Abstract
111In-labeled trastuzumab modified with nuclear localizing signal (NLS) peptides (111In-trastuzumab-NLS) efficiently delivers an Auger electron (AE) emitter 111In into the cell nucleus and is thus a promising radiopharmaceutical in AE radioimmunotherapy (AE-RIT) for targeted killing of HER2-positive cancer. However, further improvement of its therapeutic efficacy is required. In this study, the authors show a transcriptomic approach to identify potential targets for enhancing the cytotoxic effects of 111In-trastuzumab-NLS. They generated two types of 111In-trastuzumab-NLS harboring different numbers of NLS peptides, 111In-trastuzumab-NLS-S and -L. These radioimmunoconjugates (230 and 460 kBq) showed a significant higher cytotoxicity to SKBR3 human breast cancer cells overexpressing HER2 compared to 111In-trastuzumab. Microarray analysis revealed that NF-kB-related genes (38 genes) were significantly changed in transcription by 111In trastuzumab-NLS-L (230 kBq) treatment. Quantitative reverse transcription polymerase chain reaction confirmed the microarray data by showing transcriptional alternation of selected NF-κB target genes in cells treated with 111In-trastuzumab-NLS-L. Interestingly, bortezomib, a drug known as a NF-κB modulator, significantly enhanced the cytotoxicity of 111In-trastuzumab-NLS-L in SKBR3 cells. Taken together, the transcriptome data suggest the possibility that the modulation of NF-kB signaling activity is a molecular signature of 111In-trastuzumab-NLS and coadministration of bortezomib may be efficacious in enhancement of AE-RIT with 111In-trastuzumab-NLS.
Key words: : Auger electron radioimmunotherapy, Bortezomib, DNA microarray, NF-κB
Introduction
Auger electron (AE) is a low-energy electron from eV to keV with a very short track length in the nm to μm.1 AE emitters such as 125I and 111In are highly toxic when they decay in the nucleus or in close proximity to DNA, because the multiplicity of short-range AEs emitted results in very high local energy deposited and the linear energy transfer (LET) of them approaches that of α-emitters.2,3
The use of antibody modified with the nuclear localizing signal (NLS) has been proposed to achieve the efficient delivery of AE emitters into the tumor cell nucleus.4,5 Previous studies showed that 111In-labeled trastuzumab modified with NLS peptides (111In-trastuzumab-NLS) had greater nuclear transport of 111In and cytotoxic effect compared to trastuzumab itself and 111In-trastuzumab in human breast cancer cells overexpressing HER2 both in vitro and in vivo.6,7 Thus, 111In-trastuzumab-NLS is a promising therapeutic agent in AE radioimmunotherapy (AE-RIT) for targeted killing of HER2-positive cancer cells. However, to further improve its therapeutic efficacy, better understanding of the cytotoxic mechanism and cellular responses to 111In-trastuzumab-NLS is required.
It is well known that irradiation induces global gene expression changes in irradiated cells.8–10 These changes induce various cellular responses such as cell cycle arrest and cell death/survive through a complex molecular network and signaling pathway. DNA microarray analysis is a powerful technique to evaluate gene expression changes and is widely used for classifying cancer subtypes, predicting prognosis and therapeutic responses, and understanding the molecular mechanism of cancer.11 This technique has been applied for identifying genes and the molecular pathway associated with radiosensitivity or radioresistance of irradiated cancer cells to enhance the therapeutic efficacy of radiotherapy.12–14 However, there are few reports describing gene expression profiles in cells treated by AE-RIT.
The authors hypothesized that comprehensive analysis of gene expression using DNA microarray would allow to find gene expression signatures in 111In-trastuzumab-NLS treatment for enhancing therapeutic efficacy of this pharmaceutical. In this study, they performed microarray analyses to identify gene expression signature characteristics of 111In-trastuzumab-NLS treatment. These analyses revealed gene expression patterns induced by 111In-trastuzumab-NLS and suggested the possibility that the modulation of NF-κB pathway activity is a molecular signature of 111In-trastuzumab-NLS treatment. Furthermore, they demonstrated that bortezomib, a NF-κB modulator, enhanced the cytotoxicity of 111In-trastuzumab-NLS in human breast cancer SKBR3 cells overexpressing HER2.
Materials and Methods
Cells
SKBR3 and MCF-7 cells were obtained from ATCC (Manassas, VA). Human mammary epithelial cells (HMEpC) were purchased from Toyobo (Tokyo, Japan). SKBR3 cells were cultured in McCoy's 5a modified medium (Sigma-Aldrich, St Louis, MO). MCF-7 cells were cultured in DMEM (High Glucose) (Wako, Osaka, Japan). HMEpC were cultured in HMEpC Total Kit (Toyobo). All media, except HMEpC Total Kit, were supplemented with 10% fetal bovine serum (Nichirei Biosciences, Tokyo, Japan). All cell lines were cultured in humidified atmosphere containing 5% CO2 at 37°C.
Antibody and reagent
Trastuzumab (Herceptin®) was purchased from Chugai Pharmaceutical (Tokyo, Japan). Bortezomib (Velcade®) was commercially obtained from Janssen Pharmaceutical (Tokyo, Japan), dissolved in DMSO (5 μM), and stored at −20°C.
Preparation of chelate-conjugated trastuzumab-NLS
Trastuzumab was conjugated with CHX-A”-DTPA (DTPA) (Macrocyclics, Dallas, TX) for labeling with 111In. Briefly, trastuzumab (5 mg/mL) was reacted with a 2.5-fold molar excess of DTPA overnight at 37°C and then purified on a Sephadex G-50 column (GE Healthcare UK Ltd., Little Chalfont, England) eluted with phosphate-buffered saline (PBS; pH 7.5). Synthetic NLS peptides (CGYGPKKKRKVGG; Invitrogen, Carlsbad, CA) were then conjugated to DTPA-modified trastuzumab using the heterobifunctional crosslinking agent sulfosuccinimidy1-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC; Wako). Briefly, DTPA-trastuzumab (2–3 mg/mL in PBS, pH7.5) was reacted with a 10- to 20-fold molar excess of sulfo-SMCC (2.5 mg/mL) at room temperature for 1 hour and purified on a Sephadex G-50 column eluted with PBS (pH 7.5). Maleimide-derivatized DTPA-trastuzumab was transferred to a Ultracel-50K ultrafiltration device (Millipore, Billerica, MA) eluted with PBS (pH 7.0), concentrated to 4–7 mg/mL, and reacted with a 60-fold molar excess of NLS peptides (10 mmol/mL in PBS, pH7.0) overnight at 4°C. DTPA-trastuzumab with NLS peptides was purified on a Sephadex G-50 column eluted with PBS (pH 7.0).
Radiolabeling of DTPA-conjugated antibodies with 111In
111InCl3 (Nihon Medi-Physics, Tokyo, Japan) in 1 M acetate buffer (pH 6.0) was mixed with DTPA-conjugated trastuzumab or trastuzumab-NLS and allowed to react for 1 hour at room temperature. Radiolabeled antibodies were purified by spin filtration using a Sephadex G-50 column. The specific activity of the 111In-labeled antibodies was ∼111 kBq/μg. Radiolabeled antibodies were verified by high-performance liquid chromatography (HPLC).
Nuclear uptake of 111In
111In-labeled antibodies (∼25,000 cpm) were incubated with agitation at 37°C for 1 or 10 hours with 1 × 106 SKBR3 cells in microtubes. The tubes were centrifuged at 6500 rpm at 4°C for 1 minute. The supernatant was removed, and the pellet was washed in 1 mL of ice-cold PBS. The cell pellet was lysed with 400 μL of lysis buffer in the Nuclei EZ Prep Nuclei Isolation Kit (Sigma-Aldrich) on ice for 5 minutes and then centrifuged at 500 g at 4°C for 5 minutes. This procedure was repeated twice. Finally, nucleus was pelleted and resuspended in 800 μL of PBS to count the radioactivity in a γ-counter (ALOKA, Tokyo, Japan). The percentage of nuclear uptake relative to cell-associated 111In activity was calculated.
Cytotoxicity
The cytotoxic assay was conducted using alamarBlue cell viability reagent (Invitrogen). 111In-trastuzumab and 111In-trastuzumab-NLS-S and -L (230 and 460 kBq) were added to 2 × 103 SKBR3 cells plated in a 96-well microplate (BD Biosciences, Franklin Lakes, NJ) and incubated with those radiopharmaceuticals for 5–7 days in humidified atmosphere containing 5% CO2 at 37°C. For combination therapy, both bortezomib (5 nM) and 111In-trastuzumab-NLS-L (370 kBq) were added to 1 × 104 SKBR3 cells plated in a 96-well microplate and incubated with those agents for 5 days in humidified atmosphere containing 5% CO2 at 37°C. After treatment, 1/10 volume of alamarBlue reagent was added into cell culture media and incubated at 37°C for 2 hours. The absorbance of the wells was measured at 570 nm by a microplate reader (Bio-Rad, Hercules, CA), or the fluorescence of the wells (Ex: 530 nm, Em: 590 nm) was measured by a fluorescence microplate reader (BioTek, Tokyo, Japan).
Western blot
Ten micrograms of protein was loaded into 4%–20% SDS-PAGE gel (Bio-Rad), resolved by electrophoresis and transferred to PVDF membrane (Bio-Rad). Membranes were immunoblotted using antibodies to HER2, α-tubulin, HRP-conjugated rabbit IgG, and HRP-conjugated mouse IgG. All antibodies were purchased from Cell Signaling Technology, except an antibody to α-tubulin (Millipore). The membranes were imaged and quantified on an ImageQuant LAS500 imager (GE Healthcare) using Luminata Forte Western HRP Substrate (Millipore) and analyzed by ImageQuant TL software (GE Healthcare).
Microarray experiment
RNA samples for the microarray experiment were prepared from cells untreated or treated with unlabeled or 111In-labeled antibodies for 7 days toward to 2 × 103 SKBR3 cells plated in a 96-well microplate. Total RNA was extracted by the RNeasy Micro Kit (Qiagen, Venlo, Netherlands) and evaluated for integrity by Nanodrop 2000 (Thermo Scientific, Waltham, MA). Labeled cRNA probes were synthesized using the Low Input Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA) and subsequently hybridized to SurePrint G3 Human GE Microarray Kit 8 × 60K ver2.0 (Agilent Technologies) using Gene Expression Hybridization Kit (Agilent Technologies). Hybridization images were scanned by SureScan Microarray Scanner System (Agilent Technologies). Data were analyzed using GeneSpring GX 12.0 (Agilent Technologies) and Ingenuity Pathway Analysis (IPA) software (Qiagen). The microarray data have been deposited in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under accession number GSE67193.
Quantitative reverse transcription polymerase chain reaction
Reverse transcription was performed with 3 ng of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. The polymerase chain reaction (PCR) was carried out in a StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA) using the TaqMan Gene Expression Assay (Applied Biosystems) and TaqMan Fast Advanced Master Mix (Applied Biosystems). Data were analyzed with StepOne software version 2.1 (Applied Biosystems). The human β-actin gene was used as an endogenous control. The relative quantification of transcription was determined using the formula 2−ΔΔCt.
Statistical analysis
Statistical analysis was performed using JMP version 9 software (SAS Institute Japan, Tokyo, Japan). Analysis of variance followed by the Tukey–Kramer test was used. A p-value of <0.05 was considered significant.
Results
Characterization of 111In-trastuzumab-NLS
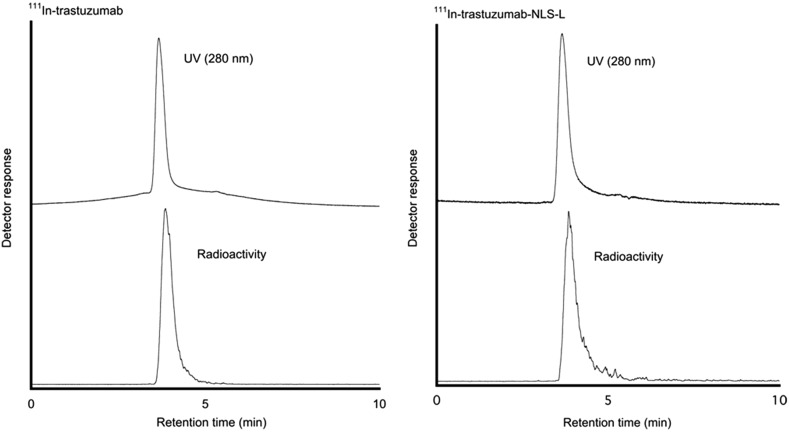
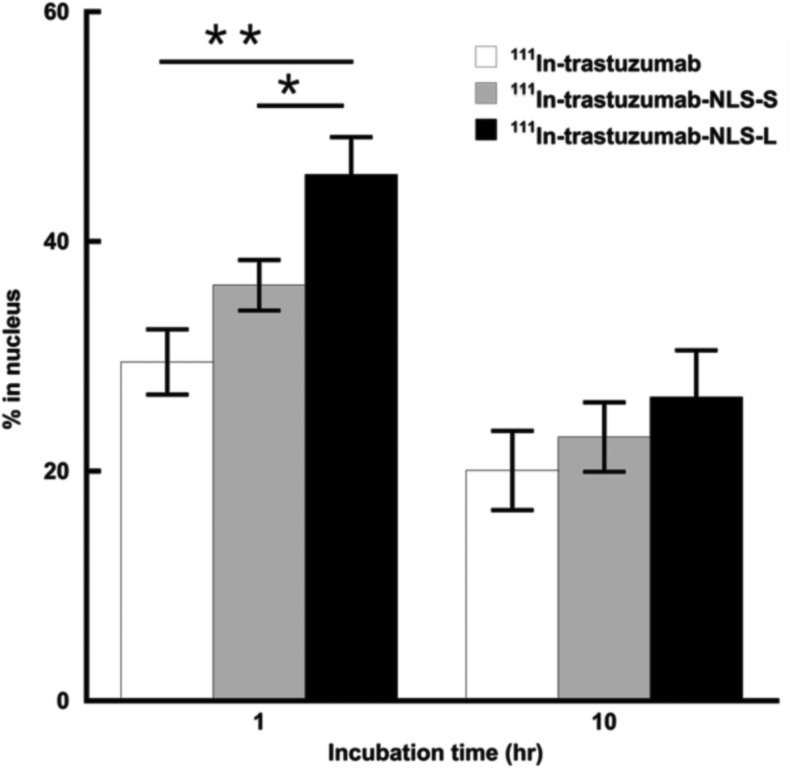
The authors generated two types of trastuzumab-NLS (trastuzumab-NLS-S and trastuzumab-NLS-L). These modified antibodies attached with DTPA were analyzed by SDS-PAGE under a nonreducing condition to determine their relative molecular mass (Fig. 1). The mean values of relative molecular mass increased relative to DTPA-trastuzumab were 5680 and 14,530 in DTPA-conjugated trastuzumab-NLS-S and trastuzumab-NLS-L, respectively. According to these values, ∼4 and 10 NLS peptides were conjugated to trastuzumab in trastuzumab-NLS-S and trastuzumab-NLS-L, respectively. 111In-labeled antibodies were verified by HPLC (Fig. 2). Figure 3 shows nuclear translocation of 111In by trastuzumab-NLS in human breast cancer SKBR3 cells. Nuclear uptake of 111In was increased in 111In-trastuzumab-NLS-L compared to 111In-transtuzumab and 111In-trastuzumab-NLS-S at 1 hour of incubation (p < 0.01 and p < 0.05, respectively). There were no significant differences in nuclear translocation of 111In among the three radiolabeled antibodies at 10 hours of incubation, although the trend was observed that nuclear uptake of 111In was increased as the number of NLS peptides conjugated increased.
FIG. 1.
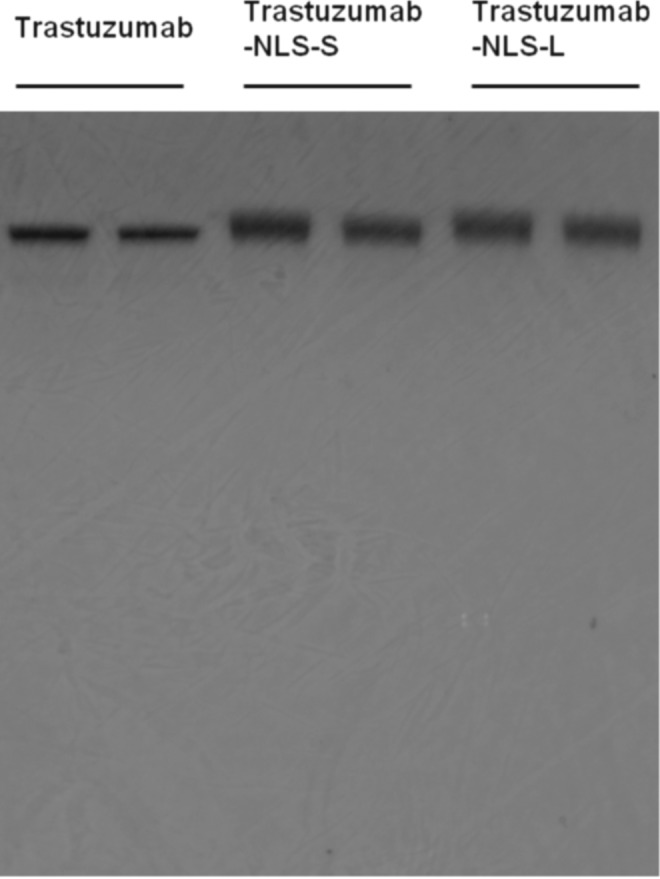
SDS-PAGE of trastuzumab, trastuzumab-NLS-S, and trastuzumab-NLS-L. Each sample has duplicate loading. Note that the size of trastuzumab-NLS antibodies increases along with the number of NLS peptides attached. NLS, nuclear localizing signal.
FIG. 2.
High-performance liquid chromatography chart of 111In-trastuzumab and 111In-trastuzumab-NLS-L.
FIG. 3.
Nuclear uptake of 111In in trastuzumab, trastuzumab-NLS-S, and trastuzumab-NLS-L. Incubation times are indicated in the graph. Percentages of 111In in cell nucleus are calculated by 111In activity in nuclear fraction relative to total 111In activity of nuclear, cytosolic, and membrane fractions. Mean ± SD. n = 3. *p < 0.05, **p < 0.01. ANOVA followed by the Tukey–Kramer test. ANOVA, analysis of variance.
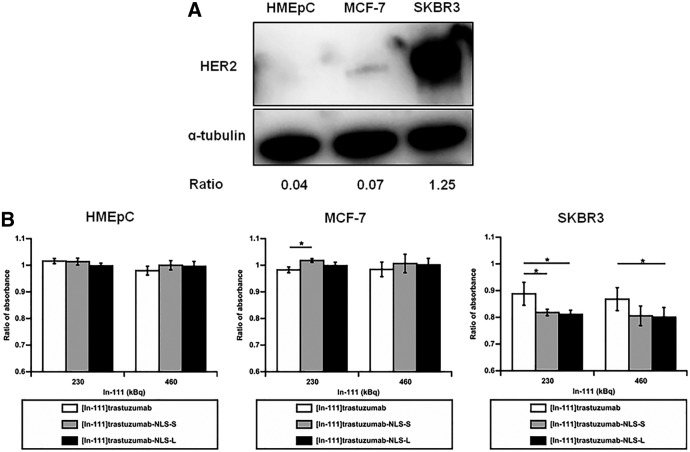
Cytotoxicity of 111In-trastuzumab-NLS in HMEpC and breast cancer cells
The cytotoxicity of 111In-trastuzumab-NLS was evaluated by the absorbance measurement of a cell viability indicator dye in HMEpC, MCF-7, and SKBR3 cells (Fig. 4). Western blot showed that the ratios of band intensity of HER 2 relative to α-tubulin in HMEpC, MCF-7, and SKBR3 cells were 0.04, 0.07, and 1.25, respectively, suggesting that the level of HER2 protein in SKBR3 cells was ∼30-fold higher than that in HMEpC and that MCF-7 cells had a very low level of HER2 expression (Fig. 4A). Thus, the authors used SKBR3 cells as HER2-overexpressing cells, whereas MCF-7 cells and HMEpC were used as human breast cancer cells with very low expression of HER2 protein and normal HMEpC, respectively. 111In-trastuzumab-NLS showed a significantly higher cytotoxic effect than 111In-trastuzumab in SKBR3 cells when incubated with 230 and 460 kBq of 111In-trastuzumab-NLS-L and 230 kBq of 111In-trastuzumab-NLS-S (p < 0.05) for 5–7 days, but not in HMEpC and MCF-7 (Fig. 4B).
FIG. 4.
HER2 expression in cells used and cytotoxic assay. (A) Western blot of HER2 expression in HMEpC, MCF-7, and SKBR3 cells. α-tubulin as loading control. Ratios of band intensity of HER2 relative to α-tubulin are indicated below the images. (B) Cytotoxicity of 111In-trastuzumab and 111In-trastuzumab-NLS in HMEpC, MCF-7, and SKBR3 cells. Bars show the ratio of absorbance of alamarBlue indicator dye when the absorbance in untreated cells considers as 1. Mean ± SD. n = 3–5. *p < 0.05. ANOVA followed by the Tukey–Kramer test. HMEpC, human mammary epithelial cells.
Microarray analyses of SKBR3 cells treated with 111In-trastuzumab-NLS-L
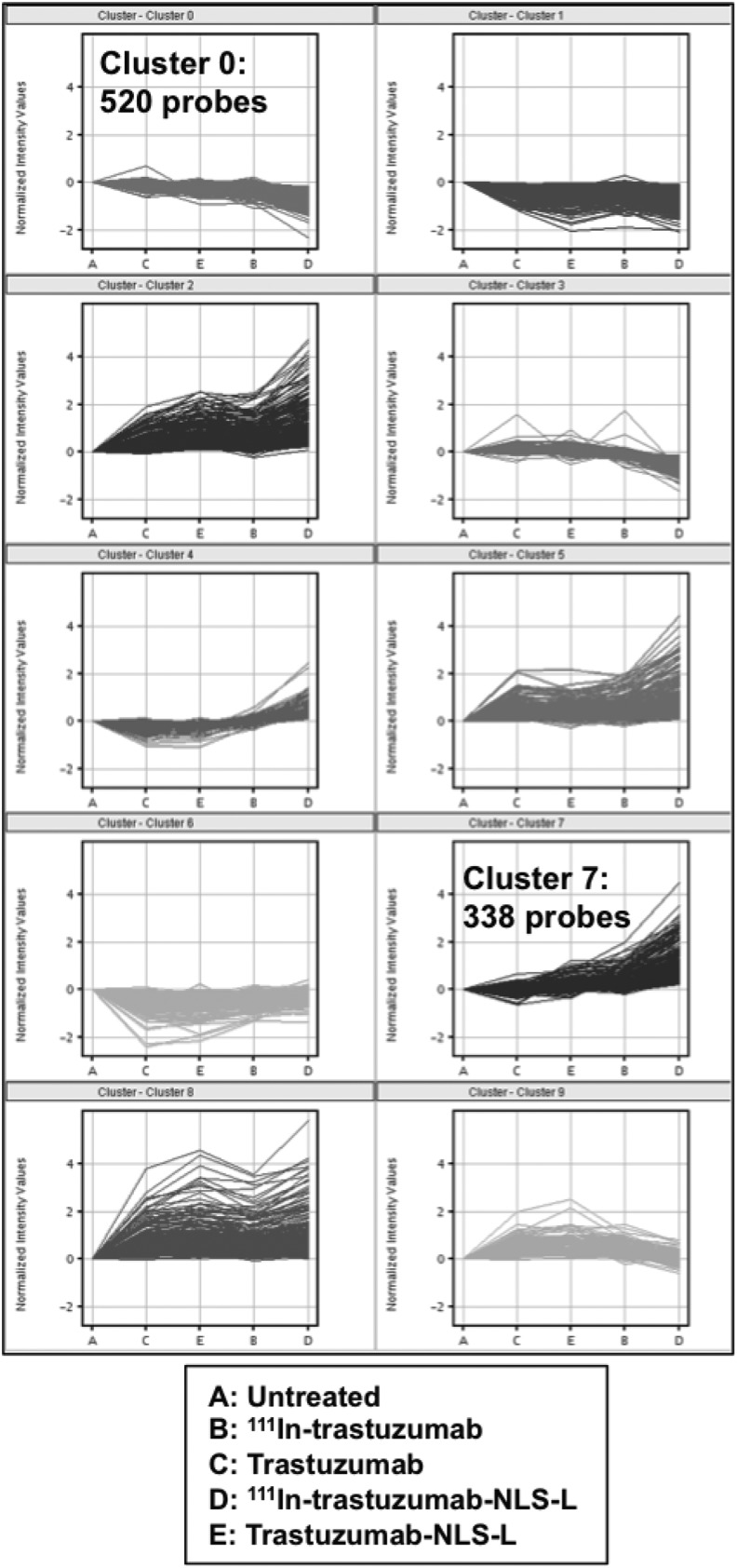
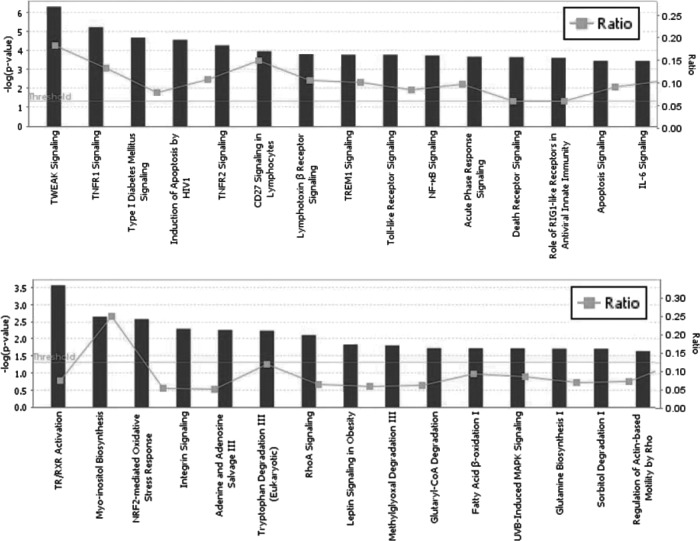
To find the genes affected by 111In-trastuzumab-NLS-L, the authors performed gene expression profiling using RNAs isolated from SKBR3 cells treated for 7 days with 230 kBq of 111In-trastuzumab-NLS-L. For comparison, RNA samples extracted from untreated SKBR3 cells or cells treated with trastuzumab, 111In-trastuzumab, and trastuzumab-NLS-L were also used. A total of 15 data from five groups were used for this study. After normalization of the raw data and correlation analyses, they performed hierarchical clustering analysis using 19,243 probes. Hierarchical clustering showed that gene expression profiles of the cells treated with 111In-trastuzumab-NLS-L were distinctly clustered, whereas other samples were not (Fig. 5). They next aimed to find significant differences of each treatment group (group B-E) relative to the untreated group (group A). The results are summarized in Table 1, suggesting that treatment of 111In-trastuzumab-NLS-L (group D) induced gene expression changes most significantly among the treatment groups. To clarify gene expression patterns among five groups, k-means clustering analysis was performed using 3190 probes that were significantly changed among the five groups. k-means clustering among five groups showed 10 patterns of classification (Fig. 6). Among them, two patterns of the clusters (cluster 7 and cluster 0) revealed that 338 and 520 probes were specifically upregulated and downregulated by treatment of 111In-trastuzumab-NLS-L (group D), respectively. To further characterize the gene expression changes induced by 111In-trastuzumab-NLS-L, they performed IPA using probe sets in cluster 0 and 7. IPA revealed the pathways affected by the treatment of 111In-trastuzumab-NLS-L (Fig. 7). The gene sets that were upregulated specifically by the treatment of 111In-trastuzumab-NLS-L were included in the signaling pathway such as TWEAK and TNFR1 signaling.
FIG. 5.
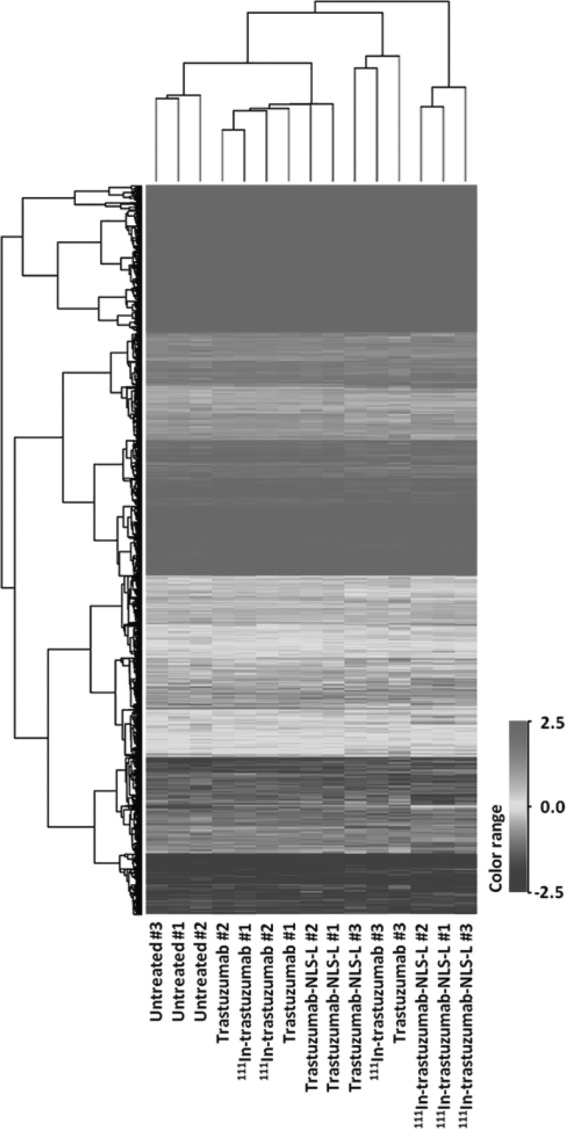
Comparison of gene expression profiles of 15 samples by hierarchical clustering.
Table 1.
Number of Probes That Were Differentially Expressed in the Treated Groups Compared with Untreated Group
| Significance | p < 0.05 | p < 0.05 | p < 0.01 | ||
|---|---|---|---|---|---|
| Group/group | Fold change | >1.5 or <0.67 | 1–1.5 or 0.67–1 | >4 or <0.25 | Total |
| B/A | Upregulation | 180 | 87 | 3 | 270 |
| Downregulation | 135 | 127 | 0 | 262 | |
| C/A | Upregulation | 146 | 86 | 2 | 234 |
| Downregulation | 114 | 87 | 0 | 201 | |
| D/A | Upregulation | 853 | 325 | 92 | 1270 |
| Downregulation | 864 | 421 | 1 | 1286 | |
| E/A | Upregulation | 285 | 93 | 17 | 395 |
| Downregulation | 188 | 131 | 0 | 319 |
Treatment groups are as follows. Group A = untreated; B = 111In-trastuzumab (230 kBq); C = trastuzumab (approximately equal amount of antibody to group B); D = 111In-trastuzumab-NLS-L (230 kBq); E = trastuzumab-NLS-L (approximately equal amount of antibody to group B). n = 3, each group.
NLS, nuclear localizing signal.
FIG. 6.
k-means clustering. X-axis denotes the different treatments.
FIG. 7.
Pathways affected by 111In-trastuzumab-NLS-L detected by Ingenuity Pathway Analysis using the information from upregulated (top) and downregulated (bottom) genes. Top 15 pathways are shown. Y-axis means an inverse of log of p-value. Gray lines show the ratio of the numbers of gene, including each pathway.
NF-κB pathway analysis by IPA
TWEAK and TNFR1 signaling are closely related to the NF-κB pathway.15,16 The NF-κB pathway plays critical roles in regulating cell survival/death and can become a potential combination therapeutic target.17–19 Therefore, the authors focused on the NF-κB pathway and further performed IPA to find NF-κB target genes affected by 111In-trastuzumab-NLS-L treatment and identified 30 upregulated and 8 downregulated genes out of 654 genes of the NF-κB pathway (Table 2).
Table 2.
NF-κB Target Genes Affected by 111In-Trastuzumab-NLS-L Treatment
| Gene symbol (log2 ratio) | |
|---|---|
| Upregulated | PLAU (2.823), SERPINA3 (2.496), CCL2 (2.382), CXCR4 (2.360), ICAM1 (2.288), BIRC3 (2.211), IL32 (2.161), TAP1 (1.764), PSMB9 (1.671), TNFAIP (1.491), MAP3K8 (1.233), RELB (1.193), IL18 (1.092), NCOA7 (1.067), MAP3K14 (1.047), IRF7 (1.003), PMAIP1 (0.994), C1R (0.977), EFNA1 (0.959), TNFAIP2 (0.949), NFKBIA (0.833), TNIP1 (0.805), CCL17 (0.684), HLA-A (0.640), SOCS1 (0.637), CASP3 (0.636), WTAP (0.627), CASP8 (0.612), NFKB1 (0.491), PEA15 (0.483) |
| Downregulated | AR (-1.410), MYLK3 (-1.160), PSMD10 (-0.808), TSC22D3 (-0.760), BCL11A (-0.645), NR3C1 (-0.645), PPARA (-0.557), BAD (-0.221) |
Validation of microarray data by quantitative reverse transcription PCR
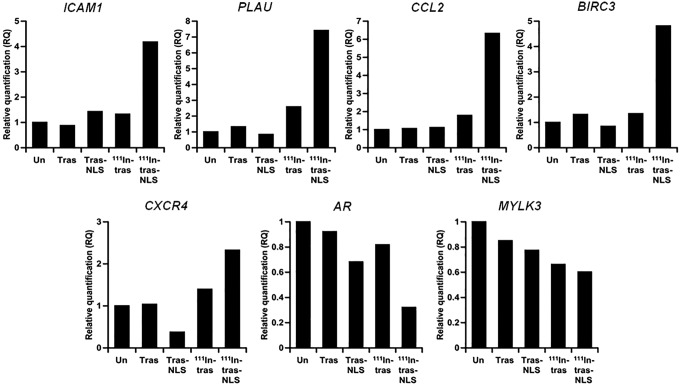
To validate the results of microarray analyses, the authors performed quantitative reverse transcription PCR for selected NF-κB target genes (Fig. 8). Consistent with the microarray results, these quantitative analyses suggested that the selected NF-κB target genes, such as ICAM1, PLAU, CCL2, BIRC3, and CXCR4, were transcriptionally upregulated by treatment with 111In-trastuzumab-NLS-L compared to other groups, whereas AR and MYLK 3 genes were downregulated. These data validated microarray data and further supported that the NF-κB pathway was remarkably modulated by 111In-trastuzumab-NLS-L.
FIG. 8.
Quantitative reverse transcription polymerase chain reaction validation of gene expression changes of selected NF-κB target genes in untreated SKBR3 cells (Un) and cells treated with unlabeled trastuzumab (Tras), unlabeled trastuzumab-NLS-L (Tras-NLS), 111In-trastuzumab (111In-tras), and 111In-trastuzumab-NLS-L (111In-tras-NLS). Values show relative quantification in each group (n = 3/group) by biological grouping analysis when the expression level in Un is 1.
Enhanced cytotoxicity of 111In-trastuzumab-NLS by bortezomib in SKBR3 cells
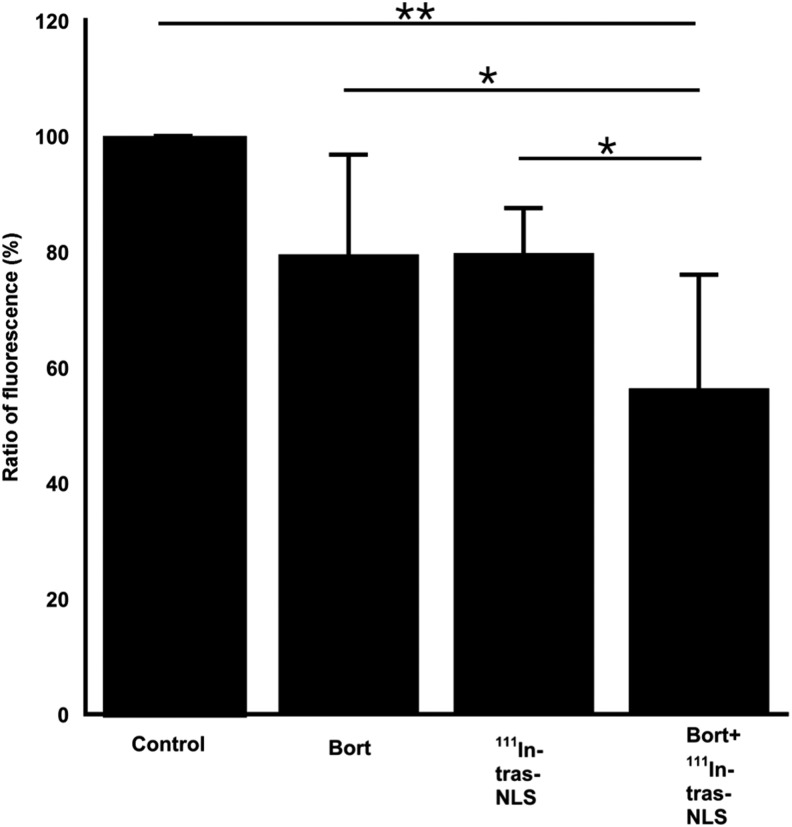
To investigate whether pharmacological intervention of NF-κB pathway enhances the cytotoxicity of 111In-trastuzumab-NLS-L, the authors examined the effects of bortezomib, a clinically approved drug that modulates the NF-κB pathway, on the cytotoxicity of 111In-trastuzumab-NLS-L in SKBR3 (Fig. 9). The cytotoxic assay using alamarBlue dye showed that bortezomib (5 nM) significantly enhanced the cytotoxicity of 111In-trastuzumab-NLS-L in SKBR3 cells (56.3% ±19.7%) compared to the monotherapy of bortezomib itself at the same concentration (79.5% ±17.2%, p < 0.05) and 370 kBq of111In-trastuzumab-NLS-L (79.8% ±7.8%, p < 0.05) and untreated control (100%, p < 0.01).
FIG. 9.
Cytotoxicity to SKBR3 cells by monotreatment with either 5 nM bortezomib (Bort) or 370 kBq of 111In-trastuzumab-NLS-L (111In-tras-NLS) and combined treatment with both (Bort+111In-tras-NLS). Bars show ratio of fluorescence of alamarBlue indicator dye when the fluorescence in control cells considers as 1. Mean ± SD. n = 6. *p < 0.05, **p < 0.01. ANOVA followed by the Tukey–Kramer test.
Discussion
In this study, the authors performed microarray analyses to find gene expression changes induced by 111In-trastuzumab-NLS. They found that NF-kB-related genes were significantly changed in transcription by 111In-trastuzumab-NLS treatment and further demonstrated that bortezomib significantly enhanced the cytotoxic effect of 111In-trastuzumab-NLS to human breast cancer SKBR3 cells.
The authors found that 111In-trastuzumab-NLS had greater nuclear delivery of 111In in SKBR3 cells when incubated for 1 hour, as the numbers of NLS peptides attached increased. These results were consistent with the previous work by Chen et al., although they used different types of antibody and cell lines.4 However, in consideration of the fact that internalization of 111In-trastuzumab increasingly continued until 12 hours after incubation6 and the result that there were no significant differences in nuclear transport of 111In at 10 hours after incubation between 111In-trastuzumab and 111In-trastuzumab-NLS, they need further studies to evaluate total nuclear accumulation of radioactivity in those radiopharmaceuticals. In agreement with the previous study,6 they found that 111In-trastuzumab-NLS had a higher cytotoxic effect in SKBR3 cells compared to 111In-trastuzumab.Furthermore, they found that the cytotoxicity of 111In-trastuzumab-NLS-S and -L was apparent in SKBR3 cells overexpressing HER2, but not in HMEpC and MCF-7 cells with very low expression of HER2. These data confirmed that 111In-trastuzumab-NLS was an effective cytotoxic agent against breast cancer cells overexpressing HER2 with excellent targeting properties and had little adverse effects to normal breast epithelium.
The microarray data showed that the gene sets involved in several signaling pathways were significantly altered by 111In-trastuzumab-NLS treatment. Among them, the authors focused on the NF-κB pathway for further detailed analyses, since the NF-κB pathway has recently become an attractive focal point for drug discovery and can become a potential combination therapeutic target.17–19 Furthermore, there are some clinically available drugs that can target the NF-κB pathway.20 qPCR results confirmed the microarray results that the modulation of NF-κB pathway was characteristic of 111In-trastuzumab-NLS treatment by showing the gene expression changes of representative NF-κB target genes. In fact, the NF-κB pathway seems to be activated by photon and particle radiation.21–24 The mechanism(s) of radiation-induced NF-κB pathway activation remain elusive. DNA damage, however, is suggested as a key factor to activate the NF-κB pathway.25,26 Therefore, DNA damage induced by AE irradiation from 111In-trastuzumab-NLS-L may trigger NF-κB activation. Based on the data, they hypothesized that the intervention of NF-κB pathway may enhance the cytotoxicity of 111In-trastuzumab-NLS-L. Bortezomib was used to test this hypothesis, as it is a NF-κB modulator drug currently approved in clinical use.27 As expected, bortezomib significantly sensitized SKBR3 cells to 111In-trastuzumab-NLS-L, supporting this hypothesis. Interestingly, bortezomib has recently been used as a combination therapy agent in clinical trials of 153Sm-lexidronam and 90Y-ibritumomab tiuxetan for the treatment of relapsed/refractory multiple myeloma and follicular non-Hodgkin lymphoma, respectively.28,29 Rae et al. recently reported that bortezomib radiosensitized tumor cells in combination with 131I-metaiodobenzylguanidine.30 These studies suggested the potential of bortezomib as the radiosensitizer for low-LET radiation. Generally, high-LET radiation tends to be less radiosensitized compared with low-LET radiation because of the higher cell killing effect, with its high relative biologic effectiveness and low oxygen enhancement ratio.31 This study raises the possibility that bortezomib could also provide radiosensitization effect to high-LET radiation and it might be an attractive new approach for the treatment of not only AE-RIT but also RIT using α-emitters and high-LET external radiation therapy.
There are several limitations of this study. Although the authors focused on the NF-κB pathway and successfully demonstrated that bortezomib significantly enhanced the cytotoxicity of 111In-trastuzumab-NLS-L, targeting signaling pathways other than the NF-κB pathway may also be effective for cell killing in combination therapy with 111In-trastuzumab-NLS-L. The mechanism(s) of altered NF-κB activity by 111In-trastuzumab-NLS-L and the molecular basis for enhanced cytotoxicity in combination of 111In-trastuzumab-NLS-L and bortezomib are not fully understood. There is a considerable interest in determining the effects of bortezomib on in vivo tumor growth in combination with 111In-trastuzumab-NLS-L. Further studies will address those issues.
Conclusions
The authors performed transcriptome analyses of AE-RIT using 111In-trastuzumab-NLS and showed that the modulation of NF-κB signaling activity is a candidate for molecular signature of 111In-trastuzumab-NLS treatment. They further demonstrated the potential of combination therapy with bortezomib and 111In-trastuzumab-NLS-L for inhibiting the growth of human breast cancer SKBR3 cells overexpressing HER2. This study will contribute to better understanding of the cytotoxic effects of 111In-trastuzumab-NLS and offer a new strategy to increase the therapeutic efficacy of AE-RIT against human cancer.
Acknowledgments
The authors thank Chemicals Evaluation and Research Institute (Tokyo, Japan) for microarray data analysis. This work was supported, in part, by JSPS KAKENHI Grant Number 24390296 and the President's grant from National Institute of Radiological Sciences, Japan.
Disclosure Statement
No conflicting financial interests exist.
References
- 1.Kassis AI, Adelstein SJ. Radiobiologic principles in radionuclide therapy. J Nucl Med 2005;46 Suppl 1:4S. [PubMed] [Google Scholar]
- 2.Humm JL, Howell RW, Rao DV. Dosimetry of Auger-electron-emitting radionuclides: Report no. 3 of AAPM Nuclear Medicine Task Group No. 6. Med Phys 1994;21:1901. [DOI] [PubMed] [Google Scholar]
- 3.Cornelissen B, Vallis KA. Targeting the nucleus: An overview of Auger-electron radionuclide therapy. Curr Drug Discov Technol 2010;7:263. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Wang J, Hope K, et al. Nuclear localizing sequences promote nuclear translocation and enhance the radiotoxicity of the anti-CD33 monoclonal antibody HuM195 labeled with 111In in human myeloid leukemia cells. J Nucl Med 2006;47:827. [PubMed] [Google Scholar]
- 5.Costantini DL, Hu M, Reilly RM. Peptide motifs for insertion of radiolabeled biomolecules into cells and routing to the nucleus for cancer imaging or radiotherapeutic applications. Cancer Biother Radiopharm 2008;23:3. [DOI] [PubMed] [Google Scholar]
- 6.Costantini DL, Chan C, Cai Z, et al. (111)In-labeled trastuzumab (Herceptin) modified with nuclear localization sequences (NLS): An Auger electron-emitting radiotherapeutic agent for HER2/neu-amplified breast cancer. J Nucl Med 2007;48:1357. [DOI] [PubMed] [Google Scholar]
- 7.Costantini DL, McLarty K, Lee H, et al. Antitumor effects and normal-tissue toxicity of 111In-nuclear localization sequence-trastuzumab in athymic mice bearing HER-positive human breast cancer xenografts. J Nucl Med 2010;51:1084. [DOI] [PubMed] [Google Scholar]
- 8.Amundson SA, Bittner M, Meltzer P, et al. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res 2001;156:657. [DOI] [PubMed] [Google Scholar]
- 9.Tsai MH, Cook JA, Chandramouli GV, et al. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res 2007;67:3845. [DOI] [PubMed] [Google Scholar]
- 10.Smirnov DA, Morley M, Shin E, et al. Genetic analysis of radiation-induced changes in human gene expression. Nature 2009;459:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: When will molecular signatures become relevant to patient care? Nat Rev Cancer 2007;7:545. [DOI] [PubMed] [Google Scholar]
- 12.Ishigami T, Uzawa K, Higo M, et al. Genes and molecular pathways related to radioresistance of oral squamous cell carcinoma cells. Int J Cancer 2007;120:2262. [DOI] [PubMed] [Google Scholar]
- 13.Amundson SA, Do KT, Vinikoor LC, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res 2008;68:415. [DOI] [PubMed] [Google Scholar]
- 14.Niu N, Qin Y, Fridley BL, et al. Radiation pharmacogenomics: A genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 2010;20:1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitoh T, Nakayama M, Nakano H, et al. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem 2003;278:36005. [DOI] [PubMed] [Google Scholar]
- 16.Varfolomeev EE, Ashkenazi A. Tumor necrosis factor: An apoptosis JuNKie? Cell 2004;116:491. [DOI] [PubMed] [Google Scholar]
- 17.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: Balancing life and death—a new approach to cancer therapy. J Clin Invest 2005;115:2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melisi D, Chiao PJ. NF-kappa B as a target for cancer therapy. Expert Opin Ther Targets 2007;11:133. [DOI] [PubMed] [Google Scholar]
- 19.Gupta SC, Sundaram C, Reuter S, et al. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta 2010;1799:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 2009;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasad AV, Mohan N, Chandrasekar B, et al. Activation of nuclear factor kappa B in human lymphoblastoid cells by low-dose ionizing radiation. Radiat Res 1994;138:367. [PubMed] [Google Scholar]
- 22.Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci U S A 1998;95:13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan M, Ahmed KM, Coleman MC, et al. Nuclear factor-kappaB and manganese superoxide dismutase mediate adaptive radioresistance in low-dose irradiated mouse skin epithelial cells. Cancer Res 2007;67:3220. [DOI] [PubMed] [Google Scholar]
- 24.Hellweg CE, Baumstark-Khan C, Schmitz C, et al. Carbon-ion-induced activation of the NF-kappaB pathway. Radiat Res 2011;175:424. [DOI] [PubMed] [Google Scholar]
- 25.Janssens S, Tschopp J. Signals from within: The DNA-damage-induced NF-kappaB response. Cell Death Differ 2006;13:773. [DOI] [PubMed] [Google Scholar]
- 26.McCool KW, Miyamoto S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol Rev 2012;246:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen D, Frezza M, Schmitt S, et al. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr Cancer Drug Targets 2011;11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berenson JR, Yellin O, Patel R, et al. A phase I study of samarium lexidronam/bortezomib combination therapy for the treatment of relapsed or refractory multiple myeloma. Clin Cancer Res 2009;15:1069. [DOI] [PubMed] [Google Scholar]
- 29.Roy R, Evens AM, Patton D, et al. Bortezomib may be safely combined with Y-90-ibritumomab tiuxetan in patients with relapsed/refractory follicular non-Hodgkin lymphoma: A phase I trial of combined induction therapy and bortezomib consolidation. Leuk Lymphoma 2013;54:497. [DOI] [PubMed] [Google Scholar]
- 30.Rae C, Tesson M, Babich JW, et al. Radiosensitization of noradrenaline transporter-expressing tumour cells by proteasome inhibitors and the role of reactive oxygen species. EJNMMI Res 2013;3:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linstadt D, Blakely E, Phillips TL, et al. Radiosensitization produced by iododeoxyuridine with high linear energy transfer heavy ion beams. Int J Radiat Oncol Biol Phys 1988;15:703. [DOI] [PubMed] [Google Scholar]