Abstract
Phosphatidylinositol 3-kinase Vps34 complexes regulate intracellular membrane trafficking in endocytic sorting, cytokinesis and autophagy. We present the 4.4 Å crystal structure of the 385 kDa endosomal complex II (PIK3C3-CII), consisting of Vps34, Vps15 (p150), Vps30/Atg6 (Beclin 1) and Vps38 (UVRAG). The subunits form a Y-shaped complex, centered on the Vps34 C2 domain. Vps34 and Vps15 intertwine in one arm where the Vps15 kinase domain engages the Vps34 activation loop to regulate its activity. Vps30 and Vps38 form the other arm that brackets the Vps15/Vps34 heterodimer, suggesting a path for complex assembly. Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) revealed conformational changes accompanying membrane binding and identified a Vps30 loop that is critical for the ability of complex II to phosphorylate giant liposomes on which complex I is inactive.
The class III phosphatidylinositol-3-kinase (PI3K) known as Vps34 (vacuolar protein sorting 34, encoded by PIK3C3) is present in all eukaryotes. It phosphorylates phosphatidylinositol (PI) to yield phosphatidylinositol 3-phosphate (PtdIns3P) (1), which is important in all vesicle sorting pathways to lysosomes, including phagocytosis, autophagy, multivesicular body formation and cytoplasm-to vacuole targeting (Cvt). In yeast, PtdIns3P production is totally dependent on Vps34 (2, 3), whereas in mouse embryonic fibroblasts, a VPS34 knockout suggests that 65% of PtdIns3P derives from Vps34, with a class II PI3K producing the remainder (4). Vps34 forms complexes with other proteins (Fig. 1A): Vps15 (encoded by PIK3R4, known as p150 in mammalian cells), Vps30 (encoded by VPS30/ATG6 in yeast, equivalent to mammalian Beclin 1, encoded by BECN1) and either Vps38 (UVRAG) or Atg14 (ATG14L). Atg14/ATG14L defines complex I, involved in autophagy, which also includes Atg38 (5) as a fifth subunit (NRBF2 in mammalian cells), whereas Vps38 is characteristic of complex II and essential for vacuolar protein sorting (3). In mammalian cells, complex II is also involved in autophagy, receptor degradation and cytokinesis (6) as well as signalling (7, 8), recycling (9) and lysosomal tubulation (10). Independently from complex I and II, Beclin 1 and UVRAG also play separate roles in endosome function and neuron viability (11). Elevated expression of PIK3C3, PIK3R4 and BECN1 is associated with more aggressive forms of chronic lymphocytic leukemia (CLL) (12), consistent with autophagy being involved in tumour maintenance. The crucial roles of complex I and complex II in stress response, nutrition and disease have led to efforts to develop both activators and inhibitors, like the recently developed ATP-mimetic inhibitors of Vps34 kinase domain (7, 13, 14).
Fig. 1.
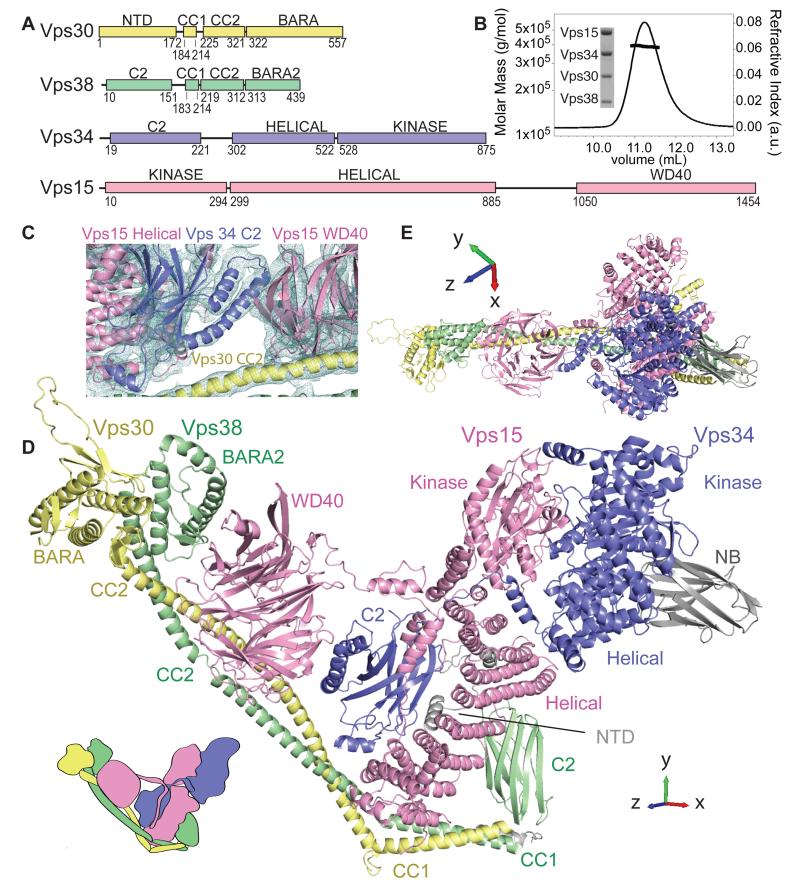
Complex II structure. (A) Domain organisation of complex II subunits. (B) MALS and SDS-PAGE analyses of complex II show a 390 kDa heterotetramer with 1:1:1:1 stoichiometry. (C) Experimental electron density contoured at 1.1σ for part of the model. (D) Complex II has a Y shape with two arms and a base. NB denotes nanobody. (E) Rotated view of the complex.
Important questions concerning Vps34 complexes remain, such as the nature of the relationship between Vps15 and Vps34, and the roles of Vps30 and Vps38/Atg14 in the functions of these complexes and how the complexes recognize membranes. To address these questions and assist the development of complex-specific drugs, we determined the crystal structure of complex II, characterised its dynamics and membrane binding.
The X-ray crystal structure of complex II
Saccharomyces cerevisiae complex II displayed a 1:1:1:1 ratio of four subunits, (Fig. 1B). Crystallization required a nanobody (15) that recognized the Vps34 helical domain, as determined by HDX-MS (residues 386-406, fig. S1). Data from seven native crystals and phases from two Ta6Br12 derivative crystals (Table S1) produced a high quality 4.4 Å resolution experimental electron density map (Fig. 1C). Building the structure was challenging at this resolution. Initial models for several domains of the complex derived from previous structures and distant homologues (16-18) were fitted first and the remainder of the structure was built directly into the density. The final model consists of 2834 residues out of the 3469, with most of the missing residues predicted to be disordered. Although at this resolution side chains were not visible, the sequence register was inferred from previously determined structures for most of Vps34, the WD40 domain of Vps15 and the C-terminal domain of Vps30. An approximate sequence register was assigned for the remainder of the structure. The real-space correlation of the model with the density suggests that the fit is reasonable for most of the structure. The poorest fit to the density is in the Vps38 N-terminal C2 domain (fig. S2).
Overall architecture of complex II
The complex has a Y shape, with two long arms and a short hook-like base (Fig. 1D, E). The base is built entirely of the Vps30 and Vps38 N-terminal domains and coiled-coil 1 (CC1) domains. One of the arms (15 nm in length) consists of Vps15 and Vps34 (Fig. 1E) while the other arm (18 nm) includes domains from all four subunits arranged along the Vps30 and Vps38 coiled-coil 2 (CC2). Interestingly, Vps30 and Vps38 show similar architectures except for their N-terminal domains, where Vps38 has a C2 domain, while Vps30 is mostly unstructured (fig. S3, A and B). At the C-terminus, Vps30 has a BARA domain that binds side-by-side to the C-terminal domain of Vps38, which we named BARA2. The two arms of complex II correspond to the V-shape seen in the low-resolution EM structure of complex I (19). It is thus likely that most of the details seen in complex II are preserved in complex I.
Vps15/Vps34 catalytic heterodimer
Vps34 and Vps15 intertwine in an anti-parallel fashion, with each of the three domains of Vps15 [kinase and helical (KINHEAT), and WD40] interacting with at least one domain of Vps34 [C2, helical and kinase (HELCAT)] (Fig. 1D, fig. S3, C and D). This network of interactions explains the co-dependent relationship of the two proteins: Vps34 is essential for Vps15 integrity (3) whereas Vps15 is necessary for Vps34 membrane recruitment and activity in vivo (20).
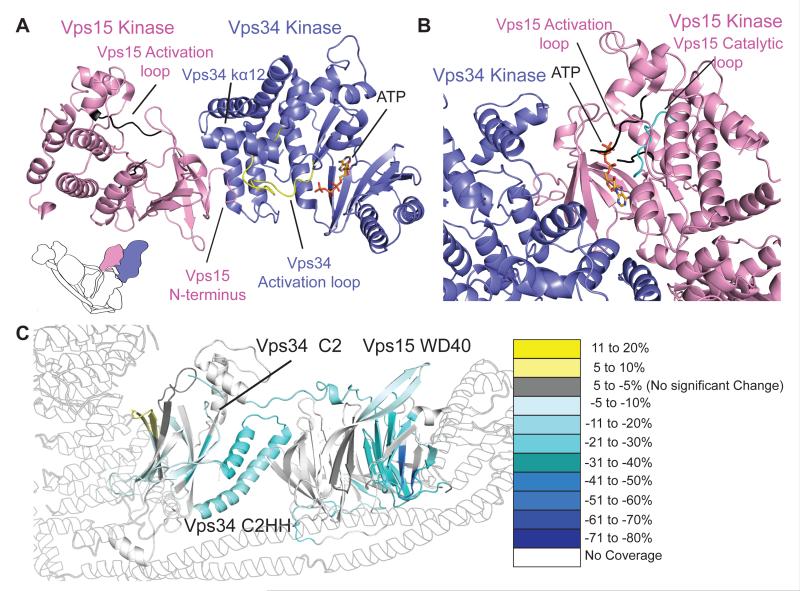
The N-lobe of the Vps15 kinase domain lies at the tip of the right arm, interacting with the C-lobe of the Vps34 kinase domain (Fig. 2A). It is not certain whether Vps15 is an active kinase or a pseudokinase. Wild-type Vps15 is phosphorylated whereas kinase-dead variants are not, suggesting autophosphorylation (21). Vps15 exhibits non-typical residues in critical catalytic elements: a 145-HGD sequence motif instead of HRD in the catalytic loop; a 165-DFA sequence motif instead of DFG in the activation segment and the absence of a GxGxxG motif (P-loop) that normally binds phosphates in ATP (22) (fig. S3C). Although these substitutions are quite rare in the human kinome, they are found both in active and pseudo kinases (23). The Vps15 kinase domain in our structure is probably in an inactive conformation since the long, partially ordered “activation loop” intrudes into the ATP binding site and would prevent ATP binding (Fig. 2B).
Fig. 2.
The Vps15/Vps34 heterodimer. (A) Vps15 kinase domain N-lobe interacts with Vps34 activation loop (yellow) and C-lobe helices kα10 and kα12 . ATP in Vps34 was modelled based on other lipid kinases. (B) The activation loop (black) in Vps15 active site would prevent ATP binding (ATP model based on other protein kinases). (C) Vps15-WD40 and Vps34-C2 (including C2HH) peptides are showing less HDX in complex II than in the Vps15/Vps34. HDX for the rest of the complex (grey outline) is not shown.
The central part of Vps15 forms 9 helical pairs wound into a right-handed helical solenoid stretching from the Vps15 kinase domain to the Vps38-CC1/CC2 (Fig. 1D). The first eight helical pairs are HEAT-type repeats. The helical solenoid has a concave face that nestles Vps34-C2. The Vps15 solenoid is followed by four more helices in a bundle (residues 788-885) that caps the hydrophobic end of the solenoid. Most of the Vps15 linker between the solenoid and WD40 is disordered (residues 889-985), except for an extended section (residues 986-1050) threading along the surface of Vps34-C2.
The C-terminal domain of Vps15 is a seven-bladed WD40 domain (16) that bridges the Vps15/Vps34 and the Vps30/Vps38 pairs. On one side, conserved residues in the WD40 β3/β4 loop of blade 4 (1261-ArgPhe, referred to as RF loop) reach Vps34-C2, while on the other side the only two other invariant residues, Glu1077 and Ser1121 (between blades 1 and 7) contact Vps38-BARA2. Finally, one surface of the WD40 straddles Vps30-CC2. Due to this interaction, the greatest HDX differences in WD40 between the heterotetramer and the Vps15/Vps34 heterodimer are for peptides in blades six and seven of WD40, which contact Vps30-CC2 (Fig. 2C). A cluster of Beclin 1 phosphorylation sites map to this CC2/WD40 interface (fig. S4), where they could affect assembly of the active heterotetramer. The region of Vps30 in contact with the WD40 coincides with a region in mammalian Beclin 1 that was referred to as the evolutionary conserved domain (ECD) (24). Instead of binding Vps34 directly as previously proposed (24), ECD binds to Vps15-WD40 and therefore only indirectly to Vps34.
Vps15 as a potential regulator of Vps34
Several catalytically important elements of Vps34 contact the kinase domain of Vps15: the activation loop (residues 749-767), helix kα10 and the kα11/kα12 elbow (residues 860-864) (Fig. 2A). These interactions are consistent with the observation that deletions in the kα11/kα12 region prevented association between Vps34 and Vps15 (25). In protein kinases, the dynamics of the activation loop regulates catalytic efficiency (26). The proximity of Vps15 could modulate the active conformation of the Vps34 lipid-binding site, including the activation loop. SUMOylation of residue K840 in helix kα10 of hsVPS34 increases kinase activity (27). This modification could regulate both the conformation of the Vps34 activation loop and its interaction with Vps15.
Vps15 could influence Vps34 in multiple ways: stabilizing the enzyme, inhibiting basal activity by restricting the activation loop and contributing to membrane recruitment. The roles of Vps15 would be akin to the multiple roles of regulatory subunits of class IA PI3Ks. Oncogenic mutations of class I PI3Ks unmask the inhibitory role of the regulatory subunit (28). In a human metastatic melanoma, a Vps15 R107Q mutation in the kinase domain (29) is in close proximity to the C2/HELCAT Linker Helix and could affect Vps34 activity by re-orientating the Vps15/Vps34 kinase domains.
VPS34 C2 domain as a hub for complex II formation
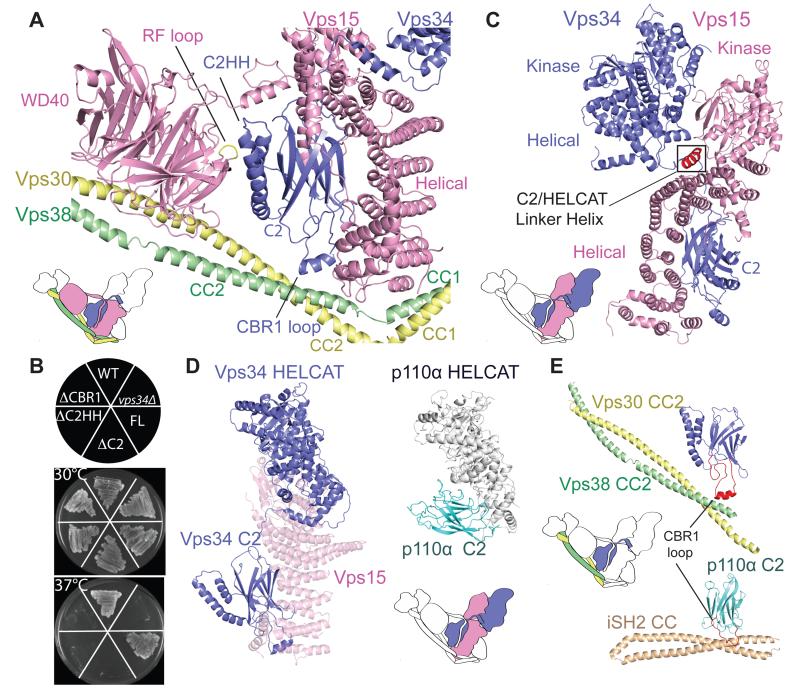
Although each Vps34 domain interacts with Vps15, the most extensive contacts involve Vps34-C2, which rests between the arms of the Y, engaging all subunits (Fig. 3A). This domain was crucial for the function of Vps34 in cells (Fig. 3B), and it was important for tight association with Vps15 (fig. S5). C2 domains often serve as membrane interaction modules, however, given the dense protein-protein interaction network centered on Vps34-C2, this domain is unlikely to also bind membranes.
Fig. 3.
Vps34-C2 is at the heart of complex II. (A) C2 β-sandwich and its distinct structural features (C2HH and CBR1 loop). (B) Effects of Vps34-C2 mutants on temperature-sensitive growth. Deletion of either the Vps34-C2 (ΔC2), the C2HH (ΔC2HH) or CBR1 (ΔCBR1) prevents growth at 37°C, similar to vps34Δ. Full-length Vps34 (FL) grows at 37°C, just as the wild-type strain (WT). (C) The helix (red) in the Vps34 C2/HELCAT linker fits between Vp15 kinase and helical domains. (D) Vps34-C2 does not contact HELCAT (left), in contrast to class I PI3Ks (right). In complex II, Vps15 bridges Vps34-C2 and Vps34-HELCAT. (E) The Vps34-C2 CBR1 loop contacts Vps30/Vps38 CC2. This is similar to the CBR1 loop of class IA PI3Ks contacting the coiled-coil (iSH2 CC) of the regulatory subunit.
The relationship of Vps34-C2 to Vps34-HELCAT is completely different than what has been observed for class I PI3Ks. The C2 domain of class I PI3Ks interacts extensively with the helical domain, while Vps34-C2 is remote from the rest of the enzyme, cradled by Vps15-HEAT (Fig. 3, C and D). This relationship of Vps34-C2 to the Vps34-HELCAT may imply substantial flexibility around the linker between these two regions of Vps34, as suggested by a relatively high global HDX (fig. S6). Long range motions of Vps34-HELCAT have been observed in the EM study of mammalian complexes I and II (19). Here, most of the C2/HELCAT linker appeared disordered, however a conserved Linker Helix (Fig. 3C) nestles in a crook between the Vps15 and Vps34, where it could provide a pivot for Vps34-HELCAT motions.
In addition to a β-sandwich scaffold, Vps34-C2 deploys two distinct features assisting its role as a nucleus for complex II. The extended loop between β1/β2 strands (CBR1) reaches the Vps30/Vps38 CC2 (Fig. 3E), analogous to the critical interaction of the equivalent loop in class I PI3K C2 domains with the iSH2 coiled-coil of the p85 regulatory subunit (28). This constitutes the only contact of Vps34 with the Vps30/Vps38 unit and explains the previously reported binding of hsVPS34-C2 to a Beclin 1 region encompassing CC2 (30). Deletion of CBR1 resulted in a phenotype similar to deletion of VPS34 (Fig. 3B), suggesting that this is an important interaction in the complex.
A helical hairpin insertion (C2HH, for C2 Helical Hairpin) in the β7/β8 loop reaches the RF loop in Vps15-WD40 (Fig. 3A). This insertion showed less HDX in complex II compared to the Vps15/Vps34 dimer (Fig. 2C), suggesting a stabilizing role for C2HH. The global exchange for C2HH was, nevertheless, higher than the rest of Vps34-C2, indicating flexibility (fig. S6), which might be in turn regulated by phosphorylations (fig. S4). Several of these modifications were affect the activity of mammalian VPS34 (31, 32). Deletion of C2HH in yeast Vps34 resulted in a temperature sensitive phenotype (Fig. 3B).
Organization of the Vps30/Vps38 pair
The Vps30 and Vps38 interact along their entire lengths, forming an elongated structure that fits like a bracket around the Vps15/Vps34 heterodimer, along which Vps38 and, to a smaller extent, Vps30, establish multiple contacts with Vps15/Vps34. The two central coiled-coils (a short CC1 and a longer CC2) of Vps30/Vps38 form a parallel heterodimer. A parallel arrangement of these two proteins was also proposed for the mammalian complexes I and II based on a low-resolution EM model (19) while the homodimeric CC2 of Beclin 1 forms an anti-parallel homodimer (33). The bracket-like organisation of the Vps30/Vps38 pair relative to Vps15/Vps34 suggests that the complex might assemble from these two pairs. This is supported by a pulldown experiment showing that purified Vps30/Vps38 heterodimer can capture purified Vps15/Vps34 (fig. S7).
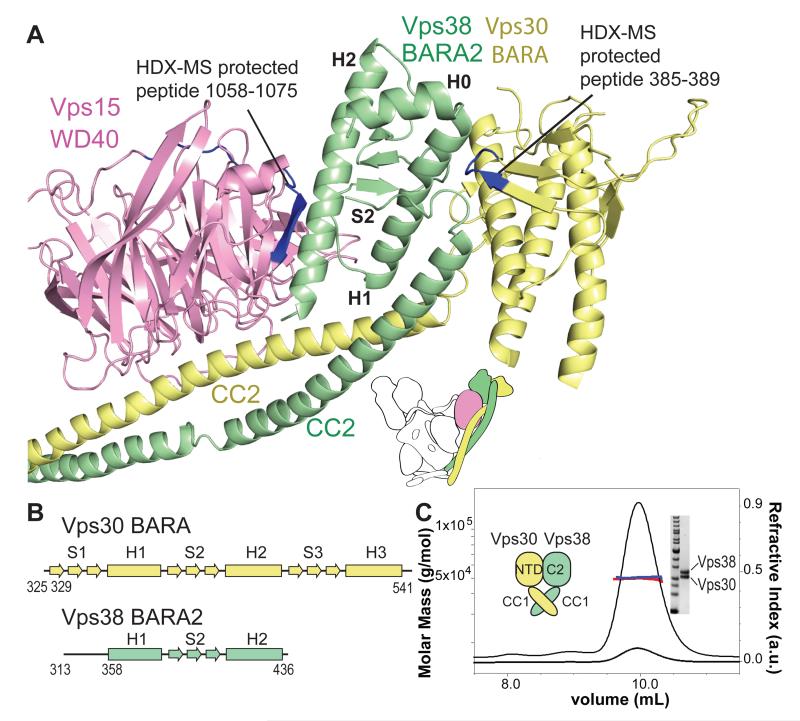
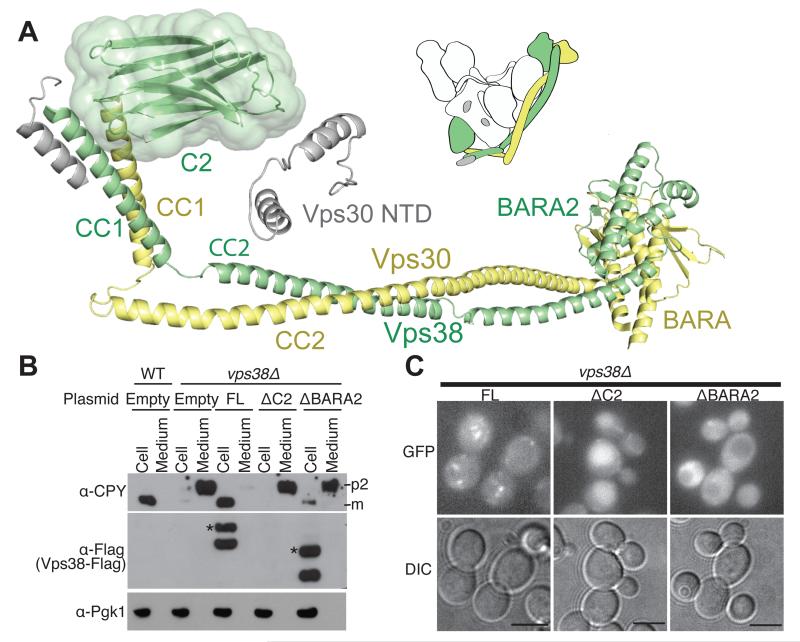
The C-terminal Vps30-BARA and Vps38-BARA2 are at the tip of the left arm, with Vps30-BARA interacting exclusively with Vps38, and Vps38-BARA2 contacting both Vps30 and Vps15-WD40. Vps30-BARA in complex II agrees closely with the structure of the isolated BARA (17, 34) (Fig. 4A). While Vps30-BARA has three β-α repeats, Vps38-BARA2 has two incomplete BARA-type repeats. The first repeat includes a helix preceded by an extended structure but not forming a clear β-sheet whereas the second repeat is complete (Fig. 4B).
Fig. 4.
Organisation of the Vps30/Vps38 pair. (A) Vps38-BARA2 bridges Vps30-BARA to Vps15-WD40. (B) Vps38-BARA2 fold is related to Vps30-BARA. (C) MALS shows that Vps30-NTD and Vps38-C2 together with CC1 regions, form a stable 50 kDa complex, as expected for a 1:1 heterodimer.
The most conserved part of Vps30-BARA forms an extensive interface with Vps38-BARA2, which is consistent with HDX data (Vps30 peptide 385-389 at the BARA/BARA2 interface being more protected in complex II then in free Vps30, Fig. 4A). The Vps30-NTD, CC1 and CC2 also contribute to interaction with Vps38. Indeed, the NTD, C2 and CC1 domains of Vps30 and Vps38 are sufficient to form a stable heterodimer in solution (Fig. 4C).
The N-terminal domains of Vps30 and Vps38 meet at the base of the complex, where they interact primarily with Vps15. Vps30-NTD is not well defined, but we could build three disconnected helices that could be part of it (Fig. 5A). In our interpretation, one of these helices forms a triple helical bundle with CC1 of Vps38/Vps30. Furthermore, the density assigned to Vps30-NTD is adjacent to Vps38-C2. This close spatial relationship is supported by our HDX-MS that identified a single nanobody binding to both Vps38-C2 (residues 45-55) and Vps30-NTD (residues 63-76). HDX suggested additional contacts, not apparent at this resolution, between these domains, based on the large protection of peptide 82-86 in Vps30-NTD in complex II versus Vps30 alone (Table S2A).
Fig. 5.
Vps30 and Vps38 N-terminal domains are important for complex II stability and function. (A) Vps30 and Vps38 parallel arrangement brings Vps30-NTD close to Vps38-C2. For Vps30-NTD (grey), a partial model consisting of three helices is suggested. (B) CPY assay in WT and vps38Δ strains expressing Vps38 truncations. Protein bands for CPY, Vps38, and Pgk1 were detected with anti-CPY, anti-Flag and anti-Pgkl antibodies. Full-length (FL) and ΔBARA2 show two Vps38 bands. Asterisks indicate Vps38 expected size. (C) Fluorescence microscopy of the vps38Δ strain expressing the same truncations as (B) fused to GFP. FL showed a punctate pattern whereas ΔC2 was in the vacuole, and ΔBARA2 was cytosolic with empty vacuoles. Scale bars are 5μm.
Given that Vps38 defines complex II and is essential for vacuolar protein sorting (17), we characterised the role of Vps38-C2 and BARA2 using a CPY sorting assay in a vps38Δ mutant (Fig. 5B). ΔBARA2 showed only a partial rescue of CPY sorting whereas ΔC2 did not rescue carboxypeptidase Y (CPY) sorting due to Vps38 degradation (Fig. 5C). Thus, Vps38-BARA2 is important for vacuolar protein sorting and Vps38-C2 is essential for the stability of Vps38 and consequently for complex II formation.
Implications for complex I
In complex I, Vps38/UVRAG is substituted with Atg14/ATG14L. Although the N-terminal domains of Vps30, Vps38 and Atg14 differ, the overall similarity of their domain organizations suggests that these proteins may have evolved from a common ancestor. The lengths of the Atg14 and Vps38 coiled-coil regions are comparable, which suggests that Atg14 probably interacts with the CC1 and CC2 domains of Vps30 similarly to Vps38.
The C-terminal extension of human Atg14L contains a BATS domain previously shown to interact with membranes. In our model, the BATS domain would share proximity with other elements of the complex involved in membrane binding (see next section). In yeast, the amphiphilic helix at the C-terminus of Atg14 (residues 240-261) could act as its functional counterpart. The absence of a C2 domain in Atg14 to interact with Vps30-NTD may explain why Vps30-NTD is dispensable for autophagy, in contrast to the critical role of the Vps30 C-terminal domain (17).
Membrane interactions of complex II
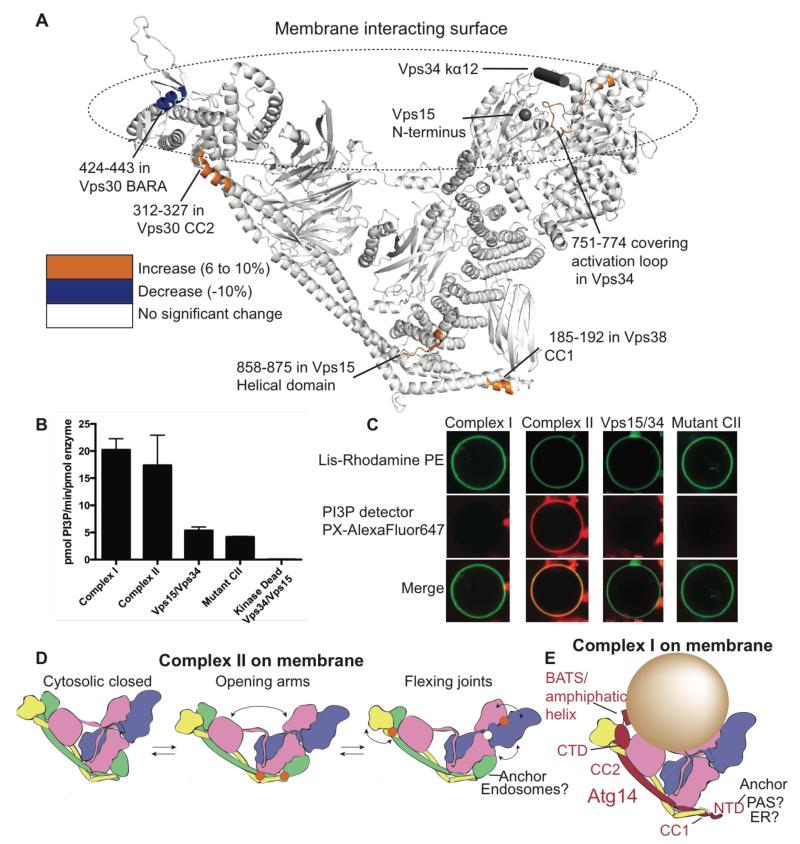
Because the activity of all PI3Ks is controlled by their association with membranes, we examined this interaction for intact complex II. Comparing HDX for complex II in the presence and absence of membranes showed that localized changes in exchange for all four subunits accompany membrane binding (Fig. 6A, Table S3). Vps30-BARA (residues 424-443) shows decreased HDX, suggesting that this region directly interacts with membranes, consistent with Vps30-BARA greatly contributing to endosomal localisation of complex II (17). The analogous region (359-FFW) in Beclin 1 BARA contains the membrane binding “aromatic finger” (34). For simplicity, we also will refer to Vps30 residues 430-FRK-432 as the “aromatic finger” (fig. S3A). Two known membrane-binding elements in Vps34, the activation loop and the C-terminal kα12 helix, did not display a decrease in HDX. Increased HDX in the Vps34 activation loop likely reflects a disruption of the contact between this loop and the N-terminus of Vps15 enabling the activation loop to bind lipids, while contacts made by kα12 helix in the closed conformation observed in complex II are replaced by membrane interactions (18). Thus, the net HDX that we observe for the Vps34 kinase domain upon membrane binding is presumably a sum of increases in HDX due to conformational changes and decreases associated with direct membrane interaction.
Fig. 6.
Activity and dynamics of Vps34 complexes on membranes. (A) Vps30-BARA binds membranes as shown by decreased HDX (blue). Regions showing increased HDX are coloured orange. The membrane-protected region of Vps30-BARA, the activation loop, kα12 helix and the Vps15 N-terminus (grey) suggest a plausible membrane-binding surface. (B) Activities of the Vps34 complexes on SUVs. Kinase-dead Vps15/Vps34 (Vps34-D731N), is used as a negative control. Error bars represent the standard deviation from the mean for triplicate measurements. (C) Complex II is active on GUVs, in sharp contrast to complex I, Vps15/Vps34 and complex II “aromatic finger” mutant. 50 GUVs were counted per condition. 50/50 GUVs showed PI3P production (red) for complex II, whereas 0/50 were positive for complex I, Vps15/Vps34 or the mutant. (D) A model for conformational changes on membranes. Regions showing increased HDX upon membrane binding (orange circles) and the dynamic C2/HELCAT Linker Helix (white circle) suggest pivoting points. (E) A model of complex I on a highly curved membrane. The N-terminal domains of Atg14 (NTD) and Vps38-C2 could contribute to anchoring the complexes at specific cellular sites.
In our model for Vps34 complexes on membranes (Fig. 6A), the tips of the two arms contact the membrane: one arm via the Vps34 activation loop, the Vps34 kα12 helix and the Vps15 N-terminal myristoylation site, and the other arm via the Vps30/Beclin 1 BARA domain. Both Vps15-WD40 and Vps34 interact with the yeast lipidated Gα subunit Gpa1 on endosomes (35) and VPS34 interacts with Gαi at the midbody (36). It is plausible that membrane-localized Gpa1 would fit into the notch between the two arms of complex II. Mammalian Vps34 complexes are regulated by Rab5 (37) and Rab7 (38), and these G-proteins may occupy the same crevice between the two arms of the complexes.
Activity of Vps34 complexes on small and large vesicles
To evaluate the influence of the Vps30-containing arm on Vps34 activity, we compared activities of complex I, complex II and the Vps15/Vps34 heterodimer. Using small unilamellar vesicles (SUVs), complexes I and II had similar activities that are higher than the heterodimer (Fig. 6B). Complex II with a mutation in the Vps30 “aromatic finger” (430-FRK-432 mutated to DDD) showed lower activity, similar to the Vps15/Vps34 dimer (Fig. 6B). Remarkably, complexes I and II greatly differed in their activities on giant unilamellar vesicles (GUVs). Complex I and the Vps15/Vps34 heterodimer were inactive on GUVs, whereas complex II efficiently phosphorylated these relatively low-curvature membranes (Fig. 6C). This ability depended critically on the Vps30 aromatic finger, since the FRK/DDD mutant had an activity indistinguishable from Vps15/Vps34. It is noteworthy that a cluster of post-translational modifications maps to loops in the Vps30-BARA (fig. S4). These inhibitory modifications may prevent the aromatic finger from interacting with membranes. Atg14 might block the basal membrane-binding ability of Vps30 in complex I on GUVs, and in cells this block could be relieved by starvation. Membrane binding was accompanied by exposure of peptides in both the arms and base of the complex (Fig. 6A), indicating conformational changes compatible with global opening/closing motions such as suggested by a normal mode analysis (Movie S1). These data allow us to devise a model for how complex II adapts to membranes (Fig. 6D). Conformational shifts in the base would enable opening of the arms to adjust to different membrane curvatures. Flexing joints at the junction between Vps30-CC2 and Vps30-BARA and between the Vps15/Vps34 kinase domains would then enhance productive membrane binding. In contrast to complex II, which operates on relatively flat endosomal membranes, complex I is associated with highly curved membranes. Accordingly, human ATG14L has a C-terminal BATS domain that senses membrane curvature (39). Highly curved membranes could better fit into the notch between the two arms (Fig. 6E). PtdIns3P recognition and curvature sensitivity conferred by the BATS domain could set up a positive feedback loop for rapid, localized build-up of PtdIns3P on highly curved membranes such as the tubular/vesicular structures associated with the tips of the autophagic isolation membrane (40) originating at the omegasome (41).
The Y-shaped complexes I/II constitute bipartite structures with catalytic activity in the Vps15/Vps34 arm and functional specificity arising from the Vps30/Vps38 (or Vps30/Atg14) arm and base. These assemblies constitute recruitment platforms for regulatory proteins and downstream factors, fine-tuning Vps34 activity. This detailed view of the whole complex provides a framework for designing selective activators and inhibitors of Vps34 complexes targeting not only the active site, but also complex-specific membrane- and protein-binding interfaces. The structure should enable a better understanding of the regulation of Vps34 autophagic and endosomal complexes by post-translational modifications, binding partners and membranes.
Materials and methods
Constructs
All DNA constructs used in this study are listed in Table S5.
Yeast strains and cell growth
Yeast strains used in this study are listed in Table S6. Cell growth for protein expression is described under Protein purification. For cell biology, yeast cells carrying plasmids were grown at 30 °C in –URA medium (0.67% yeast nitrogen base without amino acids, 0.5% casamino acids and 2% glucose, supplemented with 0.002% adenine sulfate, 0.002% tryptophan, 0.002% tyrosine and 0.002% leucine).
Protein purification – complex II
Plasmids carrying genes encoding Saccharomyces cerevisiae PI3K complex II subunits were transformed into BCY123 or YOY193 yeast cells (see Tables S5 and S6 for details). All coding fragments were under the GAL1 promoter (Somatogen Inc.). Cells were grown in YM4 medium (0.67% yeast nitrogen base without amino acids, 0.5% casamino acids and 2% glucose, supplemented with 0.002% adenine sulfate, and 0.002% tyrosine) for double-plasmid expression using pRS424- and pRS426-backbone plasmids, or –URA medium for pRS426-backbone plasmids. Flasks containing a total of 10 L of cells were grown at 30 °C in the presence of 2% galactose for 24 h. Cells were lysed in lysis buffer (50 mM Tris pH 8.8, 300 mM NaCl, 1 mM DTT, 1% Triton-X, 0.5 mM PMSF and protease inhibitor tablet (Roche, 05056489001) with glass beads (Sigma G8772), using a cell disruptor (FastPrep-24, MP Biomedicals). IgG beads (GE Healthcare 17096902) were added to the lysate, incubated for 3 h at 4 °C, then washed once with Wash 1 buffer (50 mM Tris pH 8.0, 300 mM NaCl, 1 mM DTT, 1% Triton-X, and 0.5 mM PMSF), once with Wash 2 buffer (50 mM Tris pH 8.0, 300 mM NaCl, 1 mM DTT, and 0.5 mM PMSF). The protein A (ZZ) tags were cleaved by TEV protease in Wash 2 buffer overnight. The elution fractions were concentrated and subjected to gel filtration on Superdex 200 16/60 equilibrated in 20 mM Tris pH8.0, 300 mM NaCl, 1 mM TCEP. The “aromatic finger” mutant complex II (with Vps30 residues 430-FRK-432 mutated to DDD) was purified in the same way as the wild type complex II.
Protein purification – complex I
Saccharomyces cerevisiae complex I was expressed as described above. The protocol for complex I purification was essentially the same as for complex II, with three differences. Firstly, 150 mM NaCl was used in all buffers, secondly, 0.5 % CHAPS was used as a detergent, and thirdly, 20 mM Tris pH8.8, 150 mM NaCl, 1 mM TCEP was used for gel filtration.
Protein purification – Vps15/Vps34 dimer
Saccharomyces cerevisiae Vps15/Vps34 heterodimer was expressed as described above. The purification protocol was the same as for Complex II, with some diffrences. Wash 1 and Wash 2 buffers contained 500 mM NaCl. Once the protein A tags were cleaved off with TEV protease overnight, the sample was diluted with three volumes of HEP-A buffer (20 mM Tris pH8.0, 0.5 mM DTT) and then loaded on the pre-equilibrated 5 ml Heparin HP column. The protein was then eluted with a NaCl gradient (HEP-B buffer, 2M NaCl and 0.5 mM DTT).
Kinase dead vps34 (D731N) dimer was expressed and purified as wild type.
Production of nanobodies
Nanobodies were generated against cross-linked complex II. Protein for generating nanobodies was produced from 10 L of yeast cell culture. Complex II was purified as described above except that the Wash 2 buffer was 50 mM HEPES pH 7.4, 300 mM NaCl, 1 mM TCEP, and 0.5 mM PMSF. Proteins on IgG beads were incubated overnight with TEV protease in 10 mL of Wash 2 buffer at 4 °C. After the first elution, 5 mL of Wash 2 buffer without TEV protease was added and eluted again. The two TEV elution fractions were combined and CHAPS was added to a concentration of 0.1%. Protein was concentrated for gel filtration (Amicon Ultra 100 kDa cutoff, Millipore). Gel filtration was done on Superdex 200 16/60 (GE Healthcare), which was pre-equilibrated with gel filtration buffer (20 mM HEPES pH 7.4, 300 mM NaCl, and 1 mM TCEP). The peak fractions were combined and concentrated to 400 μL at 6.3 mg/mL (16 μM). The whole amount was used for the cross-linking. Complex II at 10 μM was cross-linked using CovalX K200 (CovalX AG, Zurich, Switzerland) by following the manufacture’s protocol. After removing precipitates by centrifugation, the cross-linked material was frozen in liquid nitrogen and shipped on dry ice to the Steyaert laboratory for immunization. Complex II-specific nanobodies were generated following the previously described protocol (15). Briefly, one Ilama was immunized six times with 100 μg of cross-linked Complex II. Peripheral blood lymphocytes were extracted 4 days after the final antigen boost. RNA was purified and converted into cDNA via RT-PCR. The cDNA was cloned into phage display vector pMESy4 containing a C-terminal 6X His tag. For the phage selection, Complex II, both cross-linked and native, was directly coated on the solid phase. Antigen-bound phages were recovered by proteolysis with trypsin. 53 clones were sequenced after selections, out of which 38 tested positive in an ELISA. Nanobody 8780, which was crystallized with Complex II for structure determination, was selected using non-cross linked material.
Expression and purification of nanobodies for crystallization
WK6 Su- cells were transformed with the plasmids carrying VHH constructs genes from the selections described above. Cells were grown at 37 °C to OD600 = 0.7 then induced with 1M IPTG for 20 h at 25 °C. For the initial screening, 1 L of cell culture was grown for each nanobody. For further characterisation and crystallization trials, 6 L of cell culture were grown. Nanobodies are purified from the cell periplasm. Cells from 1 L culture were resuspended on ice with 15 mL of TES buffer (200 mM Tris-HCl pH8, 0.5 mM EDTA, 500 mM sucrose) and then rotated for 1 h at 4 °C. After incubation, 30 mL of 4X diluted TES buffer (50 mM Tris-HCl pH8, 0.125 mM EDTA, 125 mM sucrose) buffer was added to the cells followed by further rotation for 45 min at 4 °C. After ultracentrifugation, the supernatant was loaded onto Ni/NTA beads (Qiagen) equilibrated in Wash buffer (50 mM Phosphate pH7, 1 M NaCl), washed extensively in the same buffer, followed by low Imidazole wash (50 mM Phosphate pH7.4, 1 M NaCl, 15 mM Imidazole pH8). The protein was eluted with 200 mM Imidazole pH 8, 20 mM Phosphate pH 7.4, 1 M NaCl and then concentrated in Amicon Ultra 5 kDa cut-off concentrator, before being loaded on S75 10/30 gel filtration column (for small scale 1 L expression) or S75 16/60 (for large scale 6 L expression) equilibrated in 20 mM Tris-HCL pH7.5, 150 mM NaCl.
Crystallization
Purified complex II was mixed with an excess of a purified nanobody and the mixture was gel-filtered on a 16/60 Superdex 200 column. The relevant fractions were pooled, concentrated to 6-7 mg/mL and screened for crystallization in over 1900 conditions using LMB Innovadyne robotic crystallization setup (42), with 100 nL of protein and 100 nL of the reservoir solution in 96-well plates. Initial crystals of complex II were obtained using Crystal Screen 1 reagent 3 (Hampton Research), containing 0.4 M mono-ammonium dihydrogen phosphate. The crystals were optimized in grids varying the concentration of reagents, adjusting pH, screening different salts, cryogenic protectants, detergents and other additives. For data collection, crystals were produced in 96 well plates, using 200 nL of protein and 200 nL of the reservoir solution containing 0.35 M sodium acetate and 3% 1,5–diaminopentanedihydrochloride. Crystals were grown at 17 °C and reached the maximum size after 5-7 days. Crystals were cryo-protected by adding to the drop a solution containing 0.35 M sodium acetate, 3% 1,5–diaminopentanedihydrochloride and 25% ethylene glycol. Crystals were mounted in loops and flash cooled in liquid nitrogen.
Structure solution
Diffraction data were collected at the European Synchrotron Radiation Facility (ESRF) on beamline ID29 and at the Diamond light Source on beamlines I04 and I24. Native crystals of Complex II bound to a nanobody diffracted to approximately 4.4 Å resolution, some of these were soaked from 1 to 10 min in 2 mM tantalum bromide (Jena Biosciences PK-103) dissolved in the crystallization solution. We optimized data collection for tantalum-derivatized crystals by estimating the total dose absorbed using RADDOSE (43). Datasets for tantalum soaked crystals were collected at the peak absorption edge and at the two different inflection points for this element (see Table S1). Data were processed with XDS and scaled using XSCALE (44). Experimental phasing was carried out using either Autosharp or Sharp (45). The signal over noise for native crystals could be enhanced by combining seven isomorphous datasets together with XSCALE, to allow us to adopt a 4.4 Å resolution cut off. A list of initial sites was obtained using SHELX C/D (46), enabling us to obtain a first experimental map in which previously solved models of the WD40 domain of VPS15, the BARA and coiled-coil domains of VPS30/38 as well as the HELCAT of VPS34 could be readily placed. At this stage, the electron density was clear enough to build most of the helices of the helical domain of VPS15. Missing parts of the complex, such as the kinase domain of VPS15 and the C2 domains of VPS34 and VPS15 were subsequently found using homemade scripts for serial molecular replacement with PHASER (47), using model libraries obtained from HHPRED secondary structure profile matching hits (48) or BALBES (49). Improvement of the initial model was possible by successive rounds of model building and refinement followed by density modification with SOLOMON (50) from the SHARP server interface using the updated model every time as a solvent mask to generate a better experimental map. Refinement was performed using tight secondary structure and Ramachandran restraints with PHENIX (51). Given the low resolution, no side chains were incorporated, the model contains however the yeast sequence register.
HDX sample preparation: dynamics and protein contacts
For determination of the complex dynamics and protein contacts within the complex HDX-MS experiments were conducted. The D2O used in all experiments was of a ≥99.75% isotopic purity (Acros Organics). Protein samples (10 μL of 2.5 μM protein in Dilution Buffer (20 mM Tris pH 8.0, 50 mM NaCl, and 2 mM TCEP)) were mixed rapidly with 35 μL of D2O Buffer (20 mM Tris pH 8.0, 50 mM NaCl, 2 mM TCEP, 96.6% D2O), to produce a sample with a final concentration of 75% D2O. Four time points of exchange (3 s on ice, 3 s, 30 s and 300 s at 23 °C) were produced in triplicate. For each time point, the reaction was quenched by the addition of 20 μL of Quench Buffer (8.4% formic acid, 5 M guanidine-HCl), and immediate freezing in liquid nitrogen. Samples were then stored at −80 °C.
HDX sample preparation: nanobody mapping
For nanobody binding experiments, 10 μL of either 2.5 μM complex II, or 10 μL of a 2.5 μM complex II/ 3 μM nanobody solution were mixed rapidly with 40 μL of D2O buffer (producing a final concentration of 77% D2O). Reactions were incubated for 3 s on ice, before being quenched with 20 μL of Quench Buffer, and frozen in liquid nitrogen as above.
HDX sample preparation: membrane binding
Membrane binding experiments were carried out using liposomes produced as described previously (28). Briefly, phospholipids were mixed in organic solution in the desired ratios (18% (w/v) liver phosphatidylinositol (Avanti 840042C), 45% brain phosphatidylserine (Avanti 840032C), 20% brain phosphatidylethanolamine (Avanti 840022C), and 17% brain phosphatidylcholine (Avanti 840053C), (grossly mimicking yeast plasma membrane, partly modified from (52)), desiccated, resuspended in Lipid Buffer (20 mM Tris pH 8.0, 100 mM KCl, 1 mM EGTA), sonicated, freeze-thawed ten times, and then extruded eleven times through a 100 nM filter. For HDX-MS experiments, complex II (2.5 μM final concentration) was incubated with either 1 mg/mL liposomes or the equivalent volume of Lipid Buffer for 30 min prior to the addition of any deuterated buffer. HDX reactions were then conducted as described above, with 10 μL of protein or protein/lipid solution being mixed with 40 μL D2O Buffer supplemented with either 1 mg/mL or the equivalent volume Lipid Buffer (76.6% D2O). The final concentration of D2O was 61%. Four incubation times were conducted (3 s on ice, 3, 30 and 300 s at 23 °C), and the samples were quenched and stored as above.
Measurement of deuterium Incorporation
All samples were analysed using the same chromatography system and mass spectrometry instruments. Protein samples were thawed and immediately injected onto an ice-immersed ultra performance liquid chromatography (UPLC) system. The protein was initially passed through two immobilized pepsin columns (Applied Biosystems; poroszyme) at 150 μL/min for 3 min in order to digest the protein, and the resulting deuterated peptides were collected on a van-guard pre-column trap (Waters). The trap was then switched in-line to an Acquity 1.7 μm particle, 100 mm × 1 mm C18 UPLC column (Waters), and the peptides were eluted from the column using a 20 min 5 - 45% gradient of buffer A (0.1% formic acid) and buffer B (100% acetronitrile), followed by a four min 76% buffer B wash. The mass of the eluted peptides was then determined using a Xevo QTOF (Waters) acquiring for 30 min over a mass range of 350 to 1500 m/z, using an electrospray ionisation (ESI) source operated at a capillary temperature of 225 °C, and a spray voltage of 3.5 kV.
Peptide identification
The identification of non-deuterated peptides was determined by running tandem MS/MS experiments with a non-deuterated sample digested as above but with 60 min gradient over the reverse-phase column. This was repeated with a 20 min gradient, and identical peptides observed in both gradients were used to produce a linear extrapolation to correct for the retention times of peptides observed only in the 60 min gradient. A tolerance of 15 ppm was used in the MS observations, and an MS/MS tolerance of 0.2 Da was also used. The resultant MS/MS datasets were analysed using Mascot Distiller (Matrix Science). Peptides with a Mascot score less than 10 were excluded from all further analysis. Centroid values were determined using HD-Examiner software (Sierra Analytics). Every non-deuterated peptide was validated manually by searching the protein sample’s MS scan and checking for the correct charge state, the presence of any overlapping peptide envelopes, and a correct retention time (peptides failing any of these criteria were excluded from further analysis). Peptide coverage was as follows: Vps38 – 51.9%, Vps34 - 83.6%, Vps30 - 82.9%, Vps15 – 36.8%.
Mass analysis of peptide centroids
Validated non-deuterated peptides were subsequently analysed for their deuterium incorporation under various conditions. Due to the shift in isotopic envelope observed upon deuterium incorporation, additional peptides were discarded from the data set due to overlap. All peptides were once again checked for correct charge state and retention time. All deuterium levels quoted in both figures and the text are all relative levels of deuterium as no fully deuterated sample was obtained. A mathematical correction was applied using HD Examiner to compensate for the differences in the level of the deuterium in the various D2O buffers. The average error was ≤0.5 Da for any two replicates. Only changes that were greater than 0.9 Da for both observations were considered significant for our analysis. All results quoted both in the figures and the text are the greatest observed difference for that peptide at any single time point, unless otherwise explicitly stated.
CPY assay
CPY assay was modified from Noda et al. (17). Cells were grown at 30 °C to mid-log phase (A600 = 0.8 - 1.0) in –URA medium and an equivalent of 5 A600 units of cells was collected and resupended in 500 μL of –URA medium with 50 mM KPO4 pH 5.7 and 0.5 mg/mL BSA followed by incubation at 30 °C for 1 h. Cells and extracellular fraction were separated by centrifugation at 4,000 g for 1 min. Cellular fractions were subjected to the cell wall loosening procedure (2 M LiAc on ice for 2 min, followed by 0.4 M NaOH on ice for 2 min), resuspended in 50 mM Tris pH 8.0, 1% SDS, 8 M urea, 2 mM DTT and 5 mM EDTA with glass beads, and disrupted at 3000 rpm for 30 s with PowerLyzer (Mo Bio) at 4 °C. One in four volume of 4X sample buffer was added to the lysates and loaded on gel without boiling. Extracellular fractions were precipitated with final 10% TCA (from 100% stock) on ice for 30 min and centrifuged at 20,000 g for 10 min. Pellets were washed two times with 90% cold acetone, air-dried, then resuspended in 2X sample buffer, and loaded on gel without boiling. CPY is synthesized as a premature form (p1), transported to the Golgi to be glycosylated (p2), and then delivered to the vacuole, where it is cleaved to a mature form (m). The p2 and m forms of CPY were detected both in cellular (Cell) and medium (Medium) fractions using anti-CPY (1/200; Invitrogen A6428). Vps38 proteins, which were all C-terminally tagged with 3xFlag were detected by anti-Flag-HRP (1/1000; Sigma A8592). Of note, both full length Vps38 (FL) and ΔBARA2 show two protein bands. The higher molecular weight bands correspond to the sizes of interest (asterisks in Fig. 5B). Pgk1 was detected by anti-Pgk1 (1/2000; Invitrogen 459250). Anti-mouse HRP (1/2000; Sigma A9917) was used as a secondary antibody.
Protein A pull-down assay
Plasmids carrying VPS15-ZZ (pYO225) were cotransformed with an empty vector (pRS424), untagged VPS34 (pYO69) or HELCAT (pYO361) into a yeast strain (YOY193). Protein expression was induced by 2 % galactose for 24 h at 30 °C in 500 mL of YM4 medium in the presence of 2 % glycerol and 3 % lactic acid. Cells were harvested in 50 mL tubes, then 5 mL of lysis buffer (50 mM Tris pH 8.8, 300 mM NaCl, 1 mM DTT, 1 % triton, 0.5 mM PMSF, and inhibitor tablet) and 3 g of glass beads were added. Cells were disrupted with FastPrep-24 at 6.5 intensity for 45 s once. Cell debris and glass beads were removed by centrifuging at 4000 rpm for 2 min. Supernatants were transferred to ultracentrifuge tubes, and spun at 20,000 g for 10 min. IgG beads (100 μL of 50 % slurry) were added to supernatants, and rotated at 4 °C for 2 h. The protein-bound beads were transferred to Poly-Prep chromatography columns (Bio-Rad), and washed with 10 mL of wash buffer (50 mM Tris pH 8.0, 300 mM NaCl, 1 mM DTT, 1 % Triton X-100, and 0.5 mM PMSF) and 10 mL of Wash 2 buffer (50 mM Tris pH 8.0, 300 mM NaCl, and 1 mM DTT) by gravity flow. After draining Wash 2 buffer, IgG beads were resuspended in 500 μL of Wash 2 buffer. A fraction of lysate (0.04 %) and beads (2 %) were loaded on a gel.
Microscopy
Cells were grown to mid-log phase (A600 = 0.8 - 1.0), then examined by microscopy with an inverted microscope (Nikon Eclipse TE2000) equipped with a CCD camera (CoolSNAP-HQ2, Roper Scientific, Tucson, AZ), a GFP filter (Chroma Technology, Rockingham, UT) and a DIC channel. Images were analyzed using ImageJ, and levels adjusted with Adobe Photoshop.
Vps15/Vps34-Vps30/Vps38 reconstitution
A yeast plasmid carrying VPS30 and ZZ-VPS38 (pYO734) was transformed into the YOY193 strain. The transformed cells were grown overnight at 30 °C in –URA medium, then shifted to 1 L induction medium (YM4, 2 % glycerol, 3 % lactic acid, 2 % galactose, 5 μM ZnCl2, 55mg /L leucine, and 55mg /L tryptophan) at 30 °C for 24 h. About 10 g cell were obtained. Cells were split into two aliquots of 5 g in 50 ml tubes. Cells/tube were disrupted in 5 ml lysis buffer (50 mM Tris pH 8.8, 150 mM NaCl, 1 mM DTT, 1 % Triton X-100, 0.5 mM PMSF, and EDTA-free protease inhibitor tablet) and 3 g glass beads using a FastPrep24 at 6.5 intensity for 45 s. Cell debris and glass beads were spun down at 2,500 g for 2 min. Supernatants were combined, and spun at 20,000 g for 10 min. IgG beads (100 μL) were added to the supernatant, and rotated at 4 °C for 2h. For the IgG beads only control, 50 μl of IgG beads was added in 5 ml lysis buffer, and rotated as above. The control and the protein-bound IgG beads were transferred to Poly-Prep chromatography columns, and washed with 10 mL of wash buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT, 1 % Triton X-100, 5 mM ATP, 50 mM MgCl2, 2 mg/ml RNase A) and 10 mL of reaction buffer (50 mM Tris pH 8.0, 300 mM NaCl, and 1 mM DTT) by gravity flow. The protein-bound IgG beads were split into two halves, one for the control without Vps15/Vps34, the other for the reconstitution with Vps15/Vps34. After draining the buffer, 225 μl of reaction buffer was added to each column, then 25 μl of purified Vps15/Vps34 was added to a final concentration of 1 μM. Binding reaction was done on ice for 1h. Beads were washed once with 10 ml wash2 buffer (50 mM Tris pH 8.0, 300 mM NaCl, 1 mM DTT, 1 % Triton X-100), and once with 10 ml reaction buffer. 2.5% of total volume was loaded on a SDS-PAGE gel for input and IgG beads fractions.
Lipid kinase assays
Lipid kinase assays were carried out using liposomes produced as described above. Phospholipids were mixed in organic solution in the desired ratios (18% (w/v) phosphatidylinositol, 10% phosphatidylserine, 17% phosphatidylethanolamine, and 55% phosphatidylcholine), desiccated in a nitrogen stream, then resuspended in Lipid Buffer (25 mM HEPES pH 8.0, 100 mM NaCl, 1 mM EGTA) to make a 1 mg/ml stock. The stock was sonicated in a bath sonicator, freeze-thawed ten times, and then extruded eleven times through a 100 nm filter.
Lipid kinase assays were performed using the Echelon K-3000 kit, a 96- well ELISA assay for detection of PI3P. Protein dilutions and lipid stocks were made in Reaction buffer (25 mM Tris pH8.0, 150 mM NaCl, 2 mM EGTA, 0.1% CHAPS, 4 mM MnCl2, 2mM TCEP). First, 5 μl of 20 nM enzyme was aliquoted into PCR tubes. The reaction was initiated by addition of 5 μl of liposomes in Reaction buffer with 100 μM ATP to give a final enzyme concentration of 10 nM and final PI concentration of 50 μM. Control reactions without ATP were also set up for all proteins. The reaction was incubated for 1 hour at room temperature without agitation and was the stopped by addition of 16.6 mM EDTA and diluted to 80 μl in the 2x PI3P Detection Buffer (K-3004). 50 μl of this reaction was then transferred to the Detection Plate (K-3001). PI3P was then detected on a plate reader as per manufacturer’s instructions. Kinase dead Vps34 (D731N) dimer was used as a negative control. Error bars represent the standard deviation from the mean for triplicate measurements. The results shown are representative of at least three independent experiments.
Giant Unilamellar Vesicle (GUV) assay
Two concentric greased 16 mm/18 mm O-rings were placed on the metal side of an indium tin-oxide coated glass slide. An aliquot (10 μl) of the 1 mg/ml lipid mixture of 18% (w/v) liver phosphatidylinositol, 10% brain phosphatidylserine, 17% brain phosphatidylethanolamine, 55% brain phosphatidylcholine (catalogue numbers as above) and 0.1% Lisamine Rhodamine-PE (Avanti 810850P) was pipetted into the centre of the rings and dried under vacuum for 1 h. Swelling Buffer (500 mM glucose, 220 μl) was added to each ring. The slide was then placed into the Vesicle Prep Pro GUV generator (Nanion) and covered with another indium tin-oxide coated glass slide. An AC-field (1V, 10 Hz) was applied to the slides for 4 h at 60 °C. The wells of the observation chamber (Lab Tek chamber 1, Borosilicate, 155411) were pre-equilibrated with 5 mg/ml of BSA of 4 h and then washed with the Observation buffer (50 mM HEPES pH8.0, 150 mM NaCl). GUVs (50 μl) were added to a solution with enzyme and PI3P detection probe (p40PX domain labelled with AlexaFluor647) to give 200 μL of solution of Observation buffer supplemented with 1 mM EGTA, 2 mM MnCl2, 1 mM TCEP and 50 μM ATP, with an enzyme concentration of 0.5 μM and a probe concentration of 7 μM. The plasmid used for p40PX domain expression was a kind gift from Michael Wilson, Babraham Institute, Cambridge. Confocal images were acquired using Zeiss LSM 780, with an x63 (NA 1.4) oil objective lens. There was no evidence of PI3P generation on GUVs incubated with complex I, the Vps15/Vps34 heterodimer or the mutant complex II. PI3P generation appeared exclusively on GUVs incubated with complex II.
Temperature sensitivity experiment
The plasmids were transformed into SEY6210 (WT) or vps34Δ. After 2 days at 30 °C on –URA plates, newly growing colonies were repatched onto new –URA plates, and grown at 30 °C or 37 °C for 2 days. Protein expression was also confirmed by western blotting using anti-Flag-HRP (data not shown).
Supplementary Material
Acknowledgments
We are grateful to O. Perisic for help with the project and with writing the manuscript. We thank Sean Munro for helpful advice and comments on the manuscript. We thank S. Maslen, M. Skehel and S.-Y. Peak-Chew for help with mass spectrometry and K. Willibal for help with Nanobody production. We are grateful to O. Kovalevskiy and G. Murshodov for advice on refinement, A. McCoy for guidance on molecular replacement, F. Rutaganira and K. Shokat (UCSF) for Vps34 inhibitors, and K. Nagai for providing yeast strain and expression backbone plasmids. We thank the beamline scientists from ESRF beamlines ID29, ID23-1, BM30A, DLS beamlines I24, I04, I04-1, and SLS beamline PXIII. We thank INSTRUCT, part of the European Strategy Forum on Research Infrastructures (ESFRI), the Hercules Foundation Flanders and FWO (G0C8514N) for their support to Nanobody discovery. KR was supported by Cambridge Cancer Center PhD fellowship, LZ by MRC and the Cambridge Overseas Trust and JB by a grant from the British Heart Foundation (PG11/109/29247). This work was funded by the BBSRC (BB/K019155/1) and the Medical Research Council (U105184308 to RLW). Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 5DFZ.
Footnotes
References
- 1.Herman P, Emr S. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schu P, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 3.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devereaux K, et al. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS ONE. 2013;8:e76405. doi: 10.1371/journal.pone.0076405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araki Y, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thoresen SB, Pedersen NM, Liestøl K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 2010;316:3368–3378. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Bago R, et al. Characterisation of VPS34-IN1, a selective inhibitor of Vps34 reveals that the phosphatidylinositol 3-phosphate binding SGK3 protein kinase is a downstream target of Class III PI-3 kinase. Biochem J. 2014 doi: 10.1042/BJ20140889. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohatgi RA, et al. Beclin 1 regulates growth factor receptor signaling in breast cancer. Oncogene. 2015 doi: 10.1038/onc.2014.454. doi:10.1038/onc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLeod IX, Zhou X, Li Q-J, Wang F, He Y-W. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Rα surface expression. J. Immunol. 2011;187:5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munson MJ, et al. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 2015 doi: 10.15252/embj.201590992. doi:10.15252/embj.201590992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKnight NC, et al. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet. 2014;10:e1004626. doi: 10.1371/journal.pgen.1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristensen L, et al. High expression of PI3K core complex genes is associated with poor prognosis in chronic lymphocytic leukemia. Leuk. Res. 2015 doi: 10.1016/j.leukres.2015.02.008. doi:10.1016/j.leukres.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Dowdle WE, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 14.Ronan B, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nature Chemical Biology. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 15.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heenan EJ, et al. Structure and Function of Vps15 in the Endosomal G Protein Signaling Pathway. Biochemistry. 2009;48:6390–6401. doi: 10.1021/bi900621w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noda NN, et al. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J Biol Chem. 2012;287:16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller S, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baskaran S, et al. Elife. in press, doi:10.7554/eLife.05115. [Google Scholar]
- 20.Stack J, Herman P, Schu P, Emr S. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman PK, Stack JH, Emr SD. A genetic and structural analysis of the yeast Vps15 protein kinase: evidence for a direct role of Vps15p in vacuolar protein delivery. EMBO J. 1991;10:4049–4060. doi: 10.1002/j.1460-2075.1991.tb04981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor SS, Keshwani MM, Steichen JM, Kornev AP. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2012;367:2517–2528. doi: 10.1098/rstb.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reese ML, Boothroyd JC. A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J Biol Chem. 2011;286:29366–29375. doi: 10.1074/jbc.M111.253435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 25.Budovskaya Y, Hama H, DeWald D, Herman P. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277:287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- 26.Joseph RE, et al. Activation loop dynamics determine the different catalytic efficiencies of B cell- and T cell-specific tec kinases. Science Signaling. 2013;6:ra76–ra76. doi: 10.1126/scisignal.2004298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, et al. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. 2013;110:6841–6846. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc. Natl. Acad. Sci. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shull AY, et al. Novel Somatic Mutations to PI3K Pathway Genes in Metastatic Melanoma. PLoS ONE. 2012;7:e43369. doi: 10.1371/journal.pone.0043369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuya T, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slessareva J, Routt S, Temple B, Bankaitis V, Dohlman H. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 36.Boularan C, Kamenyeva O, Cho H, Kehrl JH. Resistance to inhibitors of cholinesterase (Ric)-8A and Gαi contribute to cytokinesis abscission by controlling vacuolar protein-sorting (Vps)34 activity. PLoS ONE. 2014;9:e86680. doi: 10.1371/journal.pone.0086680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 2002;3:416–427. doi: 10.1034/j.1600-0854.2002.30605.x. [DOI] [PubMed] [Google Scholar]
- 38.Stein M-P, Feng Y, Cooper K, Welford A, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 39.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uemura T, et al. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 2014;34:1695–1706. doi: 10.1128/MCB.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axe E, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog. Biophys. Mol. Biol. 2005;88:311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Paithankar KS, Garman EF. Know your dose: RADDOSE. Acta Crystallogr. D Biol. Crystallogr. 2010;66:381–388. doi: 10.1107/S0907444910006724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick G. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy A, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long F, Vagin A, Young P, Murshudov G. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zinser E, et al. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei Y, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife. 2015;4 doi: 10.7554/eLife.05289. doi:10.7554/eLife.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei Y, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun T, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. doi: 10.1038/ncomms8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang RC, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi C-S, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. autophagy. 2010;6:986–987. doi: 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia P, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012;19:788–797. doi: 10.1038/cdd.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J, Guan K-L. AMPK connects energy stress to PIK3C3/VPS34 regulation. autophagy. 2014;9:1110–1111. doi: 10.4161/auto.24877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirsch DS, et al. Insulin activation of vacuolar protein sorting 34 mediates localized phosphatidylinositol 3-phosphate production at lamellipodia and activation of mTOR/S6K1. Cell. Signal. 2014;26:1258–1268. doi: 10.1016/j.cellsig.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 64.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Molecular and Cellular Biology. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.