ABSTRACT
A major unsolved issue of prion biology is the existence of multiple strains with distinct phenotypes and this strain phenomenon is postulated to be associated with the conformational diversity of the abnormal prion protein (PrPSc). Real-time quaking-induced conversion (RT-QUIC) assay that uses Escherichia coli-derived recombinant prion protein (rPrP) for the sensitive detection of PrPSc results in the formation of rPrP-fibrils seeded with various strains. We demonstrated that there are differences in the secondary structures, especially in the β-sheets, and conformational stability between 2 rPrP-fibrils seeded with either Chandler or 22L strains in the first round of RT-QUIC. In particular, the differences in conformational properties of these 2 rPrP-fibrils were common to those of the original PrPSc. However, the strain specificities of rPrP-fibrils seen in the first round were lost in subsequent rounds. Instead, our findings suggest that nonspecific fibrils became the major species, probable owing to their selective growth advantage in the RT-QUIC. This study shows that at least some strain-specific conformational properties of the original PrPSc can be transmitted to rPrP-fibrils in vitro, but further conservation appears to require unknown cofactors or environmental conditions or both.
Keywords: fibril, prion, real-time quaking-induced conversion (RT-QUIC), recombinant prion protein (rPrP), strain, transmission
In vitro Conversion of Recombinant PrP into the Proteinase K (PK) Resistant Amyloid Fibrils
Prion diseases, or transmissible spongiform encephalopathies, are infectious and fatal neurodegenerative disorders that include Creutzfeldt-Jakob disease in humans, and scrapie and bovine spongiform encephalopathy in animals. The infectious agent, prion, is assumed to be formed mainly or exclusively by abnormal prion protein, designated PrPSc, which is partially protease-resistant1 and a β-sheet-rich conformer,2,3 frequently resulting in amyloid fibril formation. Although the pathogenesis has not been clarified fully, it is widely accepted that prion disease occurs through autocatalytic conformational conversion of the ubiquitous normal form of prion protein (PrPC) to PrPSc in a “protein only” manner.4
Studies using Escherichia coli-derived purified recombinant PrP (rPrP) has contributed to solving the controversial protein-only hypothesis. It has been demonstrated that rPrP fibrils (rPrP-fibrils) formed in vitro cause the accumulation of PrPSc in the brains of PrP-overexpressing transgenic (Tg) mice5–7 and some wild-type hamsters.8 These studies suggest that rPrP can be converted into a PrPSc-like form in vitro; however, the infectious titers seem to be much lower than that of authentic PrPSc. In contrast, prion infectivity could be propagated when brain-derived PrPC or baculovirus-derived PrPC was used as substrates for protein misfolding cyclic amplification (PMCA) in the presence of certain cofactors such as nucleic acids.9,10 Relatively high levels of prion infectivity was demonstrated by injection of PK-resistant rPrP-fibrils generated by unseeded PMCA in the presence of 1-palmitoyl-2-oleoylphosphatidylglycerol and total liver RNA into wild-type mice. Subsequently, these mice developed prion disease with an incubation period of approximately 150 days.11 However, other group failed to show infectivity of rPrP-fibrils generated by the same methods.12
Transmission of Conformational Properties of Prion Strains to rPrP-fibrils in RT-QUIC
Prion is known to provide extensive strain diversity showing different phenotypic and pathological states in mammalian species. The strain-specific characteristics can usually be serially passaged stably in the same species. Furthermore, PrPSc generated by PMCA using brain homogenate from normal animals as a source of PrPC (BH-PMCA) seeded with different mouse prion strains retained the strain-specific properties, such as incubation time, neuropathology, and biochemical characteristics from original PrPSc.13 This result indicates that the intracellular mechanisms and cell-to-cell transmission are dispensable for the maintenance and propagation of strain characteristics. The finding that PrPSc from different strains have distinct secondary structures and biochemical properties supports the notion that prion strains are manifested by conformational variations of the PrPSc.14 For example, strain-dependent differences in β-sheet-rich structures of PrPSc have been demonstrated by infrared spectroscopy.15–18 In addition, the conformational stability of PrPSc differed among prion strains, as demonstrated by guanidine hydrochloride denaturation assay followed by protease digestion.19,20 However, the mechanistic relationship between PrPSc conformational differences and the molecular basis of prion strains remains poorly understood.
The recently developed “real-time quaking-induced conversion” (RT-QUIC) is a sensitive prion detection method, in which intermittent shaking enhances the conversion of soluble rPrP into amyloid fibrils in the presence of PrPSc.21 Recent studies show that RT-QUIC assays allow highly sensitive detection of PrPSc in most species and strains, including Creutzfeldt-Jakob disease in humans,21–24 scrapie in rodents,25,26 and chronic wasting disease in cervids.27
We generated the amyloid fibrils seeded with 100 pg of PrPSc derived from either the Chandler or 22L strain in the first round of RT-QUIC (1st-rPrP-fibSc).28 Spontaneous formation of rPrP-fibrils (rPrP-fibspon) was observed by decreasing the concentration of rPrP, because there was an inverse correlation between the rate of fibril formation and the concentration of rPrP. Previous studies using FTIR and hydrogen/deuterium exchange have shown that there are structural differences between PrPSc-seeded and spontaneous rPrP-fibrils generated by PMCA.29,30 We found that the PK-resistant band pattern, structural morphology, secondary structure, and conformational stability distinguish 1st-rPrP-fibSc from rPrP-fibspon. Although there were no differences in the PK-resistant band pattern and structural morphology between Chandler-seeded (1st-rPrP-fibCh) and 22L-seeded rPrP-fibrils (1st-rPrP-fib22L), we observed significant differences in the secondary structure and conformational stability between strains. FTIR analysis showed that native rPrP had an abundance of α-helical structures, whereas 1st-rPrP-fibCh and 1st-rPrP-fib22L were substantially enriched in β-sheets. While the 1st-rPrP-fibCh was characterized by a major band at 1624 cm−1 in the β-sheet region of second-derivative spectra, the 1st-rPrP-fib22L was characterized by 2 absorbance bands at 1629 and 1617 cm−1, indicating that there were conformational differences in β-sheet structures between the 2 1st-rPrP-fibSc. Similarly, purified Chandler-PrPSc from brains of mice displayed the spectrum with a peak at 1630 cm−1, whereas purified 22L-PrPSc had 2 major maxima at 1631 and 1616 cm−1, as previously reported. Thus, the differences in β-sheet spectrum shape between strains were common to both PrPSc and 1st-rPrP-fibSc. The conformational stability of 1st-rPrP-fib22L was significantly lower than that of 1st-rPrP-fibCh, as with Chandler- and 22L-PrPSc. Furthermore, wild-type mice inoculated with the 1st-rPrP-fibSc showed an increased attack rate and a significantly shorter survival period compared with those inoculated with mock preparations. The infectious titers (per 40 μl) of 1st-rPrP-fibCh and 1st-rPrP-fib22L were estimated to be 407.2 ± 226.6 and 1067.0 ± 678.7 LD50, respectively, whereas the titers of Chandler and 22L prion were 20.2 and 28.9 LD50 units/40 pg of PrPSc, respectively, indicating that QUIC reaction in the first round resulted in a 20- to 37-fold increase in the infectious titer. These results suggest that strain features of PrPSc can be transmitted to rPrP-fibrils in a simple system solely consisting of pure rPrP. However, it is clear that the conformation of 1st-rPrP-fibSc is not identical to that of authentic PrPSc. It should be noted that the degrees of vacuolation of mice inoculated with 1st-rPrP-fibSc were significantly lower in the hippocampus and cerebellum than those of inoculated with mock preparations. The different lesion profiles may result from the conformational differences between 1st-rPrP-fibSc and authentic PrPSc.
What is Required for Maintaining Strain-Specific Conformations?
We found that the strain-specific conformational features and the infectivity disappeared in rPrP-fibrils during and after the second round,28 suggesting that RT-QUIC has the limitation of technology with respect of reproducing the prion propagation. One possible reason for the loss of the prion strain-specific traits is that E. coli-derived rPrP lacks post-translational modifications. PrP bears 2 N-linked glycosylation sites at amino acids 180 and 196 that can produce di-, mono-, and unglycosylated forms. PrPSc has varying degrees of glycosylation among strains14,31,32 and therefore the glycosylation pattern is postulated to confer strain specificity. Studies using Tg mice expressing glycosylation site mutants revealed that the strain properties of strain 79A were altered by the glycosylation state of PrPC, but the strain properties of strains ME7 and 301C were not affected.33 Moreover, the glycosylation-deficient PrPC as a substrate of PMCA by treatment with PNGase F did not affect strain-dependent neurotropisms in the 2 murine strains RML and 301C.34 Furthermore, the cell tropisms determined by the cell panel assay were altered in strains RML, 139A, 79A, and ME7 but not in strain 22L when the strains were propagated in Tg mice expressing PrP devoid of a GPI anchor.35 These results suggest that the necessity of glycans and the GPI-anchor for the transmission and preservation of strain-specific properties is dependent on the strains.
An additional reason for the loss of strain specificity from rPrP-fibrils might be because of a decrease in cofactor(s) over serial passages. Strains have been reported to differ in their RNA requirements for propagation in BH-PMCA, although RNA is not essential for maintaining strain-specific characteristics in mice.36 Moreover, another study showed that phosphatidylethanolamine is a cofactor required for the propagation of prion infectivity in seeded rPrP-PMCA but not for the transmission of strain-specific properties, because 3 different prion strains changed into a single new strain after the serial passages of rPrP-PMCA reactions.37,38 Thus, crucial cofactors or environmental conditions for maintaining strain-specific properties remain to be determined.
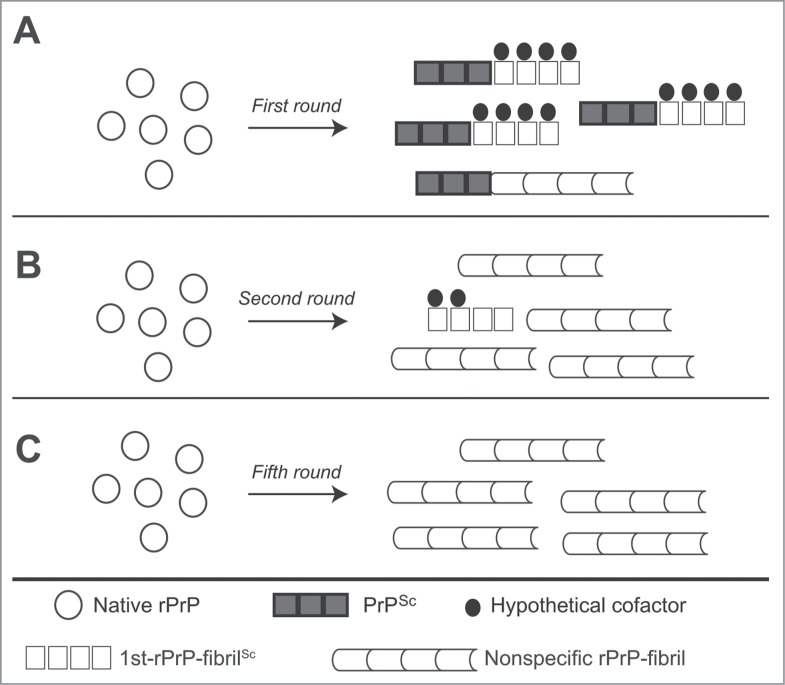
We observed the “nonspecific rPrP-fibrils” displayed no strain-specific differences in IR spectra and conformational stability after 5 serial rounds of RT-QUIC, which have the ability to cause the conversion of rPrP but failed to induce clinical signs of prion disease in the wild-type mice.28 Additionally, we found that the β-sheet spectra of rPrP-fibrils generated in the presence of small amount (1 pg) of PrPSc or generated at pH4 in the first round were similar to nonspecific rPrP-fibrils.28 These observations raise the possibility that nonspecific rPrP-fibrils lacking prion infectivity can be generated even in the first round and may interrupt the formation of the fibrils with strain-specific conformations, because of a selective growth advantage of nonspecific fibrils (Fig. 1). Of note, the formation of quasi-species that is consisting of a variety of conformational variants has been reported in prion-infected cultured cells under different environmental conditions.39,40 Furthermore, different prion strains can interfere with each other, and this is known as prion strain interference.41–44 The competition for substrates among the variants is thought to act as a selection pressure in Darwinian evolution and to cause the phenomenon of prion strain interference. Previous work showed that 2 conformational variants of rPrP-fibrils are mutually exclusive and compete for monomeric rPrP as a substrate in the rPrP-PMCA.29 Likewise, competitive amplification of 2 prion strains was observed in BH-PMCA.45 We postulate that PrPSc predominantly leads to strain-specific conformational conversion of rPrP, particularly in the presence of hypothetical cofactors, while some quantity of nonspecific fibrils could be generated simultaneously in the first round (Fig. 1A). The conditions of subsequent rounds would favor growth of nonspecific species (Fig. 1B and C). The fact that prion infectivity was often diminished in serial rPrP-PMCA46 or BH-PMCA47–49 support the hypothesis that the amplification of nonspecific rPrP fibrils is accelerated by certain conditions. Further studies are needed to ascertain the key factors responsible for maintaining the infectious and strain-specific conformations in vitro.
Figure 1.
Hypothetical models for the formation of rPrP-fibrils in sequential RT-QUIC reactions. (A) The formation of 1st-rPrP-fibSc is induced predominantly in the presence of hypothetical cofactors and brain-derived PrPSc in the first round. However, a small amount of nonspecific rPrP-fibrils may be concomitantly generated. (B) In the second round, the nonspecific rPrP-fibrils become predominant because of the paucity of hypothetical cofactors and/or a selective growth advantage of nonspecific fibrils. (C) The formation of nonspecific rPrP-fibrils occupies almost the whole reaction in the fifth round.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
REFERENCES
- 1. Meyer RK, McKinley MP, Bowman KA, Braunfeld MB, Barry RA, Prusiner SB. Separation and properties of cellular and scrapie prion proteins. Proc Natl Acad Sci U S A 1986; 83:2310–4; PMID:3085093; http://dx.doi.org/ 10.1073/pnas.83.8.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry 1991; 30:7672–80; PMID:1678278; http://dx.doi.org/ 10.1021/bi00245a003 [DOI] [PubMed] [Google Scholar]
- 3. Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 1993; 90:10962–6; PMID:7902575; http://dx.doi.org/ 10.1073/pnas.90.23.10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prusiner SB. Molecular biology of prion diseases. Science 1991; 252:1515–22; PMID:1675487; http://dx.doi.org/ 10.1126/science.1675487 [DOI] [PubMed] [Google Scholar]
- 5. Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science 2004; 305:673–6; PMID:15286374; http://dx.doi.org/ 10.1126/science.1100195 [DOI] [PubMed] [Google Scholar]
- 6. Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci U S A 2009; 106:20417–22; PMID:19915150; http://dx.doi.org/ 10.1073/pnas.0910350106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raymond GJ, Race B, Hollister JR, Offerdahl DK, Moore RA, Kodali R, Raymond LD, Hughson AG, Rosenke R, Long D, et al. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J Virol 2012; 86:11763–78; PMID:22915801; http://dx.doi.org/ 10.1128/JVI.01353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol 2010; 119:177–87; PMID:20052481; http://dx.doi.org/ 10.1007/s00401-009-0633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A 2007; 104:9741–6; PMID:17535913; http://dx.doi.org/ 10.1073/pnas.0702662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imamura M, Kato N, Yoshioka M, Okada H, Iwamaru Y, Shimizu Y, Shimizu Y, Mohri S, Yokoyama T, Murayama Y. Glycosylphosphatidylinositol anchor-dependent stimulation pathway required for generation of baculovirus-derived recombinant scrapie prion protein. J Virol 2011; 85:2582–8; PMID:21228241; http://dx.doi.org/ 10.1128/JVI.02098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132–5; PMID:20110469; http://dx.doi.org/ 10.1126/science.1183748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Timmes AG, Moore RA, Fischer ER, Priola SA. Recombinant prion protein refolded with lipid and RNA has the biochemical hallmarks of a prion but lacks in vivo infectivity. PLoS One 2013; 8:e71081; PMID:23936256; http://dx.doi.org/ 10.1371/journal.pone.0071081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J 2008; 27:2557–66; PMID:18800058; http://dx.doi.org/ 10.1038/emboj.2008.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol 2007; 8:552–61; PMID:17585315; http://dx.doi.org/ 10.1038/nrm2204 [DOI] [PubMed] [Google Scholar]
- 15. Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem 1998; 273:32230–5; PMID:9822701; http://dx.doi.org/ 10.1074/jbc.273.48.32230 [DOI] [PubMed] [Google Scholar]
- 16. Thomzig A, Spassov S, Friedrich M, Naumann D, Beekes M. Discriminating scrapie and bovine spongiform encephalopathy isolates by infrared spectroscopy of pathological prion protein. J Biol Chem 2004; 279:33847–54; PMID:15155741; http://dx.doi.org/ 10.1074/jbc.M403730200 [DOI] [PubMed] [Google Scholar]
- 17. Atarashi R, Sim VL, Nishida N, Caughey B, Katamine S. Prion strain-dependent differences in conversion of mutant prion proteins in cell culture. J Virol 2006; 80:7854–62; PMID:16873242; http://dx.doi.org/ 10.1128/JVI.00424-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baron GS, Hughson AG, Raymond GJ, Offerdahl DK, Barton KA, Raymond LD, Dorward DW, Caughey B. Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra. Biochemistry 2011; 50:4479–90; PMID:21539311; http://dx.doi.org/ 10.1021/bi2003907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci 2001; 10:854–63; PMID:11274476; http://dx.doi.org/ 10.1110/ps.39201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shindoh R, Kim CL, Song CH, Hasebe R, Horiuchi M. The region approximately between amino acids 81 and 137 of proteinase K-resistant PrPSc is critical for the infectivity of the Chandler prion strain. J Virol 2009; 83:3852–60; PMID:19176630; http://dx.doi.org/ 10.1128/JVI.01740-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, Matsubara T, Nakagaki T, Yamanaka H, Shirabe S, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med 2011; 17:175–8; PMID:21278748; http://dx.doi.org/ 10.1038/nm.2294 [DOI] [PubMed] [Google Scholar]
- 22. McGuire LI, Peden AH, Orru CD, Wilham JM, Appleford NE, Mallinson G, Andrews M, Head MW, Caughey B, Will RG, et al. Real time quaking-induced conversion analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann Neurol 2012; 72:278–85; PMID:22926858; http://dx.doi.org/ 10.1002/ana.23589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sano K, Satoh K, Atarashi R, Takashima H, Iwasaki Y, Yoshida M, Sanjo N, Murai H, Mizusawa H, Schmitz M, et al. Early detection of abnormal prion protein in genetic human prion diseases now possible using real-time QUIC assay. PLoS One 2013; 8:e54915; PMID:23372790; http://dx.doi.org/ 10.1371/journal.pone.0054915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orru CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, Fiorini M, Pocchiari M, Monaco S, Caughey B, et al. A test for Creutzfeldt-Jakob disease using nasal brushings. N Engl J Med 2014; 371:519–29; PMID:25099576; http://dx.doi.org/ 10.1056/NEJMoa1315200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 2010; 6:e1001217; PMID:21152012; http://dx.doi.org/ 10.1371/journal.ppat.1001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vascellari S, Orru CD, Hughson AG, King D, Barron R, Wilham JM, Baron GS, Race B, Pani A, Caughey B. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS One 2012; 7:e48969; PMID:23139828; http://dx.doi.org/ 10.1371/journal.pone.0048969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B, Hoover EA. Rapid antemortem detection of CWD prions in deer saliva. PLoS One 2013; 8:e74377; PMID:24040235; http://dx.doi.org/ 10.1371/journal.pone.0074377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sano K, Atarashi R, Ishibashi D, Nakagaki T, Satoh K, Nishida N. Conformational properties of prion strains can be transmitted to recombinant prion protein fibrils in real-time quaking-induced conversion. J Virol 2014; 88:11791–801; PMID:25078700; http://dx.doi.org/ 10.1128/JVI.00585-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Onwubiko HA, Priola SA, Caughey B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods 2007; 4:645–50; PMID:17643109; http://dx.doi.org/ 10.1038/nmeth1066 [DOI] [PubMed] [Google Scholar]
- 30. Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol 2011; 18:504–6; PMID:21441913; http://dx.doi.org/ 10.1038/nsmb.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clarke AR, Jackson GS, Collinge J. The molecular biology of prion propagation. Philos Trans R Soc Lond B Biol Sci 2001; 356:185–95; PMID:11260799; http://dx.doi.org/ 10.1098/rstb.2000.0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lawson VA, Collins SJ, Masters CL, Hill AF. Prion protein glycosylation. J Neurochem 2005; 93:793–801; PMID:15857383; http://dx.doi.org/ 10.1111/j.1471-4159.2005.03104.x [DOI] [PubMed] [Google Scholar]
- 33. Cancellotti E, Mahal SP, Somerville R, Diack A, Brown D, Piccardo P, Weissmann C, Manson JC. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J 2013; 32:756–69; PMID:23395905; http://dx.doi.org/ 10.1038/emboj.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piro JR, Harris BT, Nishina K, Soto C, Morales R, Rees JR, Supattapone S. Prion protein glycosylation is not required for strain-specific neurotropism. J Virol 2009; 83:5321–8; PMID:19297485; http://dx.doi.org/ 10.1128/JVI.02502-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahal SP, Jablonski J, Suponitsky-Kroyter I, Oelschlegel AM, Herva ME, Oldstone M, Weissmann C. Propagation of RML prions in mice expressing PrP devoid of GPI anchor leads to formation of a novel, stable prion strain. PLoS Pathog 2012; 8:e1002746; PMID:22685404; http://dx.doi.org/ 10.1371/journal.ppat.1002746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saa P, Sferrazza GF, Ottenberg G, Oelschlegel AM, Dorsey K, Lasmezas CI. Strain-specific role of RNAs in prion replication. J Virol 2012; 86:10494–504; PMID:22811520; http://dx.doi.org/ 10.1128/JVI.01286-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci U S A 2012; 109:8546–51; PMID:22586108; http://dx.doi.org/ 10.1073/pnas.1204498109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci U S A 2012; 109:E1938–46; PMID:22711839; http://dx.doi.org/ 10.1073/pnas.1206999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science 2010; 327:869–72; PMID:20044542; http://dx.doi.org/ 10.1126/science.1183218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weissmann C, Li J, Mahal SP, Browning S. Prions on the move. EMBO Rep 2011; 12:1109–17; PMID:21997298; http://dx.doi.org/ 10.1038/embor.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dickinson AG, Fraser H, Meikle VM, Outram GW. Competition between different scrapie agents in mice. Nat New Biol 1972; 237:244–5; PMID:4624846; http://dx.doi.org/ 10.1038/newbio237244a0 [DOI] [PubMed] [Google Scholar]
- 42. Manuelidis L. Vaccination with an attenuated Creutzfeldt-Jakob disease strain prevents expression of a virulent agent. Proc Natl Acad Sci U S A 1998; 95:2520–5; PMID:9482918; http://dx.doi.org/ 10.1073/pnas.95.5.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schutt CR, Bartz JC. Prion interference with multiple prion isolates. Prion 2008; 2:61–3; PMID:19098442; http://dx.doi.org/ 10.4161/pri.2.2.6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nishida N, Katamine S, Manuelidis L. Reciprocal interference between specific CJD and scrapie agents in neural cell cultures. Science 2005; 310:493–6; PMID:16239476; http://dx.doi.org/ 10.1126/science.1118155 [DOI] [PubMed] [Google Scholar]
- 45. Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Coinfecting prion strains compete for a limiting cellular resource. J Virol 2010; 84:5706–14; PMID:20237082; http://dx.doi.org/ 10.1128/JVI.00243-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem 2010; 285:14083–7; PMID:20304915; http://dx.doi.org/ 10.1074/jbc.C110.113464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bieschke J, Weber P, Sarafoff N, Beekes M, Giese A, Kretzschmar H. Autocatalytic self-propagation of misfolded prion protein. Proc Natl Acad Sci U S A 2004; 101:12207–11; PMID:15297610; http://dx.doi.org/ 10.1073/pnas.0404650101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klingeborn M, Race B, Meade-White KD, Chesebro B. Lower specific infectivity of protease-resistant prion protein generated in cell-free reactions. Proc Natl Acad Sci U S A 2011; 108:E1244–53; PMID:22065744; http://dx.doi.org/ 10.1073/pnas.1111255108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gonzalez-Montalban N, Lee YJ, Makarava N, Savtchenko R, Baskakov IV. Changes in prion replication environment cause prion strain mutation. FASEB J 2013; 27:3702–10; PMID:23729586; http://dx.doi.org/ 10.1096/fj.13-230466 [DOI] [PMC free article] [PubMed] [Google Scholar]