Nicotinamide mononucleotide adenylyltransferase (NMNAT) catalyzes the biosynthesis of NAD+ and NaAD+. Surprisingly, when expressed in E. coli, NMNAT mutants trapped a molecule of NADP+ in their active sites. This NADP+ was bound in a conformation that was quite different from that displayed by NAD+ in the native enzyme complex.
Keywords: structural biology, NAD biosynthesis, NMN adenylyltransferase, protein crystallography, Methanobacterium thermoautotrophicum
Abstract
Nicotinamide mononucleotide adenylyltransferase (NMNAT) catalyzes the biosynthesis of NAD+ and NaAD+. The crystal structure of NMNAT from Methanobacterium thermoautotrophicum complexed with NAD+ and SO4 2− revealed the active-site residues involved in binding and catalysis. Site-directed mutagenesis was used to further characterize the roles played by several of these residues. Arg11 and Arg136 were implicated in binding the phosphate groups of the ATP substrate. Both of these residues were mutated to lysine individually. Arg47 does not interact with either NMN or ATP substrates directly, but was deemed to play a role in binding as it is proximal to Arg11 and Arg136. Arg47 was mutated to lysine and glutamic acid. Surprisingly, when expressed in Escherichia coli all of these NMNAT mutants trapped a molecule of NADP+ in their active sites. This NADP+ was bound in a conformation that was quite different from that displayed by NAD+ in the native enzyme complex. When NADP+ was co-crystallized with wild-type NMNAT, the same structural arrangement was observed. These studies revealed a different conformation of NADP+ in the active site of NMNAT, indicating plasticity of the active site.
1. Introduction
Methanobacterium thermoautotrophicum (Mt) nicotinamide mononucleotide adenylyltransferase (NMNAT; EC 2.7.7.1) reversibly catalyzes two closely related reactions: the biosyntheses of NAD+ and its nicotinic acid analogue (NaAD+) from their respective mononucleotide precursors and ATP. The gene encoding NMNAT has been identified in bacteria, archaea and eukaryotes (Emanuelli et al., 1999 ▸, 2001 ▸; Raffaelli, Emanuelli et al., 1999 ▸; Raffaelli, Lorenzi, Amici et al., 1999 ▸; Raffaelli, Lorenzi, Mariani et al., 1999 ▸; Raffaelli et al., 1997 ▸; Schweiger et al., 2001 ▸). Crystal structures of two enzymes from archaeal organisms have been determined: one from Mt complexed with NAD+ (Saridakis et al., 2001 ▸) and one from Methanococcus jannaschii (Mj) complexed with ATP (D’Angelo et al., 2000 ▸). The structures revealed the overall dinucleotide-binding fold of these enzymes, allowed their classification as members of the T/HXGH nucleotidyltransferase superfamily of α/β phosphodiesterases and identified the residues involved in substrate and product binding (D’Angelo et al., 2000 ▸; Saridakis et al., 2001 ▸). The crystal structures of the human enzymes as well as those of the closely related nicotinic acid mononucleotide adenylyltransferases from various organisms have also been reported, confirming a high degree of conservation in fold and active-site architecture (Garavaglia et al., 2002 ▸; Han et al., 2006 ▸; Olland et al., 2002 ▸; Singh et al., 2002 ▸; Werner et al., 2002 ▸; Yoon et al., 2005 ▸; Zhang et al., 2002 ▸; Zhou et al., 2002 ▸).
The synthesis, degradation and regulation of NAD+ play a crucial role in many cellular processes; the dinucleotide functions as a coenzyme in reduction–oxidation reactions and as a substrate in DNA-ligation, protein ADP-ribosylation and protein-deacetylation reactions (Denu, 2005 ▸; Ziegler, 2000 ▸). The role of NAD+ in histone and nonhistone deacetylation reactions has recently been shown to be crucial not only in lifespan extension in yeast, worms and flies, but also in apoptosis, cell survival, transcriptional silencing and metabolism (Denu, 2005 ▸). The enzymatic conversions of NAD+ to cyclic ADP-ribose or nicotinic acid adenine dinucleotide phosphate, two calcium-mobilizing agents, suggest a role in cell signalling (Ziegler, 2000 ▸). Another transformation of NAD+ is the phosphorylation of its 2′-hydroxyl group to give NADP+ (Denicola-Seoane & Anderson, 1990 ▸). The biosynthesis of NAD+ occurs via both de novo and salvage pathways, which have been well studied in both prokaryotes and eukaryotes (Magni et al., 1999 ▸). The starting point of NAD+ biosynthesis in eukaryotes is the oxidation of tryptophan, whereas in prokaryotes it is the conversion of l-aspartic acid to iminoaspartate. The eukaryotic and prokaryotic de novo pathways converge at quinolinic acid. Subsequently, similar pathways are followed in many organisms for the conversion of quinolinic acid to NAD+. NAD+ degradation proceeds via the breakage of either the N-glycosidic or the pyrophosphate bond, resulting in the generation of either nicotinamide or nicotinamide mononucleotide. These molecules are also the products of the DNA-ligation and protein ADP-ribosylation reactions and can be converted back to NAD+ via the salvage biosynthetic pathways.
The NAD+-biosynthetic pathway provides promising targets for antibacterial therapeutics because the synthesis of NAD+ is essential in these organisms (Magni et al., 2009 ▸). More importantly, structural and functional studies have shown that human NMNATs are substantially different from prokaryotic NaMN/NMNATs, indicating that these enzymes should be valid drug targets (Sorci et al., 2009 ▸).
Sequence alignment, together with the results of several crystallographic studies, identified a number of conserved residues that are located in the active site of NMNAT and are well placed to interact with the substrates of the enzyme (Saridakis et al., 2001 ▸; Saridakis & Pai, 2003 ▸). Site-directed mutagenesis studies assigned roles in substrate binding to a number of them, including Arg11 and Arg136. Kinetic assays indicated that mutating these arginine residues to lysines had no effect on enzymatic activity (Saridakis & Pai, 2003 ▸). However, while determining the crystal structures of these and other NMNAT mutants, we were surprised to find that they specifically bound NADP+ at their active sites, a molecule scavenged from the cytosol of the Escherichia coli cells used to overexpress the protein. The native NMNAT protein had always been purified as the NAD+ complex (Saridakis et al., 2001 ▸); however, it could also be co-crystallized with NADP+. This report describes the results of our efforts to characterize the interaction of NADP+ with NMNAT at the molecular level.
2. Materials and methods
2.1. Site-directed mutagenesis
Site-directed mutagenesis was carried out using the QuikChange kit. DNA encoding native NMNAT cloned into pET-15b was used as the template for polymerase chain reaction mutagenesis. Briefly, 25 ng template DNA was incubated with the appropriate mutagenic primers, dNTPs and Pfu DNA polymerase using the temperature-cycling parameters recommended in the supplier’s manual. Following this, DpnI was added to each amplification reaction and incubated at 37°C for 6 h followed by transformation of the mutagenized plasmid into XL2 Blue cells. The mutations were verified by DNA sequencing.
2.2. Protein expression and purification
Recombinant NMNAT was overexpressed in E. coli BL21 Gold (DE3) cells harbouring a plasmid encoding three rare E. coli tRNA genes. The cells were grown at 37°C in Luria–Bertani broth with carbenicillin (50 µg ml−1) and kanamycin (50 µg ml−1) to an OD600 nm of 0.7 and were induced overnight with 0.4 mM isopropyl β-d-1-thiogalactopyranoside at 24°C. The bacteria were harvested by centrifugation and resuspended in binding buffer (50 mM Tris, 500 mM NaCl, 5% glycerol, 5 mM imidazole) supplemented with 2 mM phenylmethylsulfonyl fluoride. The bacteria were lysed by five rounds of sonication (each for 1 min at 50% maximum output, Branson Sonifier 450). Cell debris was removed by centrifugation for 30 min at 20 000g and 4°C. The supernatant was applied by gravity onto a DE52 column (Whatman) immediately coupled to a Ni–NTA column (Qiagen). The Ni–NTA column was washed with 20 volumes of binding buffer containing 30 mM imidazole. The bound NMNAT was eluted from the Ni–NTA column with 500 mM imidazole in binding buffer. The N-terminal hexahistidine tag was cleaved off with thrombin for 16 h (1 µg thrombin per milligram of recombinant protein) at 4°C in binding buffer containing 2.5 mM CaCl2. NMNAT was then dialyzed against 500 mM NaCl in 10 mM HEPES pH 7.5 and concentrated to 10 mg ml−1 using Biomax concentrators (Millipore).
2.3. Crystallization
Screening for crystallization conditions was performed using Crystal Screen (Hampton Research) and a modified ammonium sulfate grid screen at room temperature in VDX plates in a hanging-drop vapour-diffusion setup. 2 µl protein solution (10 mg ml−1) was mixed with 2 µl of the various reservoir solutions and equilibrated against 0.5 ml of these solutions. Hexagonal bipyramidal-shaped crystals appeared after 48–72 h in Crystal Screen setups that contained ammonium sulfate or lithium sulfate as precipitants and in most of the conditions with a pH higher than 6 in the modified ammonium sulfate grid screen. The crystals selected for data collection were grown in 1.5–1.6 M ammonium sulfate, 5% glycerol, 100 mM Tris pH 8.0 at 20°C. They were flash-cooled with crystallization buffer containing 30% glycerol as a cryoprotectant. In order to grow co-crystals of native NMNAT complexed with NADP+, the protein (10 mg ml−1) was incubated at 65°C with 5 mM NADP+ for 5 min to replace the NAD+ carried in native NMNAT purified from E. coli (Saridakis et al., 2001 ▸). Crystals of the resulting complex were produced as described above.
2.4. X-ray diffraction data collection and structure determination
Diffraction data from NADP+-complexed wild-type (WT) and mutant NMNAT crystals were collected on BioCARS beamline BM14C at the Advanced Photon Source, Argonne National Laboratory, USA and on beamline X8C at the National Synchrotron Light Source, Brookhaven National Laboratory, USA at 100 K using a Q4 CCD detector (ADSC). All of the X-ray data were processed and scaled with the DENZO/SCALEPACK suite of programs (Otwinowski & Minor, 1997 ▸). NADP+-complexed WT and mutant structures were solved employing the AMoRe program package using the WT NMNAT structure (PDB entry 1ej2; Saridakis et al., 2001 ▸) as the search model (Navaza, 2001 ▸; Winn et al., 2011 ▸). Model visualization and rebuilding were performed with Coot (Emsley et al., 2010 ▸), and REFMAC (Murshudov et al., 2011 ▸) was used for model refinement. Crystallographic and refinement statistics can be found in Table 1 ▸. The programs MolScript (Kraulis, 1991 ▸), Raster3D (Merritt & Murphy, 1994 ▸), PyMOL (v.1.3r1; Schrödinger) and LIGPLOT (Wallace et al., 1995 ▸) were used in the production of the figures.
Table 1. X-ray data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell (2.312.21 for wild-type Mt-NMNAT , 2.001.90 for the Arg11Lys mutant and 2.402.30 for the Arg47Glu mutant).
| Wild-type Mt-NMNAT | Arg11Lys Mt-NMNAT | Arg47Glu Mt-NMNAT | |
|---|---|---|---|
| PDB code | 4yp5 | 4yp6 | 4yp7 |
| Bound ligand | NADP | NADP | NADP |
| Crystal data | |||
| Space group | P3121 [No. 152] | P3121 [No. 152] | P3121 [No. 152] |
| Unit-cell parameters | |||
| a = b () | 124.59 | 124.47 | 124.45 |
| c () | 111.96 | 112.38 | 111.94 |
| = () | 90 | 90 | 90 |
| () | 120 | 120 | 120 |
| Diffraction data | |||
| Wavelength () | 1.000 | 1.100 | 1.000 |
| Resolution limit () | 2.21 | 1.90 | 2.30 |
| R merge (%) | 5.2 (25.5) | 7.8 (47.1) | 6.0 (21.7) |
| I/(I) | 15.7 (4.8) | 11.8 (2.4) | 18.8 (6.1) |
| Completeness (%) | 89.3 (87.0) | 98.2 (97.2) | 93.1 (93.3) |
| Model refinement | |||
| Unique reflections used | 43.177 | 74.179 | 39.652 |
| Resolution range () | 20.02.21 | 20.01.90 | 15.02.30 |
| R work (%) | 18.1 | 17.5 | 19.0 |
| R free (%) | 19.5 | 18.8 | 20.8 |
| Ramachandran plot | |||
| Outlier residues | 0 | 0 | 0 |
| Allowed residues | 3 | 3 | 0 |
| Favoured residues | 496 | 498 | 494 |
| No. of water molecules | 152 | 497 | 155 |
| R.m.s.d., bond lengths () | 0.012 | 0.012 | 0.012 |
| R.m.s.d., bond angles () | 1.59 | 1.58 | 1.57 |
2.5. Kinetic assays and data analysis
NMNATase activity was measured in a continuous coupled assay as described previously (Saridakis & Pai, 2003 ▸). Briefly, NMNATase (2 µg) was incubated with varying amounts of NMN+ and ATP together with 5 mM MgCl2, 1% ethanol, one unit of alcohol dehydrogenase, 50 mM HEPES pH 7.5 and increasing amounts of NADP+ at 65°C for 2 min. The reaction was monitored at 340 nm using a double-beam spectrophotometer (GBC Scientific Cintra 20) equipped with a temperature-controlled cell holder. Kinetic data were fitted to the appropriate rate equation using GraFit (Erathicus Software).
3. Results
3.1. NMNAT crystallization and structure determination
Crystals of the WT and mutant NMNAT enzymes complexed with NADP+ grew under conditions similar to those for WT NMNAT–NAD+ (Saridakis et al., 2001 ▸; Saridakis & Pai, 2003 ▸); however, the morphologies of the crystals were quite different. The crystals of the NADP+ complexes of the WT and the Arg11Lys and Arg47Glu mutant enzymes were hexagonal bipyramids instead of hexagonal rods. The crystals of the Arg47Lys and Arg136Lys NMNATs also had a bipyramidal shape but did not diffract well enough for further analysis. The crystal structures of the NADP+ complexes of the WT, Arg11Lys and Arg47Glu NMNATs were determined to between 2.3 and 1.9 Å resolution using molecular-replacement techniques.
3.2. Structural analysis
A trimer of NMNAT is found in the asymmetric unit of these structures, compared with a monomer in the original NMNAT structure, which was a complex with NAD+. The overall structures of the WT, Arg11Lys and Arg47Glu NMNAT proteins complexed with NADP+ are similar to that of WT NMNAT–NAD+. The r.m.s.d. of 167 equivalent Cα-atom positions of each monomeric structure within the trimer compared with WT NMNAT–NAD+ is 1.2 Å for subunit A and 1.1 Å for subunits B and C (Fig. 1 ▸). These rather large values are owing to an opening of the active site, which is necessary to accommodate the different nicotinamide nucleotide. The hexameric structures of the enzymes complexed with NADP+ are quite similar to that of the WT NMNAT–NAD+ complex. However, there are some slight differences when the structure of WT NMNAT–NAD+ is compared with that of the WT NMNAT–NADP+ complex, which are most probably caused by NADP+, although changed crystallographic contacts cannot be completely ruled out as the cause of these differences. Helices 4, 5 and 6 have moved slightly, resulting in an enlargement of the active site, which now accommodates the changed conformation of the adenylyl moiety of NADP+ (Fig. 1 ▸). In the structure of WT NMNAT–NAD+, the loop incorporating residues 123–129 was quite disordered, as indicated by high temperature factors. However, the same loop is well ordered in the WT NMNAT–NADP+ structure. There are also some subtle differences between the subunits of the trimer in the asymmetric unit. In contrast to subunits B and C, subunit A does not form any crystallographic contacts, leading to higher average B factors compared with the other two subunits. The crystallographic contacts of subunits B and C occur close to the loop comprising residues 123–129, and these crystallographic contacts are one of the reasons why this loop is more ordered in subunits B and C in the structure of the WT NMNAT–NADP+ complex.
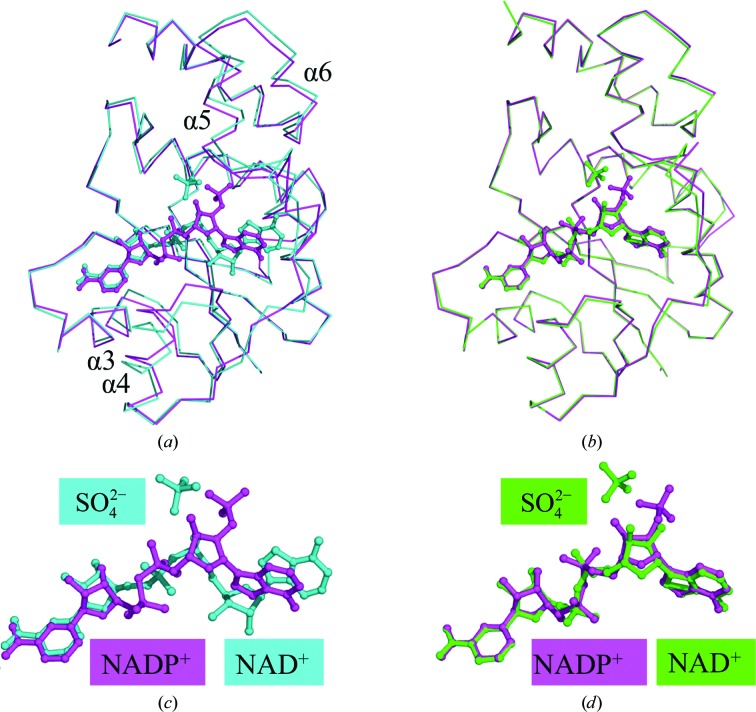
Figure 1.
(a) Superposition of ribbon diagrams of WT NMNAT complexed with NAD+ (cyan) compared with NADP+ (magenta). The largest deviation in WT NMNAT to accommodate NADP+ is seen in helices 4, 5 and 6. There is an overall opening of the active site to fit the larger NADP+ molecule. (b) Superposition of ribbon diagrams of His19Ala NMNAT complexed with NAD+ (green) compared with WT NMNAT with NADP+ (magenta). The NAD+ and NADP+ molecules are completely superposed in this alternative mode of dinucleotide binding to NMNAT. (c) Close-up of the superposition of NAD+ (cyan) and NADP+ (magenta) in WT NMNAT. The nicotinyl moiety is very closely aligned between NAD+ and NADP+, whereas the adenylyl moiety is completely misaligned. The adenylyl moiety has undergone a rotation to allow its adenine ring to interact with the side chain of Tyr126 of NMNAT. The ribose of the adenylyl moiety has also undergone a rotation to allow the 2′-phosphate to interact with His16 and His19 of NMNAT. (d) Close-up of the superposition of NAD+ in His19Ala NMNAT (green) and NADP+ in WT NMNAT (magenta). An identical rotation around the adenylyl moiety is observed between NAD+ and NADP+. A sulfate molecule mimicking the γ-phosphate of ATP was identified in the active sites of the WT and H19A NMNAT–NAD+ complexes and is shown for clarity.
The WT NMNAT–NADP+ complex was also compared with the His19Ala NMNAT–NAD+ complex (Saridakis & Pai, 2003 ▸). His19Ala NMNAT contains a mutation at a critical catalytic residue. This mutant, when purified and crystallized, also contained a molecule of NAD+ in its active site, which adopted a conformation similar to that of NADP+ in the mutants described in this work. The r.m.s.d between 164 equivalent Cα-atom positions within WT NMNAT–NADP+ and His19Ala NMNAT–NAD+ is 0.3 Å, indicating that the overall structure of WT NMNAT–NADP+ is similar to that of the His19Ala NMNAT–NAD+ complex (Fig. 1 ▸).
3.3. NADP+ analysis
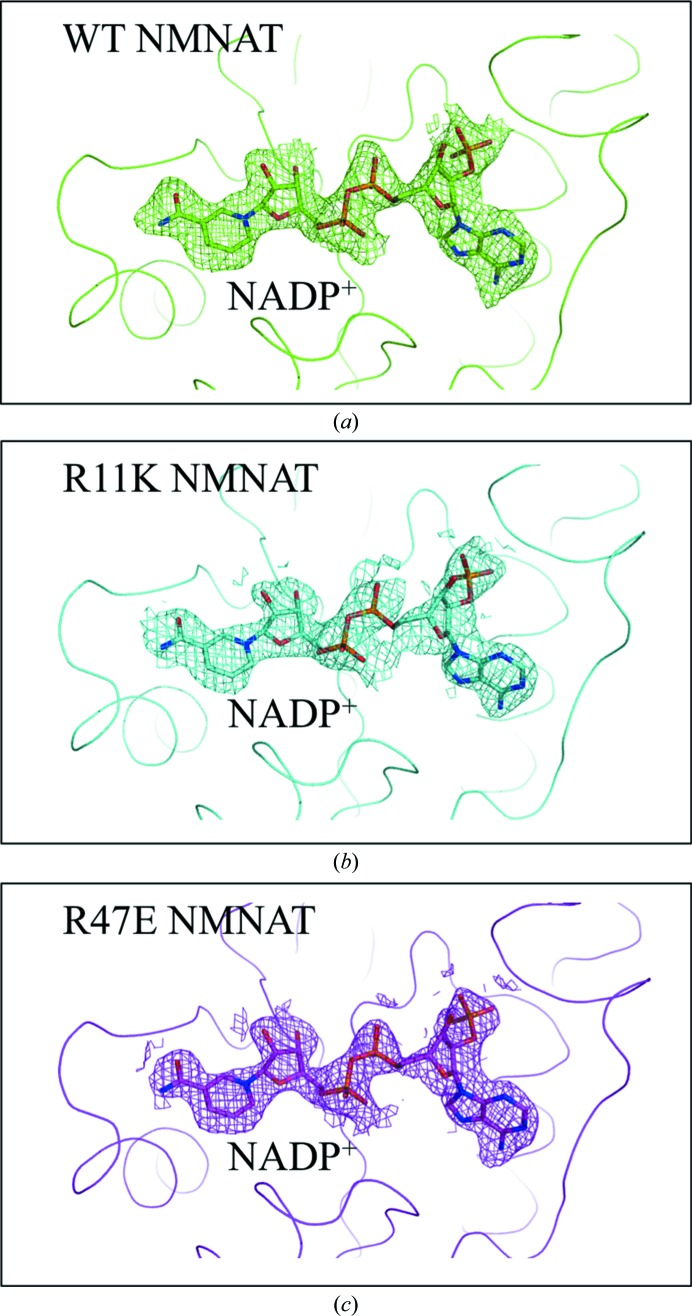
A molecule of NADP+ was initially observed in the difference Fourier maps of Arg47Glu and Arg11Lys NMNAT (Fig. 2 ▸). As no NADP+ was added during the purification or crystallization of these mutant NMNAT enzymes, we propose that each protein molecule trapped a molecule of NADP+ in its active site during expression in E. coli. This was reminiscent of WT NMNAT scavenging a molecule of its product, NAD+, from the bacterial cytosol (Saridakis et al., 2001 ▸). Once alerted to this possibility, we also succeeded in exchanging the NAD+ molecules bound in WT NMNAT with NADP+ (Fig. 2 ▸).
Figure 2.
Difference Fourier maps of WT and mutant NMNAT enzymes. Difference Fourier maps of (a) WT NMNAT, (b) Arg11Lys NMNAT and (c) Arg47Glu NMNAT after the first round of refinement, indicating the presence of NADP+ in the active site. The refined model of NADP+ is shown. All of the maps are contoured at 1σ.
The position and orientation of NADP+ in the active sites of the Arg47Glu, Arg11Lys and WT NMNATs are similar (Fig. 2 ▸) and are almost identical to the conformation of NAD+ trapped in the His19Ala mutant of NMNAT (Fig. 1 ▸). However, they are quite different from those of NAD+ in WT NMNAT (Fig. 1 ▸; Saridakis & Pai, 2003 ▸). In all structures, the NADP+ is in its extended form and its adenine ring is in the anti conformation. The nicotinamide ring is the only part of the molecule of NADP+ that closely overlays with its corresponding part of NAD+ in the conformation found in WT NMNAT. The remainder of the molecule interacts quite differently with the enzyme, with the distances between corresponding atoms increasing as one moves from the nicotinamide to the adenine ring. The nicotinylyl ribose rotates by 0.9 Å towards the cap of helix 4. The nicotinylyl phosphate has moved 2.6 Å away from the position that it occupies in WT NMNAT–NAD+ and now sits closer to the cap of helix 4, but still interacts with Asn105, the Cα atom of which (which is in helix 4) has moved by 2 Å. NADP+ makes the following contacts with NMNAT (Fig. 3 ▸), several of which are identical to those formed between NMNAT and NAD+. The nicotinamide ring is held in place by contacts made with Asn84 and stacking with Trp87. The nicotinyl ribose interacts with both Ser39 and Asp80. The O atoms on the phosphates directly bind to Asn105 and in addition make several water-mediated contacts to NMNAT. The adenylyl ribose does not engage in any hydrogen bonds. The 2′-phosphate interacts directly with His16 and His19 and makes many water-mediated contacts with NMNAT. The adenine ring stacks with the aromatic ring of Tyr126.
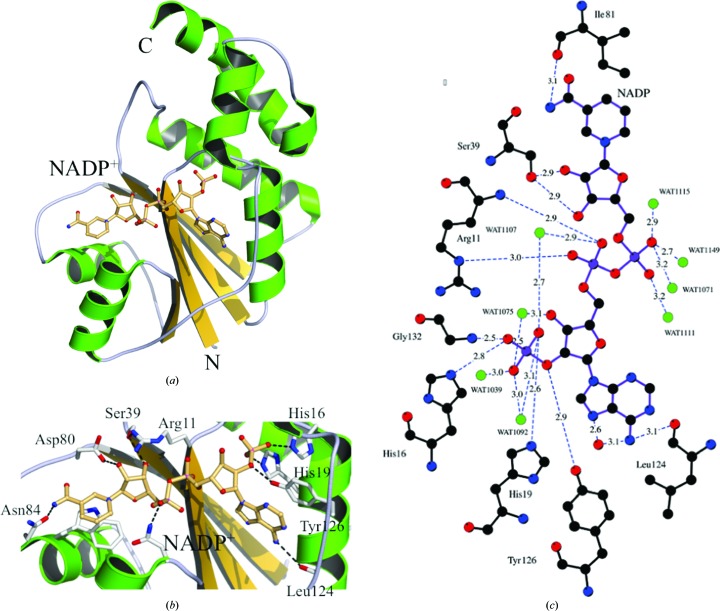
Figure 3.
Active-site interactions in the WT NMNAT–NADP+ complex. (a) Ribbon diagram of NMNAT with helices shown in green and strands in yellow. The molecule of NADP+ is shown in ball-and-stick representation. (b) Interacting NMNAT residues and NADP+ are shown in ball-and-stick representation. The most notable interactions are between the 2′-phosphate of NADP+ and NMNAT residues His16 and His19. The NADP+ adenine ring also stacks with the side chain of Tyr126. (c) Schematic representation of the interactions between WT NMNAT and NADP+. The distances and identities of the polar interactions are shown and are described in the text. Identical interactions occur between Arg11Lys and Arg47Glu NMNAT with NADP+ but have not been shown.
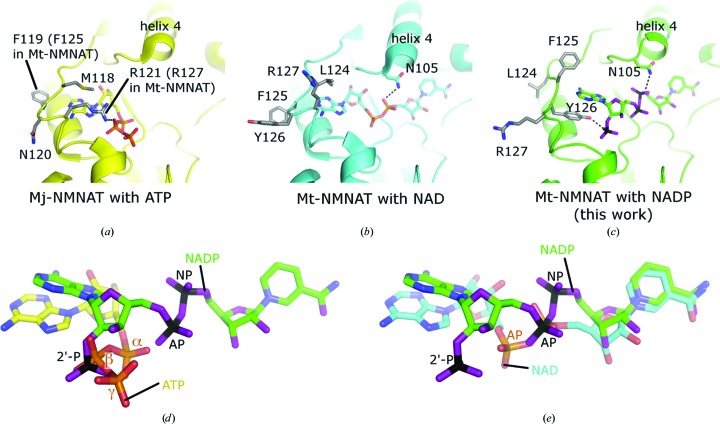
As mentioned previously, helix 4 has been pushed away from its original position in the structure of WT NMNAT–NAD+, forming a slightly wider active site to accommodate the larger NADP+ molecule (Fig. 4 ▸). The adenylyl phosphate has moved 3.4 Å from its position in the NAD+ complex. The adenylyl ribose has rotated by approximately 140° around an axis connecting the adenylyl phosphorus and the adenine-ring system, and the 3′-hydroxyl, having shifted 7.7 Å from its original position, no longer interacts with the amide of Gly104. The adenine ring is also rotated 140° and now forms a favourable stacking interaction with the aromatic ring of Tyr126, the side-chain hydroxyl group of which binds to the 2′-phosphate. In this process, the N6 amide of the adenine ring has moved approximately 4.6 Å away from its original position. In the structure of WT NMNAT–NAD+, the side chain of Tyr126 is pointing in the opposite direction (away from the active site), where it is surrounded by solvent and is therefore quite dynamic (Fig. 4 ▸). A similar arrangement is found in Mj-NMNAT–ATP, where Arg121 (Arg127 in Mt-NMNAT) rather than Tyr126 stacks with the adenine ring of ATP and again forms a hydrogen bond to the β-phosphate of ATP. In our structure, the 2′-phosphate of NADP+ superimposes well with the β-phosphate of ATP in Mj-NMNAT (Fig. 4 ▸).
Figure 4.
Adjustments of the NMNAT-binding pocket to different nucleotides and alternative conformations. (a) NMNAT with the substrate ATP (PDB entry 1f9a; D’Angelo et al., 2000 ▸). (b) NMNAT with the reaction product NAD+ (PDB entry 1ej2; Saridakis et al., 2001 ▸). (c) NMNAT with NADP+ (PDB entry 4h6l; this work). (d) Position of NADP+ compared with that of ATP (PDB entry 1f9a). (e) Position of NADP+ compared with that of NAD+ (PDB entry 1ej2). (d) and (e) were obtained by superposing the entire NMNAT protein structures onto each other while omitting the ligands. In NADP+ the nucleotide is flipped by about 180° compared with NAD+ and ATP, thereby positioning the 2′-phosphate group at the position of the β-phosphate site of ATP. The positions of the central phosphate groups in NADP+ (AP and NP) have also slightly shifted compared with NAD+, whereas the position of the nicotinamide group is almost unchanged.
3.4. NADP+ inhibition
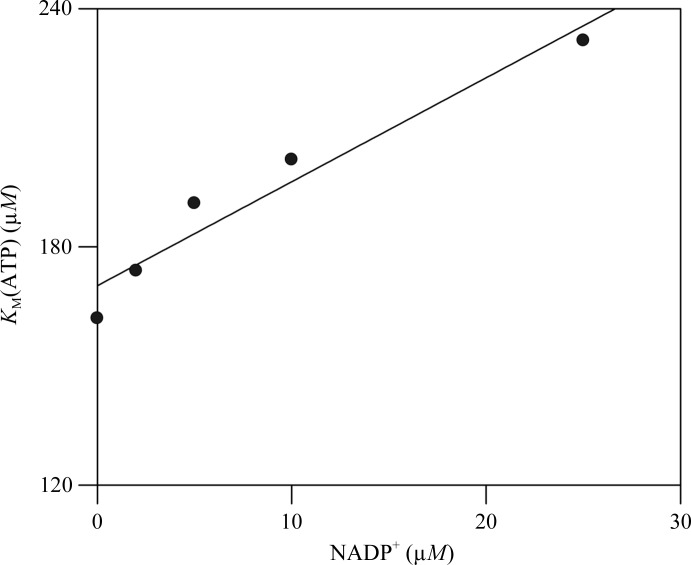
Inhibition of WT NMNAT by NADP+ was analyzed using the forward reaction (synthesis of NAD+). The K m values for ATP and NMN+ were determined by varying the concentration of NADP+. These assays demonstrated that NADP+ inhibits the NMNAT reaction in a manner competitive with ATP (K i = 65 ± 5 µM) but not NMN+ (Fig. 5 ▸).
Figure 5.
NADP+ inhibition of the NMNAT-catalyzed reaction.
4. Discussion
The biosynthesis of NADP+ can be considered as a natural extension of NAD+ biosynthesis and a single enzyme, NAD+ kinase, catalyzes its synthesis from NAD+ and ATP. Feedback inhibition of NMNAT by NADP+ would reduce the rate of synthesis of NAD+ should large amounts of NADP+ accumulate in the cell. Most importantly, NADP+ was shown to inhibit two of the three human NMNAT isoforms, NMNAT1 and NMNAT2 (Sorci et al., 2007 ▸). The NMNAT1-catalyzed and NMNAT2-catalyzed reactions were 30 and 70% inhibited by NADP+. However, NADP+ was unable to inhibit NMNAT3 under the assayed conditions. Our studies revealed that NADP+ inhibits enzyme activity by selectively competing for the ATP site with an inhibition constant of 65 ± 5 µM but not for the NMN+-binding site. This is consistent with all of our structural studies of NAD+ and NADP+ complexes of WT and mutant NMNAT enzymes.
Our analysis of NMNAT–NADP+ complexes revealed a remarkably close fit of the dinucleotide into the active site of the enzyme. NMNAT was able to readily accept the larger NADP+ molecule with only small changes necessary in the positioning of helices 4, 5 and 6. Also, by rotating about the adenylyl C′4—C′5 bond, NADP+ placed its 2′-phosphate in the position usually occupied by the β-phosphate of the substrate ATP. In this orientation, the 2′-phosphate forms hydrogen bonds to both His16 and His19. The same conformational change places the adenine ring in an orientation that allows it to stack with the aromatic ring of Tyr126. Even though Tyr126 is not conserved in NMNATs from other organisms, we propose that these enzymes should still be able to bind NADP+ in a similar conformation owing to the dominance of the energetically favourable 2′-phosphate–protein interactions (Figs. 4d and 4e ▸). We thus propose that extension of the 2′-phosphate of NADP+ to mimic the γ-phosphate group of ATP will enhance the binding affinity to NMNAT.
Comparing the crystal structures of Mt- and Mj-NMNATs complexed with NAD+, NMN+ and ATP, respectively, one can easily visualize an inline attack of NMN+ on the α-phosphate of ATP, with NAD+ and PPi as products (D’Angelo et al., 2000 ▸; Saridakis et al., 2001 ▸). As isolated from E. coli, WT NMNAT had the product NAD+ trapped in its active site in a conformation that is fully compatible with such a mechanism (Saridakis et al., 2001 ▸). Structural analysis of the His19Ala mutant of NMNAT also revealed a bound NAD+ molecule, albeit in a conformation corresponding to that now described for NADP+ (Saridakis & Pai, 2003 ▸). An NMNAT-catalyzed reaction would not be possible in this latter conformation, since it would be impossible to orient the ATP substrate in the manner seen in Mj-NMNAT (D’Angelo et al., 2000 ▸). The β- and γ-phosphates of ATP would no longer be able to interact with Arg11, His16, His19 and Arg136, residues which are involved in substrate binding and catalysis (Saridakis et al., 2001 ▸; Saridakis & Pai, 2003 ▸). There also are no other polar residues in the active site that could substitute for these lost interactions. Even if it were able to bind, ATP could not adopt a catalytically competent position that would allow an inline attack by the phosphate group of NMN+. Therefore, we postulate that NADP+ binding to NMNAT leads to inhibition of the catalytic activity of the enzyme.
The Arg11Lys and Arg136Lys NMNAT mutants both retained almost wild-type levels of enzymatic activity and bound both substrates with similar Michaelis constants to WT NMNAT (Saridakis & Pai, 2003 ▸). Therefore, it was rather unexpected to identify NADP+ in the active site of the Arg11Lys mutant, which was the first structure of the hexagonal bipyramidal crystal form that was determined. In the case of the Arg11Lys NMNAT mutant, the crystallization drop contained a mixture of the two crystal forms (hexagonal rods and hexagonal bipyramids), indicating that Arg11Lys NMNAT was capable of trapping both NAD+ and NADP+ in its active site. In contrast, the Arg47Glu, Arg47Lys and Arg136Lys NMNAT crystallization drops only contained hexagonal bipyramidal crystals, indicating that these mutants preferred to bind NADP+ rather than NAD+.
Organisms must possess the ability to regulate the intracellular concentrations of NAD+/NADP+, given the importance of these coenzymes in many cellular processes. These NMNAT–NADP+ complex structures are the first to depict the interactions between NMNAT and NADP+. The active site of Mt-NMNAT displays significant plasticity, which is important for its ability to accommodate the molecule of NADP+ in this alternate conformation. It would be interesting to determine whether NMNAT proteins from other organisms possess the same inherent plasticity and permit the binding of molecules in similar conformations.
Supplementary Material
PDB reference: NMNAT, wild type, complex with NADP, 4yp5
PDB reference: Arg11Lys mutant, complex with NADP, 4yp6
PDB reference: Arg47Glu mutant, complex with NADP, s4yp7
Acknowledgments
We thank the staff of BioCARS for help during data collection at Sector 14 of the Advanced Photon Source, as well as L. Flaks and J. Berendzen for help during data collection on beamline X8C of the National Synchrotron Light Source. We also thank Dinesh Christendat and Alexey Bochkarev for help with analysis of the diffraction data. EFP acknowledges funding from the Natural Sciences and Engineering Research Council of Canada and support through the Canada Research Chairs Program. This research was also funded in part by the Ontario Ministry of Health and Long Term Care (OMHTLC). VS acknowledges funding from the Canadian Institutes of Health Research (CIHR; grant No. 106583). The views expressed do not necessarily reflect those of the OMHLTC. Use of the Advanced Photon Source was supported by the Basic Energy Sciences, Office of Science, United States Department of Energy under Contract W-31-109-Eng-38. Use of BioCARS Sector 14 was supported by the National Center for Research Resources, National Institutes of Health under grant RR07707. A joint grant from the CIHR and NSERC of Canada enabled the use of beamline X8C at the National Synchrotron Light Source, Brookhaven National Laboratory.
References
- D’Angelo, I., Raffaelli, N., Dabusti, V., Lorenzi, T., Magni, G. & Rizzi, M. (2000). Structure, 8, 993–1004. [DOI] [PubMed]
- Denicola-Seoane, A. & Anderson, B. M. (1990). J. Gen. Microbiol. 136, 425–430. [DOI] [PubMed]
- Denu, J. M. (2005). Curr. Opin. Chem. Biol. 9, 431–440. [DOI] [PubMed]
- Emanuelli, M., Carnevali, F., Lorenzi, M., Raffaelli, N., Amici, A., Ruggieri, S. & Magni, G. (1999). FEBS Lett. 455, 13–17. [DOI] [PubMed]
- Emanuelli, M., Carnevali, F., Saccucci, F., Pierella, F., Amici, A., Raffaelli, N. & Magni, G. (2001). J. Biol. Chem. 276, 406–412. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Garavaglia, S., D’Angelo, I., Emanuelli, M., Carnevali, F., Pierella, F., Magni, G. & Rizzi, M. (2002). J. Biol. Chem. 277, 8524–8530. [DOI] [PubMed]
- Han, S., Forman, M. D., Loulakis, P., Rosner, M. H., Xie, Z., Wang, H., Danley, D. E., Yuan, W., Schafer, J. & Xu, Z. (2006). J. Mol. Biol. 360, 814–825. [DOI] [PubMed]
- Kraulis, P. J. (1991). J. Appl. Cryst. 24, 946–950.
- Magni, G., Amici, A., Emanuelli, M., Raffaelli, N. & Ruggieri, S. (1999). Adv. Enzymol. Relat. Areas Mol. Biol. 73, 135–182. [DOI] [PubMed]
- Magni, G., Di Stefano, M., Orsomando, G., Raffaelli, N. & Ruggieri, S. (2009). Curr. Med. Chem. 16, 1372–1390. [DOI] [PubMed]
- Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst. D50, 869–873. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed]
- Olland, A. M., Underwood, K. W., Czerwinski, R. M., Lo, M.-C., Aulabaugh, A., Bard, J., Stahl, M. L., Somers, W. S., Sullivan, F. X. & Chopra, R. (2002). J. Biol. Chem. 277, 3698–3707. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Raffaelli, N., Emanuelli, M., Pisani, F. M., Amici, A., Lorenzi, T., Ruggueru, S. & Magni, G. (1999). Mol. Cell. Biochem. 193, 99–102. [PubMed]
- Raffaelli, N., Lorenzi, T., Amici, A., Emanuelli, M., Ruggieri, S. & Magni, G. (1999). FEBS Lett. 444, 222–226. [DOI] [PubMed]
- Raffaelli, N., Lorenzi, T., Mariani, P. L., Emanuelli, M., Amici, A., Ruggieri, S. & Magni, G. (1999). J. Bacteriol. 181, 5509–5511. [DOI] [PMC free article] [PubMed]
- Raffaelli, N., Pisani, F. M., Lorenzi, T., Emanuelli, M., Amici, A., Ruggieri, S. & Magni, G. (1997). J. Bacteriol. 179, 7718–7723. [DOI] [PMC free article] [PubMed]
- Saridakis, V., Christendat, D., Kimber, M. S., Dharamsi, A., Edwards, A. M. & Pai, E. F. (2001). J. Biol. Chem. 276, 7225–7232. [DOI] [PubMed]
- Saridakis, V. & Pai, E. F. (2003). J. Biol. Chem. 278, 34356–34363. [DOI] [PubMed]
- Schweiger, M., Hennig, K., Lerner, F., Niere, M., Hirsch-Kauffmann, M., Specht, T., Weise, C., Oei, S. L. & Ziegler, M. (2001). FEBS Lett. 492, 95–100. [DOI] [PubMed]
- Singh, S. K., Kurnasov, O. V., Chen, B., Robinson, H., Grishin, N. V., Osterman, A. L. & Zhang, H. (2002). J. Biol. Chem. 277, 33291–33299. [DOI] [PubMed]
- Sorci, L., Cimadamore, F., Scotti, S., Petrelli, R., Cappellacci, L., Franchetti, P., Orsomando, G. & Magni, G. (2007). Biochemistry, 46, 4912–4922. [DOI] [PubMed]
- Sorci, L., Pan, Y., Eyobo, Y., Rodionova, I., Huang, N., Kurnasov, O., Zhong, S., MacKerell, A. D. Jr, Zhang, H. & Osterman, A. L. (2009). Chem. Biol. 16, 849–861. [DOI] [PMC free article] [PubMed]
- Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995). Protein Eng. Des. Sel. 8, 127–134. [DOI] [PubMed]
- Werner, E., Ziegler, M., Lerner, F., Schweiger, M. & Heinemann, U. (2002). FEBS Lett. 516, 239–244. [DOI] [PubMed]
- Winn, M. D. (2011). Acta Cryst. D67, 235–242.
- Yoon, H.-J., Kim, H. L., Mikami, B. & Suh, S. W. (2005). J. Mol. Biol. 351, 258–265. [DOI] [PubMed]
- Zhang, H., Zhou, T., Kurnasov, O., Cheek, S., Grishin, N. V. & Osterman, A. (2002). Structure, 10, 69–79. [DOI] [PubMed]
- Zhou, T., Kurnasov, O., Tomchick, D. R., Binns, D. D., Grishin, N. V., Marquez, V. E., Osterman, A. L. & Zhang, H. (2002). J. Biol. Chem. 277, 13148–13154. [DOI] [PubMed]
- Ziegler, M. (2000). Eur. J. Biochem. 267, 1550–1564. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: NMNAT, wild type, complex with NADP, 4yp5
PDB reference: Arg11Lys mutant, complex with NADP, 4yp6
PDB reference: Arg47Glu mutant, complex with NADP, s4yp7