Abstract
To assess factors influencing the success of whole genome sequencing for mainstream clinical diagnosis, we sequenced 217 individuals from 156 independent cases across a broad spectrum of disorders in whom prior screening had identified no pathogenic variants. We quantified the number of candidate variants identified using different strategies for variant calling, filtering, annotation and prioritisation. We found that jointly calling variants across samples, filtering against both local and external databases, deploying multiple annotation tools and using familial transmission above biological plausibility contributed to accuracy. Overall, we identified disease causing variants in 21% of cases, rising to 34% (23/68) for Mendelian disorders and 57% (8/14) in trios. We also discovered 32 potentially clinically actionable variants in 18 genes unrelated to the referral disorder, though only four were ultimately considered reportable. Our results demonstrate the value of genome sequencing for routine clinical diagnosis, but also highlight many outstanding challenges.
Introduction
The mainstream application of whole genome sequencing (WGS) in clinical diagnosis holds much promise. In contrast to existing genetic tools, such as targeted gene sequencing, array CGH, and exome sequencing1-5, only WGS can characterise all types of genetic variant in all parts of the genome. Such completeness, coupled with efforts to chart the distribution of genetic variation in populations6,7, will enable the identification of pathogenic variants and hence influence diagnosis, genetic counselling and treatment.
Nevertheless, clinical adoption of WGS faces many challenges including cost, speed of delivery, sensitivity, specificity and heterogeneity of variant detection, ambiguities and errors in variant annotation, a substantial informatics burden, and the difficulties of incidental findings 8,9. Consequently, while technological improvements to improve speed in critical situations such as neonatal intensive care10 and detailed evaluations of WGS and whole exome sequencing (WES) in specific disorders11 are opening the opportunity for its wider use, its reach into the clinic is, to date, limited12. In order for WGS to be adopted as a routine clinical platform, we would need to demonstrate its diagnostic yield for patients with likely genetic disorders identified by clinicians across a broad range of medical specialties, within a hospital setting. Furthermore, the challenges of reliably identifying and validating potential pathogenic variants at scale across such a disease spectrum would need to be met.
In order to address these challenges we undertook the WGS500 programme to sequence 500 patient genomes from diverse genetic disorders referred by a range of medical specialists. For all disorders, study leaders had access to additional samples and /or could follow up with functional studies for validation. Some results from this study have already been published13-19. Here, we report an overview of the results from the Mendelian and immunological disorders, representing 156 independent individual cases or families, selected because a strong genetic component was suspected (family history, early-onset, severe disorder) but prior genetic screening had failed to identify any pathogenic variants (Fig. 1). The disorders varied substantially in the number of independent cases recruited, the availability of additional family members and the likely disease model. Here, we identify and quantify the effect on success of factors relating to the genetic architecture of a disease, experimental design and analytical strategy.
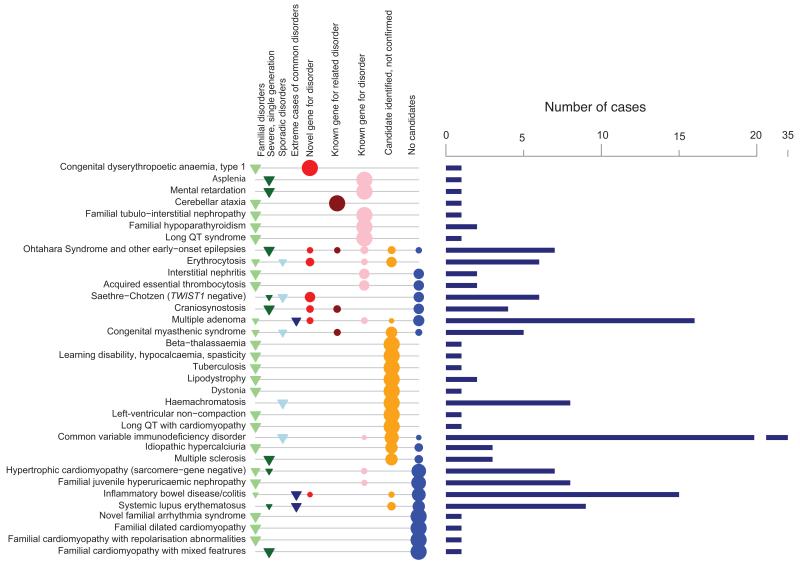
Figure 1. Overview of projects and results.
For each disorder, the number of independent cases (bars) studied is shown alongside information about the nature of the disorder: familial disorders (category 1, light green triangles), severe single-generation disorders suspected to be caused by de novo or recessive mutations (category 2, dark green), unrelated sporadic disorders (category 3, light blue) and extreme cases of common complex diseases (category 4, dark blue). The proportion of cases with each outcome class A-E is also shown (see Online Methods): pathogenic variant in novel gene for disorder (A, red circles), pathogenic variant in gene for related disorder (B, brown), pathogenic variant in known gene for disorder (C, pink), candidate pathogenic variant with validation studies underway (D, orange) and no single candidate variant, or negative results for validation of top candidate/s (blue). Size of points proportional to outcome fraction. Disorders are ranked by fraction of cases with confirmed pathogenic variants (class A to C).
Results
Variant calling, filtering and annotation
Individuals were sequenced to an average of 31.8× (range 22.7 – 60.8×) such that on average 82.7% of the genome (88.2% of the exome) was covered to at least 20× (Supplementary Fig. 1). We find no significant correlation between sequencing coverage and diagnostic success (Pearson r = −0.1, P = 0.13), indicating that, at this depth, fluctuations in coverage in WGS play a minor a role in determining success for germline disorders. For the few samples with low levels of contaminating DNA (Supplementary Fig. 1), we took additional care over interpretation of candidate pathogenic variants rather than returning to the patient for additional material; one individual with substantial contamination was excluded.
All samples were processed with the same pipelines for sequencing, variant calling and annotation (Online Methods). Concordance between the WGS data and genotypes from SNP arrays was over 99.9% at heterozygous sites (Supplementary Tables 1 and 2; Supplementary Fig. 2). Our pipeline included two key steps. First, we used a two-stage variant calling procedure with an initial round of independent calling followed by a second round which revisits the evidence in each individual for any variant called across all samples. This approach improves genotype accuracy by, for example, using strong evidence for a variant in a child to enhance support for the same variant in a parent (and vice versa). Joint calling substantially increases the accuracy of de novo mutation detection in families. For example, the number of candidate coding de novo mutations was reduced from a mean of 32.1 after independent variant calling (filtering against 1000 Genomes and the NHLBI Exome Sequencing Project, ESP) to 2.6 after joint calling of the parents and proband (Supplementary Table 3).
The second key step was that when identifying likely pathogenic variants, in addition to filtering against external data sources, we also filtered against other WGS500 samples. For example, when filtering against external data sources only, individuals had an average of 80.8 rare or novel (frequency < 0.5%) homozygous coding variants, but only 1.5 if, in addition, variants present above this frequency in other WGS500 samples were excluded (Supplementary Table 4). Using control samples sequenced using the same technology and processed through the same pipeline reduced the impact of systematic differences between our studies and others in coverage, sequencing technology, experimental protocol and data processing (Supplementary Fig. 3, Supplementary Table 3).
Finally, we found that the choice of transcript set and annotation software can affect variant annotations20. Comparison of annotation using the RefSeq and Ensembl transcript sets revealed only 44% agreement for putative loss-of-function variants. Similarly, we found agreement of only 66% for loss-of-function annotations and 87% for all exonic annotations between the tools Annovar21 and Ensembl’s Variant Effect Predictor22. In both comparisons, the greatest discrepancy was for splicing annotations (agreement of 25% between transcript sets and 57% between software tools). Such heterogeneity in how variants are annotated can substantially reduce the efficacy of WGS for clinical analysis. We therefore used multiple annotation approaches to identify candidate variants.
Evaluating biological candidacy of variants
To identify candidate disease-causing variants we used a combination of predicted functional impact, frequency in the population, transmission within a family (where appropriate) and, when multiple independent cases were available, statistical evidence for association (Online Methods). Beccause most genes harbour large numbers of rare variants6,23 many of which are absent from existing databases and affect the protein produced but only a fraction of which may influence disease risk, care has to be taken in interpreting novel variants in known disease genes. To assess the burden of such ‘variants of unknown significance’ across a range of disorders we defined candidate genes for early-onset epilepsy (EOE), X-linked mental retardation (XLMR) and craniosynostosis (CRS). For EOE we used a semi-automated approach on the basis of a three-tiered system according to medical genetic and biological information (Online Methods; Supplementary Table 5). Tier 1 comprises the set of known genes for the disorder (from the Human Gene Mutation Database, HGMD24), Tier 2 adds genes known for related disorders (from HGMD) or whose products interact directly with those in Tier 1 (from the Mammalian Protein-Protein Interaction Database, MIPS25), and Tier 3 adds genes in relevant biological pathways (from HGMD and the Gene Ontology database). For XLMR we only examined Tier 1 genes. For CRS we used lists generated by expert curation. For those individuals with the disorder, additional family members were used to identify the most likely pathogenic variants (Supplementary Table 6).
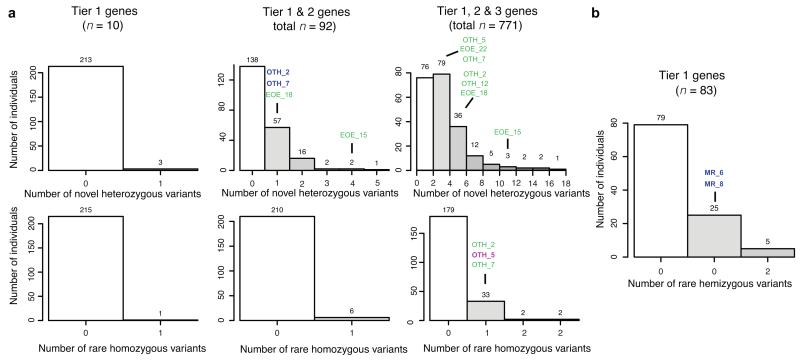
For each disorder, we found multiple unaffected individuals in WGS500 with variants in the candidate genes for XLMR, CRS and EOE that would be interpreted as potentially pathogenic had those individuals presented with the disorder in question. Within the ten known genes for EOE (Tier 1), we found that 3/216 individuals (1.3% of the sample) carried a novel heterozygous candidate variant and one (0.5%) carried a rare homozygous candidate (Fig. 2a); none of these individuals had epilepsy. As the strength of gene candidacy reduced, the effect was increased; 36% of individuals carried at least one heterozygous candidate among the Tier 1 genes or the additional 82 genes implicated in milder forms of epilepsy (Tier 2) and 96% of individuals carried one such variant in a Tier 1 or 2 gene or in one of the 771 genes involved in brain development or function (Tier 3). The proportions for homozygous candidates were 3% and 17% for Tier 1-2 and 1-3 respectively. We found no enrichment for either heterozygous or homozygous candidates in Tier 1-2 genes among the six EOE patients (Supplementary Table 7) and only 2/10 Tier 1-2 variants found in EOE samples are thought to be pathogenic based on family information; for homozygous variants in Tiers 1-3, the figure is 1/315 (Supplementary Table 6).
Figure 2. The burden of variants of unknown significance.
(a) Histograms of the number of previously unreported coding variants at conserved positions in different sets of candidate gene (Tiers 1, 1+2 and 1+2+3 for columns left to right) for early-onset epilepsy, under different inheritance models, across 216 WGS500 samples. (b) Histogram of the number of previously unreported coding variants at conserved positions in known X-linked mental retardation genes (XLMR), for the 99 male WGS500 samples. The candidate genes were chosen by high-throughput searches (Online Methods). Sample identifiers indicate individuals with the disorder in question. Sample names in green text indicate that the variant is not likely to be pathogenic (since it does not fit a plausible inheritance model or is less functionally compelling than another candidate); blue text indicates that the variant is thought to be causal (see Supplementary Table 6). OTH: Ohtahara syndrome; EOE: nonsyndromic early onset epilepsy; MR: mental retardation. See Supplementary Fig. 4 for the analysis of craniosynostosis.
Similar results were found in genes for other disorders. For CRS genes, 57/216 (26%) samples carried at least one novel heterozygous coding variant in the 38 expert-curated known causative genes, though no sample had any rare homozygous coding variants (Supplementary Fig. 4). Five CRS samples carried Tier 1-2 variants, but none are thought to be pathogenic since they were not of de novo origin. For X-linked mental retardation (XLMR) genes, the effect is striking: 30/109 male samples (28%) carry at least one previously unreported missense variant at a conserved residue within the 83 known XLMR genes (Fig. 2b). In only two of these (two brothers with MR) was the variant thought to be pathogenic.
We also investigated the burden of potentially pathogenic regulatory variants, focusing on conserved positions in regulatory regions defined by the Ensembl Regulatory Build that are less than 50 kb away from candidate genes (Supplementary Fig. 5). The mean number of novel heterozygous variants per individual was 203 (standard deviation = 102; range 102 - 614), more than twice as many as the equivalent number of novel coding variants (mean = 75, Supplementary Fig. 3), although we note that this number is inflated because there are fewer control individuals in publicly available datasets for regulatory rather than exonic variants. Many individuals had novel or rare variants at conserved sites in regulatory regions close to candidate genes for EOE and CRS (Supplementary Fig. 6). Moreover, in samples from patients with the disorder, there were typically multiple potentially regulatory variants that were consistent with a plausible inheritance model, although none of these are considered likely pathogenic because stronger candidates were present (Supplementary Table 6).
These results demonstrate that the combined use of gene candidacy, predicted functional consequence, variant frequency and evolutionary conservation, though widely used filters within pipelines for identifying pathogenic candidates, will not, by themselves, differentiate between pathogenic and non-pathogenic variants. Naïve application of such rules will lead to a high rate of false positive diagnosis, even in rare disorders within limited numbers of known genes. Moreover, focusing only on candidate genes will lead to a high false negative rate; of the eight EOE, CRS or XLMR families for which a strong candidate (Class A-C) for the pathogenic variant has been identified (Supplementary Table 6), only four of these variants are in candidate genes found using automated database searches (Tier 1, 2 or 3). In this study, as in others, additional evidence, such as functional data, familial transmission, de novo status and screens of other patients, was needed to establish pathogenicity.
Overview of findings
In 33 of the 156 cases (21%), we identified at least one variant with a high level of evidence of pathogenicity (Class A, B or C as described in Online Methods; Figure 1; Table 1; Supplementary Table 8). These comprised five nonsense variants, fifteen missense, three noncoding, two frameshifts, one in-frame indel, five variants that disrupt splicing, and two compound heterozygotes, each with one missense and one either nonsense or splicing variant (and additionally one variant that was reported independently of WGS500). Together, we identified twelve cases with variants in novel genes for which we found additional compelling genetic and/or functional evidence of pathogenicity (Class A), four cases with variants in genes known for other phenotypes but not for the disorder in question, supported by additional genetic and/or functional evidence (Class B), and seventeen cases with variants in genes already known for that phenotype (Class C). This rate of success is comparable to recent exome sequencing studies in various disorders3,5,8,26. Below we describe the range of the findings and some of the outstanding challenges identified.
Table 1. Summary of conditions for which pathogenic genes were identified (class A, B or C).
| Disease | Project category | Result class | Genea | Coding consequence | Inheritance (Zygosity)b | Variant |
|---|---|---|---|---|---|---|
|
Acquired essential thrombocytosis |
1.1 | C | THPO | splicing | D (het) | NM_001177598: c.13+1G>C |
| Asplenia | 2.2 | C | RPSAc | splicing | D (het) | NM_002295.4:c.-34+5G>C |
| Cerebellar ataxia | 1.2 | B | SPTBN2 | nonsense | AR (hom) | NM_006946:c.1881G>A:p.C627* |
|
Common variable immunodeficiency disorder |
3 | C | d | missense | d | d |
|
Congenital dyserythropoietic anaemia, type 1 |
1.2 | A | C15ORF41 | missense | AR (hom) | NM_001130010:c.533T>A:p.L178Q |
| Congenital myasthenic syndrome | 3 | B | ALG2 | missense | AR (hom) | NM_033087:c.203T>G:p.V68G |
| Craniosynostosis | 2.1 | A | ZIC1 | nonsense | DN (het) | NM_003412.3: c.1163C>A: p.S388* |
| 2.1 | B | HUWE1 | missense | DN (het) | NM_031407.6: c.329G>A:p.R110Q | |
| Erythrocytosis | 1.1 | A | EPO | noncoding | D (het) | NM_000799.2:c.-136G>A |
| 1.1 | A | EPO | noncoding | D (het) | NM_000799.2:c.-136G>A | |
| 3 | C | BPGM | missense | D (het) | NM_001724:c.269G>A:p.R90H | |
| Familial hypoparathyroidism | 1.3 | C | SOX3 | noncoding | XL (hemi) | deletion of chrX:139,502,946-139,504,327, 1.5kb downstream of SOX3 |
| 1.1 | C | CASR | missense | D (het) | NM_000388:c.2299G>C:p.E767Q | |
|
Familial juvenile hyperuricaemic nephropathy |
1.4 | C | UMOD | missense | D (het) | NM_001008389:c.410G>A:p.C137Y |
|
Familial tubulo-interstitial nephropathy |
1.1 | C | UMOD | missense (inframe insertion/deletion) | D (het) |
NM_001008389:c.279_289del:p.93_97del; NM_001008389:c.278_279insCCGCCTCC:p.V9 3fs |
| Hypertrophic cardiomyopathy (sarcomere gene-negative) | 1.1 | C | MYBPC3 | nonsense | e | NM_000256:c.1303C>T:p.Q435* |
|
Inflammatory bowel syndrome/colitis |
4 | A | d | missense | d | d |
| Interstitial nephritis | 1.4 | C | MUC1c | - | D (het) | d |
| Long QT syndrome | 1.1 | C | KCNQ1 | missense | D (het) | NM_000218:c.1195_1196insC:p.A399fs |
| Mental retardation | 2.1 | C | GRIA3 | missense | XL (hemi) | a |
|
Ohtahara syndrome and other early-onset epilepsies |
2.1 | A | PIGQ | splicing | SR (hom) | NM_004204:c.690-2A>G |
| 2.1 | B | KCNT1 | missense | UPID (hom) | NM_020822: c. 2896G>A:p.A966T | |
| 2.1 | C | KCNQ2 | missense | DN (het) | NM_004518:c.827C>T:p.T276I | |
| 2.1 | C | SCN2A | missense | DN (het) | NM_001040143:c.5558A>G:p.H1853R | |
| Multiple adenoma | 1.1 | A | POLD1 | missense | D (het) | NM_002691:c.1433G>A:p.S478N |
| 1.1 | A | POLD1 | missense | D (het) | NM_002691:c.1433G>A:p.S478N | |
| 4 | A | POLE | missense | D (het) | NM_006231:c.1270C>G:p.L424V | |
| 4 | C | MSH6 | missense and nonsense | CR (het; het) | NM_000179:c.2315G>A:p.R772Q | |
| 4 | C | BMPR1A | frameshift | AR (hom) | NM_004329.2:c.142_143insT:p.Thr49Asnfs*2 2 | |
| 4 | C | APC | splicing | D (het) | NM_001127511:c.251-2A>G | |
|
Saethre-Chotzen syndrome (TWIST1 negative) |
3 | A | TCF12 | nonsense | DN (het) | NM_207037.1:c.1283T>G; p.L428* |
| 3 | A | TCF12 | splicing | DN (het) | NM_207037.1:c.1035+3G>C | |
| 2.1 | A | CDC45 | synonymous (splicing) and missense | CR (het; het) |
NM_001178010.2:c.318C>T;p.V106=; NM_001178010.2:c.773A>G;p.D258G |
Each line represents a separate case or family, so if the same gene is reported on two lines, this signifies that the gene is thought to be pathogenic in both cases. Some genes have two mutations in the same affected individual, likely representing compound heterozygous inheritance, which is indicated in the Inheritance column.
D(het): dominant - affected individual/s heterozygous; AR (hom): autosomal recessive – affected individual/s homozygous; DN (het) :de novo – affected individual/s heterozygous; XL (hemi): X-linked recessive – affected male/s hemizygous, affected female/s homozygous; UPD (hom): uniparental isodisomy–affected individual homozygous; CR (het; het): compound recessive – affected individual /s heterozygous for two different variants in the same gene
Causal variant discovered independently of WGS500.
Details will be reported in an independent publication.
Form of inheritance not clear. See Supplementary Table 8.
Variants missed by prior genetic testing
We identified four cases where a candidate variant lay within a gene that had previously been screened by a clinical or research genetic laboratory (UK or overseas) but had been missed. These were variants in UMOD in familial juvenile hyperuricemic nephropathy (FJHN), in KCNQ1 in long QT syndrome and in APC and MSH6 in multiple adenoma. The rate of false negative results from Sanger sequencing is likely to vary considerably between genes and types of variant. Nevertheless, across the samples studied here, a relatively small fraction (2.5%) of cases resulted from false negative tests in standard clinical genetics testing.
Challenges in establishing pathogenicity
For several disorders, likely pathogenic variants were identified in genes not reported previously for those conditions or related phenotypes. When additional variants of major coding consequence were found in screens of other cases (and not controls), the evidence for pathogenicity was considered strong, including for POLE and POLD1 in multiple adenomas/colorectal cancer19, TCF12 in Saethre-Chotzen-like syndrome16, ALG2 for congenital myasthenic syndrome17, and C15ORF41 in congenital dyserythropoetic anaemia, type 114. In some cases, mouse models provided supportive evidence (e.g. confirming the role of SPTBN2 in cerebellar ataxia18), and/or functional work demonstrated that the variant affects protein function (e.g. a KCNT1 de novo mutation found in an Ohtahara syndrome patient was shown to affect potassium channel activity15).
In six cases, likely pathogenic variants were identified in genes where variants cause disorders with related phenotypes. For example, a de novo mutation that disrupts CBL splicing (NM_005188:exon9:c.1228-1G>A) was identified in a patient with severe epilepsy, microcephaly, and developmental delay15. Cbl is a ubiquitin ligase that regulates the Ras/MAPK pathway27,and heterozygous missense variantsin CBL cause facial, cutaneous and cardiac abnormalities, hypotonia and developmental delay28,29, as well as microcephaly and a predisposition to juvenile myelomonocytic leukemia30,31. However, whilst our patient had unusual cutaneous and cardiac features, these were not typical of NCFC syndrome, and review by clinicians did not alter the original diagnosis. CBL variants have previously been noted for their variable phenotypes and incomplete penetrance31. Thus, the CBL variant is a strong candidate, but no other likely pathogenic variants in CBL were identified in a panel of over 500 other epilepsy patients15.
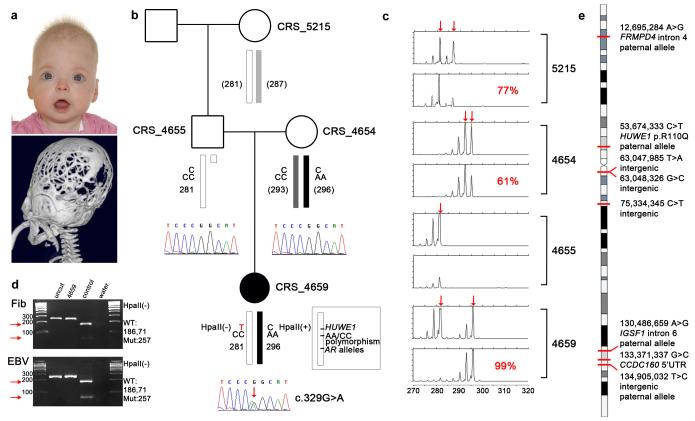
The difficulties in establishing pathogenicity are also illustrated by a de novo missense mutation (NM_031407:c.329G>A:p.R110Q) in HUWE1 identified in a girl with craniosynostosis and learning difficulties (Fig. 3a; Supplementary Note). Mutations in HUWE1 are reported to cause X-linked mental retardation and macrocephaly32-34, though not previously CRS. The mutation affects a highly conserved residue in a domain of unknown function (DUF908; Supplementary Fig. 7). The gene spans 154,641 bp and comprises 84 exons, and, because of extensive heterogeneity in CRS, the contribution to the disease is likely to be low. Thus, it was not surprising that no other HUWE1 mutations were found in a cohort of 47 unrelated cases with complex CRS. The mutation originated on the paternal X chromosome (Fig. 3b; Supplementary Fig. 7). Unexpectedly, cells from the patient show preferential inactivation of the maternally inherited, wild-type X (Fig. 3c) and, consistent with these two observations, only the mutant allele was expressed in the tissues available (fibroblast and transformed lymphoblasts) (Fig. 3d). Seven other X-linked de novo point mutations (three in genes: a 5′UTR change in CCDC160 and intronic changes in FRMPD4 and IGSF1) were identified in the same individual (Fig. 3e), though none were considered pathogenic. The finding of a substitution at a highly conserved residue in a known XLMR gene, combined with exclusive expression of the mutant allele, suggested that this mutation contributed at least to the learning difficulties in this child, but that this is a highly unusual case, and hence it was challenging to establish true pathogenicity. Recently, however, we identified, using WES, a different de novo hemizygous mutation altering the same amino acid of HUWE1 (c.328C>T encoding p.R110W) in a boy presenting with metopic craniosynostosis, moderate-severe learning disability and other dysmorphic features, supporting the evidence for causality.
Figure 3. Identification of de novo HUWE1 mutation associated with severe craniosynostosis.
(a) Upper panel, the proband (CRS_4659; female, aged 6 months) presented with an abnormal skull shape. Lower panel, three-dimensional CT scan aged 5 months shows multisuture synostosis with multiple craniolacunae. (b) Family pedigree showing dideoxy sequence chromatograms with de novo G>A mutation of the X-linked HUWE1 gene in the proband (red arrow). Schematic X chromosomes are annotated from top to bottom with the HUWE1 alleles, haplotype of AA/CC polymorphisms located 1.15 kb away from mutation and used to deduce paternal origin, and androgen receptor (AR) trinucleotide repeat allele size (allele sizes in CRS_4654 and CRS_5215 are in brackets to emphasize that phase is unknown relative to other parts of the two X chromosomes). Note that the HUWE1 mutation abolishes a HpaII restriction site. (c) Analysis of X-inactivation in whole blood samples at AR locus. For each individual, AR alleles are indicated by arrows in the upper panel, while the lower panel shows proportions of methylated alleles and percentage representation of the more highly inactivated X chromosome. (d) Exclusive expression of cDNA from the HUWE1 mutant allele in both fibroblast (Fib) and Epstein Barr virus (EBV)-transformed lymphoblastoid cells from the proband. Arrows highlight absence of expression of the normal allele in either cell type. Product sizes (bp) from different alleles are shown on the right. WT: wild-type, Mut: mutant. (e) X chromosome ideogram showing eight de novo mutations identified. Where known, the parental allele on which the variant arose is indicated.
Candidates for pathogenic regulatory variants
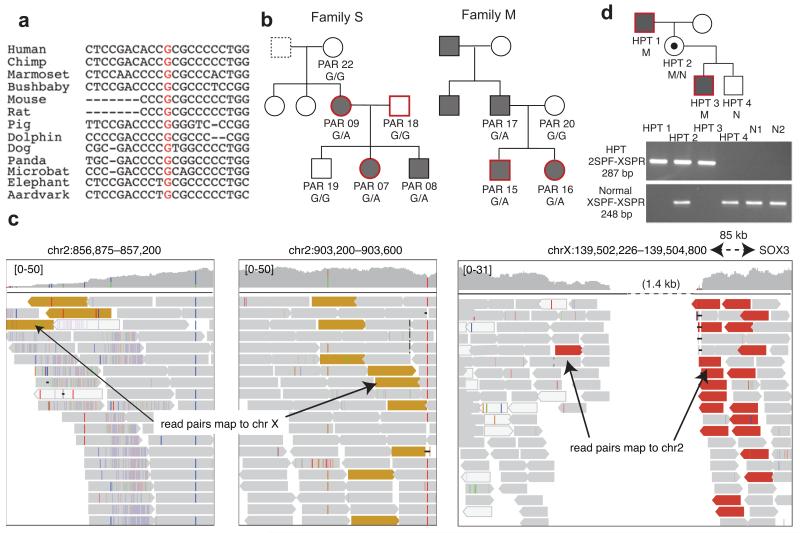
Strong candidate pathogenic variants were detected outside the coding fraction of the genome in two conditions. The same variant at a highly conserved base (chr7:100318468 G>A; Fig. 4a) within the 5′ UTR of the erythropoietin gene EPO was identified in two independent families with erythrocytosis and co-segregated with the disease (Fig. 4b; Supplementary Note). Moreover, this is the only rare exonic variant found in an 8 Mb region that is identical-by-descent in the affected individuals in these two unrelated families (the only such region), suggesting that it had a single, and probably recent, mutational origin. EPO is a strong candidate gene as erythropoietin is essential for red cell production and increased levels cause increased red cell mass, the hallmark of erythrocytosis35,36. However, genetic variation in EPO has not been linked previously with erythrocytosis and further functional data would be necessary to prove causality definitively.
Figure 4. Candidate pathogenic noncoding variants.
(a) Multi-species alignment of a region of the 5′ UTR of EPO in which a variant was identified at a conserved position (red text) in two families with erythrocytosis. (b) Erythrocytosis pedigrees studied, showing affected individuals (shaded grey), those sequenced (red borders), and genotypes of all individuals for whom we had DNA. We had no information about the father of PAR09 (dotted box).(c) Summary of read mapping in an individual with hypoparathyroidism showing evidence for an interstitial insertion-deletion event in which a ~ 50 kb region of chromosome 2p25.3 (top panel) has been duplicated and inserted into chromosome X, resulting in a 1.4 kb deletion 81.5 kb downstream of SOX3 (bottom panel). Yellow reads: mate maps to chrX; red reads: mate maps to chr2; grey reads: read and mate map to the same chromosome; white reads: read has mapping quality 0. (d) Pedigree showing segregation of the complex variant within the affected pedigree, with PCR validation below. M: mutation; N: normal. Primers 2SPF and XSPR flank the distal breakpoint of the deletion-insertion and are shown in Supplementary Figure 8. Primers XSPF and XSPR detect the normal allele. The mutation was not seen in 150 alleles from 100 unrelated normocalcemic individuals (50 males and 50 females, including N1 and N2, who are shown).
In another case, a complex event leading to deletion of 1.4 kb of the X chromosome and insertion of 50 kb from chromosome 2p (Fig. 4c; Supplementary Fig. 8) was discovered in a patient with X-linked hypoparathyroidism (Supplementary Note). This variant lies 81.5 kb downstream of SOX3, segregates with the disease (Fig. 4d) and is similar to an event reported previously in an independent kindred37. SOX3 is a strong candidate as it influences the development of the parathyroid gland38. Although the pathogenicity for these variants is not proven, that such candidates can be identified using WGS demonstrates the value of screening the noncoding genome.
Incidental findings
The identification of variants unrelated to the referred condition, but which have potential clinical and actionable significance, is a major challenge for WGS. To evaluate the burden of such incidental findings, we followed the American College of Medical Genetics and Genomics’ recommendations39 and used HGMD24 assignments of pathogenic status to identify 32 variants in 18 genes of possible clinical significance (four nonsense, three splice-site and 25 nonsynonymous variants). After detailed and lengthy review of the literature and curated variant databases, 26 could be eliminated (Supplementary Table 9), leaving six variants in four genes, each present in a single case (Table 2). Although the majority of these variants have been published in association with a relevant disease, major doubts remain about their clinical interpretation due to incomplete information on (1) variant frequencies in large populations of healthy people, (2) phenotypes when segregating within families and (3) corroborative functional studies40. Where the variant occurs at significant frequency in public databases (e.g. the RYR1 variants), the rarity of associated case reports suggests that penetrance is low or indeed zero. Even in the most apparently clear-cut case of a nonsense variant in BRCA2, the actual disease risk is likely to be reduced in the absence of a documented family history41.
Table 2. Incidental findings with potentially actionable consequences.
| Incidental finding condition | Gene | AA change | UK10K | EVS_EA | Comments |
|---|---|---|---|---|---|
| I. Reportable Incidental Findings | |||||
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | DSG2 | NM_001943: c.2397T>G:p.Y799* | absent | absent | Stop gain mutation, not previously reported, but mutation class considered pathogenic44 |
| DSG2 | NM_001943: c.2554G>T:p.E852* | absent | absent | As above | |
| Breast & ovarian cancer | BRCA2 | NM_000059: c.7558C>T:p.R2520* | absent | 0.0001 | Stop gain mutation; 5 independent R2520* in affected patients45-49. 4 reports of variant being pathogenic in ClinVar50 (submitted by independent clinical labs) and 7 records rated as causal in UMD-BRCA2 database51. Mutations of this class described in Brohet, et al. 52. |
| Long QT syndrome | KCNQ1 | NM_000218: c.877C>T:p.R293C | absent | absent | 2 independent reports in literature: i) 4 /2500 independent cases from FAMILION cohort referred for LQT genetic testing53 and ii) 1 case /388 consecutive unrelated patients with swimming triggered arrhythmia syndromes54, as compound heterozygote with G269D. Location of mutation in pore suggestive of pathogenicity. |
| II. Incidental Findings of Uncertain Significance | |||||
| Long QT syndrome | KCNQ1 | NM_000218: c.1189C>T:p.R397W | absent | 0.0006 | 3 independent reports in literature, including 3/2500 independent cases referred for long QT testing55, 5/600 cases in LQT registry53 and 1/91 independent cases of intrauterine foetal death56. Functional data from heterologous expression of mutation i) in HEK293 cells which shows markedly reduced current on whole cell patch clamp compared with WT56 and ii) in inside-out membrane patches from Xenopus oocytes which showed markedly reduced ATP binding57. Taken together, this suggests that the mutation should not be disregarded clinically as it may be weakly pathogenic, albeit with low absolute risk |
| Malignant hyperthermia | RYR1 | NM_000540: c.5036G>A:p.R1679H | 0.0006 | 0.0014 | Variant observed in single subject with complication, and positive functional testing58 but no independent replication. |
Variants deemed to be reportable and clinically actionable are listed Section I of the table. Those for which the evidence was not considered sufficient to be clinically actionable are reported or are uncertain are listed in Section II.
AA: amino acid; VUS: variant of unknown significance; UK10K: frequency in the UK10K twin cohort; EVS: Exome Variant Server; EVS_EA: frequency in European Americans in the EVS; HGMD: Human Gene Mutation Database; UMD: Universal Mutation Database (see URLs).
Any decision on clinical action must balance multiple potential harms (invasion of personal autonomy, the severity of proposed preventive intervention, associated healthcare costs) against the anticipated benefits to health. We propose that only four variants are clinically reportable (Table 2), whilst a further two variants are of uncertain significance and warrant further investigation (Table 2). For example, the R397W variant in KCNQ1 is potentially associated with long QT syndrome (LQTS) and sudden death. The frequency of this single variant in EVS exceeds that of LQTS overall, suggesting very low absolute risk; nevertheless it is probably reasonable to recommend avoidance of certain classes of medication (even if the subject does not have any obvious ECG abnormality) as this intervention can, very occasionally, be lifesaving. By contrast, we do not believe that intensive electrophysiological investigation or clinical cascade screening of the extended family are indicated. These observations highlight the urgent need for unbiased data from large biobanks to support clinical decision making.
Discussion
The goal of the WGS500 study was to evaluate the potential value of WGS in mainstream genetic diagnosis. In routine clinical settings the opportunities for time-consuming investigation of multiple variants emerging from WGS are limited. We identified multiple strategies in analysis (joint variant calling, filtering of variants against local databases and the use of multiple annotation algorithms) that improve the reliability of variants called and improved sensitivity and specificity in detecting candidate disease-causing variants.
With these innovations, WGS proved to be effective for molecular diagnosis of severe disorders for which a strong genetic component was suspected, but where screening of known genes had failed previously to identify candidates. Overall, WGS identified a pathogenic variant in 33/156 cases (21%), 23/68 (33.8%) Mendelian cases (class A, B or C in category 1 or 2), increasing to 57% (8/14) in cases where de novo or recessive models were suspected and both parents sequenced (category 2.1) (Supplementary Table 8). The majority of these variants lie within genes, hence are typically accessible through WES. However, in an independent study of 141 exomes, 3/33 sites reported in Table 1 lay outside the exome target and a further six lay within the target, but had low coverage (median < 20×; Supplementary Fig. 9). If a minimum of six reads, three of which support the variant, are required for detecting a novel heterozygous variant, we estimate that 15% of causal variants identified in this WGS study (including coding and non-coding changes) would likely have been missed by WES (0.5% in WGS500). Conversely, using 20 variant sites identified as causal from an independent exome sequencing project, we estimate that WGS at this coverage has 99.6% power to identify a novel heterozygous variant (compared with 96.1% in WES; Supplementary Fig. 9).
Moreover, WGS has additional benefits. For example, in the CRS case discussed above, WGS was important for a) identifying the HUWE1 mutation, b) identifying nearby variants for establishing parent of origin, so we could subsequently show that only the mutant chromosome was being expressed, and c) for assessing other de novo mutations on the X that might affect X chromosome inactivation. The latter two points could not have been addressed with WES data. Moreover, WGS identified, in other cases, two likely pathogenic non-coding variants and unusual chromosomal features including large deletions (for example, a 30 Mb deletion on the X chromosome of a patient with congenital myasthenia, though this is not thought to be relevant to the disorder), distant consanguinity (Supplementary Fig. 10) and uniparental disomy (as in the case of a child with Ohtahara Syndrome15).
In other types of disorder, WGS proved less successful. The number of candidate variants in families with dominantly inherited disorders makes functional validation time-consuming, and many such cases remain in active follow up. Furthermore, our hypothesis that extreme forms of complex disorders (young onset, severe disease) would enrich for monogenic forms was not confirmed. In only two cases out of 49 (one case of common variable immunodeficiency disorder and one in inflammatory bowel disease) did WGS on unrelated individuals with extreme immune-related disorders identify strong candidates for pathogenic variants, despite substantial sample sizes (n = 34 for CVID, n = 15 for IBD). Several other candidates have been identified, but pathogenicity has not been confirmed. This low success rate likely reflects the influence of multiple genetic factors, even in extreme cases. Only very large patient cohorts are well-powered for identifying novel genes with a modest contribution to the phenotype42,43 and, in any specific case, it will be difficult to assign pathogenicity to any particular variant.
Our results also highlight the outstanding challenges of WGS interpretation. Every individual carries multiple rare variants that could potentially be assessed as pathogenic for a given disorder on the basis of biological information about the gene, the coding consequence of the variant and its frequency within the population. Such variants may be benign, or have variable penetrance, making their clinical interpretation without additional information (such as de novo status or co-segregation with the disease within a family) challenging. Conversely, rigid application of biological candidacy filters will lead to false negatives. Ultimately, WGS will only be able to reliably assess the diagnostic and predictive value of any specific variant if it, or another variant in the same gene, is identified in other individuals with the same disorder for whom detailed phenotypic and clinical data are available.
Finally, the identification of pathogenic variants, exclusion of potential candidate variants and identification of incidental findings relied on close collaboration between analysts, the scientists knowledgeable about the disease and genes and clinicians expert in the specific disorders. The availability of resources and expertise for functional validation studies were critical to the assignment of causality. Provision of this network may be challenging to establish in a clinical setting but it will be an important aspect of successful translation of WGS.
Online Methods
Overview of the WGS500 Project Consortium
WGS was carried out as part of a collaboration between the Wellcome Trust Centre for Human Genetics at the University of Oxford, the Oxford NIHR Biomedical Research Centre and Illumina Inc. We sought to sequence 500 whole genomes from patients in whom findings could have immediate clinical utility in terms of diagnosis, prognosis, treatment selection, or genetic counselling and reproductive choices.
Process and criteria for sample inclusion
Proposals were invited from clinicians and researchers in Oxford, commencing in December 2010, and were reviewed by a scientific Steering Committee. Known candidate genes and large chromosomal copy number changes had to have been excluded for the patient to be included in the study, to maximise the likelihood of identifying variants in novel disease genes.
Projects were categorised as follows:
- Families with suspected Mendelian conditions with affected individuals across multiple generations (dominant or recessive). In these cases, we usually sequenced one or a few family members (chosen to maximise power for exclusion analysis) and obtained SNP array data on all available others, in order to identify regions identical by descent between affected individuals. These cases were further subdivided into:
-
1.1.1.Dominant model suspected
-
1.1.2.Recessive model suspected (often due to consanguinity)
-
1.1.3.X-linked model suspected
-
1.1.4.Multiple unrelated families with linkage to the same region/s
-
1.1.1.
- Families with suspected Mendelian conditions with one or more affected individuals in a single generation. For these, we hypothesised that they were due to de novo or recessive mutations. We used these sub-classifications:
-
2.1.Affected offspring and both parents sequenced.
-
2.2.Only affected offspring sequenced, not parents.
-
2.1.
Cohort of unrelated sporadic patients with no known family history.
Individuals with extreme forms of common disorders (e.g. early onset or severe forms).
Ethics
Individual researchers had explicit research consent to undertake genetic investigation into the cause of the relevant disease, and/or samples were obtained with clinical consent as part of efforts to identify the cause of the patient’s disease. Ethics committee reference numbers for every individual research project have been provided to the journal editors.
Sequencing library Preparation
3-5 μg of DNA was obtained from each individual, usually from blood, or otherwise from saliva or immortalised cell lines. Samples were diluted to 80 ng/μl in 10 mM Tris-Cl, pH 8.5, then quantified using the High Sensitivity Qubit system (Invitrogen). Sample integrity was assessed using 1% E-Gel EX (Invitrogen). Focused ultrasonication was carried out to fragment 2μg of DNA using the Covaris S2 system with the following settings: duty cycle = 10%, intensity = 5%, cycles/burst = 200 and time = 60 s. Libraries were constructed using the NEBNext DNA Sample Prep Master Mix Set 1 Kit (New England Biolabs), with minor modifications. Ligation of adapters was performed using 6 μl of Illumina Adapters (Multiplexing Sample Preparation Oliogonucleotide Kit). Ligated libraries were size-selected using 2% E-Gel EX (Invitrogen) and the distribution of fragments in the purified fraction was determined using Tapestation 1DK system (Agilent/Lab901). Each library was PCR-enriched with 25 μM each of the following custom primers:
Multiplex PCR primer 1.0:
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′
Index primer:
5′-CAAGCAGAAGACGGCATACGAGAT[INDEX]CAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′
Indexes were 8 bp long and formed part of an indexing system developed in-house59. Four independent PCRs were prepared per sample using 25% volume of the pre-PCR library each. After 8 cycles of PCR (cycling conditions as per Illumina recommendations) the four reactions were pooled and purified with AMPure XP beads (Beckman Coulter, Inc.). The final size distribution was determined using a Tapestation 1DK system (Agilent/Lab901). The concentration of each library was determined by Real-time PCR using the Agilent qPCR Library Quantification Kit and an MX3005P instrument (Agilent).
Whole genome sequencing and quality control
WGS was performed on either the Illumina HiSeq2000 or the HighSeq2500 run in standard mode, either by the Oxford Genomics Centre at the Wellcome Trust Centre for Human Genetics, or by Illumina Cambridge Ltd.. We generated 100 bp reads and used v2.5 or v3 clustering and sequencing chemistry. A PhiX control was spiked into the libraries. We aimed for a mean coverage of 30× and obtained a minimum of 22.7×. The mode number of lanes required to reach the desired coverage was 2⅓.
We used the recommended quality metrics in the Illumina Sequence Analysis Viewer in analysing each lane. Additionally, we generated our own quality metrics for each lane (or, in the case of multiplexes, each part of a lane), and required the following criteria to be met: <2% duplicate pairs; most frequent kmer <2%; >99% mapped; <2.5% read pairs mapping to different chromosomes; mean insert size between 340 bp and 440 bp, with a median absolute deviation of <30bp; approximately uniform genomic coverage by GC content; ~1% exonic coverage; <2% N bases at any cycle; approximately equal number of reads per tag (three samples multiplexed per lane), standard deviation <25%.
Read mapping
Sequence reads were generated using the Illumina offline basecaller (OLB; v1.9.3) and mapped to the GRCh37d5 human reference sequence. This reference genome was obtained from the 1000 Genomes Project and is based on hg19 but contains a 35.48 Mb decoy chromosome that reduces misalignment of repetitive sequence and improves accuracy of SNP discovery. Mapping was performed using BWA (v0.5.6)60 and Stampy (versions 1.0.12 - 1.0.22; see URLs)61, and merging and deduplication using Picard (v1.67; see URLs).
SNP and indel calling and genotyping
Variant calling was performed with Platypus62 (version 0.1.9; see URLs) using the default settings. This algorithm can detect SNPs and short indels (<50 bp), and is sensitive to somatic mosaic mutations at low allele frequencies63,64.
The variant calling included two stages. First, we used Platypus to identify SNPs and short indels in all samples individually (raw calling). We then ran it a second time to genotype the union of all variants in all samples. We did the raw calling on groups of related samples together (“joint calling”), so that the same sites were interrogated for all samples in the family (i.e. it was possible to distinguish homozygous reference from a missing call), and so that the observation of a variant in one of the individuals in the family reduced the required threshold for calling it in the others (see Rimmer, et al. 62).
We retained variants with a PASS flag that had a posterior probability (phred scale) of >20 that the variant segregates. Variants with a “clustered” flag (within 25 bp of another variant) were manually checked in IGV65 but not discounted.
To check for sample contamination, we plotted the distribution of the ratio of the number of reads containing the alternate allele to the total number of reads (ALT:TOTAL) for known SNPs (from dbSNP) and novel SNPs for each sample (Supplementary Figure 1D). To check for duplicates and cryptically related individuals, we ran principal components analysis on the WGS500 data. We included the three ethnic groups (CEU, YRI and JPT/CHB) from the HapMap project, which allowed us to identify a few individuals with a particularly high number of variants as ancestry outliers.
Filtering variants in trios
There were fifteen families in WGS500 from which both the parents and one or more affected children were sequenced: six trios and one quartet with early-onset epilepsy (EOE), a trio with hypertrophic cardiomyopathy (HCM), a trio with erythrocytosis (ERY) (with mother and daughter affected), four trios with craniosynostosis (CRS), a trio with Saethre-Chotzen syndrome (SC; a type of CRS), and a quartet with X-linked mental retardation (XLMR). If the parents were unaffected and only one child was affected, the initial hypothesis was that the causal mutation was de novo. To screen for these variants, we searched for variants that were absent from all public databases (1000 Genomes, dbSNP, ESP) and from other WGS500 samples, and that had been confidently called as heterozygous in the child and as homozygous reference in the parents (genotype log likelihood ratio, GLLR < −5). (This latter criterion on the GLLR was not always applied when analyzing data for specific projects.) Supplementary Table 3 demonstrates the value of applying these different filters.
We also investigated a simple recessive model in these families. Homozygous variants in the affected child (or children) had to have a frequency less than 0.5% in 1000 Genomes and ESP (corresponding to an expected homozygous frequency of 1 in 40,000), and there had to be 0 homozygotes and ≤ 2 heterozygotes amongst the unrelated WGS500 samples. We required the parents to be called as heterozygous. Results of these filters are shown in Supplementary Table 4. When filtering for compound recessive candidates (in which the child had two rare heterozygous coding variants in the same gene, one inherited from each parent) and X-linked recessive candidates, we used the same frequency thresholds as for the simple recessive case.
Annotation of variants
The functional consequences of variants were predicted using several programmes. We used ANNOVAR (February 2013 version)66 to annotate variants with respect to RefSeq genes, adding information about segmental duplications, conservation (based on the UCSC alignment of 46 mammalian genomes), GERP, SIFT, MutationTaster, phyloP and PolyPhen2 scores, dbSNP identifiers (version 1.35), and frequency in the NHLBI Exome Sequencing Project (see URLs) and Phase 1 of the 1000 Genomes Project 6. We also annotated all variants using the Variant Effect Predictor from EnsEMBL (version 69) and the nonsynonymous SNPs using PolyPhen2 (version 2.2.2r405).
Detection of copy number variants and extended homozygosity
We used several different methods to search for copy number variants (CNVs). 1) We generated count profiles for each individual by dividing each chromosome into 10 kb bins and counting number of reads in each bin. For each chromosome, we applied principal components analysis to the log of the counts (training set - one per family), and then plotted the residuals of the predicted PCs along the chromosome. This procedure served to remove noise in the data due to biases in sequence composition. Candidate CNVs (down to about 10 kb) were identified as outliers visually. 2) We applied OncoSNP-SEQ67 in germline mode to a subset of 300,000 reliable SNPs across the genome. Coverage and read counts at these locations were used as proxy for the intensity and B-allelic-frequency under a specific model of the Hidden Markov Model intended for next-generation sequencing data. 3) To identify exon-level CNVs, we used ExomeDepth68.
To search for long regions of homozygosity (LROHs), we calculated the fraction of heterozygous SNPs in 10 kb bins along the chromosomes for each individual, averaged these over 1 Mb regions, and plotted them, ignoring centromeric and other repetitive regions. We classed as “homozygous” any segments with a heterozygous/homozygous ratio < 0.2; this empirically chosen threshold avoided large homozygous regions being interrupted by genotyping errors in difficult regions, but clearly distinguished them from the rest of the genome. Homozygous regions up to 4 Mb size are common in demonstrably outbred individuals69. Consanguinity had already been reported for many of the thirty-nine individuals who had LROHs larger than 4 Mb (Supplementary Figure 10), but in cases for whom it had not, this finding prompted analysts to search for rare homozygous variants within these regions.
SNP array data
Illumina SNP arrays were run on some WGS500 samples and other relatives. This was to check the genotyping accuracy of our sequencing pipeline, to refine linkage regions, to confirm familial relationships, and, in two cases, to investigate whether large stretches of homozygosity were likely due to uniparental disomy or unreported consanguinity. We ran 200 ng of DNA on the Illumina Human CytoSNP12 array or on the 1M array (Illumina Inc.), following the manufacturer’s guidelines. Concordance between the CytoSNP12 genotypes and the WGS data is shown in Supplementary Tables 1 and 2, and the dependence on coverage in Supplementary Figure 2. In most cases, array-CGH had already been performed prior to submission of samples, but we also used QuantiSNP70 to check for CNVs, as well as Nexus Copy Number version 7 (BioDiscovery, Hawthorn, CA). We used MERLIN71 in familial studies to identify regions identical-by-descent.
Assessment of coverage in WGS500 and exome sequence data
After removing duplicate reads, we used bedtools72 to measure coverage in all WGS500 samples, and examined the cumulative distributions across the genome and exome (CCDS transcripts) (Supplementary Figure 1A,B). In order to compare this with a typical exome sequencing experiment, we took 141 whole exome datasets that had been captured using the Roche Nimblegen SeqCap EZ v.2.0 kit at the Oxford Biomedical Research Centre, removed duplicate reads, and measured coverage at each position in the CCDS transcripts. We compared the exome-wide coverage distributions between WGS500 and these exomes (Supplementary Figure 1), as well as the coverage at specific variants thought to be causal (Supplementary Figure 9).
Identifying variants of clinical relevance
Following variant calling and annotation, cases from specific projects were investigated by different analysts, and variants prioritised on the basis of mode of inheritance, functional consequence, population frequency, evolutionary consequence of position and biological relevance. Where available, parental data, linkage information and repeated occurrence across multiple independent cases with a disorder were used to aid prioritisation. Validation of biological consequence and/or screening of additional cohorts was used to confirm pathogenicity. Where a previously described variant or variant class (e.g. loss of function, frameshift) was observed in a known gene for a disorder, it was assumed to be pathogenic and missed by prior screening. All putatively causative variants were confirmed using Sanger sequencing. Information was returned to clinicians responsible for managing individual patients, who decided whether and how information was reported to them.
Classification system for results
We categorised the results for each independent case into five classes, as follows:
Class A: Mutation found in a novel gene, with additional genetic evidence (in unrelated cases) and/or functional data supporting causality.
Class B: Mutation found in a gene known for a different phenotype, with additional genetic evidence and/or functional data supporting causality.
Class C: Mutation found in a known gene for this phenotype.
Class D: Mutation found in a novel gene, with further genetic and functional validation studies in progress.
Class E: No single candidate yet, or negative results for validation of original candidates.
The results for all projects are summarised in Figure 1. Note that one of the CVID cases recovered antibody production, and was thus found to have been misdiagnosed.
Analysis of gene/variant tiering strategy
For eight of the thirteen families mentioned above from which we sequenced the affected child/children and healthy parents, we have identified the causal mutation with a reasonable level of confidence (class A, B or C in Supplementary Table 6). Thus, we used these families as test cases for the analysis in this section.
We compiled tiered lists of candidate genes for each of the three diseases: EOE, CRS and XLMR. For EOE (Supplementary Table 5), Tier 1 contains genes that are recorded in the Human Gene Mutation Database (HGMD)24 as causing Ohtahara syndrome or epileptic encephalopathy, Tier 2 contains genes that interact with Tier 1 genes (according to the Mammalian Protein-Protein Interaction Database (MIPS) database25) or that are listed in HGMD as causing more general epilepsy, and Tier 3 contains genes that are involved in biological pathways known to be involved in pathogenesis. For CRS, Tier 1 is a hand-curated list of genes mentioned in the literature as causing CRS in two or more cases, Tier 2 is a list of additional genes associated with the term “craniosynostosis” in the the Copenhagen disease gene association list (see URLs), and Tier 3 comprises additional orthologs of 270 mouse genes that are expressed in the skull73 (Eurexpress database; see URLs). For XLMR, we compiled the Tier 1 list by searching HGMD for “mental retardation” and “intellectual disability” and then restricting to chrX; we did not consider additional tiers since Tier 1 already contained 83 genes.
We analysed the mutational burden in these genes for all 216 samples, having excluded the contaminated sample HCM_2361 (Supplementary Figure 1. Specifically, we screened for coding variants in these genes that would appear to fit a de novo dominant (DN), simple recessive (SR) or X-linked (XL) model in the absence of parental information, according to the following criteria:
De novo model: novel variant heterozygous in proband and absent from 1000 Genomes, ESP and other unrelated WGS500 samples
Simple recessive model: novel or very rare variant homozygous in proband, frequency <0.5% in 1000 Genomes and ESP, with no other homozygotes and ≤ 2 heterozygotes amongst the unrelated WGS500 samples
X-linked recessive model: novel or very rare variant hemizygous in male proband, frequency <0.5% in 1000 Genomes and ESP, and no other homozygous females or hemizygous males and ≤ 2 heterozygotes amongst the unrelated WGS500 samples
A variant was considered “coding” if it was annotated by ANNOVAR66 as missense, stop gain or stop loss, an indel, or within a splice site, for one or more transcripts, and “conserved” if it had a GERP or phyloP score greater than 2, or was in a constrained element as defined by the UCSC 46-way alignment. We also used VEP to add information about regulatory regions from the Ensembl V65 Regulatory Build (see URLs).
Actionable, pathogenic incidental findings
We followed the guidelines of the American College of Medical Genetics and Genomics39 for reporting incidental findings. To identify potentially disease-causing mutations, we first took all the mutations in HGMD24, left-aligned the indels, and removed variants with erroneous reference alleles. We retained those classed as “DM” (disease mutation) that were within 10 bp of an exon of a gene in Table 1 of Green et al.39. We then searched all WGS500 samples for these mutations, and for nonsense or frameshift mutations in the same genes, which would be expected to be pathogenic. We removed any variants with a frequency >1% in 1000 Genomes or the Exome Variant Server. Variants that were i) classified as variants of unknown significance in curated, disease-specific mutation databases ii) had high frequency in Exome Variant Server iii) had negative functional data iv) showed lack of segregation in literature reports or v) were unlikely to be of significance from their sequence context (e.g. splicing variants without clear splicing signatures) were removed. These are listed in Supplementary Table 9. The remaining variants (Table 2) were then scrutinized by a panel of clinical experts, who decided which variants had enough data supporting pathogenicity to report.
Supplementary Material
Acknowledgements
Funded by a Wellcome Trust Core Award (090532/Z/09/Z) and Medical Research Council Hub grant (G0900747 91070) to PD, the National Institute for Health Research (NIHR) Biomedical Research Centre Oxford, the Department of Health’s NIHR Biomedical Research Centres funding scheme, and Illumina Inc. Additional support is acknowledged from the BBSRC (BB/I02593X/1) to GL and GMV; Wellcome Trust grants 093329, 091182, and 102731 to AOMW, and 100308 to LF; the Newlife Foundation for Disabled Children (10-11/04) to AOMW; AtaxiaUK to AHN; the Haemochromatosis Society to KR; the European Research Council (FP7/2007-2013) Grant agreement no. 281824 to JCK and no. 305608 to OD, the Jeffrey Modell Foundation NYC and Baxter Healthcare to SYP and H. Chapel; Action de Recherche Concertée (ARC10/15-029, Communauté Française de Belgique) to OD; FNRS, FRSM and Inter-University Attraction Pole (IUAP, Belgium Federal Government) to OD; the NCCR Kidney.CH program (Swiss National Science Foundation) to OD; the Gebert Rüf Stiftung (Project GRS-038/12) to OD; the Swiss National Science Foundation 310030-146490 to OD; the Shriners Hospitals for Children (grant 15958) to MW; the Medical Research Council grants G9825289 and G1000467 to RVT, G1000801 to DH, and MC_UC_12010/3 to LF. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health.
We would like to thank the patients and their families who consented to these studies and the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics for the generation of the sequencing data. Additionally, we are grateful to F. Harrington, C. Mignion, V. Sharma, I. Taylor and I. Westbury for assistance with molecular genetic analysis, and the staff of the OUH NHS Immunology Laboratory for DNA preparation.
URLs
Stampy read mapper, http://www.well.ox.ac.uk/project-stampy
Platypus variant caller, https://github.com/andyrimmer/Platypus
Picard, www.picard.sourceforge.net
Ensembl regulatory build, http://www.ensembl.org/info/docs/funcgen/regulatory_build.html
NHLBI Exome Variant Server, http://evs.gs.washington.edu/EVS
Copenhagen disease gene association list, http://diseases.jensenlab.org/Search
Eurexpress database, http://discovery.lifemapsc.com/gene-expression-signals/high-throughput/ish-large-scale-dataset-eurexpress
Universal Mutation Database for BRCA2: http://www.umd.be/BRCA2
Footnotes
Accession Codes
The majority of samples studied in WGS500 were consented for clinical investigation only. A small number of samples were collected for general research and WGS data for these samples are available from the ENA under accession number XXX.
Competing Financial Interests
All authors at Illumina (see affiliations) are employees of Illumina Inc., a public company that develops and markets systems for genetic analysis. GM, GL and PD are founders and shareholders of Genomics Ltd, a company that develops genome analytics.
References for main text
- 1.Need AC, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–61. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon-Salazar TJ, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaulieu CL, et al. FORGE Canada Consortium: Outcomes of a 2-Year National Rare-Disease Gene-Discovery Project. Am J Hum Genet. 2014;94:809–17. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–25. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 10.Saunders CJ, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilissen C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 12.Jacob HJ, et al. Genomics in clinical practice: lessons from the front lines. Sci Transl Med. 2013;5:194cm5. doi: 10.1126/scitranslmed.3006468. [DOI] [PubMed] [Google Scholar]
- 13.Cazier JB, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014;5:3756. doi: 10.1038/ncomms4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babbs C, et al. Homozygous mutations in a predicted endonuclease are a novel cause of congenital dyserythropoietic anemia type I. Haematologica. 2013;98:1383–7. doi: 10.3324/haematol.2013.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin HC, et al. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma VP, et al. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet. 2013;45:304–7. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cossins J, et al. Congenital myasthenic syndromes due to mutations in ALG2 and ALG14. Brain. 2013;136:944–56. doi: 10.1093/brain/awt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lise S, et al. Recessive mutations in SPTBN2 implicate beta-III spectrin in both cognitive and motor development. PLoS Genet. 2012;8:e1003074. doi: 10.1371/journal.pgen.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2012;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy DJ, et al. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6:26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–4. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 26.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 28.Denayer E, Legius E. What’s new in the neuro-cardio-facial-cutaneous syndromes? Eur J Pediatr. 2007;166:1091–8. doi: 10.1007/s00431-007-0535-7. [DOI] [PubMed] [Google Scholar]
- 29.Martinelli S, et al. Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet. 2010;87:250–7. doi: 10.1016/j.ajhg.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemeyer CM, et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez B, et al. Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J Med Genet. 2010;47:686–91. doi: 10.1136/jmg.2010.076836. [DOI] [PubMed] [Google Scholar]
- 32.Nava C, et al. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry. 2012;2:e179. doi: 10.1038/tp.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isrie M, et al. HUWE1 mutation explains phenotypic severity in a case of familial idiopathic intellectual disability. Eur J Med Genet. 2013;56:379–82. doi: 10.1016/j.ejmg.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Froyen G, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–43. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMullin MF. The classification and diagnosis of erythrocytosis. Int J Lab Hematol. 2008;30:447–59. doi: 10.1111/j.1751-553X.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 36.Jelkmann W. Regulation of erythropoietin production. Journal of Physiology. 2011;589:1251–8. doi: 10.1113/jphysiol.2010.195057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowl MR, et al. An interstitial deletion-insertion involving chromosomes 2p25.3 and Xq27.1, near SOX3, causes X-linked recessive hypoparathyroidism. J. Clin. Invest. 2005;115:2822–31. doi: 10.1172/JCI24156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zajac JD, Danks JA. The development of the parathyroid gland: from fish to human. Current Opinion in Nephrology and Hypertension. 2008;17:353–6. doi: 10.1097/MNH.0b013e328304651c. [DOI] [PubMed] [Google Scholar]
- 39.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in Medicine. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metcalfe K, et al. Family history of cancer and cancer risks in women with BRCA1 or BRCA2 mutations. J Natl Cancer Inst. 2010;102:1874–8. doi: 10.1093/jnci/djq443. [DOI] [PubMed] [Google Scholar]
- 42.Zuk O, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–64. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moutsianas L, et al. The Power of Gene-based Rare Variant Methods to Detect Disease-associated Variation and Test Hypotheses about Complex Disease. PLoS Genet. doi: 10.1371/journal.pgen.1005165. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kapplinger JD, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57:2317–27. doi: 10.1016/j.jacc.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castera L, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet. 2014;22:1305–13. doi: 10.1038/ejhg.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chong HK, et al. The validation and clinical implementation of BRCAplus: a comprehensive high-risk breast cancer diagnostic assay. PLoS One. 2014;9:e97408. doi: 10.1371/journal.pone.0097408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borg A, et al. Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE study. Hum Mutat. 2010;31:E1200–40. doi: 10.1002/humu.21202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebbeck TR, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2004;22:1055–62. doi: 10.1200/JCO.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 49.Hakansson S, et al. Moderate frequency of BRCA1 and BRCA2 germ-line mutations in Scandinavian familial breast cancer. Am J Hum Genet. 1997;60:1068–78. [PMC free article] [PubMed] [Google Scholar]
- 50.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caputo S, et al. Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012;40:D992–1002. doi: 10.1093/nar/gkr1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brohet RM, et al. Breast and ovarian cancer risks in a large series of clinically ascertained families with a high proportion of BRCA1 and BRCA2 Dutch founder mutations. J Med Genet. 2014;51:98–107. doi: 10.1136/jmedgenet-2013-101974. [DOI] [PubMed] [Google Scholar]
- 53.Moss AJ, et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–9. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi G, et al. Spectrum and frequency of cardiac channel defects in swimming-triggered arrhythmia syndromes. Circulation. 2004;110:2119–24. doi: 10.1161/01.CIR.0000144471.98080.CA. [DOI] [PubMed] [Google Scholar]
- 55.Kapplinger JD, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crotti L, et al. Long QT syndrome-associated mutations in intrauterine fetal death. JAMA. 2013;309:1473–82. doi: 10.1001/jama.2013.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, et al. Intracellular ATP binding is required to activate the slowly activating K+ channel I(Ks) Proc Natl Acad Sci U S A. 2013;110:18922–7. doi: 10.1073/pnas.1315649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vukcevic M, et al. Functional properties of RYR1 mutations identified in Swedish patients with malignant hyperthermia and central core disease. Anesth Analg. 2010;111:185–90. doi: 10.1213/ANE.0b013e3181cbd815. [DOI] [PubMed] [Google Scholar]
Methods only references
- 59.Lamble S, et al. Improved workflows for high throughput library preparation using the transposome-based Nextera system. BMC Biotechnol. 2013;13:104. doi: 10.1186/1472-6750-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–8. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagnamenta AT, et al. Exome sequencing can detect pathogenic mosaic mutations present at low allele frequencies. J Hum Genet. 2012;57:70–2. doi: 10.1038/jhg.2011.128. [DOI] [PubMed] [Google Scholar]
- 64.Ruark E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–10. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012 doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yau C. OncoSNP-SEQ: a statistical approach for the identification of somatic copy number alterations from next-generation sequencing of cancer genomes. Bioinformatics. 2013;29:2482–4. doi: 10.1093/bioinformatics/btt416. [DOI] [PubMed] [Google Scholar]
- 68.Plagnol V, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–54. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McQuillan R, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Colella S, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–25. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 72.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diez-Roux G, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.