Abstract
The miniaturization of gene transfer assays to either 384 or 1536-well plates greatly economizes the expense and allows much higher throughput when transfecting immortalized and primary cells compared to more conventional 96-well assays. To validate the approach, luciferase and GFP reporter gene transfer assays were developed to determine the influence of cell seeding number, transfection reagent to DNA ratios, transfection time, DNA dose, and luciferin dose on linearity and sensitivity. HepG2, CHO and 3T3 cells were transfected with polyethylenimine (PEI)-DNA in both 384 and 1536 well plates. The results established optimal transfection parameters in 384-well plates in an total assay volume of 35 μl and in 1536-well plates in a total volume of 8 μl. A luciferase assay performed in 384-well plates produced a Z' score of 0.53, making it acceptable for high throughput screening. Primary hepatocytes were harvested from mouse liver and transfected with PEI-DNA and calcium phosphate DNA nanoparticles in 384-well plates. Optimal transfection of primary hepatocytes was achieved on as few as 250 cells per well in 384-well plates, with CaPO4 proving to be 10-fold more potent than PEI.
Keywords: gene transfection, gene delivery, 384-well, 1536-well, microplate, mammalian cell culture, Luciferase assay
Introduction
High throughput screening (HTS) has become an important technique in screening compound libraries to find new targets for drug discovery. Advanced robotics have allowed miniaturization of 6-well and 96-well cell based assays to 384-well and 1536-well, thereby increasing throughput to more efficiently perform thousands of assays [1, 2]. This decreased size in microplate format is achieved by sophisticated liquid handling pipets and sensitive plate reading detectors, that reduce assay volume while retaining high accuracy and precision.
In vitro gene transfer assays have been used for over two decades to screen for novel nonviral vectors to improve gene transfer efficiency. In vitro transfection studies have led to the discovery of potent gene transfer cationic lipids [3-5], polyethylenimine (PEI) and its analogues[6, 7], cyclodextrin derivatives[8, 9], chitosan and polysaccharide based polymers[10], dendrimers[11-13], polylysines[14, 15], and disulfide cross-linking polypeptides[16-18]. In vitro gene assays typically use plasmids expressing luciferase or green fluorescent protein (GFP) as reporter genes, which are most often performed in either 6-well or 96-well formats.
Combinatorial and parallel chemistry has made it possible to generate large compound libraries that have accelerated the identification of new nonviral gene delivery vectors. As chemical libraries grow, the miniaturization of gene transfer assays are necessary to better manage time, materials and cost. Gene transfer assays have been previously used in 384-well plate formats [6, 11, 19-27] but the full optimization of all parameters have not been reported and these have not been adapted to 1536 well plates [28-31]. In addition to transient transfection of cells, the use of cell lines stably transfected with luciferase or GFP have been used in a miniaturized assay format to facilitated screening for new siRNA delivery vectors [6, 11, 19-27].
Most in vitro transfection assays aimed at developing new vectors, resort to using rapidly dividing immortal mammalian cancer cell lines [32]. The rapid cell division of cancer cell lines allows growth of large numbers of cells needed for scaled-up 6-well and 96-well gene transfer assays. In addition, nonviral gene transfer agents are more efficient at transforming rapidly dividing cells due to the presence of a transient nuclear membrane barrier [33]. Thereby, gene transfer agents developed for transforming rapidly dividing cells often fail to mediate significant expression in non-dividing cells [34]. This contributes to the lack of correlation between in vitro and in vivo efficacy for nonviral gene delivery agents.
The transfection of primary hepatocytes has been reported using calcium phosphate nanoparticles (CaPO4) [35, 36], cationic lipids [37-39] and galactose based cationic glycolipids[40, 41]. However, non-dividing mouse or rat primary hepatocytes are laborious to prepare making their transfection in either 6-well or 96-well plates low throughput. Therefore, miniaturized gene transfer assays that allow much greater throughput are a significant advantage when transforming primary cells [42].
In the present report, we have optimized in vitro gene transfer assays for use in both 384-well plate and 1536-well plate formats. Mammalian cancer cell lines and mouse primary hepatocytes were compared. Assays were optimized for cell density, DNA dose, and transfection time. The results demonstrate a highly optimized and efficient assay to screen large libraries for gene transfer in vitro.
Materials and methods
ONE-Glo Luciferase assay system was purchased from Promega. gWiz-Luc, a 6732bp plasmid with luciferase reporter gene driven by a CMV promoter, and gWiz-GFP, a 5757bp plasmid with green fluorescent protein reporter gene driven by a CMV promoter, were obtained from Aldevron. gWiz-Luc and gWiz-GFP were amplified in a DH5α strain of Escherichia coli and purified with QIAGEN Endofree plasmid kit according to the manufacturer's instructions. Firefly luciferase was purchased from Roche Applied Science (Indianapolis, IN). Dulbecco Modified Eagle Medium (DMEM/F12) without phenol red and William's E medium were purchased from Gibco Life Technologies. Fetal bovine serum (FBS) was obtained from Gibco Life Technologies and was inactivated by incubation at 50°C for 30 min. Penicillin/streptomycin was purchased from Gibco Life Technologies, containing 10,000 units/mL penicillin and 10,000 μg/mL streptomycin. L-glutamine was purchased from Sigma-Aldrich. Nonessential amino acids were obtained from Gibco Life Technologies. HepG2, CHO and NIH 3T3 cells were acquired from the American Type Culture Collection (Manassas, VA). Cell lines were maintained routinely in cell culture media (DMEM/F12 supplemented with 10% FBS and 1% penicillin/streptomycin). Primary hepatocytes were isolated by collagenase perfusion method from mouse as reported [43, 44], and were cultured in William's E medium, containing 1% penicillin/streptomycin, 1% 200 mM L-glutamine, 1% nonessential amino acids amd 10% FBS. Anhydrous 25 KDa PEI, was obtained from Sigma Aldrich. Black solid wall 384-well and 1536-well cell culture plates were purchased from VWR.
Luciferase Calibration Curve
A luciferase calibration curve was constructed to establish linearity of response. HepG2 cells were plated as described below, and after 24 hr the cells were aspirated using a Janus 384-pin head. Luciferase (30 μL of 0.64 -10,000 pg per μL) was pipetted onto cells in triplicate followed immediately by the addition of 10-30 μL of ONE-Glo. Alternatively, HepG2 cells were plated in 1536-well plates and luciferase (2 μL of 4.6-10,000 pg per μL) was added to triplicate wells followed by 1-3 μL of ONE-Glo. Immediately following the addition of luciferin, both 384 and 1536 plates were centrifuged at 1,000 RPM for 1 min, followed by incubation at room temperature for 4 min, with subsequent bioluminescence measurement on the Envision plate reader.
In Vitro Gene Transfection of HepG2 Cells in 384 and 1536 Well Plates
Liquid handling was performed on a Perkin-Elmer Janus automated workstation, using WinPREP® software for Janus 4.8. A 384-pin head loaded with disposable pipette tips was used to transfer liquid in 384-well and 1536-well microplates. Bioluminescence and fluorescence intensities were measured using a Perkin-Elmer Wallac Envision 2104-0010 multilabel reader, using Envision manager software version 1.12. Luciferase bioluminescence was measured with an emission filter of 700 nm at a height of 6.5 mm. GFP fluorescence was measured using an excitation wavelength at 480 nm and emission wavelength at 510 nm, and measurement height of 6.5 mm.
HepG2 cells were plated using a BioTek Multiflo equipped with a 5 μL cassette to dispense cells into 384-well plates and a 1 μL cassette to dispense into 1536-well plates. Prior to use, the dispensing cassettes were washed with 70% ethanol and dry autoclaved, then primed with cell suspension. A uniform cell density was achieved by gently stirring cell suspensions to prevent sedimentation during plating. HepG2 cells were suspended in DMEM phenol red free culture medium at a concentration ranging from 100-400 cells per μL determined by hemocytometer. HepG2 cells were plated at varying density into 384-well plates by dispensing 25 μL per well whereas 6 μL per well was dispensed for 1536-well plates. Plated HepG2 cells were cultured at 37°C in a humidified 5% CO2 incubator for 24 hr prior to transfection.
PEI DNA polyplexes were prepared at N:P (nitrogen to phosphate) ratio of 9 by mixing equal volumes of gWiz-Luc (0.5-8 μg in 100 μL ) or gWiz-GFP with PEI (0.6-9.3 μg in 100 μL) in HBM buffer (5 mM HEPES, 2.7 M mannitol pH 7.5), followed by incubation at RT for 30 min prior to transfection of cells. CaPO4 DNA nanoparticles were prepared according to Olton et al [45]. CaCl2 (13 μL of 2.5 M) was added to gWiz-Luc (0.5-9.3 μg in a total volume of 117 μL of water) followed by incubation at RT for 15 min. The DNA (130μL) was added to an equal volume of 280 mM NaCl, 10 mM KCl, 12 mM dextrose, 50 mM HEPES free acid and 1.25 mM Na3PO4 pH 7.5, at a controlled rate of 13.4 μL per sec. The resulting CaPO4 DNA nanoparticles were incubated at RT for 15 min before addition to cells. CaPO4 DNA or PEI DNA were added to HepG2 cells in 384-wells (5 μL) or 1536-wells (2 μL) using a Hamilton Microlab Star liquid handling system. Following transfection, plates were incubated at 37°C in humidified 5% CO2 for times ranging from 24-96 hr. Plates were then cooled to RT, centrifuged at 1,000 RPM for 1 min, ONE-Glo was added to each well (384-well 10 μL, 1536-well 2 μL), and the bioluminescence was measured after 5 min on an Envision plate reader.
In Vitro Transfection of Primary Hepatocyes in 384-Well plates
Primary hepatocytes were isolated from ICR mice by collagenase perfusion as previously described [50] with an average cell viability of 85-90 % as determined by trypan blue exclusion. Primary hepatocyes were prepared in William's essential media at 4-200 cells per μL. The cell suspensions were plated onto a 384-well plate (45 μL per well) manually with a multichannel pipetter. Cells were allowed to sediment for 15 min at 37°C in a humidified 5 % CO2 incubator prior to transfection. CaPO4 DNA or PEI DNA (5 μL) prepared as described above were added to cells using a Hamilton Microlab Star liquid handling system. Primary hepatocytes were transfected for 24 h at 37°C in a humidified 5 % CO2 incubator. Plates were cooled to RT, ONE-Glo (10 μL) was added to each well, and the bioluminescence was measured as described above. Alternatively, gWiz-GFP transfections were analyzed by measuring fluorescence on an Envision plate reader. The results were analyzed for statistical significance using two-way ANOVA analysis.
Results and Discussion
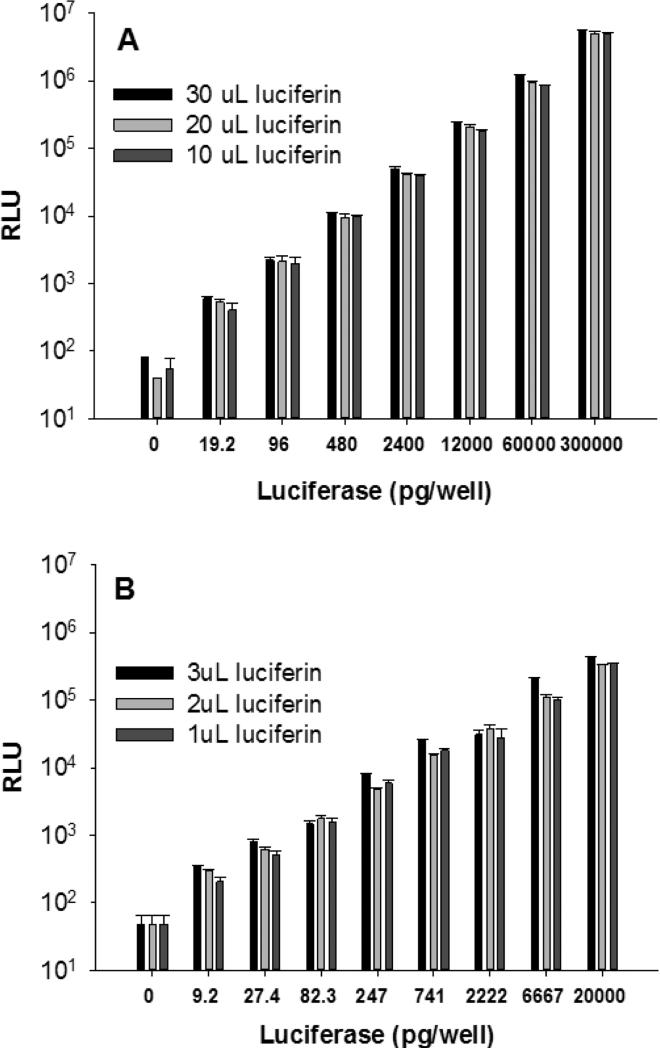
The goal of this study was to develop optimal transfection parameters for primary and cultured hepatocytes, as well as other cell lines in 384 and 1536-well plates. A standard curve was generated to establish the linear dynamic range of the bioluminescence response (relative light unit, RLU) in both plate formats. The addition of ONE-Glo luciferin produces stable luminescence for approximately 30 min when 30 μL is added to 30 μL of media containing luciferase. A linear response of RLU covering 5-orders of magnitude is generated when increasing luciferase across the range of 0-300,000 pg, establishing the lack of detector saturation at 107 RLU. However, considering the cost of ONE-Glo, the linearity of the assay was also examined with the addition of 10 and 20 μL per well. The signal intensity and linearity were nearly identical, such that 10 μL was chosen for all subsequent experiments (Fig. 1A).
Figure 1.
Luciferase Standard Curve in 384 and 1536-Well Plates. Panel A illustrates the linearity of response as a function of increasing amounts of luciferase and increasing luciferin (ONE-Glo) added to plated HepG2 cells in a 384-well plate. The results are the mean and standard deviation of n = 3. Panel B illustrates the linearity of response and dynamic range in a 1536-well plate (n = 4).
Similar experiments were performed in 1536-well plates, where 1-3 μL of ONE-Glo was added to serial diluted luciferase on HepG2 cells in a total volume of media of 6 μL (Fig. 1B). A linear response covering 4-orders of magnitude was determined with only a slight increase in standard deviation when using 1 μL of ONE-Glo. Consequently, 2 μL of ONE-Glo was added to 1536-well plates throughout the study.
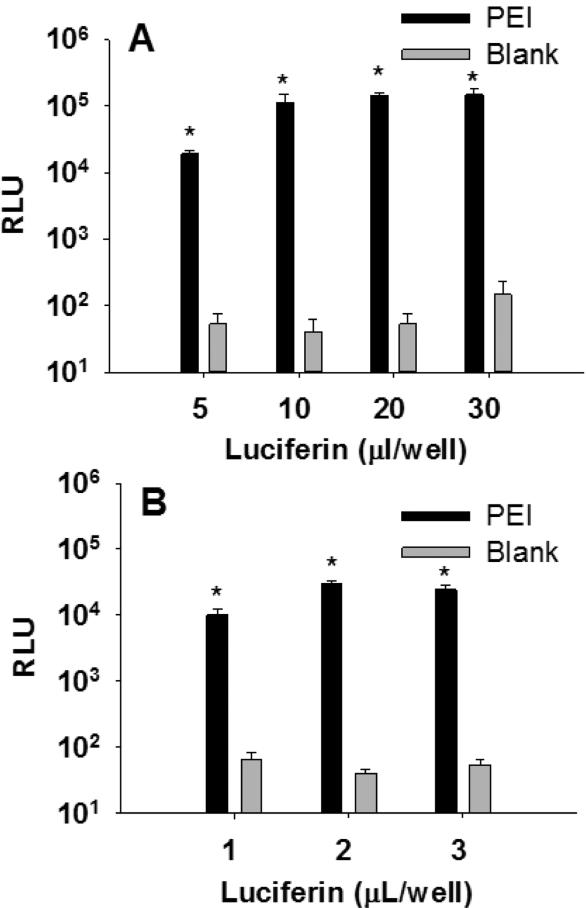
To verify a linear response when diminished amounts of ONE-Glo was added to transfected cells, PEI was used to transfect gWiz-Luc, a luciferase expressing plasmid. The transfection of 5000 HepG2 cells per well, plated in 25 μl in 384-well plates, was performed by adding 250 ng of gWiz-Luc in complex with 1 μg of PEI (nitrogen to phosphate (N:P) ratio of 9). At 48 hr post-transfection, 5-30 μL of ONE-Glo was added, and the RLU were measured. The resulting RLU intensities in 384-well plate format were equivalent for 10-30 μL of ONE-Glo, but were 5-fold lower when 5 μL was used. The magnitude of expression (105 RLU), established a greater than 3,000 signal-to-background (S:B) ratio relative to mock transfected cells (Fig. 2A) .
Figure 2.
Transfection of HepG2 Cells in 384 and 1536-Well Format. Panel A illustrates the result of 384-well transfection of 5000 HepG2 cells in 25 μL media. At 48 hr following the addition of PEI gWiz-Luc (250 ng, N:P = 9:1) in 5 μL, 5-30 μL of ONE-Glo was added, and RLU were determined after 5 min (n = 3). Panel B illustrates transfection of 1200 HepG2 cells in 6 μL media in 1536-well plates, with 80 ng (2 μL) of PEI/gWiz-Luc (N:P = 9). At 48 hr after post-transfection, 1-3 μL of ONE-Glo was added prior to measuring RLU after 5 min (n = 4). * indicates p ≤ 0.05 relative to blank.
In 1536-well plates, 1,200 HepG2 cells per well were plated and dosed with 80 ng of gWiz-Luc polyplex in a total volume of 8 μL. After 48 hr, 3 μL of media was removed followed by the addition of 1-3 μL of ONE-Glo. The luciferase response of 104 RLU established an S:B ratio of 455, which was insensitive to the volume of ONE-Glo added (Fig. 2B).
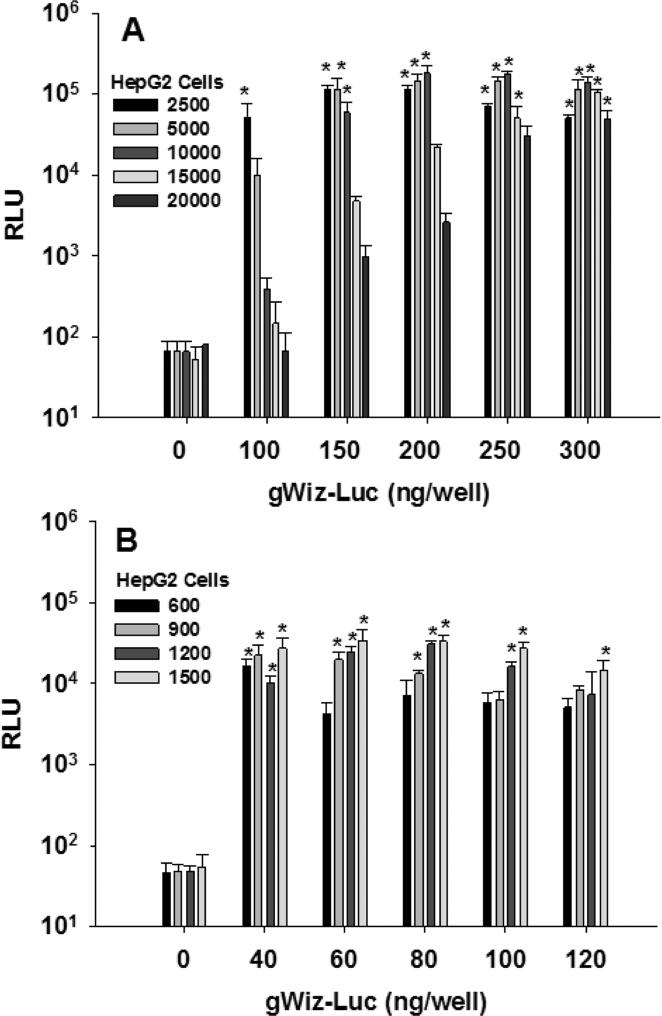
To establish the influence of cell density on transfection efficiency, HepG2 cells were cultured at plating densities ranging from 2,500-20,000 cells per well in 384-well plates and transfected with 0-300 ng of PEI gWiz-Luc (N:P = 9). After 48 hr, the addition of 10 μL of ONE-Glo luciferin, followed by bioluminescence analysis, revealed the cell plating number strongly influences gene transfer at 100 ng DNA doses, with highest expression at low (2500 cells per well) seeding density. The effect of cell seeding density on expression progressively diminished with increasing DNA dose, such that a 300 ng dose resulted in a constant gene transfer efficiency as a function of cell seeding number (Fig. 3A). This result is consistent with low seeding numbers resulting in greater gene transfer efficiency due to rapid cell division, which diminishes as cells reach confluency [46]. The difference in expression is diminished as the PEI gWiz-Luc dose increases, which is likely the result of increased PEI concentrations.
Figure 3.
Influence of Cell Plating Number on Transfection in 384 and 1536-Well Format. Panel A illustrates the result of plating HepG2 cells (25 μL, 2500-20000 cells) in 384-well plates and transfecting with 5 μL (0-300 ng, PEI gWiz-Luc, N:P = 9). At 48 hr post-transfection, 10 μL of ONE-Glo was added and the RLU was measured after 5 min (n = 3). Panel B illustrates the result of transfection of 0-120 ng (2 μl) of PEI gWiz-Luc (N:P 9) into HepG2 cells (600-1500 cells in 6 μL per well) in 1536-well plates. At 48 hr, 3 μL was removed and 2 μL of ONE-Glo was added the bioluminescence data was measured at 25 min (n = 4). * indicates p ≤ 0.05 relative to 0 DNA dose.
When the cell seeding number was varied from 600-1,500 cells per well in 1536-well plates, and transfection was performed with PEI gWiz-Luc varying from 40-120 ng per well, the luciferase expression (RLU) was insensitive to cell seeding number across the dosing range, which indicates maintenance of rapid cell division at cell seeding numbers that remain below confluency. (Fig. 3B). Transfection of 600 cells per well with 40 ng per well provided an optimal transfection efficiency. Increases in PEI-gWiz-Luc dose were tolerated without decreases in expression, indicating the lack of PEI toxicity at the highest doses.
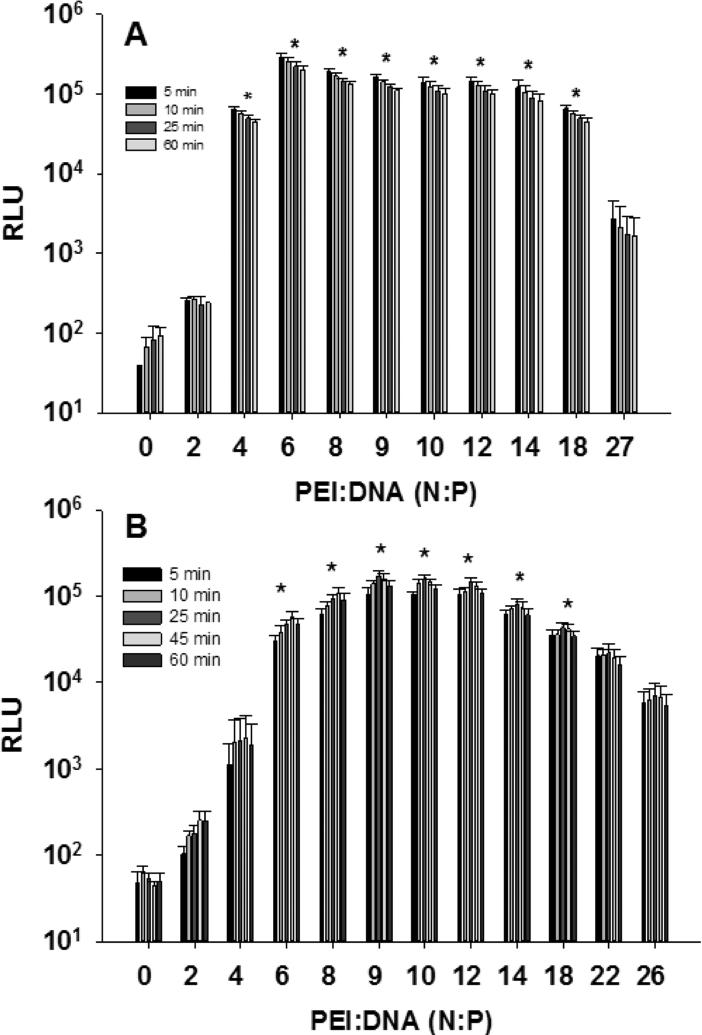
Using optimized cell densities and DNA doses, the PEI gWiz-Luc ratio was varied across a broad range. The PEI:DNA ratio is known to strongly influence gene transfer efficiency and has been most often reported as the nitrogen to phosphate (N:P) ratio [47]. As the ratio of PEI to DNA increases to beyond that necessary to fully compact DNA, which is typically N:P 2:1 (1:1 weight ratio of PEI and DNA), the excess unbound PEI serves as an endosomal buffering agent to facilitate gene transfer [47]. However, at excessively high concentrations, PEI is toxic to cells resulting in diminished expression [47]. As illustrated in Fig 4, transfection using an N:P of 6 results in the most efficient transfection in 384-well whereas an N:P of 9 was optimal in 1536-well plates. The transfection efficiency was relatively insensitive to N:P in 384-well plates across a broad N:P ratio ranging from 6-18, whereas a narrow range of 9-12 was optimal in 1536-well plates (Fig. 4). However, while the optimal ranges of N:P ratios may serve as a general guide, optimized expression based on varying N:P is also dependent on cell seeding number, PEI DNA doses, and the type of cell being transformed.
Figure 4.
Influence of Varying PEI to DNA Ratio and Bioluminescence Acquisition Time on Gene Transfer Efficiency. Panel A illustrates the result of transfecting 250 ng PEI gWiz-Luc with N:P ranging from 0-27 onto 5000 HepG2 cells per well in 384-well plates. Following the addition of 10 μL of ONE-Glo, bioluminescence was measured at a time varying from 5-60 min (n = 3). Panel B illustrates the result of transfecting 75 ng per well of PEI gWiz-Luc (2 μl) with N:P ranging from 0-26 onto 1200 HepG2 cells per well in 1536-well plates. At 48 hr following transfection, 3 μL was removed and 2 μL ONE-Glo was added followed by bioluminescence measurement at 5, 10, 25, 45 and 60 min (n = 4). An N:P of 0 indicates a transfection control with naked DNA which fails to mediate luciferase expression. * indicates p ≤ 0.05 relative to 0 N:P.
Throughout this study the luciferase expression was determined by measuring bioluminescence at 5 min following the addition of ONE-Glo. The Envision plate reader measures RLU from an entire 384-well plate in 1 min, whereas reading a 1536-well plate requires 15 min. Likewise, the addition of ONE-Glo to all 384-wells of a plate is accomplished in a single pipetting event using a 384-well head. Whereas, the addition of ONE-Glo to a 1536-well plate requires four pipetting events from the 384-well head resulting in a time delay of several min. Thereby, if the bioluminescence signal is not sufficiently stable, significant differences in RLUs could arise from time delays when reading 1536-well plates.
In the 384-well plates, RLU intensities acquired at 5 min after the addition of ONE-Glo were consistently higher and progressively decreased as the time delay to read bioluminescence was increased up to 1 hr (Fig. 4A). However, the differences in intensities were not statistically significant, indicating that ONE-Glo produces stable bioluminescence for up to 1 hr in 384-well plates. In contrast, in 1536-well plates, RLU intensities increased with read delays of 5-25 min following addition of ONE-Glo, and reached a stable maximum between 25-45 min, before decreasing at 60 min. While these small differences were also not statistically significant, they most likely arise from the combination of longer read-time across 1536-well plates and from the slower mixing of ONE-Glo in the small total volume of 8 μL in 1536-well plates.
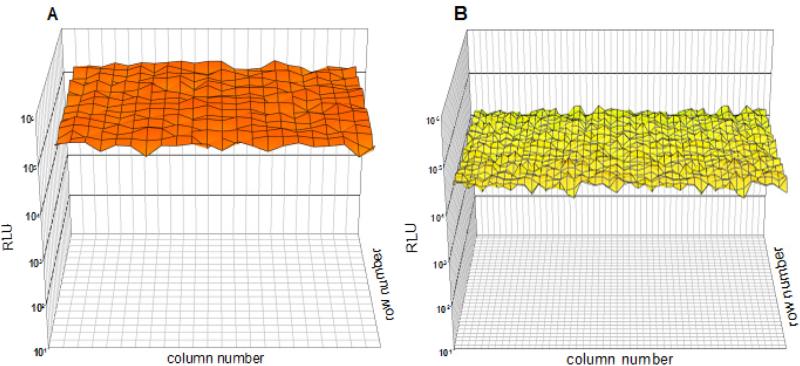
Whole plate transfections were carried out in both 384 and 1536-well format to examine the precision of the gene transfer assay and its suitability for HTS studies (Fig. 5). All 384 wells were seeded with 5000 HepG2 cells per well and all 22 columns of wells were transfected with PEI gWiz-Luc (250 ng, N:P 9), whereas 2 columns were used as controls by omitting PEI gWiz-Luc. In 1536-well plates, 44 columns were loaded with HepG2 cells (1200 cells per well) and PEI gWiz-Luc (80 ng per well, N:P 9), whereas 4 columns were used as controls.
Figure 5.
Whole Plate Transfection in 384 and 1536-Well Plates. The result of applying an optimized transfection protocol to HepG2 cells in 384 (Panel A) and 1536-well (Panel B) format are illustrated.
The RLU intensity across the 384-well plate was constant, with a coefficient of variation (CV) of 16 %, and a Z’ factor at 0.53. In the 1536 format, RLU intensities were more variable, with a CV of 19 %, and Z’ factor of 0.42, which is below an excellent assay threshold of 0.5. This greater variation in 1536-well format is caused by an edge effect that decreases signal intensities in the outer wells due to dehydration during the 48 hr transfection period. Consequently, the transfection in the outer wells of 1536-well plate should be omitted to improve the precision of the assay.
The optimized parameters for HepG2 cell transfection in 384 and 1536-well plates are included in Table 1. Overall, approximately four-fold fewer cells and DNA were needed for 1536-well transfections compared to 384-well transfections. However, this also resulted in a five-fold lower S:B ratio. A similar transfection optimization conducted for NIH-3T3 and CHO cells also resulted in comparable reductions in S:B ratio (Table 1).
Table 1.
Summary of Optimized Transfection Parameters for Cells on 384 and 1536-Well Plates.
| Plate | 384-Well Plate | 1536-Well Plate | ||||
|---|---|---|---|---|---|---|
| Cells/well | DNA dose ng/well | S:Ba Ratio | Cells/well | DNA dose ng/well | S:Ba Ratio | |
| HepG2 | 5000-15000 | 150-300 | 2000 | 900-1200 | 40-100 | 400 |
| NIH 3T3 | 2500-5000 | 200-300 | 3800 | 400-900 | 60-120 | 750 |
| CHO | 2500-10000 | 100-300 | 3200 | 600-1200 | 40-120 | 500 |
| Volumeb | 25 μL | 6 μL | ||||
| Polyplex | 5 μL | 2 μL | ||||
| ONE-Glo | 10 μL | 2 μL | ||||
Signal to Baseline ratio for luciferase expression.
Represents the volume of media in the assay.
Optimized transfection parameters described above, were also used to express green fluorescent protein by substituting gWiz-GFP for gWiz-Luc. GFP expression offers the advantage of direct fluorescence analysis without the addition of substrate, and the cells can be repeatedly analyzed over time. In addition, GFP transfection is frequently monitored by FACS and confocal fluorescence microscopy [8, 22, 26, 27, 30, 35].
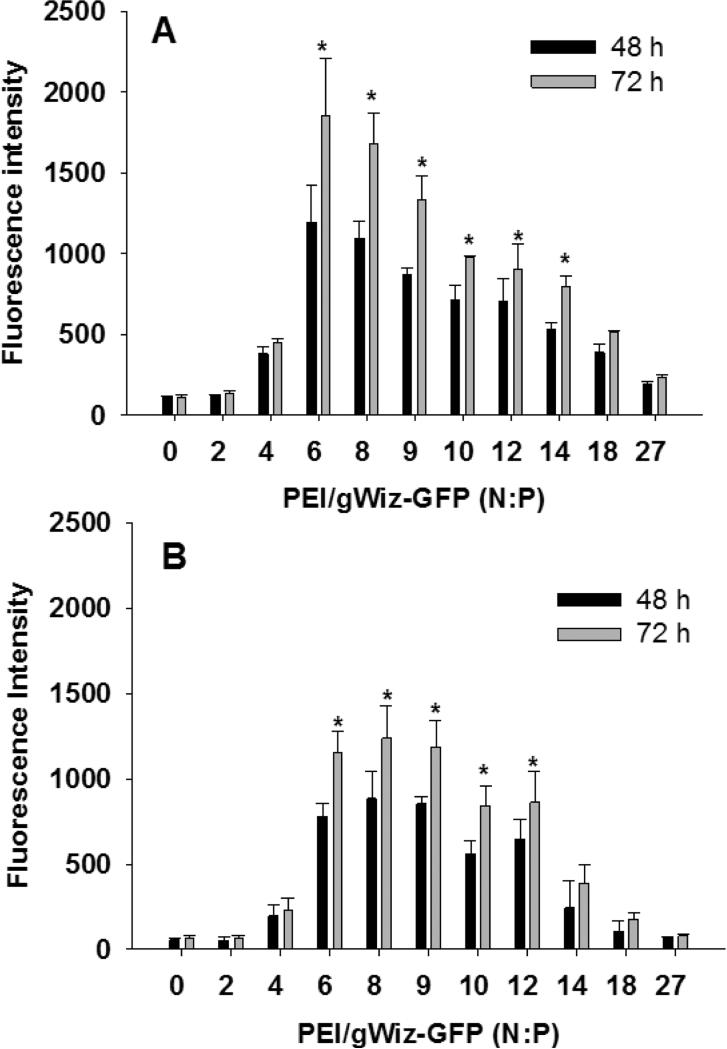
HepG2 cells (10,000 cells seeded per well) were transfected in 384-well plates with 250 ng of PEI gWiz-GFP at N:P ratios ranging from 0-27. At 48 and 72 hr the cells were analyzed by fluorescence with excitation of 480 nm and emission of 510 nm. Maximum expression at 48 hr occurred at an N:P ratio of 6, which provided a fluorescent intensity of 1200 units and an S:B of approximately 10 relative to background fluorescence intensity (120 units) for untransfected control wells at N:P of 0 (Fig. 6A). The fluorescence intensity increased to 1800 units at 72 hr, resulting in an S:B of approximately 15, but the standard deviation of the analysis also increased.
Figure 6.
Transfection of HepG2 Cells with GFP in 384 and 1536-Well Plates. Panel A illustrates the result of transfection of 10,000 cells (25 μL) per well in 384-well plates with transfection with 250 ng (5 μL) of PEI gWiz-GFP (N:P 0-27). At 48 and 72 hr the fluorescence was measured at excitation 480 and emission 510 nm. Panel B illustrates the result of transfecting 1500 HepG2 cells (6 μl) in 1536-well plates with 60 ng (2 μL) of PEI gWiz-GFP (N:P 0-27) followed by fluorescence intensity analysis at 48 and 72 hr. An N:P of 0 indicates a transfection control with naked DNA which fails to mediate GFP expression.* indicates p ≤ 0.05 relative to 0 N:P.
Similar results were determined from PEI gWiz-GFP (60 ng ranging in N:P from 0-27) transfection of HepG2 cells (1500 cells per well) in 1536-well plate. A maximal fluorescence intensity of 870 units at N:P of 8 resulted in an S:B of 15. Increasing the transfection time to 72 hr resulted in an increased fluorescence intensity of 1100, with an increase in the standard deviation. gWiz-GFP transfection offers the main advantages of repeated analysis of cells by direct fluorescence measurement without the need to lyse or add substrate, in addition to being compatible with FACS and confocal. However, compared to luciferase expression with an S:B of 2000 in 384-well and 400 in 1536-well plates, GFP results in S:B values of 10-15 in both plate formats.
Miniaturization and robotic automation significantly increases the speed and decreases the amount of DNA, cells, media, and transfection reagents needed to optimize and routinely perform transfection assays with greater precision, relative to 6-well and 96-well formats. However, it is essential to miniaturize when transforming primary mammalian cells due to the limited number of most cells isolated from animal sources. The in vitro transfection of primary hepatocytes has been reported using calcium phosphate nanoparticles [42,43] and commercially available cationic lipids [44-46].
Prior studies were performed on 6-well or 96-well plates, with cell densities of 106 to 105 per well. Primary hepatocytes isolated from mouse liver by collagenase perfusion vary with both the strain and the age of mice. The number of cells that can be obtained from a mouse liver is also limited, so the number of cells per well must be kept low in order to test a reasonable number of samples, which favors the 384 or 1536-well formats.
Primary hepatocytes were determined to be more fragile than HepG2 cells during plating. At 24 hr following automated pipetting by Biotek Multiflo, primary hepatocytes were 60% (5 μL cassette) or 50% viable (1 μL cassette), whereas manual seeding in 384-well plates with an 8-channel pipet resulted in 80% viability. Primary hepatocytes also sediment faster than HepG2 cells, requiring continuous gentle shaking to maintain cell suspensions. Primary hepatocytes were plated on 384-well plates with or without collagen coating. Cell viabilities were similar in both conditions with approximately 80% viable at 24 hr, 70% viable at 48 hr, and less than 40% viable at 72 hr. Therefore, transfections were performed 45 min following manually pipetting primary hepatocytes into non-collagen coated 384-well plates.
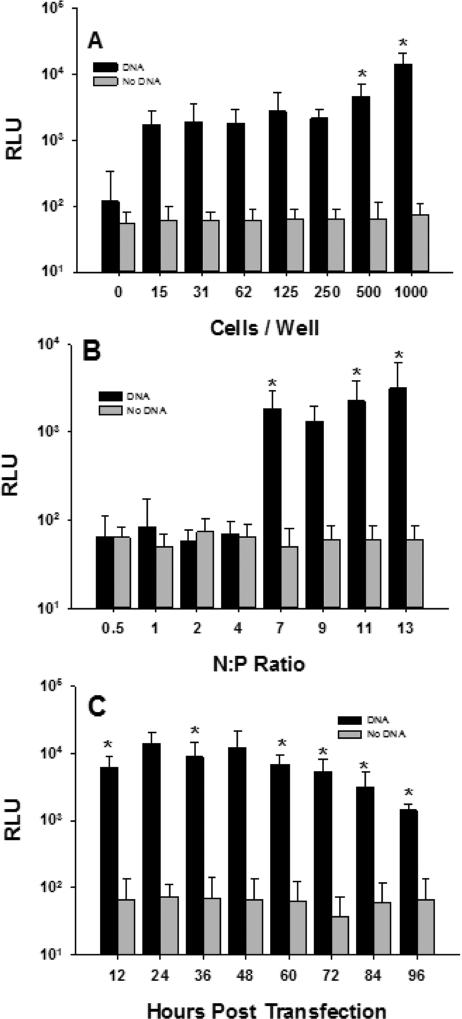
Optimal PEI mediated transfection of primary hepatocytes was determined by plating 0-1,000 cells per well on 384-well plates and transfecting with 400 ng PEI gWiz-Luc at N:P of 7. A cell seeding number of 250 cells per well resulted in a S:B of 10. However, lower cell seeding numbers down to as few as 15 cells per well resulted in comparable signal intensity (Fig. 7A). An optimal PEI gWiz-Luc N:P ratio was 7, below which no expression was observed and above which the expression was constant (Fig. 7B). Under optimal conditions, the transfection of primary hepatocytes with PEI gWiz-Luc established maximal expression at 12-48 hr followed by a decrease during 48-96 hours (Fig. 7C).
Figure 7.
Transfection of Primary Hepatocytes in 384-Well Plates. Panel A illustrates the transfection of 0-1000 primary hepatocytes in 45 μL in a 384-well plate with 400 ng (5 μL) of PEI gWiz-Luc (N:P 7). At 24 hr, 10 μL of ONE-Glo was added, followed by measurement of bioluminescence at 5 min. (n = 9). Panel B illustrates transfection of 1000 primary hepatocytes as described in panel A, with 400 ng (5 μl) of PEI gWiz at (N:P 0.5 to 13). Panel C illustrates the transfection of 250 primary hepatocytes per well with 400 ng of PEI gWiz-Luc (N:P 7) with bioluminescence measured every 12 hours for 96 hours (n = 9). * indicates p ≤ 0.05 relative to 0 DNA dose.
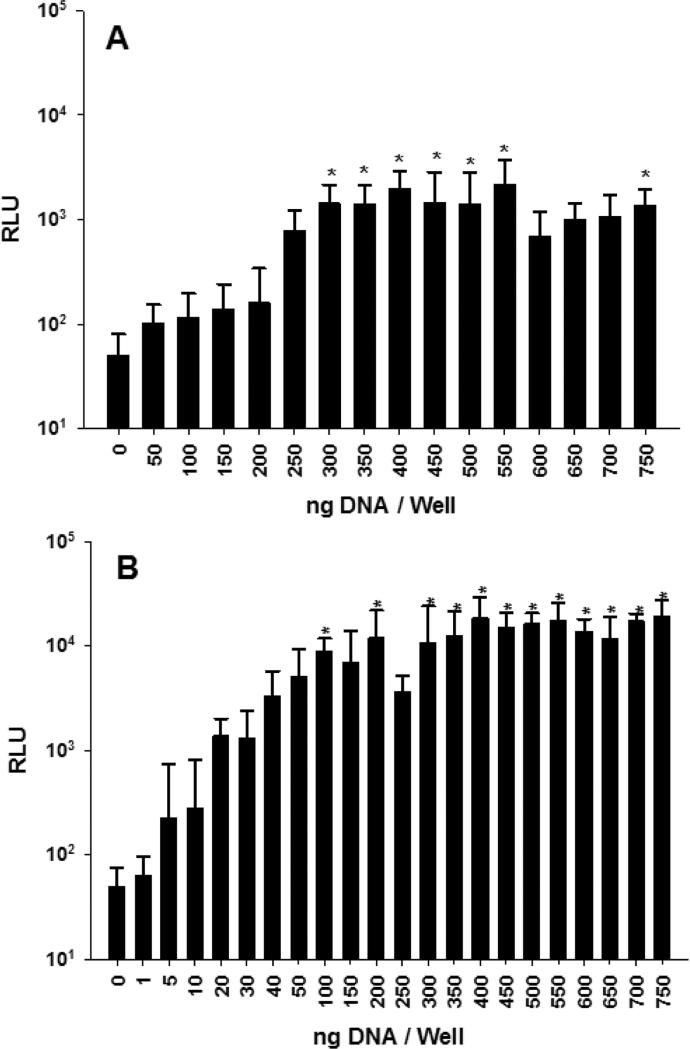
PEI and CaPO4 nanoparticles were compared as transfection agents for primary hepatocytes on 384-well plates. Based on prior studies, a calcium to phosphate ratio of 200 was selected for highest transfection efficiency [42, 49]. Primary hepatocytes (250 per well) were plated in 384-well plates and transfected with 0 to 750 ng of PEI gWiz-Luc (N:P 7) or CaPO4 gWiz-Luc at (Ca:P, 200) and analyzed for luciferase expression at 24 hr. PEI mediated highest expression at 300-550 ng DNA dose whereas CaPO4 transfection was maximal at 200-750 ng DNA dose (Fig 8A and B). The miniaturization of 384-well gene transfer assays to 250 primary hepatocytes per well produced low Z' values of −1.9 for PEI and −0.9 for CaPO4 mediated gene transfer. This result is primarily due to the higher CV (80% for PEI and 62% for CaPO4) caused by greater variability in primary hepatocytes compared to HepG2 cells (16%). The finding that CaPO4 consistently produced a more efficient transfection than PEI in primary hepatocytes at all doses is most likely a reflection of the different cell uptake processes and endosomal escape mechanisms for PEI versus CaPO4.
Figure 8.
PEI and CaPO4 Transfection of Primary Hepatocytes in 384-Well Plates. Panel A illustrates the transfection of 250 primary hepatocytes (45 μL) per well with 0-750 ng of PEI gWiz-Luc (N:P 7). At 24 hr 10 μL of ONE-Glo was added and bioluminescence was measured at 5 min (n = 9). Panel B illustrates the transfection of primary hepatocytes (45 μL) plated in 384-well plates at 250 cells per well with 250 ng of CaPO4 calcium phosphate. At 24 hr 10 μL of ONE-Glo was added and bioluminescence was measured at 5 min (n = 9). * indicates p ≤ 0.05 relative to 0 DNA dose.
Conclusion
The parameters examined serve as a guide to arrive at optimal transfection assays for other cell types in 384 and 1536-well microplates. Of the parameters explored, the cell seeding number and PEI-DNA dose most strongly influence the S:B ratio. High doses at low cell seeding result in cell toxicity and low S:B ratio. The number of cells required to produce acceptable S:B is significantly reduced in 384 and 1536-well format compared to more traditional 96 and 6-well plates. The number of primary hepatocytes required to produce acceptable S:B is also approximately 10-fold less than required when transforming HepG2 cells. CaPO4 is a more potent gene transfer agent than PEI when used to transform primary hepatocytes. Optimized miniature transfection assays provide the most efficient format to explore the potency of new transfer agents on varying cell types as a predictor of in vivo efficacy.
Acknowledgement
The authors gratefully acknowledge support from NIH Grant GM097093, NIH T32GM008365 predoctoral training grant (SC), and a fellowship from the American Foundation for Pharmaceutical Education (SC). NIH supports for UIHTS facility from S10 RR029274-01 is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Current Opinion in Pharmacology. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Sterling J, Bojanic D, Eglen RM, Heyse S, Strulovici B. Current trends in high-throughput screening. Assay and drug development technologies. 2008;6:491–504. doi: 10.1089/adt.2008.9989. [DOI] [PubMed] [Google Scholar]
- 3.Gao X, Kim K-S, Liu D. Nonviral gene delivery: What we know and what is next. AAPS Journal. 2007;9:E92–E104. doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Midoux P, Pichon C, Yaouanc J-J, Jaffres P-A. Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. British J Pharmacol. 2009;157:166–178. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barua S, Ramos J, Potta T, Taylor D, Huang H-C, Montanez G, Rege K. Discovery of Cationic Polymers for Non-Viral Gene Delivery Using Combinatorial Approaches. Combin Chem & High Throughput Screening. 2011;14:908–924. doi: 10.2174/138620711797537076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner DK, Zhao X, Buss H, Langer R, Hammond PT. Crosslinked linear polyethylenimine enhances delivery of DNA to the cytoplasm. J Cont Rel. 2013;167:101–107. doi: 10.1016/j.jconrel.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo - polyethylenimine. Proc Natl Acad of Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez H, Hwang SJ, Davis ME. New class of polymers for the delivery of macromolecular therapeutics. Bioconj Chem. 1999;10:1068–1074. doi: 10.1021/bc990072j. [DOI] [PubMed] [Google Scholar]
- 9.Cryan SA, Holohan A, Donohue R, Darcy R, O'Driscoll CM. Cell transfection with polycationic cyclodextrin vectors. Euro J Pharmaceut Sci. 2004;21:625–633. doi: 10.1016/j.ejps.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Liu YM, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. J Amer Chem Soc. 2005;127:3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 11.Kwok A, Eggimann GA, Reymond J-L, Darbre T, Hollfelder F. Peptide dendrimer/lipid hybrid systems are efficient DNA transfection reagents: structure-activity relationships highlight the role of charge distribution across dendrimer generations. ACS Nano. 2013;7:4668–4682. doi: 10.1021/nn400343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayele HK, Sakthivel T, O'Donell M, Pasi KJ, Wilderspin AF, Lee CA, Toth I, Florence AT. Versatile peptide dendrimers for nucleic acid delivery. J Pharmaceut Sci. 2005;94:446–457. doi: 10.1002/jps.20230. [DOI] [PubMed] [Google Scholar]
- 13.Sanclimens G, Shen H, Giralt E, Albericio F, Saltzman MW, Royo M. Synthesis and screening of a small library of proline-based biodendrimers for use as delivery agents. Biopolymers. 2005;80:800–814. doi: 10.1002/bip.20301. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli UK. Characterization of DNA condensates induced by poly(ethylene oxide) and polylysine. Proc Natl Acad Sci USA. 1975;72:4288–4292. doi: 10.1073/pnas.72.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine pDNA complexes. Bioconj Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 16.McKenzie DL, Kwok KY, Rice KG. A potent new class of reductively activated peptide gene delivery agents. J Biol Chem. 2000;275:9970–9977. doi: 10.1074/jbc.275.14.9970. [DOI] [PubMed] [Google Scholar]
- 17.McKenzie DL, Smiley E, Kwok KY, Rice KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconj Chem. 2000;11:901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 18.Baumhover NJ, Anderson K, Fernandez CA, Rice KG. Synthesis and In Vitro Testing of New Potent Polyacridine-Melittin Gene Delivery Peptides. Bioconj Chem. 2010;21:74–83. doi: 10.1021/bc9003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Raad M, Teunissen EA, Lelieveld D, Egan DA, Mastrobattista E. High-content screening of peptide-based non-viral gene delivery systems. J Cont Rel. 2012;158:433–442. doi: 10.1016/j.jconrel.2011.09.078. [DOI] [PubMed] [Google Scholar]
- 20.Van Vliet LD, Chapman MR, Avenier F, Kitson CZ, Hollfelder F. Relating chemical and biological diversity space: A tunable system for efficient gene transfection. Chembiochem. 2008;9:1960–1967. doi: 10.1002/cbic.200800003. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Zahner D, Su Y, Gruen C, Davidson G, Levkin PA. A biomimetic lipid library for gene delivery through thiol-yne click chemistry. Biomaterials. 2012;33:8160–8166. doi: 10.1016/j.biomaterials.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 22.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angewandte Chemie-International Edition. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 23.Akinc A, Lynn DM, Anderson DG, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Amer Chem Soc. 2003;125:5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 24.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang Y-H, Langer R, Sawicki JA, Anderson DG. Rapid optimization of gene delivery by parallel end-modification of poly(beta-amino ester)s. Mol Ther. 2007;15:1306–1312. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]
- 25.Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K, Brown D. High-throughput RNAi screening in vitro: From cell lines to primary cells. RNA-A. 2005;11:985–993. doi: 10.1261/rna.7288405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borawski J, Lindeman A, Buxton F, Labow M, Gaither LA. Optimization procedure for small interfering RNA transfection in a 384-well format. J Biomol Screening. 2007;12:546–559. doi: 10.1177/1087057107300172. [DOI] [PubMed] [Google Scholar]
- 27.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Effects of Base Polymer Hydrophobicity and End-Group Modification on Polymeric Gene Delivery. Biomacromolecules. 2011;12:3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garyantes TK. 1536-well assay plates: when do they make sense? Drug Discovery Today. 2002;7:489–490. doi: 10.1016/s1359-6446(02)02246-8. [DOI] [PubMed] [Google Scholar]
- 29.Kariv I, Cao H, Marvil PD, Bobkova EV, Bukhtiyarov YE, Yan YP, Patel U, Coudurier L, Chung TDY, Oldenburg KR. Identification of inhibitors of bacterial transcription/translation machinery utilizing a miniaturized 1536-well format screen. J Biomol Screening. 2001;6:233–243. doi: 10.1177/108705710100600405. [DOI] [PubMed] [Google Scholar]
- 30.Maffia AM, Kariv I, Oldenburg KR. Miniaturization of a mammalian cell-based assay: Luciferase reporter gene readout in a 3 microliter 1536-well plate. J Biomol Screening. 1999;4:137–142. doi: 10.1177/108705719900400307. [DOI] [PubMed] [Google Scholar]
- 31.Rajasarkka J, Virta M. Miniaturization of a Panel of High Throughput Yeast-Cell-Based Nuclear Receptor Assays in 384-and 1536-Well Microplates. Combin Chem & High Throughput Screening. 2011;14:47–54. doi: 10.2174/1386207311107010047. [DOI] [PubMed] [Google Scholar]
- 32.Donato MT, Lahoz A, Castell JV, Gomez-Lechon MJ. Cell lines: A tool for in vitro drug metabolism studies. Current Drug Metabolism. 2008;9:1–11. doi: 10.2174/138920008783331086. [DOI] [PubMed] [Google Scholar]
- 33.Zhou R, Geiger RC, Dean DA. Intracellular trafficking of nucleic acids. Expert Opin Drug Deliv. 2004;1:127–140. doi: 10.1517/17425247.1.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichon C, Billiet L, Midoux P. Chemical vectors for gene delivery: uptake and intracellular trafficking. Curr Opin Biotechnol. 21:640–645. doi: 10.1016/j.copbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Gaunitz F, Papke M, Gebhardt R. Transient transfection of primary cultured hepatocytes using CaPO4/DNA precipitation. Biotechniques. 1996;20:826. doi: 10.2144/96205st01. [DOI] [PubMed] [Google Scholar]
- 36.Edwards M, Wong SC, Chotpadiwetkul R, Smirlis D, Phillips l.R., Shephard EA. Transfection of primary cultures of rat hepatocytes. Methods in molecular biology (Clifton, N.J.) 2006;320:273–282. doi: 10.1385/1-59259-998-2:273. [DOI] [PubMed] [Google Scholar]
- 37.Park J-S, Surendran S, Kamendulis LM, Morral N. Comparative nucleic acid transfection efficacy in primary hepatocytes for gene silencing and functional studies. BMC research notes. 2011;4:8–8. doi: 10.1186/1756-0500-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe Y, Nomoto H, Takezawa R, Miyoshi N, Akaike T. Highly efficient transfection into primary cultured mouse hepatocytes by use of cationic-liposomes - an application for immunization. J of Biochem. 1994;116:1220–1226. doi: 10.1093/oxfordjournals.jbchem.a124667. [DOI] [PubMed] [Google Scholar]
- 39.Jarnagin WR, Debs RJ, Wang SS, Bissell DM. Cationic lipid-mediated transfection of liver cells in primary culture. Nucleic Acids Res. 1992;20:4205–4211. doi: 10.1093/nar/20.16.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letrou-Bonneval E, Chevre R, Lambert O, Costet P, Andre C, Tellier C, Pitard B. Galactosylated multimodular lipoplexes for specific gene transfer into primary hepatocytes. J Gene Med. 2008;10:1198–1209. doi: 10.1002/jgm.1212. [DOI] [PubMed] [Google Scholar]
- 41.Mukthavaram R, Marepally S, Venkata MY, Vegi GN, Sistla R, Chaudhuri A. Cationic glycolipids with cyclic and open galactose head groups for the selective targeting of genes to mouse liver. Biomaterials. 2009;30:2369–2384. doi: 10.1016/j.biomaterials.2008.12.074. [DOI] [PubMed] [Google Scholar]
- 42.Gardmo C, Kotokorpi P, Helander H, Mode A. Transfection of adult primary rat hepatocytes in culture. Biochem Pharmacol. 2005;69:1805–1813. doi: 10.1016/j.bcp.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Park Y, Man S, Liu D, Rice KG. Cross-linked low molecular weight glycopeptide-mediated gene delivery: relationship between DNA metabolic stability and the level of transient gene expression in vivo. J Pharm Sci. 2001;90:2010–2022. doi: 10.1002/jps.1152. [DOI] [PubMed] [Google Scholar]
- 44.Collard WT, Yang Y, Kwok KY, Park Y, Rice KG. Biodistribution, metabolism, and in vivo gene expression of low molecular weight glycopeptide polyethylene glycol peptide DNA co-condensates. J Pharm Sci. 2000;89:499–512. doi: 10.1002/(SICI)1520-6017(200004)89:4<499::AID-JPS7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 45.Olton D, Li J, Wilson ME, Rogers T, Close J, Huang L, Kumta PN, Sfeir C. Nanostructured calcium phosphates (NanoCaPs) for non-viral gene delivery: Influence of the synthesis parameters on transfection efficiency. Biomaterials. 2007;28:1267–1279. doi: 10.1016/j.biomaterials.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 46.Fasbender A, Zabner J, Zeiher BG, Welsh MJ. A low rate of cell proliferation and reduced DNA uptake limit cationic lipid-mediated gene transfer to primary cultures of ciliated human airway epithelia. Gene therapy. 1997;4:1173–1180. doi: 10.1038/sj.gt.3300524. [DOI] [PubMed] [Google Scholar]
- 47.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Cont Rel. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]