The crystal structure of the response regulator RPA3017 from the photosynthetic bacterium R. palustris has been determined at 1.9 Å resolution. RPA3017 is part of a two-component system that involves two tandem bacteriophytochromes involved in red-light signaling.
Keywords: two-component signal transduction, response regulator, red-light signaling, crystal structure, phosphotransfer
Abstract
Two-component signal transduction is the major signaling mechanism that enables bacteria to survive and thrive in complex environmental conditions. The photosynthetic bacterium R. palustris employs two tandem bacteriophytochromes, RpBphP2 and RpBphP3, to perceive red-light signals that regulate the synthesis of light-harvesting complexes under low-light conditions. Both RpBphP2 and RpBphP3 are photosensory histidine kinases coupled to the same response regulator RPA3017. Together, they constitute a two-component system that converts a red-light signal into a biological signal. In this work, the crystal structure of RPA3017 in the unphosphorylated form at 1.9 Å resolution is presented. This structure reveals a tightly associated dimer arrangement that is conserved among phytochrome-related response regulators. The conserved active-site architecture provides structural insight into the phosphotransfer reaction between RpBphP2/RpBphP3 and RPA3017. Based on structural comparisons and homology modeling, how specific recognition between RpBphP2/RpBphP3 and RPA3017 is achieved at the molecular level is further explored.
1. Introduction
Two-component systems (TCSs) are a common signal transduction mechanism widely found in prokaryotes, fungi, yeast and some plants (Capra & Laub, 2012 ▸; Gao & Stock, 2009 ▸; Stock et al., 2000 ▸). Bacteria use TCSs extensively to detect, perceive and respond to environmental stress and signals such as light, oxygen, population density, nutrients and other signals (Capra & Laub, 2012 ▸; Stock et al., 2000 ▸). At the core of a TCS is a phosphotransfer reaction between a sensory histidine kinase (HK) and its cognate response regulator (RR). Sensory HKs typically exhibit a modular architecture in a parallel dimer framework, where the sensory and effector domains are arranged in a ‘beans-on-a-stalk’ fashion. Sensory HKs are membrane-bound in many TCS pathways, in which extracellular sensory domains are connected to 1–2 transmembrane helices that transmit signals across the cell membrane so as to regulate the autophosphorylation of an HK inside the cell. The phosphoryl group of histidine is further relayed to a conserved aspartate of the cognate response regulator to provoke downstream responses (Capra & Laub, 2012 ▸).
Bacterial light signaling is also mediated by two-component systems. Most photoreceptors are soluble sensory HKs since light, unlike chemical signals, can readily penetrate the cell membrane. In the photosynthetic bacterium Rhodopseudomonas palustris, two red-light photoreceptors, bacteriophytochromes RpBphP2 and RpBphP3, regulate the synthesis of light-harvesting complexes under low-light conditions (Giraud et al., 2005 ▸). Both RpBphP2 and RpBphP3 are red-light-sensitive HKs that are highly homologous in sequence and tertiary structure (Bellini & Papiz, 2012 ▸; Yang et al., 2007 ▸, 2015 ▸). Their N-terminal photosensory core modules consist of three domains, namely PAS (Per–ARNT–Sim), GAF (cGMP phosphodiesterase/adenyl cyclase/FhlA) and PHY (phytochrome). The red-light-absorbing chromophore biliverdin (BV) is primarily embedded in the GAF domain and forms a covalent linkage with a cysteine residue (Essen et al., 2008 ▸; Yang et al., 2008 ▸; Fig. 1 ▸ a). At the C-terminus is the HK domain that undergoes light-dependent autophosphorylation (Anders et al., 2011 ▸; Rockwell et al., 2006 ▸; Yang et al., 2011 ▸). RpBphP2 and RpBphP3 are encoded by tandem genes in the same operon next to the gene for the response regulator RPA3017 (Giraud et al., 2005 ▸). In vitro biochemical experiments showed that both RpBphP3 and RpBphP3 are paired with RPA3017 in the cognate HK–RR phosphotransfer reaction, although they exhibit distinct photoconversion properties (Giraud et al., 2005 ▸). This established RPA3017 as the cognate response regulator that is paired with RpBphP2/RpBphP3 in red-light signaling.
Figure 1.
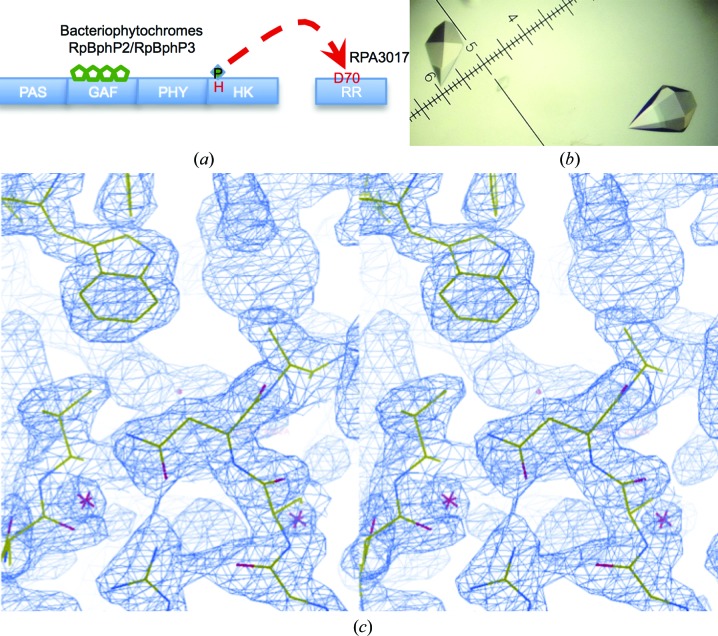
Structure determination of RPA3017. (a) The phosphorelay scheme of the two-component system between bacteriophytochromes RpBphP2/RpBphP3 and RR3017. (b) Typical RPA3017 crystals. (c) Stereoview of a representative region in the figure-of-merit-weighted experimental map following SAD phasing and density modification (contoured at 2.5σ).
The genome of R. palustris contains 63 putative sensory HKs and 79 response-regulator receiver domains (Larimer et al., 2003 ▸). Together, they regulate a wide range of cellular functions under diverse environmental conditions for adaptation and growth. Since most RRs are not covalently linked to sensory HKs, cognate pairing between HKs and RRs is essential for precise signal transmission to avoid unwanted cross-talk between different signaling pathways (Capra & Laub, 2012 ▸). Although much is known about the structures of RpBphP2 and RpBphP3 (Bellini & Papiz, 2012 ▸; Yang et al., 2007 ▸, 2015 ▸), it is still unclear how phosphotransfer between RpBphP2/RpBphP3 and RPA3017 is achieved at the molecular level and which structural elements of RpBphP2/RpBphP3 and RPA3017 contribute to the phosphotransfer reaction and specificity. In this work, we present the crystal structure of RPA3017 in the unphosphorylated state at 1.9 Å resolution determined by the single-wavelength anomalous dispersion method. We examine the dimerization mode of RPA3017 as well as protein–protein interactions between RpBphP2/RpBphP3 and RPA3017 using structural analyses and homology modeling. We also examine the active site and the reaction mechanism of the phosphotransfer reaction in RPA3017. This work sheds light on the molecular events involved in red-light signaling pathways in R. palustris.
2. Materials and methods
2.1. Cloning, expression and purification
The rpa3017 gene was amplified using the genomic DNA of R. palustris CGA009 (ATCC accession No. BAA-98D-5) as a template. The forward and reverse primers are 5′-CAGCCATATGATGAACCGCCAGCGCACACTGCC-3′ and 5′-CCACCTCGAGTCCCGTTCGATAAGCCTCGGTGG-3′, respectively. The PCR product was then inserted into the expression vector pET-24c between NdeI and XhoI restriction sites. The resulting construct was used to overexpress RPA3017, which carries an additional three residues (GLE) at the C-terminus followed by a 6×His affinity tag.
Escherichia coli BL21 (DE3) cells were transformed with the RPA3017 construct and grown in LB medium at 37°C until the OD600 reached ∼0.4–0.6. Following induction with 1 mM IPTG, the cells were grown overnight at 17°C before harvesting via centrifugation. The cell pellet was resuspended in lysis buffer (500 mM NaCl, 100 mM Tris–HCl pH 8.0) using a homogenizer and then subjected to cell lysis by sonication on ice. Following centrifugation, the supernatant of the cell lysate was collected, filtered and loaded onto a HisTrap Ni2+ column (GE Healthcare) for purification of His-tagged RPA3017 via immobilized metal ion-affinity chromatography. The RPA3017 protein was eluted using 250 mM imidazole, which was removed during the subsequent concentration process. Similar protocols were used to express and purify selenomethionine-substituted (SeMet) RPA3017 except that a minimal medium containing a cocktail of six normal amino acids and l-selenomethionine was used in the cell culture (Doublié, 1997 ▸).
2.2. Crystallization, X-ray data collection and structure determination
The purified native and SeMet RPA3017 were crystallized at 20°C using the hanging-drop vapor-diffusion method. A protein sample at a concentration of 3–5 mg ml−1 was mixed in a 1:1 ratio with a precipitant consisting of 0.2 M sodium citrate tribasic, 0.1 M sodium cacodylate pH 6.5, 4%(v/v) 2-propanol. Crystals appeared in 1 d and grew to full size (0.4 × 0.4 × 0.6 mm) in 3–7 d (Fig. 1 ▸ b). The crystals were cryoprotected in liquid nitrogen using the precipitant solution containing ∼25% glycerol.
X-ray diffraction data for native and SeMet RPA3017 were collected on the LS-CAT 21-ID-G beamline at the Advanced Photon Source, Argonne National Laboratory. The initial crystal structure of SeMet RPA3017 was determined in space group P6522 (unit-cell parameters a = b = 60.35, c = 203.03 Å) at 2.15 Å resolution using the single-wavelength anomalous diffraction (SAD) method in PHENIX (Adams et al., 2010 ▸). The native RPA3017 structure was determined in space group P65 (unit-cell parameters a = b = 60.98, c = 200.37 Å) using molecular replacement in PHENIX with the SeMet RPA3017 structure as a search model; each asymmetric unit contained two RPA3017 molecules. All X-ray data were indexed, integrated and scaled using the HKL-2000 software package (Otwinowski & Minor, 1997 ▸). Coot (Emsley & Cowtan, 2004 ▸) was used in model building. All structural illustrations were generated using PyMOL (http://www.pymol.org). The coordinates of the RPA3017 structure have been deposited in the Protein Data Bank as entry 4zyl.
3. Results and discussion
3.1. Crystal structure of RPA3017
The crystal structure of the response regulator RPA3017 was determined at 1.9 Å resolution using the single-wavelength anomalous dispersion (SAD) method and was refined to a final R factor and R free of 0.186 and 0.226, respectively (Table 1 ▸). The final model includes residues 7–149 with well resolved electron density (Fig. 1 ▸ c). The N-terminal six residues and the C-terminal 13 residues including the 6×His affinity tag were disordered and were not visible in the electron-density map.
Table 1. Statistics of X-ray data collection and structure refinement.
| Native RPA3017 | SeMet RPA3017 | |
|---|---|---|
| Resolution () | 501.90 (1.931.90) | 502.15 (2.192.15) |
| Completeness (%) | 99.5 (100) | 99.4 (99.7) |
| R merge | 0.044 (0.545) | 0.057 (0.775) |
| Multiplicity | 11.3 (11.4) | 20.3 (18.7) |
| I/(I) | 56.0 (3.83) | 61.1 (2.9) |
| Space group | P65 | P6522 |
| Unit-cell parameters () | a = b = 60.98, c = 200.37 | a = b = 60.35, c = 203.02 |
| Beamline | 21-ID-G, APS | 21-ID-G, APS |
| Refinement | ||
| R factor | 0.186 (0.224) | |
| Free R factor | 0.226 (0.251) | |
| Resolution () | 321.90 (1.961.90) | |
| Mean B factor (2) | 56.0 | |
| R.m.s.d., bond lengths () | 0.014 | |
| R.m.s.d., bond angles () | 1.361 | |
| Protein structure | 2 molecules [residues 7149] | |
| No. of waters | 148 | |
| Ramachandran plot (%) | ||
| Favored | 99 | |
| Allowed | 1 | |
| Disallowed | 0 | |
| PDB code | 4zyl | |
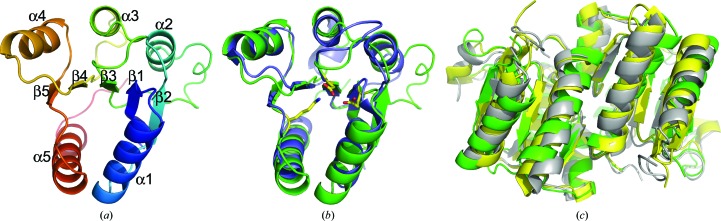
The RPA3017 structure adopts a Rossmann fold with five alternating β-strands and α-helices, a structural motif typical of RR structures (Gao & Stock, 2009 ▸). The highly conserved catalytic residues (Glu14, Asp70 and Lys122) are clustered at one end of the (α/β)5 structure and identify the active site of RPA3017 (Fig. 2 ▸ a). The RPA3017 structure reveals two tightly associated monomers that are related by noncrystallographic twofold symmetry perpendicular to the crystallographic 65 axis. The C-terminal structural segments (α4–β5–α5) constitute the dimer interface and bury a large surface area of 1165 Å2 (calculated using the PISA server; Krissinel & Henrick, 2007 ▸; Fig. 3 ▸). This mode of dimerization is nearly identical to those of RRs involved in phytochrome signaling such as Rcp1 from Synechocystis PCC6803 (PDB entries 1jlk and 1i3c; Im et al., 2002 ▸) and RcpA/RcpB from Calothrix PCC7601 (PDB entries 1k66 and 1k68; Benda et al., 2004 ▸). Since these RR structures were determined independently under various crystallization conditions in different space groups, we postulate that the RPA3017 structure represents a biologically relevant dimer rather than a crystallization artifact.
Figure 2.
Crystal structure of RPA3017. (a) Ribbon diagram of the RPA3017 monomer. Topology from the N-terminus to the C-terminus is represented by rainbow colors from blue to red. (b) Structure superposition of RPA3017 (green) and the response regulator RR468 (blue; PDB entry 3dgf). Residues in yellow mark the active site. (c) Structural alignment of the dimer structures of RPA3017 (green), Rcp1 (yellow; PDB entry 1i3c) and RcpB (grey; PDB entry 1k66) according to one subunit (chain A).
Figure 3.
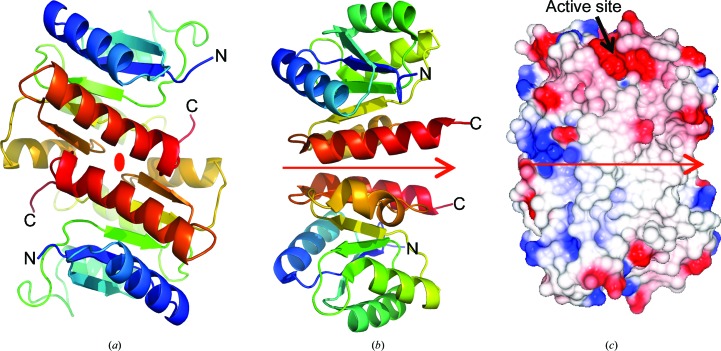
The dimer scaffold of RPA3017. (a) Two RPA3017 subunits are related by a noncrystallographic twofold symmetry axis marked by the oval-shaped symbol in red. The tracing in each subunit is colored in a rainbow from blue to red from the N-terminus to the C-terminus. The C-terminal structural elements (in orange/red) constitute the antiparallel dimer interface. (b) The receiver domain of the E. coli PhoP structure (PDB entry 3r0j) forms a parallel dimer with the C-terminal segments at the dimer interface. The red arrow indicates the twofold symmetry axis. (c) Surface rendering shows tight dimer association in the RPA3017 structure and the electrostatic surface reveals the active site in a negatively charged cleft.
Although response regulators are monomeric in the HK–RR phosphotransfer reaction (Casino et al., 2009 ▸; Podgornaia et al., 2013 ▸), their regulatory roles seem to depend on dimerization that is governed by the activation and/or phosphorylation state. Based on >200 RR structures in the PDB, the α4–β5–α5 surface in the RR structure emerges as the primary dimer interface (Gao & Stock, 2010 ▸). Like RPA3017, PhoP from Mycobacterium tuberculosis (PDB entry 3r0j) forms a dimer via its C-terminal structural segments (α4–β5–α5; Menon & Wang, 2011 ▸). However, their mode of dimerization differs. Specifically, PhoP adopts a parallel dimer scaffold denoted the ‘4-5-5 dimer’, while the RPA3017, Rcp1 and RcpA/B structures exhibit an antiparallel scaffold denoted the ‘inverted 4-5-5 dimer’ (Gao & Stock, 2010 ▸; Figs. 3 ▸ d and 3 ▸ e). It is noteworthy that the active sites for the phosphotransfer reaction are readily accessible in both scenarios. To date, four major modes of dimerization have been identified among RR structures and are denoted 4-5-5, inverted 4-5-5, 5-5 and 4-5 (Gao & Stock, 2010 ▸). It is plausible that the mode of dimerization is dictated by downstream regulatory functions. For example, full-length PhoP contains a C-terminal DNA-binding domain and acts as a transcriptional regulator (Menon & Wang, 2011 ▸). Upon activation, the N-terminal receiver domain of PhoP would favor parallel dimerization to promote DNA binding. In contrast, RPA3017 is a single-domain RR that is not covalently linked to any effector domains. It is expected to transfer its phosphoryl group to an aspartate of unknown downstream regulator(s) that control the synthesis of LH4 (Giraud et al., 2005 ▸).
3.2. The active site and the phosphotransfer reaction
The active site of RPA3017 resides in a shallow, negatively charged cleft found at the opposing ends of the RPA3017 dimer structure (Fig. 3 ▸). Three highly conserved residues, namely Glu14, Asp70 and Lys122, are located at the bottom of the cleft and serve as catalytic residues in the phosphotransfer reaction between the HK domains of RpBphP2/RpBphP3 and RPA3017. Asp70 is predicted to be the substrate Asp residue that receives the phosphoryl group from His532 of RpBphP2 (Fig. 4 ▸).
Figure 4.
The active site for the phosphotransfer reaction. (a) RPA3017 (green) is positioned with the DHp domain of HK (blue) according to the framework of the HK853–RR468 complex structure (PDB entry 3dge). The active-site residues of RPA3017 are shown as yellow sticks, while His532 of RpBphP2 is shown in cyan. (b) His532 of RpBphP2, the phosphoryl group (indicated by a sulfate group) and Asp70 of RPA3017 align to carry out inline nucleophilic attack in the phosphotransfer reaction.
To explore the molecular mechanism of the phosphotransfer reaction, we aligned the RPA3017 structure with that of RR468 in the HK853–RR468 complex (Casino et al., 2009 ▸; PDB entry 3dge) using a least-squares fitting procedure (Coot; Emsley & Cowtan, 2004 ▸). Structural superposition revealed a root-mean-square difference (r.m.s.d.) of 1.9 Å over 122 aligned residues with only 19% sequence identity (Fig. 2 ▸ b). The Oδ atom of the phosphoacceptor Asp70 in RPA3017 lines up with the N∊ atom of His260 of HK853 (corresponding to His532 of RpBphP2) and a sulfate group that marks the position of the phosphoryl group (Fig. 4 ▸ a). This active-site architecture supports a reaction mechanism in which phosphotransfer is achieved via nucleophilic inline attack by Asp70 (Fig. 4 ▸ b). While Glu14 may act as an activator for the nucleophilic Asp70, Lys122 is more likely to contribute to stabilization of the transition state, which involves the phosphoryl group, His532 and Asp70 in a linear pentavalent complex (Mildvan, 1997 ▸). These highly conserved residues are also expected to play important roles in the dephosphorylation reaction of RPA3017 (Immormino et al., 2015 ▸).
3.3. Putative interface between RpBphP2/RpBphP3 and RPA3017
In order to position the phosphoaccepting Asp70 for inline nucleophilic attack, RPA3017 must properly dock onto the HK domain of RpBphP2/RpBphP3. A typical HK domain contains two main parts: the helical hairpin DHp domain and the ATPase domain that catalyzes the autophosphorylation of HK via ATP hydrolysis. The phosphorylation site (His532 of RpBphP2) is located on the first helix of the DHp helical hairpin, which is connected to the PHY domain in the photosensory core module via a long helical linker denoted the PHY–HK linker. The four-helix bundle in the DHp domain promotes the formation of a parallel dimer in RpBphP2/RpBphP3, which extends the helical spine of the photosensory core module (PCM) at the dimer interface (Yang et al., 2015 ▸). Upon absorbing a photon, the helical spine of RpBphP2/RpBphP3 is thought to undergo light-induced structural rearrangements which alter the disposition of the PHY–HK linker helix relative to the ATPase domain, thereby affecting autophosphorylation (Takala et al., 2014 ▸; Yang et al., 2015 ▸).
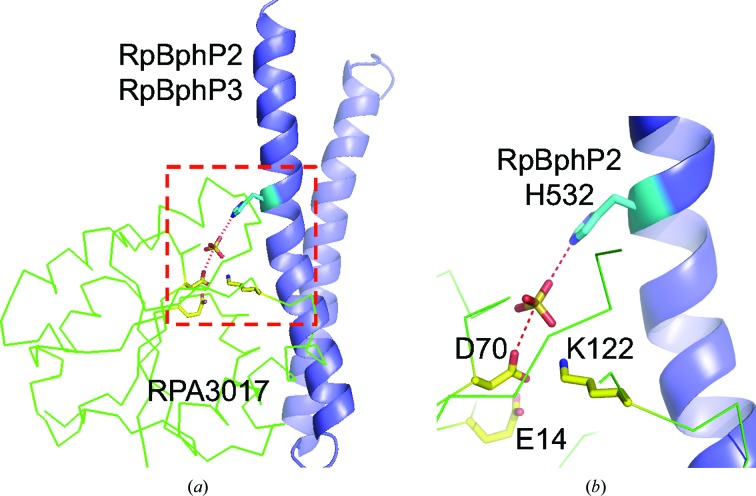
A possible docking site for RPA3017 with minimal steric clashes with the ATPase core is the lower portion of the DHp helical bundle near His532 of RpBphP2. In the absence of any crystal structure of the HK domain of a bacteriophytochrome, we built a homology model of the HK domain of RpBphP2/RpBphP3 using SWISS-MODEL (Biasini et al., 2014 ▸; Fig. 5 ▸ a). Casino and coworkers identified key positions on the DHp domain of HK and the α1 helix of RR that confer specificity based on the crystal structure of the HK853–RR468 complex (Casino et al., 2009 ▸). Their proposal was further supported by rational interface engineering that enables partner switching between two distinct HK–RR pairs: HK853–RR468 and PhoR–PhoB (Podgornaia et al., 2013 ▸). We therefore adopt the framework of the HK853–RR68 complex (PDB entry 3dge) to explore possible interfaces between RPA3017 and RpBphP2 by aligning the homology model of the HK domain of RpBphP2 (RpBphP2-HK) and the crystal structure of RPA3017 onto the HK853 and RR468 structures, respectively (using SSM in Coot). In the model of RpBphP2-HK–RPA3017, the α1 helix and the connecting loop α5–β5 of RPA3017 directly interact with RpBphP2/RpBphP3 like a clamp that holds the first helix of the DHp domain (Fig. 5 ▸ a). Additional contacts may be found between the α4–β4 loop and the ATPase domain of HK (Casino et al., 2009 ▸). Interestingly, the dimer scaffold of RPA3017 is largely compatible with the HK–RR complex. In fact, the second subunit in the RPA3017 dimer may play a role in stabilizing the complex by directly interacting with the ATPase domain of the partner subunit (Fig. 5 ▸ b).
Figure 5.
The putative interface between RpBphP2 and RPA3017. (a) Relative positioning of the homology model of the HK domain of RpBphP2 (RpBphP2-HK; DHp in blue and ATPase in green) and RPA3017 (secondary-structure elements from the N-terminus to the C-terminus identified by rainbow colors from blue to red). The α1 helix of RPA3017 joins the DHp domain to form a helix bundle. The homology model of RpBphP2-HK was built using the SWISS-MODEL server (Biasini et al., 2014 ▸). The crystal structure of the HK853–RR468 complex (PDB entry 3dge) was used as the template in homology modeling and served as a structural framework for exploring the interface between RpBphP2-HK and RPA3017. (b) Docking of the RPA3017 dimer (the two subunits are colored yellow and cyan, respectively) against the parallel dimer scaffold of the HK domain, in which one subunit is colored green and blue and the other grey. The docking was guided by a least-squares fit procedure (SSM in Coot; Emsley & Cowtan, 2004 ▸) between the structures of RR468 and subunit A of RPA3017.
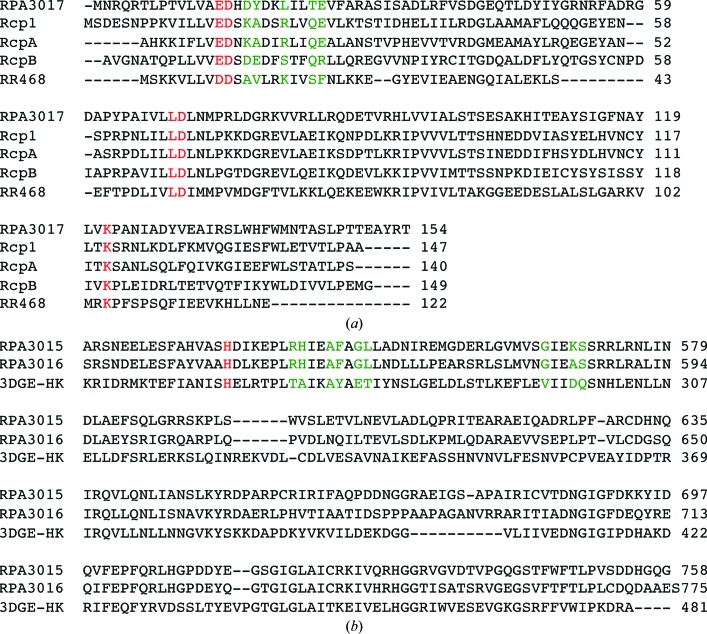
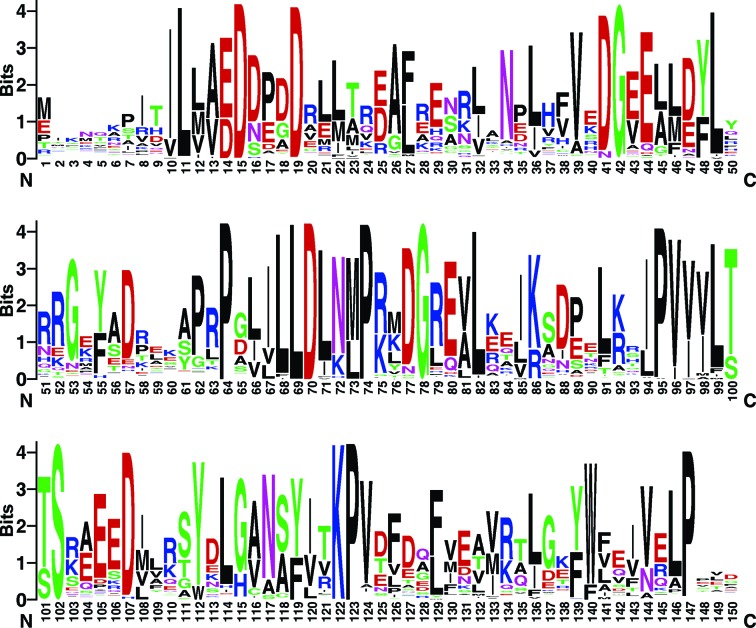
Based on sequence alignment (Fig. 6 ▸) and homology modeling, the surface area of the DHp domain of RpBphP2 (Arg540, His541, Phe545, Gly547, Leu548, Gly566 and Ser570) and the α1 helix of RPA3017 consisting of Asp17, Tyr18, Leu21, Thr24 and Glu25 are involved in specific HK–RR recognition. Not surprisingly, these residues carrying ‘specificity codes’ co-vary in the sequence space of the large families of HK and RR proteins (Capra & Laub, 2012 ▸; Podgornaia et al., 2013 ▸), so they are located in variable regions according to the sequence logo (Crooks et al., 2004 ▸) derived from 250 RRs from UniProt (Fig. 7 ▸). Consistent with this, RpBphP2 and RpBphP3 have identical residues at these equivalent positions, as they both recognize RPA3017. The nature of these putative interface residues suggests that specificity is conferred via hydrogen-bond and salt-bridge interactions together with shape complementarity. The crystal structure of the RpBphP2/RpBphP3–RPA3017 complex with additional mutational data is desirable to further explore this hypothesis.
Figure 6.
Sequence alignment of the RR and HK structures used in structural comparisons and homology modeling. (a) Sequence alignment of the five RR structures used in this work. Their PDB codes are 4zyl (RPA3017), 1i3c (Rcp1), 1k68 (RcpA), 1k66 (RcpB) and 3dge (RR468). The conserved catalytic residues are colored red. The interface residues are colored green. (b) Sequence alignment of the HK domains of RpBphP2, RpBphP3 and HK853. The His residues at the phosphorylation site are marked in red, while the variable interface residues are marked in green.
Figure 7.
The sequence logo generated based on 250 RR sequences in UniProt. The RPA3017 sequence was used in the BLAST search and in residue numbering. This figure was produced using the WebLogo server (Crooks et al., 2004 ▸).
4. Conclusions
Red-light signaling in R. palustris is mediated by a two-component system consisting of bacteriophytochromes RpBphP2/RpBphP3 and their cognate response regulator RPA3017. The high-resolution crystal structure of RPA3017 complements extensive crystallographic studies on RpBphP2/RpBphP3 and advances our understanding of how light perception is achieved at the molecular level. Firstly, this structure features a dimer scaffold that seems to be conserved among RRs related to phytochrome signaling, although its physiological implications require further investigation. Secondly, the active-site architecture suggests that RPA3017 receives the phosphoryl group via inline nucleophilic attack, a mechanism that is likely to be common to many TCSs. Thirdly, putative interfaces between RpBphP2/RpBphP3 and RPA3017 are identified. These specificity-conferring interactions ensure precise signal relays and coordinated light responses in R. palustris.
Supplementary Material
PDB reference: RPA3017 involved in red-light signaling in Rhodopseudomonas palustris, 4zyl
Acknowledgments
We thank the LS-CAT staff for beamline access and support. The use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357. Xuefei Yang is supported by the 111 Project of China Grant B07041 (to Qifa Zhang). This work was supported by NIH grant R01EY024363 (to Xiaojing Yang and Keith Moffat) and the University of Illinois at Chicago Intramural Research Fund (Xiaojing Yang).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Anders, K., von Stetten, D., Mailliet, J., Kiontke, S., Sineshchekov, V. A., Hildebrandt, P., Hughes, J. & Essen, L.-O. (2011). Photochem. Photobiol. 87, 160–173. [DOI] [PubMed]
- Bellini, D. & Papiz, M. Z. (2012). Acta Cryst. D68, 1058–1066. [DOI] [PubMed]
- Benda, C., Scheufler, C., Tandeau de Marsac, N. & Gärtner, W. (2004). Biophys. J. 87, 476–487. [DOI] [PMC free article] [PubMed]
- Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., Kiefer, F., Cassarino, T. G., Bertoni, M., Bordoli, L. & Schwede, T. (2014). Nucleic Acids Res. 42, W252–W258. [DOI] [PMC free article] [PubMed]
- Capra, E. J. & Laub, M. T. (2012). Annu. Rev. Microbiol. 66, 325–347. [DOI] [PMC free article] [PubMed]
- Casino, P., Rubio, V. & Marina, A. (2009). Cell, 139, 325–336. [DOI] [PubMed]
- Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. (2004). Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed]
- Doublié, S. (1997). Methods Enzymol. 276, 523–530. [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Essen, L.-O., Mailliet, J. & Hughes, J. (2008). Proc. Natl Acad. Sci. USA, 105, 14709–14714. [DOI] [PMC free article] [PubMed]
- Gao, R. & Stock, A. M. (2009). Annu. Rev. Microbiol. 63, 133–154. [DOI] [PMC free article] [PubMed]
- Gao, R. & Stock, A. M. (2010). Curr. Opin. Microbiol. 13, 160–167. [DOI] [PMC free article] [PubMed]
- Giraud, E., Zappa, S., Vuillet, L., Adriano, J.-M., Hannibal, L., Fardoux, J., Berthomieu, C., Bouyer, P., Pignol, D. & Verméglio, A. (2005). J. Biol. Chem. 280, 32389–32397. [DOI] [PubMed]
- Im, Y. J., Rho, S.-H., Park, C.-M., Yang, S.-S., Kang, J.-G., Lee, J. Y., Song, P.-S. & Eom, S. H. (2002). Protein Sci. 11, 614–624. [DOI] [PMC free article] [PubMed]
- Immormino, R. M., Starbird, C. A., Silversmith, R. E. & Bourret, R. B. (2015). Biochemistry, 54, 3514–3527. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Larimer, F. W. et al. (2003). Nature Biotechnol. 22, 55–61. [DOI] [PubMed]
- Menon, S. & Wang, S. (2011). Biochemistry, 50, 5948–5957. [DOI] [PMC free article] [PubMed]
- Mildvan, A. S. (1997). Proteins, 29, 401–416. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Podgornaia, A. I., Casino, P., Marina, A. & Laub, M. T. (2013). Structure, 21, 1636–1647. [DOI] [PMC free article] [PubMed]
- Rockwell, N. C., Su, Y.-S. & Lagarias, J. C. (2006). Annu. Rev. Plant Biol. 57, 837–858. [DOI] [PMC free article] [PubMed]
- Stock, A. M., Robinson, V. L. & Goudreau, P. N. (2000). Annu. Rev. Biochem. 69, 183–215. [DOI] [PubMed]
- Takala, H., Björling, A., Berntsson, O., Lehtivuori, H., Niebling, S., Hoernke, M., Kosheleva, I., Henning, R., Menzel, A., Ihalainen, J. A. & Westenhoff, S. (2014). Nature (London), 509, 245–248. [DOI] [PMC free article] [PubMed]
- Yang, X., Kuk, J. & Moffat, K. (2008). Proc. Natl Acad. Sci. USA, 105, 14715–14720. [DOI] [PMC free article] [PubMed]
- Yang, X., Ren, Z., Kuk, J. & Moffat, K. (2011). Nature (London), 479, 428–432. [DOI] [PMC free article] [PubMed]
- Yang, X., Stojković, E. A., Kuk, J. & Moffat, K. (2007). Proc. Natl Acad. Sci. USA, 104, 12571–12576. [DOI] [PMC free article] [PubMed]
- Yang, X., Stojković, E. A., Ozarowski, W. B., Kuk, J., Davydova, E. & Moffat, K. (2015). Structure, 23, 1179–1189. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: RPA3017 involved in red-light signaling in Rhodopseudomonas palustris, 4zyl