in Gln and Asn residues and contains tandem repeats.9 The carboxy-terminal domain (C-domain), with a globular structure 10, is required and sufficient for translation termination and cell viability.6,7 The middle domain (M-domain) that connects the N- and C-domains also has some influence on [PSI+] propagation.11 The N-domain alone or together with the M-domain, and also the full-length Sup35p, assemble into self-propagating amyloid fibrils.12,13 Thus, the understanding of the [PSI+] prion properties primarily requires a better appreciation of the fibril structures formed by the N-domain.
Figure 1.
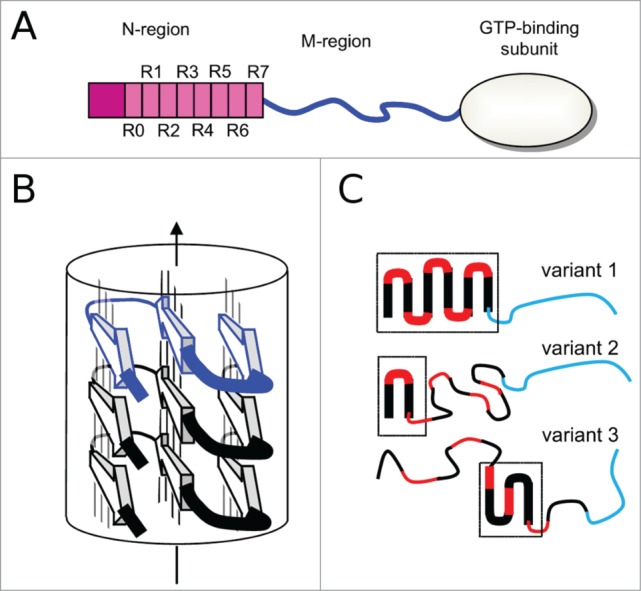
(A) Schematic representation of Sup35p domain organization: the N-terminal region (1-123 amino acids) is shown with 8 repeats (R0-R7); the naturally unfolded M-region (124-245 amino acids) is in blue; C-terminal domain (246-685 amino acids) has a globular fold. (B) A model of parallel and in-register superpleated β-structure, with H-bonded β-strand denoted as arrows linked by thin lines. As an example, a serpentine with 3 β-strands is shown. (C) Putative serpentines of Sup35p fibrils corresponding to different prion variants. A region with fibril-forming potential is shown by red and black lines, while an unfolded linker that is not able to form fibrils is in blue. Note that number of black-red units does not obligatory correspond to the number of repeats in the sequence of N-domain. Parts of Sup35p variants that are involved in the fibril structure are shown by thick lines and outlined by frames. Variant 1 has a larger region of the serpentine than variant 2, however both variants have one common structural element, but variant 3 has both different positions and the different shape of serpentine fold. It is important to mention that once formed the serpentines of each prion variant are axially stacked in the same conformation.
In recent years, there have been major advances in the study of the structural arrangement of the Sup35p prion fibrils. By negative staining, cryo-EM and scanning transmission EM (STEM), fibrils of full-length Sup35p show a ˜8-nm-wide backbone surrounded by a 65nm-wide cloud of globular C-domains connected to the fibril by the extended M-region.14 In accordance with X-ray and electron diffraction studies the fibrils have cross-β structure.5,15,16 Scanning transmission electron microscopy mass measurements for Sup35p N-fibrils, NM-fibrils and full-length Sup35p fibrils showed that filaments have one subunit per 0.47 nm.14 A number of solid-state NMR measurements of Sup35p fibrils revealed that they contain β-strands in a parallel, in-register arrangement regardless of which appendage is attached to the prion domain.17–20
It is still unclear which part(s) of the Sup35p N-domain is critical for fibril formation. Mutational analysis of the Sup35 protein (substitutions with proline, insertions of glycine, or deletions of large fragments) has shown that only the first half of the N-domain (up to ∼60 residues) may be required for the [PSI+] phenotype and fibril-formation.3,21–26 Solid state NMR studies detected the structured core within the first 30 residues.19 The other solid-state NMR data suggest that almost the entire N-domain and even a part of the M-domain have an in-register parallel β-structure.17,20 The other studies suggested that in addition to this region, a downstream region (amino acids 110–128) can also be implicated in amyloid formation.25 The discrepancy between the experimental data in the determination of the core region of the Sup35p fibrils may be linked to the polymorphism of the fibril structures. Sup35p can adopt different fibril structures depending on [PSI+] variants or “strains”.4,5,27
Two types of structural models of the ARN-region fibrils have been proposed based on experimental data: a β-helical model 16,28 and a superpleated β-structural model.29 The disagreement of the β-helical model with in-register parallel β-strand arrangement detected by solid state NMR 17–20 makes this model unlikely. Current data favor the superpleated β-structure model (for a review see ref. 30). In this structure, each polypeptide chain has a serpentine fold, and successive serpentines are stacked in register, one on top of the other (Fig. 1B). This arrangement generates an array of elongated parallel β-sheets, each composed of identical strands and aligned with the fibril axis. The β-strands form a so-called cross-β structure that runs perpendicular to the filament axis. At the same time there is no clear evidence about the exact positions of the β-strand and turns regions. This uncertainty can reflect the limits of the experimental approaches, but also may be linked to the structural polymorphism of Sup35p fibrils. Different forms of Sup35p fibrils can involve different regions of N-domain in the serpentines having different shapes (Fig. 1C). Importantly, in principle, a highly amyloidogenic sequence of the N-domain could have serpentines covering the first ˜100 residues.29 However, due to the slight twist of the axially stacked monomers in the amyloid fibrils, the real serpentines may contain from one to multiple β-arcades. These smaller serpentines can be formed in different regions within the N-domain depending on prion variant (Fig. 1C). In addition, the serpentines may vary in their positions of β-strands and turns.
Despite the progress in understanding of structural arrangement of fibrils, the relationship between the fibril structure and properties of [PSI+] still remains elusive. In the next section we will address this question, by analyzing through the prism of the fibril structure the effects of substitutions in the N-domain on aggregation of Sup35p and [PSI+] propagation. We will also provide an extended view on the results of our recent works 31,32, which demonstrated the utility of structural information for the design of the mutational analysis experiments.
2. Effect of Sup35p MUTATIONS ON [PSI+]
Deletion of either N-, M- or C-domain of Sup35p allowed to determine the roles of each of these domains, in the normal and prion states.6,8,11,23,24 To establish subtle effects, a series of studies analyzing the effects of SUP35 point mutations has been undertaken. For example, the random mutagenesis screens of SUP35 were performed in 2 independent studies aimed to find amino acid substitutions eliminating prion phenotype.3,22 A number of such point mutations have been obtained, which inhibited the nonsense-suppressor efficiency of [PSI+]. For this reason they were named Asu (from antisuppression). Some of these mutations also prevent [PSI+] propagation and were named PNM (from “[PSI+] no more” (for a review see ref. 33)). Although the screens covered the whole SUP35 gene, all substitutions were located within the first 59 residues of Sup35p, with the highest occurrence in the first 25 residue region (Fig. 2).3,22 These results suggested that the N-terminal 60 residue region is critical for prion maintenance. Almost all of these spontaneous mutations led to substitutions of neutral to charged amino acid residues. The effect of the mutations has a straightforward explanation in the model of the parallel and in-register superpleated β-structure 29 where electrostatic repulsion of the juxtaposed charged residues of the same sign destabilize the prion fibrils and, therefore, can lead to the loss of [PSI+]. It is worth noting that the β-helical model 16,28 does not provide an explanation for these experimental results because, in the β-helix, the introduced charged residues are located far from each other and cannot destabilize the fibrils by means of electrostatic repulsion.
Figure 2.
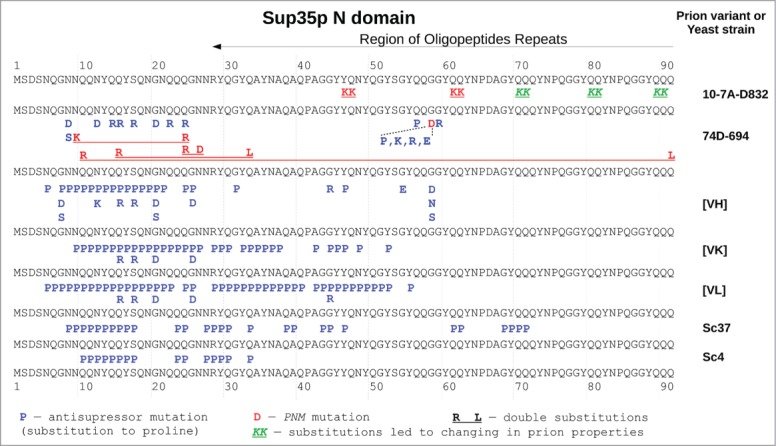
The effects of majority of substitutions in Sup35p affecting [PSI+] propagation are variant specific. Sequence of N-domain of Sup35p (in black regular font) is shown several times and separate substitutions corresponding to different prion variants. Location of the sequence repeats according to 31 is shown on top. Numbers and dashed lines mark corresponding positions in the protein. The substitutions affecting [PSI+] prion are shown by bold letters in corresponding positions. Prion variants affected by substitution are marked on the right. Red letters denote PNM mutations, blue – antisupressor mutations and/or reduced protein aggregation (part of them may be PNM, but exact data are available only for 74D-694 yeast strain), green italic – substitutions changing [PSI+] properties. Double substitutions are underlined. The data used are from Refs. 3, 21, 22, 25, 26, 31, and 34.
The agreement between the published mutations and the superpleated β-structural arrangement encouraged us to design new mutations that would provide further insight into the structure and properties of the [PSI+] prion. We constructed 5 mutant alleles, each carrying a pair of positively charged lysine residues in equivalent positions of repeats R1 to R5, to uniformly cover the repeat-containing part of the N-domain.31 The corresponding mutations were located within predicted β-strand regions at positions occupied by uncharged residues QQ or QN and expected to destabilize the in-register β-structure due to electrostatic repulsion. We demonstrated that the most N-terminal substitutions Y46K/Q47K and Q61K/Q62K led the prion loss, whereas the other alleles were able to maintain the [PSI+] prion with 100% efficiency. This suggests that the C-terminal boundary of the superpleated β-structure for the [PSI+] variant studied in our work lies between residues 63 and 69. Effect of other types of point mutations also supported a ∼60 residue long fibril-forming region. For example, substitutions of glycine 58 to proline led to efficient prion destabilization.21,34 On the contrary, insertions of hydrophobic residues (isoleucine and valine) within the first 25 residues of Sup35p N-domain led to increase the frequency of de novo [PSI+] prion induction. Even basal expression of some of these mutant alleles promoted prion formation. On the other hand, deletions of tyrosine residues in this region reduced efficiency of [PSI+] appearance and made the prion mitotically unstable.35 A subsequent study confirmed that aromatic and hydrophobic residues promote prion formation.36 In another work it was also shown that insertions of valines or isoleucines in the polyglutamine sequence that was fused to the Sup35p MC-domain instead of the N-domain, increase protein aggregation compared with the polyglutamine.37 These results are in accordance with the parallel and in-register superpleated β-structure, where the hydrophobic residues are placed one over the other along the fibril and should form energetically favorable contacts that can stabilize the fibril structure.
Interpretation of the consequences of the sup35 point mutations is hampered by the fact that their effect depends on prion variant.3,38,39 The variant specificity explains differences in lengths of Sup35p regions containing substitutions in the spontaneous mutation screens: the first 34 or 58 amino acid residues 3,22 – in these studies different yeast strains with distinct prion variants were used. Variant-specific effects were also described for PNM mutations.38,39 The structural polymorphism of prion variants may also explain the disagreement between the solid state NMR data on the location of the fibril-forming regions.17,19,25,27
Site-directed mutagenesis with a substitution to proline or an insertion of glycine was used for mapping possible β-strands within a prion domain of different variants.25,26 Proline (and, in a less extent, glycine), as a β-structure breaker, should decrease Sup35p aggregation and prion mitotic stability if it is located within the β-strands. It was confirmed that [PSI+] prion variants differ in the lengths of fibril-forming regions and in the localization of β-strands and turns. These results were supported by hydrogen/deuterium exchange data that determined protection of the corresponding β-strands.25 The observed differences can be easily explained by the polymorphism of the superpleated β-structures (Fig. 1C).
The new prion variants can also be formed by the sup35 mutant alleles that carry mutations outside of the fibril-forming region.31 For example, the mutant allele Q80K/Q81K led to formation of a new strong prion variant of [PSI+] that is characterized by an increased proportion of the prion fibrils and reduced amount of soluble Sup35p in the cell, in comparison with the wild type protein. In contrast, mutant allele Q89K/Q90K yields a weaker prion variant.
3. ArchCandy as a computational tool to predict effects of mutations
The core structural element of a majority of naturally-occurring and disease-related amyloid fibrils is a β-arcade representing a parallel and in register stacks of β-strand-loop-β-strand motifs called β-arches.40–42 Based on an assumption that protein sequences that are able to form β-arcades are amyloidogenic, a computational program “ArchCandy” to predict amyloidogenic regions in proteins has been developed.32 ArchCandy allows us to establish the relationship between the effect of mutations on the fibril forming potential of Sup35p in a more quantitative way, in comparison to the previous rather qualitative considerations (see section 2).
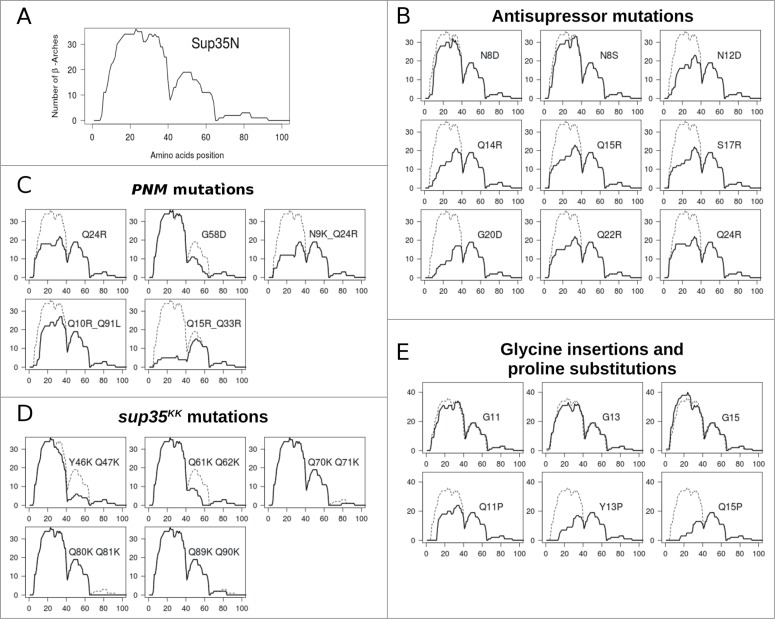
The superpleated β-structure consists of β-arcades (Fig. 1B). The Sup35p fibrils may have one or several successive β-arcades depending on prion variants (Fig. 1C). Figure 3 summarizes the ArchCandy predictions of the effects of known Sup35p mutations. In agreement with the experimental data, ArchCandy assigns the highest amyloidogenic potential of the full length Sup35p to N-domain.32 All PNM and Asu mutations of Sup35p revealed by the spontaneous mutation screens decrease the amyloidogenic potential predicted by ArchCandy (Fig. 3 B–C). In the N-domain, the amyloidogenic potential is maximal within the first 40 residues, followed by the other high-score region between 41 to 64 positions (Fig. 3A). In agreement with this prediction, most of the substitutions in Sup35p affecting [PSI+] are located within the first 40 residues.3,22,25,26 At the same time, the mutations in regions 41–64 are also important for prion propagation of some prion variants (Fig. 2).3,21,22,25,26,38 We also predicted effects of sup35KK mutant alleles published in our recent work.31 In agreement with the experimental data, ArchCandy assigns a lower amyloidogenicity score to alleles leading to prion loss (Y46K/Q47K and Q61K/Q62K) (Fig. 3D) and predicts almost no effect on the fibril-forming potential for the other downstream sup35KK alleles. Among these alleles, Q80K/Q81K leads to the strongest [PSI+] phenotype.31 To explain this data, we proposed that Q80K/Q81K substitution makes the prion-forming region shorter and this increases the strength of [PSI+] prion.31,43 This correlation between the fibril-forming region length and strength of prion phenotype was already described elsewhere.25–27 ArchCandy predicts that Q80K/Q81K substitution eliminates any traces of amyloidogenicity after position 64 (Fig. 3D).
Figure 3.
ArchCandy prediction of the effect of amino acids substitutions on Sup35p fibril-forming potential. (A) Wild type Sup35p. The fibril-forming potential is measured by the number of predicted β-arcades (with ArchCandy score above 0.575) for each amino acids in Sup35p N-domain. For example, residue 23 of wild type Sup35p can participate in 37 different β-arcades. Panels B–E compare ArchCandy predictions for modified Sup35 proteins (continuous lines) and the wild type protein (dashed line). For proline substitutions and glycine insertions only some typical results are shown. Each plot is labeled by corresponding amino acid substitution(s) or sup35 allele. The data used are from Refs. 21, 22, 26 and 31.
The observed destabilization of [PSI+] prion in the proline-containing mutant alleles 26 can be explained by the decrease of the amyloidogenic potential predicted by ArchCandy (Fig. 3E). ArchCandy was also able to predict (with a few exceptions) the increase of prion formation, related to the insertions of hydrophobic residues, within the first 25 residues of N-domain and [PSI+] prion destabilization after deletion of tyrosines.35 At the same time, ArchCandy failed to predict strong destabilization effect of some sup35 mutant alleles with glycine insertions (Fig. 3E) 26, pointing on the necessity of some improvement for the ArchCandy algorithm.
4. Conclusions
Yeast [PSI+] prion formed by aggregated Sup35 protein is one of the most suitable and well characterized system for the investigation of the prion phenomenon. Numerous studies of [PSI+] prion have greatly advanced the field, owing to their experimental tractability, which is additionally enhanced by the powerful genetics of yeast, the relatively short generation times of fungi, and, in several cases, simple metabolic assays for the presence of the prion. However, until recently, the lack of information about the 3D arrangement of Sup35p prion fibrils, along with the molecular mechanisms of prion formation and infectivity hindered progress in this area. The recent arrival in this field of new experimental techniques has provided breakthroughs in the understanding of the 3D arrangement of amyloid fibrils. As a result, the parallel and in-register superpleated β-structure has emerged as a consensus model for Sup35p fibrils. Today, the effect of the majority of published sup35 mutations can be explained within the frame of this structural model. Hence, the agreement between the effect of the published mutations and the superpleated β-structural arrangement encouraged us to design a set of new mutant alleles, which should provide us with a better insight into the location and size of the prion-forming domain.31
A recently developed computational approach, ArchCandy 32, allows us to evaluate the probability of proteins to form fibrils, with the superpleated β-structure. Application of this bioinformatics tool to the set of the known sup35 mutant alleles showed a good correlation between the ArchCandy prediction and the experimentally observed effects on Sup35p fibrollogenesis and prion properties. This opens an avenue to the interpretation of the experimental results, design of new mutations, and eventually a more complete understanding of the relationship between the effect of mutations on the fibril forming potential of Sup35p, in a more objective and automated manner. The high correlation between the ArchCandy prediction of the protein amyloidogenicity and the ability to form prions provides additional support of the notion that the propensity to aggregate underlies prionogenesis. At the same time, we are aware that amyloidogencity is not sufficient for the explanation of the prion phenomenon. The real situation may be more complicated with chaperons and several other cellular cofactors involved in prion propagation and maintenance. Further investigations of these molecular mechanisms need to be carried out prior to the development of new bioinformatics algorithms that study these aspects.
Despite progress, the prion phenomenon still has a number of unresolved questions, such as, a link between the fibril structures and the strength of the prion infectivity; interplay between different prion strains and co-aggregation of Sup35p and its mutant alleles. Many of these questions require a deep insight into the detailed 3D structure of each prion strain. One function of ArchCandy allows us to generate such 3D structural models, which can be used to advance in these complicated problems, in the context of limited capacities of the experimental structural approaches. For example, the ArchCandy prediction of possible β-arcades of the prion domains may provide further insight into the origin of new prion variants, supporting either the “cloud” hypothesis of intrinsically heterogeneous prion isolates 44 or the hypothesis of a “deformed templating” pathways.45
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Daniel Roche for critical reading of the manuscript and suggestions.
Funding
The authors acknowledge Saint-Petersburg State University for following research grants: 1.37.291.2015, 0.37.696.2013 (SB, MB, GZ), 1.50.1041.2014 (MB), 1.50.2218.2013 (GZ), 1.42.1274.2014 (SB). This work was also supported by RFBR (13-04-00645 (SB, MB, GZ), 14-04-32213(SB)) and by the grant of the President of The Russian Federation (MK-4854.2015.4 (SB)).
REFERENCES
- 1.Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012; 191:1041–72; PMID:22879407; http://dx.doi.org/ 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of ; PSI prion factor in Saccharomyces cerevisiae. Genetics 1996; 144:1375–86; PMID:8978027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King CY. Supporting the structural basis of prion strains: induction and identification of PSI variants. J Mol Biol 2001; 307:1247–60; PMID:11292339; http://dx.doi.org/ 10.1006/jmbi.2001.4542 [DOI] [PubMed] [Google Scholar]
- 4.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature 2004; 428:323–8; PMID:15029196; http://dx.doi.org/ 10.1038/nature02392 [DOI] [PubMed] [Google Scholar]
- 5.King C, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature 2004; 428:319–23; PMID:15029195; http://dx.doi.org/ 10.1038/nature02391 [DOI] [PubMed] [Google Scholar]
- 6.Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 1993; 7:683–92; PMID:8469113; http://dx.doi.org/ 10.1111/j.1365-2958.1993.tb01159.x [DOI] [PubMed] [Google Scholar]
- 7.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 1995; 14:4065–72; PMID:7664746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-mendelian determinant [ psi +] in yeast Saccharomyces cerevisiae. Genetics 1994; 137:671–6; PMID:8088512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smirnov VN, Inge-Vechtomov SG. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene 1988; 66:45–54; PMID:3047009; http://dx.doi.org/ 10.1016/0378-1119(88)90223-5 [DOI] [PubMed] [Google Scholar]
- 10.Kong C, Ito K, Walsh MA, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell 2004; 14:233–45; PMID:15099522; http://dx.doi.org/ 10.1016/S1097-2765(04)00206-0 [DOI] [PubMed] [Google Scholar]
- 11.Liu J-J, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci USA 2002; 99:16446–53; PMID:12461168; http://dx.doi.org/ 10.1073/pnas.252652099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover JR, Kowala S, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997; 89:811–9; PMID:9182769; http://dx.doi.org/ 10.1016/S0092-8674(00)80264-0 [DOI] [PubMed] [Google Scholar]
- 13.King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wüthrich K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA 1997; 94:6618–22; PMID:9192614; http://dx.doi.org/ 10.1073/pnas.94.13.6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baxa U, Keller PW, Cheng N, Wall JS, Steven AC. In Sup35p filaments (the [PSI+] prion), the globular C-terminal domains are widely offset from the amyloid fibril backbone. Mol Microbiol 2011; 79:523–32; PMID:21219467; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07466.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 2000; 289:1317–21; PMID:10958771; http://dx.doi.org/ 10.1126/science.289.5483.1317 [DOI] [PubMed] [Google Scholar]
- 16.Kishimoto A, Hasegawa K, Suzuki H, Taguchi H, Namba K, Yoshida M. beta-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem Biophys Res Commun 2004; 315:739–45; PMID:14975763; http://dx.doi.org/ 10.1016/j.bbrc.2004.01.117 [DOI] [PubMed] [Google Scholar]
- 17.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA 2006; 103:19754–9; PMID:17170131; http://dx.doi.org/ 10.1073/pnas.0609638103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel beta-sheet structures, independent of hydration. Biochemistry 2009; 48:5074–82; PMID:19408895; http://dx.doi.org/ 10.1021/bi900345q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luckgei N, Schütz AK, Bousset L, Habenstein B, Sourigues Y, Gardiennet C, Meier BH, Melki R, Böckmann A. The conformation of the prion domain of Sup35p in isolation and in the full-length protein. Angew Chem Int Ed Engl 2013; 52:12741–4; PMID:24123863; http://dx.doi.org/ 10.1002/anie.201304699 [DOI] [PubMed] [Google Scholar]
- 20.Gorkovskiy A, Thurber KR, Tycko R, Wickner RB. Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc Natl Acad Sci USA 2014; 111:E4615–E4622; PMID:25313080; http://dx.doi.org/ 10.1073/pnas.1417974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doel SM, Mccready SJ, Nierras CR, Cox BS. The dominant PNM2 mutation which eliminates the PSI factor of Saccharomyces cerevisiae is the result of missense mutation in the SUP35 gene. Genetics 1994; 137:659–70; PMID:8088511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998; 93:1241–52; PMID:9657156; http://dx.doi.org/ 10.1016/S0092-8674(00)81467-1 [DOI] [PubMed] [Google Scholar]
- 23.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol 2004; 2:E86; PMID:15045026; http://dx.doi.org/ 10.1371/journal.pbio.0020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shkundina IS, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics 2006; 172:827–35; PMID:16272413; http://dx.doi.org/ 10.1534/genetics.105.048660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature 2007; 449:233–7; PMID:17767153; http://dx.doi.org/ 10.1038/nature06108 [DOI] [PubMed] [Google Scholar]
- 26.Chang H-Y, Lin J-Y, Lee H-C, Wang H-L, King C-Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc Natl Acad Sci USA 2008; 105:13345–50; PMID:18757753; http://dx.doi.org/ 10.1073/pnas.0802215105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem Biol 2014; 21:295–305; PMID:24485763; http://dx.doi.org/ 10.1016/j.chembiol.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 2005; 435:765–72; PMID:15944694; http://dx.doi.org/ 10.1038/nature03679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure. Proc Natl Acad Sci USA 2004; 101:7885–90; PMID:15143215; http://dx.doi.org/ 10.1073/pnas.0402427101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxa U, Cassese T, Kajava AV, Steven AC. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv Protein Chem 2006; 73:125–80; PMID:17190613; http://dx.doi.org/ 10.1016/S0065-3233(06)73005-4 [DOI] [PubMed] [Google Scholar]
- 31.Bondarev SA, Shchepachev VV, Kajava AV, Zhouravleva GA. Effect of charged residues in the N-domain of Sup35 protein on prion [PSI+] stability and propagation. J Biol Chem 2013; 288:28503–13; PMID:23965990; http://dx.doi.org/ 10.1074/jbc.M113.471805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed AB, Znassi N, Château M-T, Kajava AV. A structure-based approach to predict predisposition to amyloidosis. Alzheimers Dement 2014; doi: 10.1016/j.jalz.2014.06.007 [DOI] [PubMed] [Google Scholar]
- 33.Cox BS, Tuite MF, McLauchlin C. The psi factor of yeast: a problem in inheritance. Yeast 1988; 4:159–78; PMID:3059716; http://dx.doi.org/ 10.1002/yea.320040302 [DOI] [PubMed] [Google Scholar]
- 34.Marchante R, Rowe M, Zenthon J, Howard MJ, Tuite MF. Structural definition is important for the propagation of the yeast [PSI+] prion. Mol Cell 2013; 50:675–85; PMID:23746351; http://dx.doi.org/ 10.1016/j.molcel.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED. Increasing prion propensity by hydrophobic insertion. PLoS One 2014; 9:e89286; PMID:24586661; http://dx.doi.org/ 10.1371/journal.pone.0089286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacLea KS, Paul KR, Ben-Musa Z, Waechter A, Shattuck JE, Gruca M, Ross ED. Distinct amino acid compositional requirements for formation and maintenance of the [PSI+] prion in yeast. Mol Cell Biol 2015; 35:899–911; PMID:25547291; http://dx.doi.org/ 10.1128/MCB.01020-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandrov AI, Polyanskaya AB, Serpionov GV, Ter-Avanesyan MD, Kushnirov VV. The effects of amino acid composition of glutamine-rich domains on amyloid formation and fragmentation. PLoS One 2012; 7:e46458; PMID:23071575; http://dx.doi.org/ 10.1371/journal.pone.0046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derkatch IL, Bradley ME, Zhou P, Liebman SW. The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI+] prion in yeast. Curr Genet 1999; 35:59–67; PMID:10079323; http://dx.doi.org/ 10.1007/s002940050433 [DOI] [PubMed] [Google Scholar]
- 39.Verges KJ, Smith MH, Toyama BH, Weissman JS. Strain conformation, primary structure and the propagation of the yeast prion [PSI+]. Nat Struct Mol Biol 2011; 18:493–9; PMID:21423194; http://dx.doi.org/ 10.1038/nsmb.2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D structure of Alzheimer's amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA 2005; 102:17342–7; PMID:16293696; http://dx.doi.org/ 10.1073/pnas.0506723102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petkova AT, Yau W-M, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry 2006; 45:498–512; PMID:16401079; http://dx.doi.org/ 10.1021/bi051952q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kajava AV, Baxa U, Steven AC. Beta arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. FASEB J 2010; 24:1311–9; PMID:20032312; http://dx.doi.org/ 10.1096/fj.09-145979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bondarev SA, Shirokolobova ED, Trubitsina NP, Zhouravleva GA. Modification of [PSI+] prion properties by combining amino acid changes in N-terminal domain of Sup35 protein. Mol Biol (Mosk) 2014; 48:270–7; http://dx.doi.org/ 10.1134/S0026893314020034 [DOI] [PubMed] [Google Scholar]
- 44.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science 2010; 327: 869–72; PMID:20044542; http://dx.doi.org/ 10.1126/science.1183218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makarava N, Baskakov IV. The Evolution of Transmissible Prions: The Role of Deformed Templating. PLoS Pathog 2013; 9:1–3; http://dx.doi.org/ 10.1371/journal.ppat.1003759 [DOI] [PMC free article] [PubMed] [Google Scholar]