The crystal structure of glyoxalase I from Z. mays is reported.
Keywords: glyoxalase, methylglyoxal, maize
Abstract
The glyoxalase system is ubiquitous among all forms of life owing to its central role in relieving the cell from the accumulation of methylglyoxal, a toxic metabolic byproduct. In higher plants, this system is upregulated under diverse metabolic stress conditions, such as in the defence response to infection by pathogenic microorganisms. Despite their proven fundamental role in metabolic stresses, plant glyoxalases have been poorly studied. In this work, glyoxalase I from Zea mays has been characterized both biochemically and structurally, thus reporting the first atomic model of a glyoxalase I available from plants. The results indicate that this enzyme comprises a single polypeptide with two structurally similar domains, giving rise to two lateral concavities, one of which harbours a functional nickel(II)-binding active site. The putative function of the remaining cryptic active site remains to be determined.
1. Introduction
The enzyme glyoxalase I [S-d-lactoylglutathione methylglyoxal lyase (isomerizing); EC 4.4.1.5] catalyzes the glutathione-dependent conversion of methylglyoxal (MG), a cytotoxic α-oxoaldehyde, to S-d-lactoylglutathione. The product thus formed is then converted to d-lactate by glyoxalase II, regenerating reduced glutathione. This system is ubiquitously present in all forms of life from ancient archaea to bacteria and higher eukaryotes, and is the major pathway for the detoxification of MG, which is generated spontaneously during the course of glycolysis. In humans, this system has been extensively studied, since increased levels of MG have been linked to the development of several disorders (Rabbani & Thornalley, 2011 ▸). In plants, this pathway is thought to play an important function in conferring tolerance to multiple abiotic stresses (Kaur et al., 2014 ▸; Mustafiz et al., 2014 ▸; Yadav et al., 2008 ▸; Veena et al., 1999 ▸). In addition, although less studied, glyoxalases are involved in biotic stress. Chen et al. (2004 ▸) found that glyoxalase I is upregulated in resistant maize kernels after inoculation with Aspergillus flavus. Furthermore, Lin et al. (2010 ▸) reported that an expressed sequence tag encoding a glyoxalase I was isolated from a suppression subtractive hybridization cDNA library of wheat spike inoculated with Fusarium graminearum. On the other hand, F. verticillioides (Sacc.) Nirenberg (synonym F. moniliforme Sheldon, teleomorph Gibberella moniliformis Wineland) is one of the most burdensome pathogens of maize; it is an endophytic and hemibiotrophic fungus that causes the disease known as ear rot. This microorganism not only causes severe reductions in cereal quality and yield, thus leading to major economic losses, but also produces secondary metabolites such as fumonisins, in particular fumonisin B1, which are toxic to humans (Marasas, 1995 ▸). This fungus can be found in maize fields at different stages of maize ear development (Chulze et al., 1996 ▸). The symptoms of the disease vary from asymptomatic infection to severe rotting of all parts of the plant. Under unfavourable conditions for plant growth, the fungus is not in a balanced state with the plant, resulting in degrees of pathological response (Bacon et al., 2001 ▸). Our proteomic experiments have shown that Zea mays glyoxalase I (ZmGLX1) is also upregulated in moderately resistant maize lines after inoculation with F. verticillioides compared with susceptible maize lines (unpublished work). Together, these results suggest a key role for glyoxalase I in the resistance of maize to fungal infections. Therefore, a deeper understanding of the structure–function relationship of this enzyme is expected to shed light on plausible methods of reinforcing the antimicrobial defence of the plant.
Glyoxalase I enzymes from numerous organisms have been biochemically characterized, including bacteria, plants, yeast, animals and protozoan parasites (Suttisansanee & Honek, 2011 ▸; He et al., 2000 ▸; Sukdeo et al., 2004 ▸; Deswal & Sopory, 1991 ▸; Espartero et al., 1995 ▸; Mustafiz et al., 2014 ▸; Marmstål et al., 1979 ▸; Gomes et al., 2005 ▸; Martins et al., 2001 ▸; Aronsson et al., 1978 ▸; Cameron et al., 1997 ▸; Akoachere et al., 2005 ▸; Ariza et al., 2006 ▸; Greig et al., 2006 ▸). However, only six glyoxalases I have been structurally described thus far, namely the enzymes from Homo sapiens (Aronsson et al., 1978 ▸; Cameron et al., 1997 ▸), Escherichia coli (He et al., 2000 ▸), Leishmania major (Ariza et al., 2006 ▸), Mus musculus (Kawatani et al., 2008 ▸), Clostridium acetobutylicum (Suttisansanee et al., 2011 ▸) and Pseudomonas aeruginosa (Bythell-Douglas et al., 2015 ▸). All of them display a characteristic homodimeric quaternary structure, in which each monomer comprises two βαβββ domains that interact to generate a continuous eight-stranded β-sheet with another βαβββ domain present in the opposite monomer (Fig. 1 ▸ a). The two active sites are formed at the concavity of each interdomain β-sheet, which corresponds to the interface between monomers. An important exception is the enzyme from C. acetobutylicum, in which the eight-stranded β-sheet results from the interaction between two βαβββ domains in the same monomer (Suttisansanee et al., 2011 ▸). Such a versatile topology, which is typical of the glyoxalase fold, has been suggested to be the result of gene-duplication and domain-swapping events (Cameron et al., 1997 ▸).
Figure 1.
Overall structure of glyoxalase I. (a) Three-dimensional structure of E. coli glyoxalase I (PDB entry 1f9z; He et al., 2000 ▸), depicting the typical topology of homodimeric glyoxalases, where each monomer comprises two βαβββ domains that give rise to two eight-stranded β-sheets (β1–8) with the opposite monomer, with each of them harbouring one active site. (b) Topological relationships among secondary-structure elements in monomeric ZmGLX1. Each of the N- and C-terminal domains (α1–2β1–8 and α3–5β9–16, respectively) corresponds to the monomers found in homodimeric glyoxalases I. Sites A and B are the concavities formed by interdomain β-sheets β5–12 and β1–4/13–16, respectively. The discontinuous β-sheets β6 and β14 are indicated by dotted rectangles. The cartoon and molecular-surface representation for each protein is displayed on the right for comparison.
Although currently characterized glyoxalase I enzymes are homodimeric in nature, the existence of monomeric forms of these enzymes has been suggested in some organisms based on primary-sequence comparisons, including yeast, Plasmodium, rice and wheat (Ridderström & Mannervik, 1996 ▸; Iozef et al., 2003 ▸; Mustafiz et al., 2014 ▸; Lin et al., 2010 ▸). Monomeric glyoxalases have also been suggested to contain four βαβββ domains, although their spatial organization in the protein quaternary structure has not been discerned. Yeast and P. falciparum glyoxalases are among the few characterized enzymes comprising a single polypeptide with two active sites that catalyze the same reaction (Frickel et al., 2001 ▸; Deponte et al., 2007 ▸). In addition, glyoxalase I from rice is a monomeric enzyme which appears to have only one active site (Mustafiz et al., 2014 ▸).
Biochemical and structural studies have shown that glyoxalases are metalloenzymes that require either nickel(II) or zinc(II) for catalytic activity (Sukdeo et al., 2004 ▸). It has been suggested that the metal specificity may be predicted based on the amino-acid sequence and the length, given that zinc(II)- dependent glyoxalases contain short additional regions that are absent in the nickel(II)-dependent enzymes (Suttisansanee et al., 2015 ▸). On the other hand, structural analyses indicate that only the enzymes in which the metal cofactor displays an octahedral coordination geometry are catalytically active (He et al., 2000 ▸; Suttisansanee et al., 2011 ▸, 2015 ▸). Such an octahedral geometry arises from the metal coordination to four conserved residues present in His/Glu/Gln/Glu or His/Glu/His/Glu metal-binding motifs, completed by two solvating water molecules.
In order to obtain further insights into monomeric plant glyoxalases I, we pursued the biochemical and structural characterization of the glyoxalase I from maize, ZmGLX1. We show that ZmGLX1 requires a single nickel(II) ion for catalysis. In addition, its crystallographic structure, the first atomic model of a glyoxalase I available from a plant, displays two structurally similar domains, giving rise to two lateral concavities, one of which harbours a functional nickel(II)-binding active site, as confirmed by site-directed mutagenesis. The putative function of the remaining cryptic active site remains to be determined.
2. Materials and methods
2.1. General
Methylglyoxal (MG) and glutathione (GSH) were purchased from Sigma–Aldrich. All chromatographic steps were performed on an Amersham Biosciences liquid-chromatography system operating at 277 K. Metal standards were purchased from Fisher Scientific and were diluted with ultrapure water. All other chemicals used in this study were commercially available and were of the highest quality.
2.2. Gene amplification and cloning
The coding DNA sequence for Z. mays glyoxalase I (accession No. GRMZM2G181192 for the B73 maize line, available at the Gramene database; http://www.gramene.org) was obtained from cDNA of L4637 maize grains using the primer set ZmGLX1 Fw and ZmGLX1 Rv, which include NcoI and XhoI restriction sites at the 5′ end and the 3′ end of the fragment, respectively (Supplementary Table S1). The amplified 894 bp PCR product was cloned into the pGEMT Easy vector (Promega) and transformed into E. coli DH5α cells by electroporation using a Bio-Rad apparatus. After sequence confirmation, the glxI sequence fragment was digested with the above-mentioned enzymes and cloned into pET-28b(+) expression vector (Novagen) to obtain the pET-28b-Glx1 vector. This cloning strategy resulted in the addition of a noncleavable His-tag sequence at the C-terminus of the ZmGLX1 protein.
A different cloning strategy was used to obtain the wild-type and E144Q mutant enzymes without a His-tag. In these cases, the primers used for cloning in pET-28b(+) allowed expression of the proteins as an N-terminal fusion with a thrombin-cleavable His-tag using NheI and XhoI cloning sites. The new constructs were named pET-28b-Glx1(His6-less) for the wild-type sequence and pET-28b-E144Q for the mutant sequence. To obtain the E144Q variant sequence, overlap extension PCR was performed using Phusion DNA polymerase (Thermo Scientific), following the manufacturer’s recommendations. The primers used for this PCR are described in Supplementary Table S1.
2.3. Protein overexpression and purification
ZmGLX1 was recombinantly produced from E. coli BL21 Rosetta cells using the pET-28b-Glx1 vector. This system yielded high-level expression of recombinant ZmGLX1 protein (UniProt C0PK05) fused to a hexahistidine tag at its C-terminal end. In a typical protein preparation, 400 ml of transformed E. coli BL21 Rosetta culture was grown in auto-induction medium. Optimal overexpression was achieved using auto-induction medium supplemented with trace-metal ions followed by 24 h incubation at 303 K, as described previously (Studier, 2005 ▸). The bacterial cultures were harvested by centrifugation and resuspended in 50 mM Tris–HCl pH 8.0, 1 mM phenylmethylsulfonyl fluoride, 0.01 mg ml−1 DNAse, 5 mM MgCl2. Sonication was performed six times for 30 s, followed by ultracentrifugation at 10 000 rev min−1 in the SS34 rotor of a Sorvall centrifuge. The bacterial lysate was applied onto an Ni–NTA column (Invitrogen). After washing with 50 mM Tris–HCl pH 8.0, 300 mM NaCl, 20 mM imidazole, the fusion protein was eluted with 50 mM Tris–HCl pH 8.0, 300 mM NaCl, 250 mM imidazole. Fractions containing ZmGLX1 were pooled and dialyzed against 50 mM Tris–HCl pH 7.2, 0.2 M NaCl. The protein obtained using this protocol was named ‘trace-metals ZmGLX1’. Additionally, we modified this expression protocol by adding 1 mM NiCl2 instead of trace metals, with the aim of obtaining a nickel(II)-enriched protein sample. We named this protein sample ‘Ni-only protein’.
The wild-type and E114Q variants without a His-tag were produced in E. coli BL21 Rosetta cells transformed with the pET-28b-Glx1(His6-less) or pET-28b-E144Q vectors, and were cultured using the same overexpression protocol as used for the Ni-only protein. Once obtained, the fusion proteins were treated with a 1:100 ratio of thrombin protease for 1 h at 310 K in 50 mM Tris–HCl pH 7.2, 0.2 M NaCl medium. Cleaved proteins were obtained after a new purification step of Ni–NTA chromatography, in which the His-tag and undigested protein were retained. The different overexpression strategies are summarized in Supplementary Table S2.
2.4. Oligomeric state of ZmGLX1
The molecular weight and oligomeric state of trace-metals ZmGLX1 were estimated by gel-filtration chromatography using a High Resolution HiPrep 16/60 Sephacryl S200 column equilibrated in 50 mM Tris–HCl, 0.2 M NaCl buffer at pH 8. A calibration curve was prepared using the standards carbonic anhydrase (29 kDa), albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), apoferritin (443 kDa) and thyroglobulin (669 kDa). Subsequently, the relative molecular weight of the eluted protein was interpolated from a linear calibration plot of elution volume versus log(molecular weight).
2.5. Protein concentration
The enzyme concentration was determined spectrophotometrically by measuring the A 280 nm and using an extinction coefficient Δ∊280 nm = 37 945 M −1 cm−1. This parameter was calculated from amino-acid composition information by applying a modified Edelhoch method (Gill & von Hippel, 1989 ▸). The Bradford colorimetric assay was also used to corroborate these measurements.
2.6. Metal-content analyses
The metal content of the ZmGLX1 samples was measured using atomic absorption spectroscopy in a Metrolab 250 AA instrument. A 30 µM solution of pure protein in 50 mM Tris–HCl, 0.2 M NaCl pH 7.2 was tested for zinc, nickel, copper, cobalt, calcium, magnesium and iron. The metal-content data presented in this paper represent an average from at least three preparations of each growth condition. Atomic absorption standards (Sigma) were used for the calibration curves. Dialysis buffer (50 mM Tris–HCl, 0.2 M NaCl pH 7.2) was used as a blank.
2.7. Steady-state kinetics
Parameters were determined by measuring the initial rate of formation of S-d-lactoylglutathione at 240 nm considering a Δ∊240 nm of 3370 M −1 cm−1 (Racker, 1951 ▸). The measurements were performed in a reaction medium consisting of 500 µl 100 mM phosphate buffer pH 7.2 (previously determined as the optimum pH, evaluating the activity as a function of the measured buffer pH) at 298 K in a Jasco 550 UV–visible spectrophotometer. The amount of enzyme ranged between 100 and 200 nM. The metal cofactor was at 100 µM. The GSH level was kept constant at 1.7 mM and the MG concentration was varied from 10 to 1000 µM. In an analogous experiment, the MG level was maintained constant at 1.7 mM, while the GSH concentration was varied from 10 to 1000 µM. The measurements were performed at least in triplicate. Kinetic constants were determined by fitting the initial rate data to the Michaelis–Menten equation, considering the spontaneously formed hemithioacetal to be the only substrate and to be formed with a K eq of 0.385 (Rae et al., 1990 ▸).
2.8. Metal-activation studies
A profile for metal activation of trace-metals ZmGLX1 with various divalent metal chlorides was obtained using the method described by Sukdeo & Honek (2008 ▸). Samples of ZmGLX1 dialyzed in 50 mM Tris–HCl, 0.2 M NaCl pH 7.2 were pre-incubated with 100 molar equivalents of CoCl2, NiCl2, ZnCl2, CaCl2, MnCl2, CuCl2, MgCl2 and Fe(NH4)SO4. The enzyme activity was assayed as described above. The measurements were performed at least in triplicate.
2.9. Crystallization of trace-metals ZmGLX1
Crystals were grown using the sitting-drop vapour-diffusion method in 96-3 three-well Intelli-Plate trays (Art Robbins Instruments). Drops consisted of 1.0 µl 1:1 mixtures of protein sample (20 mg ml−1 protein in 20 mM HEPES pH 8.0, 20 mM NaCl) and reservoir solution and were prepared with an Oryx8 liquid pipettor robot (Douglas Instruments). Initial hits appeared after 1–2 d of incubation at 293 K in condition No. 48 of the NeXtal Classics Suite (Qiagen), consisting of 4 M sodium formate (final pH ∼9.7). This crystallization condition was further optimized by the addition of 1% PEG 4000. Crystals were mounted in nylon loops (Hampton Research) and flash-cooled in liquid nitrogen after cryoprotection in 7 M sodium formate.
2.10. X-ray diffraction data collection, structure determination and analysis
Diffraction data were collected remotely on beamline BL7-1 of the Stanford Synchrotron Radiation Light Source (Cohen et al., 2002 ▸; Soltis et al., 2008 ▸). Reflections were indexed, integrated and scaled with XDS (Kabsch, 2010 ▸) and AIMLESS (Evans & Murshudov, 2013 ▸). The crystals were found to belong to space group P63, with one molecule per asymmetric unit. Phaser was used for molecular-replacement phasing (McCoy et al., 2007 ▸). The search model for molecular replacement was obtained with the Phyre2 server (Kelley & Sternberg, 2009 ▸) starting from the protein sequence (UniProt ID C0PK05). Structure refinement was performed with REFMAC5 (Murshudov et al., 2011 ▸) and the CCP4 suite of programs (Winn et al., 2011 ▸). Manual building was conducted with Coot (Emsley et al., 2010 ▸) using σA-weighted 2F o − F c and F o − F c Fourier difference maps. Throughout the final cycles of restrained refinement, anisotropic B factors were refined for a single Ni2+ ion partly coordinated to the C-terminal hexahistidine tag. Structure validation was performed with SFCHECK (Vaguine et al., 1999 ▸) and the built-in functions implemented in Coot. Figures were prepared with PyMOL v.1.2 (Schrödinger) and CorelDRAW X7.
Structure-based sequence alignments were calculated with Expresso (Armougom et al., 2006 ▸) and processed with Jalview (Waterhouse et al., 2009 ▸). Structural homologues of ZmGLX1 were retrieved with the DALI server (Holm & Rosenström, 2010 ▸) using our structure, PDB entry 5d7z, as a query. Phylogenetic trees were calculated with MEGA6 (Tamura et al., 2013 ▸), using the maximum-likelihood method, in which the evolutionary rate differences among all sites were estimated with five categories of a discrete gamma distribution.
3. Results
3.1. Gene cloning and protein expression
The protein encoded by a sequence identical to that of the B73 maize line (GRMZM2G181192) showed increased expression levels in kernels of resistant maize lines (L4637) compared with susceptible ones (L4674) after F. verticillioides infection (Fauguel et al., unpublished work). The protein sequence exhibits the highest similarity to lactoylglutathione lyase or glyoxalase I (EC.4.4.1.5) from different organisms and shares 89.4% sequence similarity with the recently characterized monomeric nickel-dependent glyoxalase I from Oryza sativa (Mustafiz et al., 2014 ▸).
To fully characterize this protein, we amplified its coding sequence from cDNA obtained from a maize line that is moderately resistant to F. verticillioides infection, as described in §2. Different expression and purification strategies were attempted. However, overexpression of ZmGLX1 in a soluble form was only possible through the utilization of cultures of E. coli BL21 Rosetta transformed with pET-28b-Glx1 plasmid using an auto-induction medium supplemented with trace metals (Studier, 2005 ▸). The purification procedure took advantage of the affinity of the hexahistidine tag provided by the expression vector in order to isolate the protein by immobilized metal-affinity chromatography (IMAC) using Ni–NTA resin. This successful purification procedure resulted in an average yield of 100 mg per litre of culture of what we named trace-metals ZmGLX1 (Supplementary Fig. S1). Additionally, we attempted an overexpression protocol in which we supplemented the culture medium with NiCl2 instead of trace metals. In this case we obtained a sample enriched with nickel(II), named Ni-only protein, the overexpression yield of which was significantly reduced.
In order to corroborate the molecular mass and oligomeric state of the purified protein, we performed a size-exclusion chromatography test. Based on our results, ZmGLX1 is a monomeric enzyme with a molecular mass of ∼30 kDa, which is in agreement with the predicted size based on the protein sequence (32 kDa).
3.2. Metal content of ZmGLX1
Previous studies on glyoxalase I have reported diverse metal contents for this enzyme (Suttisansanee & Honek, 2011 ▸; Frickel et al., 2001 ▸; Sukdeo et al., 2004 ▸; Yadav et al., 2008 ▸; Deswal & Sopory, 1999 ▸). In general, the recombinant forms of glyoxalase isozymes characterized so far were isolated with nickel or zinc bound to their active sites. We used atomic absorption spectroscopy to analyze the metal bound to recombinant trace-metals ZmGLX1, using commercially available atomic absorption standards for a set of the most probable metals (Table 1 ▸). Given the low metal contents, it is clear that trace-metals ZmGLX1 is obtained mainly as an apo-protein. This may be attributed to a loss of metal during purification or to limited metal availability during overexpression. To evaluate the latter hypothesis, we replaced the addition of trace metals to the bacterial cultures by supplementation with 1 mM NiCl2. From this procedure we were able to obtain Ni-only ZmGLX1, although the protein yield was significantly lower. Nevertheless, atomic absorption measurements confirmed the presence of nearly one equivalent of nickel per mole of protein (Table 1 ▸). In order to discard the possibility of this nickel equivalent binding to the His-tag instead of the active site, we generated His6-less ZmGLX1 by subcloning the enzyme construct with a cleavable His-tag (Supplementary Table S2). We determined a nickel content of 0.8 ± 0.2 per His6-less ZmGLX1, strongly suggesting that nickel(II) ion is bound to the protein.
Table 1. Metal content of as-isolated ZmGLX1 measured by atomic absorption spectroscopy.
Metal content is defined as the ratio of the metal:protein molar concentrations. The buffer is 50mM TrisHCl, 0.2M NaCl pH 7.2. ND, not detected.
| Metal content | ||||
|---|---|---|---|---|
| Metal | Trace-metals ZmGLX1 | Ni-only ZmGLX1 | His6-less ZmGLX1 | His6-less E114Q ZmGLX1 |
| Ca | 0.4 0.1 | ND | ND | ND |
| Zn | 0.10 0.05 | ND | ND | ND |
| Co | 0.08 0.03 | ND | ND | ND |
| Ni | 0.07 0.02 | 0.80 0.02 | 0.80 0.20 | 0.30 0.10 |
Additionally, to obtain insight into the metal-binding sites of ZmGLX1, we assayed the metal content of His6-less E144Q ZmGLX1 protein with a mutation in one of the putative metal ligands. Using the same overexpression protocol, this variant expressed with a significantly lower nickel content, suggesting that the only nickel(II)-binding site is that containing residue Glu144 as a metal ligand (Table 1 ▸).
3.3. Catalytic profile of ZmGLX1
We performed enzymatic assays with trace-metals ZmGLX1 using methylglyoxal (MG) and glutathione (GSH) as substrates (Fig. 2 ▸). The activity profiles as a function of pH showed an activity peak at pH 7.2 in both the absence (Fig. 2 ▸ a) and the presence of 100 µM nickel(II) or cobalt(II), which was selected as the pH for all subsequent kinetic characterization assays. The enzyme was fully inhibited by the addition of 100 µM EDTA to the reaction medium (data not shown), which is in agreement with its requirement for divalent metal ions for catalysis. To evaluate whether ZmGLX1 is capable of catalysis in the presence of different metal ions, we assayed the enzyme activity with an excess concentration of several divalent metal ions (Fig. 2 ▸ b). As previously reported for other glyoxalase I variants, such as those from E. coli, O. sativa and C. acetobutylicum (Sukdeo & Honek, 2008 ▸; Mustafiz et al., 2014 ▸; Suttisansanee et al., 2011 ▸), the highest catalytic activity was observed in the presence of nickel(II). The next metal activating trace-metals ZmGLX1 was cobalt(II), with about 35% of the maximum activity in the presence of nickel(II). In contrast, other added divalent metal ions such as iron(II), manganese(II), magnesium(II), zinc(II), copper(II) or calcium(II) had negligible effects on the activity, which was comparable with the activity of the as-isolated protein observed in the absence of any added metals (Fig. 2 ▸ b).
Figure 2.
Kinetic profile of ZmGLX1. (a) Effect of pH on the specific activity of ZmGLX1. Sodium phosphate buffer with a pH range of 5.8–8.2 was used for the enzyme assay. The maximum activity was achieved at pH 7.2. The same behaviour was witnessed in the presence of added 100 µM nickel(II) or cobalt(II). (b) Metal-dependent activation of ZmGLX1. The highest catalytic activity was observed in the presence of nickel(II). The reaction medium was 100 mM phosphate buffer pH 7.2 with 100 µM of the corresponding divalent metal ion added. The enzyme concentration was 92 µM. NA indicates the activity of ZmGLX1 as isolated, i.e. without any added metals in the reaction medium. (c) Typical Michaelis–Menten plots of ZmGLX1 in the presence of nickel(II) (triangles) or cobalt(II) (circles). The hemithioacetal (HTA) resulting from spontaneous reaction between MG and GSH was considered to be the substrate. The reaction medium was 100 mM phosphate buffer pH 7.2.
Michaelis–Menten plots for ZmGLX1 were determined by varying the MG concentration (10–100 µM) at a constant GSH concentration (1.7 mM) and also by varying the GSH concentration (10–100 µM) at a constant MG concentration (1.7 mM) (Fig. 2 ▸ c). The kinetic parameters for the trace-metals enzyme sample with 100 µM nickel(II) or cobalt(II) added are summarized in Table 2 ▸. We considered the hemithioacetal (HTA) as the only substrate, which is formed spontaneously by the reaction of GSH and MG. The reported values are lower than those obtained for glyoxalases from other organisms, probably owing to lower metal uptake during the catalytic reaction. To validate this hypothesis, we evaluated the kinetic behaviour of Ni-only ZmGLX1. We found that although it shows a similar substrate affinity to trace-metals ZmGLX1, it exhibits twice the V max, suggesting that the metal uptake from the reaction medium is not efficient enough to reach the maximum activity observed for protein that has been folded in vivo with the metal ion.
Table 2. Comparison of the kinetic parameters obtained for three representative nickel(II)-dependent enzymes.
One international unit (IU) is defined as the production of 1mol of S-D-lactoylglutathione product per minute.
| Enzyme | K m, HTA (M) | V max (IUmg1) |
|---|---|---|
| Z. mays † | ||
| Trace-metals, 100M nickel(II) | 56.0 5.0 | 43.3 1.1 |
| Trace-metals, 100M cobalt(II) | 32.3 2.3 | 12.3 0.2 |
| Ni-only, 100M nickel(II) | 40.5 3.2 | 87.5 1.2 |
| Trace-metals, as purified | 58.0 8.2 | 3.2 1.2 |
| His6-less, 100M nickel(II) | 39.2 4.2 | 90.1 2.5 |
| O. sativa ‡ | 99.8 1.3 | 131 12 |
| E. coli § | 27.0 0.4 | 676 17 |
In addition, we have determined the activity performance of His6-less ZmGLX1 and its E144Q mutant. The wild-type enzyme showed maximal activity rates of 90.1 ± 2.5 IU mg−1. In contrast, the E144Q variant does not show catalytic activity, which is in agreement with its inability to bind nickel.
3.4. Protein crystallization and structure solution
One of the preparations of trace-metals ZmGLX1 was used for crystallization assays. Crystals of recombinant histidine-tagged protein were grown in a medium consisting of 4 M sodium formate at 298 K (final pH of ∼9.7) with 1% PEG 4000. The crystals belonged to the hexagonal space group P63. The asymmetric unit contained a single polypeptide chain, in which 280 (Val15–His295) of the 302 residues are clearly defined in the electron density. Notably, the crystal packing involves a nickel(II) ion co-purified with the protein, which is revealed as a strong electron-density peak surrounded by six His N∊2 atoms corresponding to His290 and His295 belonging to the His-tag at the carboxyl-termini of three symmetry-related monomers (Fig. 3 ▸). X-ray diffraction data-collection and refinement statistics are summarized in Table 3 ▸.
Figure 3.
Crystal packing partially formed by coordination of nickel(II) (arising from Ni-IMAC purification). The side chains of His290 and His295 belong to the C-terminal hexahistidine tag (amino-acid residues labelled in parentheses; yellow) in α-helix α5 (orange, green and blue).
Table 3. X-ray crystallographic data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Data collection | |
| Space group | P63 |
| Unit-cell parameters (, ) | a = b = 90.69, c = 74.71, = = 90, = 120 |
| Resolution range () | 38.81.73 (1.771.73) |
| Unique reflections | 36195 (1895) |
| Completeness (%) | 99.7 (95.0) |
| I/(I) | 20.7 (1.8) |
| Wilson B factor (2) | 26.5 |
| Multiplicity | 4.7 (2.8) |
| R merge | 0.037 (0.461) |
| Refinement | |
| PDB code | 5d7z |
| Resolution () | 1.73 |
| R cryst (%) | 16.8 |
| R free † (%) | 20.1 |
| No. of atoms | |
| Protein | 2211 |
| Solvent | 146 |
| Ligands | 42 |
| Mean B factor (2) | 29.8 |
| R.m.s.d., bond lengths () | 0.021 |
| R.m.s.d., angles () | 2.026 |
The working and free sets contained 34334 and 1843 reflections, respectively, i.e. R free was calculated using 5.1% of the reflections.
3.5. Overall structure
As previously mentioned, glyoxalases I are usually dimeric in nature, but monomeric forms of these enzymes have been reported in several species. In these organisms the enzymes have two glyoxalase domains (Fig. 1 ▸). Z. mays glyoxalase I displays the typical fold defining the glyoxalase I family, although all four βαβββ motifs belong to a single polypeptide. A structural similarity search using the DaliLite server (Holm & Rosenström, 2010 ▸) indicates that the highest structural similarity of ZmGLX1 in the Protein Data Bank is to the human protein of unknown function GloD4 (PDB entry 3zi1; 1.04 Å all-atom fitting r.m.s.d.; Structural Genomics Consortium, unpublished work). GloD4 displays a typical glyoxalase-like fold, with two potential active sites in a single polypeptide; however, it does not seem to bind any metal ions since the metal-binding motifs are missing. The next closest structural homologues are the homodimeric glyoxalases I from E. coli and L. major, which display two identical active sites, followed by the enzymes from P. aeruginosa and H. sapiens. See Supplementary Fig. S2 for a full phylogenetic tree based on the DALI server output. The N- and C-terminal domains of ZmGLX1, Val15–Pro154 and Leu155–His295, respectively, adopt the same position as each monomer in most homodimeric glyoxalases I (Fig. 1 ▸), displaying 46% sequence identity and only 0.57 Å all-atom fitting r.m.s.d. to each other. The equivalent interdomain β-sheet concavities can also be found in ZmGLX1: site A (β-sheets 5–12) and site B (β-sheets 1–4 and 13–16) (Fig. 1 ▸ b). A tubular ring-shaped electron density present in site A was modelled as a polyethylene glycol molecule chelating a sodium ion in a mode that resembles crown-ether complexes, with O—Na distances ranging from 2.33 to 2.79 Å (Supplementary Fig. S3). Site B is partially occupied by the α-helical segment α5 in the C-terminus of the protein. Even though five of the 13 residues comprising the α5 segment are part of the His-tag, the main interactions holding this helix in place correspond to hydrophobic residues of the protein, particularly Leu26, Ala228, Val280 and Phe285 (Supplementary Fig. S4), which remarkably are also present in the human protein GloD4, the function of which is still unknown. Finally, four formate anions were found in site A, interacting electrostatically with Arg120, Arg148, Arg161, Arg182 and Lys184. Notably, the corresponding residues in site B do not show electron density indicative of formate anions, i.e. Arg31 (Arg161), Arg52 (Arg182), Arg54 (Lys184), Arg254 (Arg120) and Asn282 (Arg148) (cf. the sequence alignment in Fig. 4 ▸). The only formate ion in site B is found near Lys276, equivalent to Met142 in site A. Finally, the the inter-concavity surface is largely comprised of hydrophobic residues, particularly aromatic (Phe and Tyr) and branched (Ile, Leu and Val) amino acids. These residues, which are all located in inner secondary-structural elements (i.e. excluding β-sheets 2/10 and 6/14 and helix α5), are conserved among different proteins (Fig. 4 ▸), highlighting their key roles in maintaining the overall protein fold.
Figure 4.
Sequence alignment of selected glyoxalases I. Amino-acid sequences of glyoxalase I enzymes of known three-dimensional structure compared with ZmGLX1 (UniProt C0PK05; PDB entry 5d7z) and S. cerevisiae glyoxalase I (UniProt P50107): L. major (UniProt Q68RJ8; PDB entry 2c21; Ariza et al., 2006 ▸), E. coli (UniProt P0AC81; PDB entry 1f9z; He et al., 2000 ▸), H. sapiens (UniProt Q04760; PDB entry 1fro; Cameron et al., 1997 ▸) and M. musculus (UniProt Q9CPU0; PDB entry 2za0; Kawatani et al., 2008 ▸). The N- and C-termini of ZmGLX1 have been treated separately in order to allow direct comparisons with homodimeric glyoxalases I. The phylogenetic tree on the left is a maximum-likelihood consensus tree (1000 bootstraps). Amino-acid positions involved in metal-cofactor binding are shaded green. The position of positively charged residues lining the site A and B concavities are shaded orange. Hydrophobic residues comprising the inter-concavity inner interface are highlighted in grey. Secondary-structure assignments are based on the ZmGLX1 structure (PDB entry 5d7z) reported in this work. Sheets β6 and β14 (dotted arrow) are discontinuous (cf. Fig. 1 ▸ b).
3.6. Residues involved in metal-cofactor binding
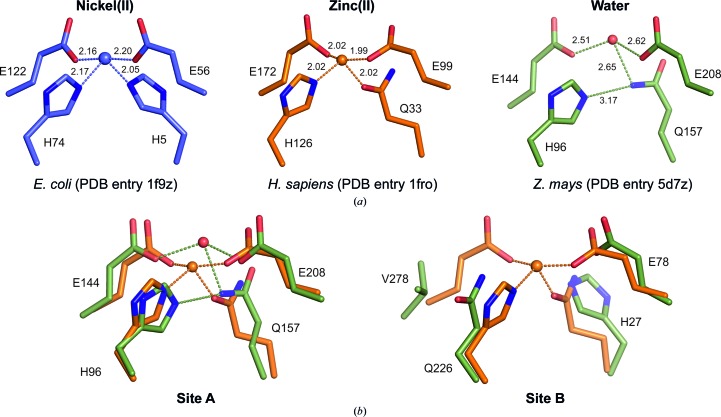
The absence of transition-metal ions in sites A or B prevents conclusive localization of the active site(s). Structure-guided sequence alignments with known glyoxalases I allow us to predict the amino-acid residues involved in metal-cofactor binding: His96, Glu144, Gln157 and Glu208 in site A and His27, Glu78, Gln226 and Val278 in site B (Figs. 4 ▸ and. 5 ▸). Mutagenesis and sequence-analysis studies of monomeric glyoxalase I from S. cerevisiae have shown that this enzyme has two functional active sites, with the conserved metal-binding motifs H25E89H269E318 and H117E163H185E242. This is not the case for ZmGLX1 owing to the presence of Val278 disrupting the site B motif. No electron-density peaks suggestive of bound metal ions were detected near the His27, Glu78 and Gln226 triad in site B, whereas a weak electron-density peak was found near Gln157 and Glu residues 144 and 208 (which are most likely to be deprotonated at pH ∼9.7), which was modelled as a water molecule. The possibility of this peak corresponding to a low-occupancy transition-metal ion can be discarded based on the longer metal–ligand distances and distorted coordination geometry (Fig. 6 ▸). However, ZmGLX1 requires divalent transition-metal ions for activity, particularly nickel(II) or cobalt(II). This unexpected lack of bound transition metals can be attributed to decreased metallation in vivo or an experimental artifact, considering that the crystals grow in a strongly chelating medium such as 4 M sodium formate pH 9.7, whereas the optimum pH for the enzyme is 7.2. Crystallization attempts in the presence of 1 mM NiSO4 resulted in a structure indistinguishable from that obtained from crystals grown without added metals, and the crystals did not withstand significant changes in the mother-liquor pH. The clear electron density for a nickel(II) ion near the carboxy-terminus, held by crystal-packing intermolecular interactions, suggests that the protein does not display a particularly high affinity towards this metal cofactor under such experimental conditions.
Figure 5.
Electron density around residues normally involved in transition-metal binding among homodimeric glyoxalases I (contoured at 1σ): His96, Glu144, Gln157 and Glu208 (site A; left) and His27, Glu78, Gln226 and Val278 (site B; right). Site A harbours a PEG molecule chelating an Na+ ion fortuitously co-crystallized with the protein.
Figure 6.
Metal-binding modes of selected glyoxalases I. (a) The metal-binding sites of most glyoxalases I, such as those from E. coli (blue) and H. sapiens (orange), display a transition-metal ion in an octahedral or trigonal geometry. In the case of ZmGLX1 (green) a transition metal was not found, but a modest side-chain reorganization seems to be likely to allow metal binding with a coordination geometry comparable to reported glyoxalases I. Dotted lines indicate metal–ligand interactions; numbers indicate distances in Å. (b) Superposition of human glyoxalase I metal-binding sites with the corresponding regions in ZmGLX1 (sites A and B). For clarity, only ZmGLX1 residues are indicated.
By superposing the structure of ZmGLX1 with available structures of glyoxalase I complexed with substrate-analogue inhibitors, several aspects of the enzyme substrate-binding mode can be discerned. H. sapiens glyoxalase I has been co-crystallized with the substrate analogue HIPC [S-(N-hydroxy-N-p-iodophenylcarbamoyl)glutathione], revealing its substrate-binding mode (Cameron et al., 1999 ▸). Upon structure superposition, HIPC appears to interact with key positively charged groups in ZmGLX1 site A, which bind formate anions in our structure (Fig. 7 ▸ a). Two formate anions bound to the Arg120 and Arg161 side chains map the positions of the respective glutathione glycyl and γ-glutamyl α-carboxyl groups. In the same way, two formate anions near Arg148, Arg182 and Lys184 map the putative location of negatively charged groups in native hemithioacetal substrates. Finally, the sodium ion, which is probably located close to the native metal cofactor, appears to be properly located near the carbamoyl group of HIPC, which emulates the glyoxal moiety of native substrates. HIPC can also be modelled in ZmGLX1 site B, despite the fact that the protein is unlikely to bind any metals in this concavity (Fig. 7 ▸ b). In this case there is only one formate anion in the vicinity of Lys276 and Arg254, which maps the position of the glutathione glycyl moiety. Residues Arg254 and Arg31 occupy the positions of Arg120 and Arg161 in site A, respectively, suggesting that site B is able to accommodate the glutathione moiety. Furthermore, Arg52 appears at the position of Arg182, which presumably interacts with negatively charged groups conjugated to glutathione. Notably, residues Arg148 in site A and Arg54 in site B are not conserved in other glyoxalases I (Fig. 4 ▸), suggesting that these residues are involved in ZmGLX1-specific ligand recognition.
Figure 7.
Inferring the ZmGLX1 ligand-binding modes. Analysis of the ZmGLX1 substrate-binding mode, as estimated through superposition with the structure of human glyoxalase I (PDB entry 1qin; Cameron et al., 1999 ▸) complexed with the substrate analogue HIPC [S-(N-hydroxy-N-p-iodophenylcarbamoyl)glutathione; orange]. (a) HIPC binding modelled in site A. (b) HIPC binding modelled in site B. Note that formate anions (FMT; yellow) in ZmGLX1 map the position of negatively charged groups in the substrate, particularly in site A.
4. Discussion
Recent reports indicate that glyoxalase I enzymes are upregulated in plants as a defence response to fungal infections, suggesting that the mechanism of methylglyoxal detoxification is part of a broader defence system. In order to shed light on the molecular properties of plant glyoxalase enzymes, we performed biochemical and structural characterization of the glyoxalase I from Z. mays, ZmGLX1.
Heterologous expression of ZmGLX1 using E. coli cultures grown in autoinducing medium yielded a protein with functional properties comparable with analogous enzymes. We found that the supplementation of culture media with trace metals gives rise to essentially metal-free ZmGLX1. However, supplementation with 1 mM NiCl2 resulted in a protein sample with exclusively nickel bound, as confirmed by atomic absorption measurements, although a lower protein yield was obtained. Besides, our in vitro metal-activation assays indicate that ZmGLX1 is maximally active in medium supplemented with nickel(II), which allowed us to confirm that ZmGLX1 is a nickel(II)-dependent metalloenzyme. Interestingly, trace-metals ZmGLX1 contained about 0.4 molar equivalents of Ca2+, despite the enzyme activity not being significantly affected by Ca2+. Even though this observation needs further investigation, the presence of Ca2+ is not unexpected, as it has been suggested that the Ca2+–calmodulin system is involved in modulation of glyoxalase I (Deswal & Sopory, 1999 ▸).
The kinetic mechanism of glyoxalase I has been a matter of intense debate for over a century, since it is still unclear whether it is a two-substrate enzyme (i.e. MG and GSH) or a one-substrate enzyme acting on the hemithioacetal formed by the spontaneous reaction of MG and GSH. We have determined the kinetic parameters for this enzyme assuming that it is a one-substrate enzyme. However, we cannot rule out that the actual substrates are MG and GSH, as proposed by Lages et al. (2012 ▸).
The overall fold of ZmGLX1, comprising an arrangement of four βαβββ motifs in two domains, is similar to that found in homodimeric surrogates of known three-dimensional structure, as revealed by our high-resolution structure. However, instead of forming two intermonomer eight-stranded β-sheets, ZmGLX1 is a single polypeptide comprising two interdomain eight-stranded β-sheets, only one of which, namely the middle eight-stranded β-sheet β5–12 (site A), harbours a functional active site. The presence of Val278 replacing a conserved glutamate metal-ligand residue in most glyoxalases I strongly suggests that site B (the eight-stranded β-sheet β1–4/13–16) in ZmGLX1 is unable to properly bind a metal cofactor and hence would lack glyoxalase activity. Besides, metal-content and activity assays of the ZmGLX1 E144Q mutant indicate that a single nickel(II) ion binds to site A. This result, together with the fact that the enzymatic activity is strictly dependent on the nickel(II) content, allows us to conclude that the protein is active with a single nickel(II) ion bound in site A. The ZmGLX1 structure shows that there are no nearby residues in site B whose side chain could compensate for the absence of a Glu278 residue in order to allow the binding of an octahedrally coordinated metal ion. Even if the protein were able to bind a metal cofactor using the His27, Gln226 and Glu78 triad, an enzyme catalyzing the same reaction with two different active sites seems to be highly unlikely. On the other hand, the observation that site B seems to be able to bind glutathione conjugates opens the possibility of ZmGLX1 activity being noncompetitively affected by such molecules, probably as part of a yet to be discovered regulatory mechanism or a different catalytic activity such as that recently discovered for Glo1 from P. aeruginosa (Bythell-Douglas et al., 2015 ▸). These features are not uncommon in this protein family. For instance, it has been suggested that the homodimeric glyoxalase I from E. coli displays distinct specificities in each of its two active sites (Su et al., 2008 ▸) and that the monomeric glyoxalases I from S. cerevisiae and P. falciparum contain two active sites with different substrate preferences and catalytic activities (Frickel et al., 2001 ▸; Deponte et al., 2007 ▸).
Detoxification of methylglyoxal, a toxic byproduct of the glycolytic pathway, is nearly as universal as glycolysis itself, and expression of proficient glyoxalase enzymes is the most widespread mechanism of defence against the harmful effects of this metabolite. Such strong selective pressure over extended evolutionary periods may have shaped glyoxalase I enzymes to optimally suite a wide variety of environments, including the biotic stresses caused by fungal infections. In this context, additional research efforts are needed in order to fully characterize Z. mays glyoxalase I, as well as other members of this highly specialized enzyme family.
Supplementary Material
PDB reference: ZmGLX1, 5d7z
supporting information. DOI: 10.1107/S1399004715015205/dw5137sup1.pdf
Acknowledgments
We thank ANPCyT (PICT 358), CONICET, SECTEI and INTA for funding. The authors gratefully acknowledge Dr R. Girolami for his help with the atomic absorption determinations. VCB is an Assistant Researcher from CONICET. JMG is the recipient of an LANL Director’s Postdoctoral Fellowship, grant DOE-LDRD 20120776PRD4. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
References
- Akoachere, M., Iozef, R., Rahlfs, S., Deponte, M., Mannervik, B., Creighton, D. J., Schirmer, H. & Becker, K. (2005). Biol. Chem. 386, 41–52. [DOI] [PubMed]
- Ariza, A., Vickers, T. J., Greig, N., Armour, K. A., Dixon, M. J., Eggleston, I. M., Fairlamb, A. H. & Bond, C. S. (2006). Mol. Microbiol. 59, 1239–1248. [DOI] [PubMed]
- Armougom, F., Moretti, S., Poirot, O., Audic, S., Dumas, P., Schaeli, B., Keduas, V. & Notredame, C. (2006). Nucleic Acids Res. 34, W604–W608. [DOI] [PMC free article] [PubMed]
- Aronsson, A. C., Marmstål, E. & Mannervik, B. (1978). Biochem. Biophys. Res. Commun. 81, 1235–1240. [DOI] [PubMed]
- Bacon, C. W., Yates, I. E., Hinton, D. M. & Meredith, F. (2001). Environ. Health Perspect. 109, 325–332. [DOI] [PMC free article] [PubMed]
- Bythell-Douglas, R., Suttisansanee, U., Flematti, G. R., Challenor, M., Lee, M., Panjikar, S., Honek, J. F. & Bond, C. S. (2015). Chem. Eur. J. 21, 541–544. [DOI] [PubMed]
- Cameron, A. D., Olin, B., Ridderström, M., Mannervik, B. & Jones, T. A. (1997). EMBO J. 16, 3386–3395. [DOI] [PMC free article] [PubMed]
- Cameron, A. D., Ridderstrom, M., Olin, B., Kavarana, M. J., Creighton, D. J. & Mannervik, B. (1999). Biochemistry 38, 13480–13490. [DOI] [PubMed]
- Chen, Z.-Y., Brown, R. L., Damann, K. E. & Cleveland, T. E. (2004). Phytopathology, 94, 938–945. [DOI] [PubMed]
- Chulze, S. N., Ramirez, M. L., Farnochi, M. C., Pascale, M., Visconti, A. & March, G. (1996). J. Agric. Food Chem. 44, 2797–2801.
- Clugston, S. L., Yajima, R. & Honek, J. F. (2004). Biochem. J. 377, 309–316. [DOI] [PMC free article] [PubMed]
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst. 35, 720–726. [DOI] [PMC free article] [PubMed]
- Deponte, M., Sturm, N., Mittler, S., Harner, M., Mack, H. & Becker, K. (2007). J. Biol. Chem. 282, 28419–28430. [DOI] [PubMed]
- Deswal, R. & Sopory, S. K. (1991). FEBS Lett. 282, 277–280. [DOI] [PubMed]
- Deswal, R. & Sopory, S. K. (1999). Biochim. Biophys. Acta, 1450, 460–467. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Espartero, J., Sánchez-Aguayo, I. & Pardo, J. M. (1995). Plant Mol. Biol. 29, 1223–1233. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Frickel, E. M., Jemth, P., Widersten, M. & Mannervik, B. (2001). J. Biol. Chem. 276, 1845–1849. [DOI] [PubMed]
- Gill, S. C. & von Hippel, P. H. (1989). Anal. Biochem. 182, 319–326. [DOI] [PubMed]
- Gomes, R. A., Sousa Silva, M., Vicente Miranda, H., Ferreira, A. E., Cordeiro, C. A. & Freire, A. P. (2005). FEBS J. 272, 4521–4531. [DOI] [PubMed]
- Greig, N., Wyllie, S., Vickers, T. J. & Fairlamb, A. H. (2006). Biochem. J. 400, 217–223. [DOI] [PMC free article] [PubMed]
- He, M. M., Clugston, S. L., Honek, J. F. & Matthews, B. W. (2000). Biochemistry 39, 8719–8727. [DOI] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Iozef, R., Rahlfs, S., Chang, T., Schirmer, H. & Becker, K. (2003). FEBS Lett. 554, 284–288. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kaur, C., Ghosh, A., Pareek, A., Sopory, S. K. & Singla-Pareek, S. L. (2014). Biochem. Soc. Trans. 42, 485–490. [DOI] [PubMed]
- Kawatani, M., Okumura, H., Honda, K., Kanoh, N., Muroi, M., Dohmae, N., Takami, M., Kitagawa, M., Futamura, Y., Imoto, M. & Osada, H. (2008). Proc. Natl Acad. Sci. USA, 105, 11691–11696. [DOI] [PMC free article] [PubMed]
- Kelley, L. A. & Sternberg, M. J. (2009). Nature Protoc. 4, 363–371. [DOI] [PubMed]
- Lages, N. F., Cordeiro, C., Sousa Silva, M., Ponces Freire, A. & Ferreira, A. E. (2012). PLoS One. 7, e32749. [DOI] [PMC free article] [PubMed]
- Lin, F., Xu, J., Shi, J., Li, H. & Li, B. (2010). Mol. Biol. Rep. 37, 729–735. [DOI] [PubMed]
- Marasas, W. F. O. (1995). Nat. Toxins, 3, 193–198. [DOI] [PubMed]
- Marmstål, E., Aronsson, A. C. & Mannervik, B. (1979). Biochem. J. 183, 23–30. [DOI] [PMC free article] [PubMed]
- Martins, A. M., Mendes, P., Cordeiro, C. & Freire, A. P. (2001). Eur. J. Biochem. 268, 3930–3936. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Mustafiz, A., Ghosh, A., Tripathi, A. K., Kaur, C., Ganguly, A. K., Bhavesh, N. S., Tripathi, J. K., Pareek, A., Sopory, S. K. & Singla-Pareek, S. L. (2014). Plant J. 78, 951–963. [DOI] [PubMed]
- Rabbani, N. & Thornalley, P. J. (2011). Semin. Cell Dev. Biol. 22, 261. [DOI] [PubMed]
- Racker, E. (1951). J. Biol. Chem. 190, 685–696. [PubMed]
- Rae, C., Berners-Price, S. J., Bulliman, B. T. & Kuchel, P. W. (1990). Eur. J. Biochem. 193, 83–90. [DOI] [PubMed]
- Ridderström, M. & Mannervik, B. (1996). Biochem. J. 316, 1005–1006. [DOI] [PMC free article] [PubMed]
- Soltis, S. M. et al. (2008). Acta Cryst. D64, 1210–1221. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Su, Z., Sukdeo, N. & Honek, J. F. (2008). Biochemistry 47, 13232–13241. [DOI] [PubMed]
- Sukdeo, N., Clugston, S. L., Daub, E. & Honek, J. F. (2004). Biochem. J. 384, 111–117. [DOI] [PMC free article] [PubMed]
- Sukdeo, N. & Honek, J. F. (2008). Drug Metabol. Drug Interact. 23, 29–50. [DOI] [PubMed]
- Suttisansanee, U. & Honek, J. F. (2011). Semin. Cell Dev. Biol. 22, 285–292. [DOI] [PubMed]
- Suttisansanee, U., Lau, K., Lagishetty, S., Rao, K. N., Swaminathan, S., Sauder, J. M., Burley, S. K. & Honek, J. F. (2011). J. Biol. Chem. 286, 38367–38374. [DOI] [PMC free article] [PubMed]
- Suttisansanee, U., Ran, Y., Mullings, K. Y., Sukdeo, N. & Honek, J. F. (2015). Metallomics, 7, 605–612. [DOI] [PubMed]
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Veena, Reddy, V. S .& Sopory, S. K. (1999). Plant J. 17, 385–395. [DOI] [PubMed]
- Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. (2009). Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yadav, S. K., Singla-Pareek, S. L. & Sopory, S. K. (2008). Drug Metabol. Drug Interact. 23, 51–68. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ZmGLX1, 5d7z
supporting information. DOI: 10.1107/S1399004715015205/dw5137sup1.pdf