Abstract
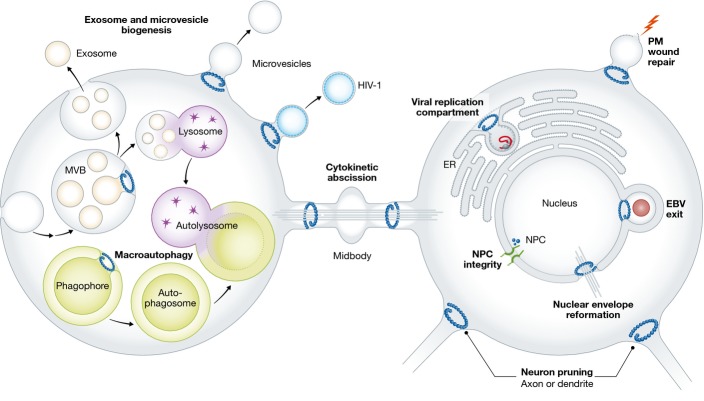
The ESCRT proteins are an ancient system that buds membranes and severs membrane necks from their inner face. Three “classical” functions of the ESCRTs have dominated research into these proteins since their discovery in 2001: the biogenesis of multivesicular bodies in endolysosomal sorting; the budding of HIV-1 and other viruses from the plasma membrane of infected cells; and the membrane abscission step in cytokinesis. The past few years have seen an explosion of novel functions: the biogenesis of microvesicles and exosomes; plasma membrane wound repair; neuron pruning; extraction of defective nuclear pore complexes; nuclear envelope reformation; plus-stranded RNA virus replication compartment formation; and micro- and macroautophagy. Most, and perhaps all, of the functions involve the conserved membrane-neck-directed activities of the ESCRTs, revealing a remarkably widespread role for this machinery through a broad swath of cell biology.
Keywords: exosome, exovesicle, nuclear envelope reformation, plasma membrane wound repair, shedding microvesicle
Introduction
The endosomal sorting complex required for transport (ESCRT) complexes comprise an ancient system for membrane remodeling and scission. The ESCRTs were first discovered and named for their central role in sorting membrane proteins from endosomes to lysosomes (Katzmann et al, 2001). The formation of multivesicular bodies (MVBs) is a key step in this pathway, and the ESCRT proteins comprise the main machinery of MVB biogenesis (Hanson & Cashikar, 2012). MVBs are formed from endosomes by the inward budding of the limiting membrane into the lumen, followed by the severing of the narrow neck connecting the bud and the limiting membrane (Fig1). Ubiquitinated membrane proteins are sorted into these buds by the ESCRT proteins. The severing event results in the release of cargo-laden intralumenal vesicles (ILVs) into the interior of the MVB.
Figure 1.
Overview of the cellular functions of the ESCRTs
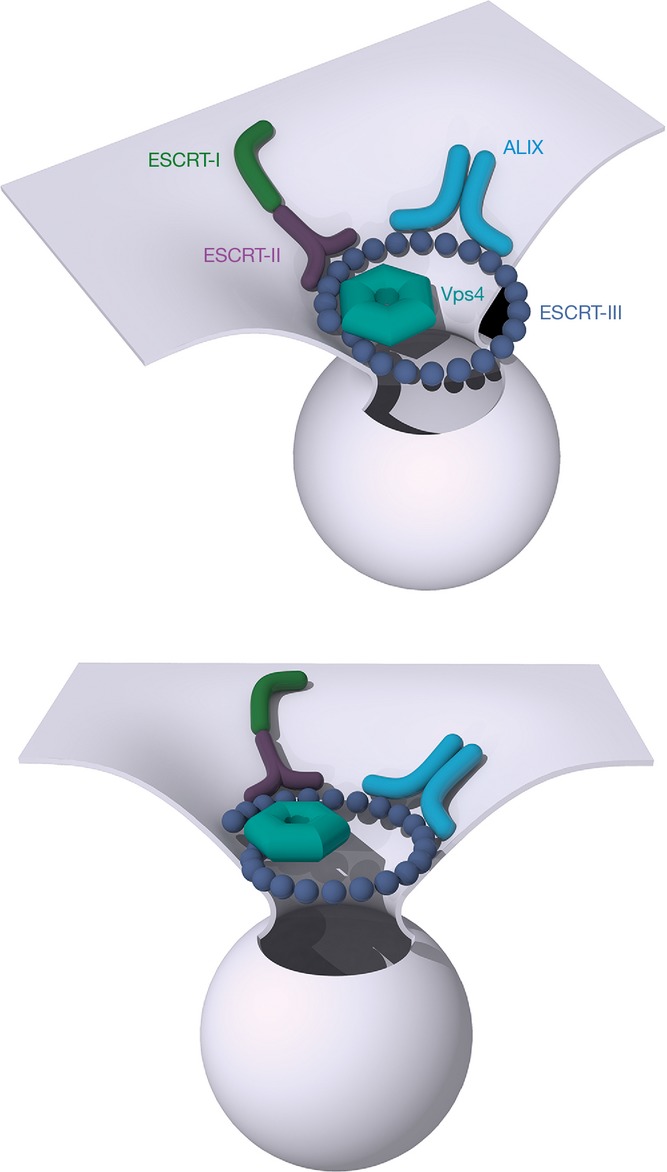
The ESCRTs first emerged in the Archaea, and their earliest function was in cell division (Lindas et al, 2008; Samson et al, 2008). The Crenarchaeota orthologs of ESCRT-III subunits and the AAA+ ATPase Vps4 sever the narrow membrane neck connecting daughter cells, a role shared from the Crenarchaea to Schizosaccharomyces pombe to mammals (Samson & Bell, 2009) (Table1). Thus, the primal biochemical function of the ESCRTs is to constrict and sever narrow membrane necks (although the biophysical mechanism of membrane scission remains unknown) (Hurley & Hanson, 2010; Peel et al, 2011; Henne et al, 2013; McCullough et al, 2013). The additional biochemical activities of the ESCRTs, such as membrane budding and the recruitment of ubiquitinated membrane proteins, are later elaborations (Samson & Bell, 2009; Wideman et al, 2014). These ubiquitin-associated activities are directed mainly by the ESCRT-I and ESCRT-II complexes and the ESCRT-associated protein ALIX. In yeast and animals, ubiquitinated cargo is organized into a flat clathrin-coated domain by ESCRT-0 prior to encountering ESCRT-I (Raiborg et al, 2001; Sachse et al, 2002). As an early endosomal clathrin adaptor and a later arrival in evolution (Wideman et al, 2014), ESCRT-0, in spite of its name, is not a core component of the membrane budding and scission machinery. The core machinery consists of ESCRT-I and ESCRT-II on the one hand, and ALIX on the other. ESCRT-I/ESCRT-II and ALIX function as two branches of the ESCRT pathway that feed into a common ESCRT-III and Vps4 membrane scission machinery (Fig2). The structures of these proteins have been exhaustively studied and are well described elsewhere (Williams & Urbe, 2007; Hurley, 2010).
Table 1.
Usage of ESCRT complexes in diverse cellular functions
| MVB | HIV-1 release | Cytokinesis | Exosome | Microvesicle | PM wound repair | Neuron pruning | EBV budding | NPC QC | NE reformation | VRRC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Targeting element | ESCRT-0 Ub | Gag p6 | CEP55 | Syntenin | ARRDC1 Ub | ALG-2/Ca2+ | ? | BFRF1 | Heh2 | UFD1 | TBSV-p33 BMV-1a |
| Upstream ESCRT | ESCRT-I | ESCRT-I ALIX | ESCRT-I ALIX | ALIX | ESCRT-I | ALIX | ESCRT-I | ALIX | None | None | ESCRT-I ALIX |
| Bridging ESCRT | ESCRT-II | ESCRT-II? | ESCRT-II? | Not needed | ? | Not needed | HD-PTP | Not needed | None | None | |
| Special ESCRT-III subunits | IST1 | CHMP7, IST1 | |||||||||
| Spastin | ✓ | ? | ✓ |
All of the pathways shown involve CHMP4/Snf7 and Vps4. Endosomal microautophagy mirrors the MVB pathway, and macroautophagy is not included due to insufficient data. The table emphasizes generalizations of typical situations that are mostly drawn from animal cells. Entries for nuclear pore complex quality control (NPC QC) and viral RNA replication compartment (VRRC) formation pertain to yeast. Requirements may vary in yeast, plants, Archaea, and in various special cases in animals. Plasma membrane budding of many animal viruses other than HIV-1 is also ESCRT dependent, but details of ESCRT usage vary.
Figure 2.
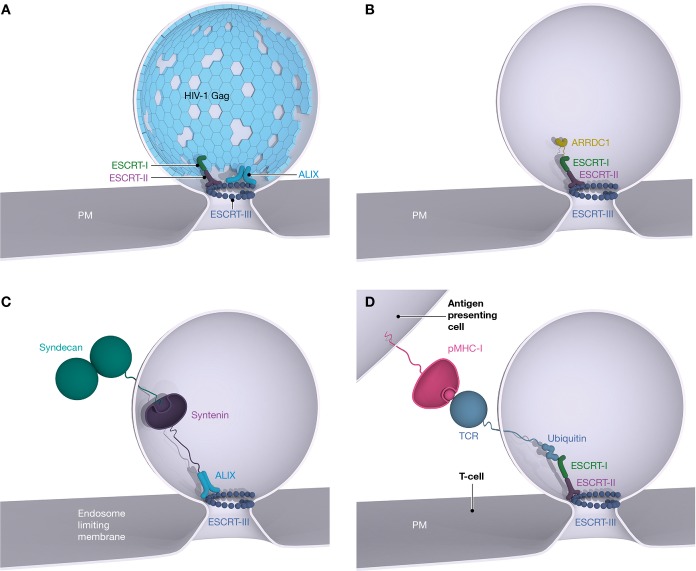
Two branches of the ESCRT pathway
The ESCRT-I/ESCRT-II and ALIX branches of the ESCRT pathway are illustrated in the context of a generic vesicle bud neck. The upstream ESCRTs (I/II and ALIX) are shown on the outer face of the bud neck with respect to ESCRT-III; however, the actual nanoscale arrangement of the molecules is unknown.
The modular organization of the ESCRT system and its versatility has made it an evolutionary success story. The initial discovery of ESCRT function in MVB biogenesis (Katzmann et al, 2001) was quickly followed by the realization that HIV and other viruses hijack the ESCRTs to bud from the plasma membrane of infected cells with the same membrane topology (Garrus et al, 2001; Martin-Serrano et al, 2001; VerPlank et al, 2001; Demirov et al, 2002) (Fig3A). Six years later, this was followed by the discovery that the ESCRTs function in cell division in mammals (Carlton & Martin-Serrano, 2007) and in a subset of Archaea (Lindas et al, 2008; Samson et al, 2008) (Fig4A). ESCRT-I and ESCRT-II are nearly as ancient as ESCRT-III and Vps4, as they are present in the genomes of the Lokiarchaeota, a proposed novel phylum of archaebacteria with eukaryote-like membrane trafficking processes (Spang et al, 2015).
Figure 3.
Viral and vesicular budding by the ESCRTs
(A) Budding of HIV-1 from the plasma membrane of an infected human cell uses both ESCRT-I and ALIX. ESCRT-II is shown despite a lack of genetic evidence for its role in HIV-1 budding, because it is capable of bridging ESCRT-I and ESCRT-III in vitro, and a bridging factor of some kind is necessary. (B) ARRDC1 mediates microvesicle budding through the ESCRT-I pathway. (C) Syntenin mediates ALIX recruitment in the biogenesis of syndecan-containing exosomes. (D) T-cell receptor-containing microvesicles are shed into the immunological synapse in an ESCRT-I-dependent process. Vps4 is required in all of these processes but not shown.
Figure 4.
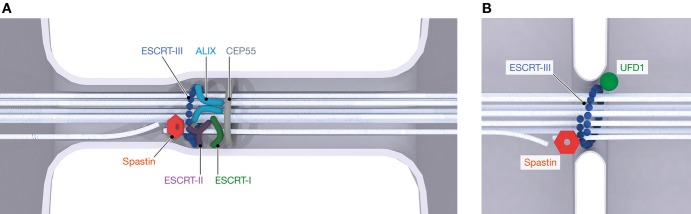
Membrane neck scission by ESCRTs in cell division
(A) The classical cytokinetic function of both the ESCRT-I/ESCRT-II and ALIX branches of the pathway in membrane abscission, and coordination with microtubule severing by spastin. (B) Resolution of gaps between fragments of reforming nuclear membrane in telophase, again coordinated with microtubule severing. Vps4 is required in both pathways but not shown.
It is deeply satisfying that all three of these pathways involve membrane neck severing with a common topology. These three cellular functions of the ESCRTs have been intensively studied and by now could be considered the “classical” functions of the ESCRTs. In the past several years, the classical repertoire of ESCRT functions has been surpassed by new roles. In this review, I take stock of the new additions to the list and place them in the context of the classical functions. I consider to what extent the new functions fit with the established mechanistic paradigms and how they highlight both the strengths and deficiencies of current tools and concepts in the field.
Diagnosing the need for an ESCRT
Saccharomyces cerevisiae strains deleted in core ESCRT genes are viable. This makes Baker’s yeast the most powerful model system for studying the classical role of ESCRTs in MVB biogenesis. Dysfunction in MVB biogenesis in yeast is readily scored using model cargoes such as carboxypeptidase S (CPS). Other model organisms, such as Drosophila melanogaster (Thompson et al, 2005; Vaccari & Bilder, 2005; Herz et al, 2006) and Arabidopsis thaliana (Spitzer et al, 2006), are tractable for ESCRT genetics and have made important contributions to understanding the dependence or independence of various pathways on the ESCRTs. The loss of core ESCRT genes, such as TSG101, VPS25, and CHMP5 which encode subunits of ESCRT-I, ESCRT-II, and ESCRT-III, respectively (see Table2 for subunit compositions), is embryonically lethal in mice (Wagner et al, 2003; Shim et al, 2006; Handschuh et al, 2014). Mouse hypomorphs such as the ESCRT-II mutant VPS25ENU are viable, but have striking developmental defects (Handschuh et al, 2014). Cells derived from hypomorphic mice promise to be useful in the future. CRISPR-Cas9-edited A549 cells genomically deleted in TSG101 are viable (Sanyal et al, 2013). Apart from this report, it is remarkable how few studies to date have used gene-edited mammalian cells in ESCRT research.
Table 2.
Composition of the ESCRT complexes
| Complex | Yeast protein | Alternative names (yeast) | Metazoan protein | Alternative names (metazoan) | Motifs and domains |
|---|---|---|---|---|---|
| ESCRT-I | Vps23 | Stp22 | TSG101 | UEV, PRD, stalk, headpiece | |
| Vps28 | VPS28 | Headpiece, Vps28 CTD | |||
| Vps37 | Srn2 | VPS37A, B, C, D | Basic helix, stalk, headpiece | ||
| Mvb12 | MVB12A, B, UBAP1 | Stalk, MAPB | |||
| ESCRT-II | Vps22 | Snf8 | EAP30 | Basic helix, Winged-helix | |
| Vps25 | EAP20 | Winged-helix | |||
| Vps36 | EAP45 | Winged-helix, GLUE, NZF1 (yeast), NZF2 (yeast) | |||
| ESCRT-III | Vps2 | Did4 Chm2 | CHMP2A, B | MIM1 | |
| Vps20 | Chm6 | CHMP6 | MIM2 | ||
| Vps24 | CHMP3 | Weak MIM1 | |||
| Snf7 | Vps32 | CHMP4A, B, C | Shrub | Weak MIM2 | |
| Vps60 | Chm5 | CHMP5 | |||
| Did2 | Chm1 Vps46 | CHMP1A, B | MIM1 | ||
| CHMP7 | |||||
| Ist1 | IST1 | MIM1, MIM2 | |||
| Vps4-Vta1 | Vps4 | VPS4A, B | SKD1 | MIT, AAA | |
| Vta1 | VTA1 | LIP5 | MIT | ||
| Bro1/ALIX | Bro1 | Vps31 | ALIX | AIP1 | Bro1, V, PRD |
UEV, ubiquitin E2 variant, a ubiquitin-binding domain with no catalytic activity; PRD, proline-rich domain; CTD, C-terminal domain; MAPB, MVB12-associated β-prism domain; GLUE, GRAM-like ubiquitin binding in EAP45, a variant PH domain that also binds to PI(3)P; NZF, neural zinc finger domain; MIM, MIT-interacting motif; MIT, microtubule interacting and trafficking; AAA, ATPases associated with various cellular activities; Bro1, N-terminal CHMP4-binding domain of Bro1, ALIX, and related proteins; V, V-shaped ubiquitin and signal binding central domain of Bro1 and ALIX.
In the great majority of studies into ESCRT function in mammalian cells, RNA interference (RNAi) has been the tool of choice. Cultured cells have been shown to survive knockdown of even multiple core ESCRT genes (Morita et al, 2011). A loss of function upon ESCRT knockdown is the closest thing to a “gold standard” for the dependence of a mammalian cell process on the ESCRTs. The converse does not hold, however. The absence of a loss-of-function phenotype on knockdown is not necessarily robust evidence for the ESCRT independence of a pathway, given variability in the efficiency and duration of the loss of the expressed protein. In some cases, RNAi knockdowns have had no phenotype even when other lines of evidence have suggested an important role for ESCRTs. For example, knockdown of ESCRT-II subunits has no effect on HIV-1 release (Morita et al, 2011) and on mammalian cytokinesis (Morita et al, 2007). Biochemical reconstitution (Carlson & Hurley, 2012), imaging (Goliand et al, 2014), and dominant-negative studies (Goliand et al, 2014) support a functional role. In this instance, where negative findings from RNAi studies differ from those of other approaches, the field has yet to reach a consensus.
Of course, none of the foregoing experiments, on their own, differentiate between direct contributions to the step under study and indirect contributions through perturbations of established functions. Identification and knockdown of specific adaptors that distinguish unique aspects of ESCRT function are invaluable in this regard. In HIV-1 release, the p6 domain of Gag has this function (Garrus et al, 2001; Martin-Serrano et al, 2001; VerPlank et al, 2001; Demirov et al, 2002), and in mammalian cytokinesis, CEP55 has this role (Carlton & Martin-Serrano, 2007; Morita et al, 2007; Lee et al, 2008). Visualization of ESCRTs at the site of action is also an important criterion for a direct role.
There are no pharmacological inhibitors available for the ESCRT pathway, although overexpression of Walker EQ mutant VPS4 (Babst et al, 1998) is widely used and usually effective. Overexpression of an ALIX mutant defective in CHMP4 binding (Kim et al, 2005; Fisher et al, 2007; Usami et al, 2007) is a popular and effective tool for diagnosing a role for the ALIX-CHMP4 branch of the ESCRT pathway. Overexpression of a fragment of ESCRT-II subunit VPS25 involved in binding to ESCRT-III inhibits cytokinesis (Im et al, 2009; Goliand et al, 2014). Overexpression of GFP-tagged ESCRT-III subunits is also inhibitory, although this is typically considered something to be avoided in imaging studies, rather than actively pursued for functional inhibition. Biochemical reconstitution addresses what components are minimally sufficient rather than what is essential, but can be used to address what is essential in the context of a minimal system. Going forward, spatially (Loncle et al, 2015) or temporally (Matusek et al, 2014) restricted deletions of essential ESCRT genes will be helpful. In weighing the literature retrospectively, and in planning strategies to move ahead, it is important to bear these issues in mind.
ESCRTs, exosomes, and microvesicles
The terms exovesicle and extracellular vesicle refer to any biological vesicle extant outside of a cell, regardless of its origin (Raposo & Stoorvogel, 2013). Here I will use the term microvesicle to indicate extracellular vesicles that directly bud from the plasma membrane. “Microvesicle” is synonymous with “ectosome”, “shedding vesicle”, and “microparticle”. In contrast, exosomes originate as ILVs within MVBs that fuse with the plasma membrane. MVBs are also referred to as multivesicular endosomes (MVEs).
The concept of an ESCRT role in exosome biogenesis is not new and seems natural given that exosomes originate in MVBs and that ESCRTs comprise the major machinery for MVB biogenesis. However, several prominent studies employing dominant-negative VPS4 (Trajkovic et al, 2008) and knockdowns (van Niel et al, 2011) reported negative or mixed findings with respect to ESCRT requirements in exosome biogenesis. The tetraspanin CD63 was consistently observed in exosomes, suggesting that tetraspanins might comprise a separate mechanism for the biogenesis of MVBs destined for fusion with the plasma membrane and exosome release (van Niel et al, 2011). Evidence began to emerge in 2011–2012 that ESCRTs play a direct role in exovesicle biogenesis. The first suggestion came from the observation that a peptide from the SP2 region of HIV-1 Gag inhibited both exovesicle release and VPS4B association with exovesicle cargoes (Gan & Gould, 2011). In C. elegans, loss of the lipid flippase TAT-5 leads to a buildup of phosphatidylethanolamine on the outside of the plasma membrane, which triggers extensive microvesicle shedding (Wehman et al, 2011). Increased shedding correlates with increased ESCRT localization at the PM, which is further increased by depletion of Vps4. RNAi against ESCRT-0 and ESCRT-I subunits partially suppresses shedding, but depletion of the key ESCRT-II and ESCRT-III subunits has no effect, leaving a mixed picture (Wehman et al, 2011). In a contemporaneous effort, the ubiquitin ligase adaptor ARRDC1 was shown to directly recruit ESCRT-I to microvesicle budding sites via its PSAP motif (Fig3B). The TSG101 knockdown, VPS4-DN expression, and PSAP mutant were shown to have robust phenotypes (Nabhan et al, 2012), strengthening the case for an ESCRT-microvesicle connection.
Syndecans are the major heparan sulfate-presenting proteins on the cell surface, and they are also found in exosomes. Syndecan-positive exosomes contain CD63 and ceramide and thus qualify as “canonical” exosomes. Syndecans are sorted into exosomes by a specific adaptor protein, syntenin. Syntenin uses a YPXL motif to bind specifically to ALIX (Baietti et al, 2012; Hurley & Odorizzi, 2012) (Fig3C). Downstream of ALIX, CHMP4, and VPS4 knockdowns block syndecan exosome biogenesis (Baietti et al, 2012). Knockdown of CD63 does not block the biogenesis of these exosomes, but the ESCRT knockdowns were shown to decrease the amount of CD63 released (Baietti et al, 2012). This portrays CD63 as a fellow traveler with syndecan rather than a core component of the exosome release pathway. The Cos transmembrane proteins of yeast have been proposed to serve as the yeast counterpart of mammalian tetraspanins (MacDonald et al, 2015). Ubiquitinated Cos proteins provide a sorting signal in trans to non-ubiquitinated cargo destined for ILVs and so facilitate their sorting by the ESCRTs (MacDonald et al, 2015). The concept that ESCRTs and CD63 could work together by similar means is attractive.
The use of temporally controllable RNAi targeted to Hedgehog (Hh)-producing cells made it possible to explore the role of ESCRTs in long range Hh signaling in wing development in Drosophila (Matusek et al, 2014). Knockdown of ALIX and subunits of ESCRT-I, ESCRT-II, and ESCRT-III block the Hh signal (Matusek et al, 2014). Hh is secreted from cells in exovesicles that contain ˜160 other proteins, including several of the ESCRTs (Matusek et al, 2014). These exovesicles appear to be shed directly from the plasma membrane. At the immunological synapse, T-cell receptors (TCR) form complexes with pathogen-bound major histocompatibility complexes (pMHC) on antigen-presenting cells. These complexes are immobilized when the TCR-containing microvesicles are secreted into the synaptic center. This process was convincingly shown to be dependent on ESCRT-I and VPS4 (Choudhuri et al, 2014) using RNAi and dominant-negative approaches. The process is closely analogous to the release of HIV-1 virions from the plasma membrane of infected T cells (Fig3D), and it is natural to infer that HIV-1 co-opted its use of the ESCRT from the TCR microvesicle budding mechanism (Choudhuri et al, 2014). To sum up, the sustained findings in the past few years of multiple examples of ESCRT-dependent exosome and microvesicle biogenesis in diverse species and contexts suggest that the ESCRTs have a ubiquitous and fundamental role in these processes.
ESCRTs and plasma membrane wound repair
When eukaryotic cells are perforated in their plasma membranes, they react to patch the holes within a matter of seconds (Andrews et al, 2014). The increase in [Ca2+] at the site of the wound signals the need for repair. Holes can be created experimentally by detergents, pore-forming toxins, mechanical injury, or a laser. In every one of these cases, ALIX, ESCRT-III, and VPS4 are recruited subsequent to the Ca2+ signal (Jimenez et al, 2014). The central ESCRT-III subunit CHMP4B is particularly important, and cell survival following wounding is compromised by its knockdown. The damaged membrane is shed outward from the plasma membrane in much the same manner as in microvesicle budding (Jimenez et al, 2014).
The initial recruitment of ESCRTs to wound sites is rapid and energy independent and thus does not require ubiquitination (Jimenez et al, 2014). Consistent with the initial ubiquitin independence of the pathway, and the key role of Ca2+, the ESCRTs are recruited via the EF hand protein ALG-2 (Scheffer et al, 2014). In its Ca2+-bound state, ALG-2 directly interacts with ALIX. ALG-2 appears to be the unique ESCRT-recruitment element in plasma membrane wound repair (Scheffer et al, 2014), analogous to CEP55 in cytokinesis, the Gag p6 domain in HIV-1 budding, or the syntenin YPXL motif in syndecan exosome biogenesis (Table1).
ESCRTs and neuron pruning
Early in their development, neurons generate a profusion of branches. These branches, both dendrites and axons, are subject to shortening. Extending the branch metaphor, this process is referred to as “pruning”. Drosophila has been a model system of choice for the study of neuron pruning. Three different laboratories have now shown that the ESCRT machinery is deeply involved in this process in Drosophila. In the first of these studies to appear, RNAi against ESCRT-I (Vps28) and ESCRT-III (Shrub, the Drosophila CHMP4 ortholog) and dominant-negative Vps4 blocked dendrite pruning. The effect was attributed to a block in the downregulation of the cell surface protein neuroglian (Nrg). It was postulated the Nrg inhibits dendrite pruning, which is relieved when Nrg is endocytosed and downregulated via the MVB pathway and lysosomal degradation (Zhang et al, 2014). This was followed by a report that ESCRT-0 is required for axon pruning (Issman-Zecharya & Schuldiner, 2014). In this case, the finding is that another cell surface receptor, Patched (Ptc), is not downregulated normally when ESCRT function is lost, leading to sustained inhibition of pruning. To the extent that the receptor downregulation mediates this effect, this is not a “new” function of the ESCRTs, rather a new instance of the ESCRTs’ classical role in the MVB pathway.
The putative roles of Nrg and Ptc as pruning inhibitors beg the question as to the molecular mechanism of pruning itself. A third study suggested that the ESCRTs are directly involved in severing the membrane necks of both axons and dendrites (Loncle et al, 2015). This function of the ESCRTs requires ESCRT-I and ESCRT-III, but not ESCRT-II or ALIX. In this instance, the ESCRT-II independence is persuasive, as homozygous deletion of vps25 in the context of mosaic analysis with a repressible cell marker (MARCM) recapitulates the RNAi phenotype. It is unusual for an ESCRT-III-dependent process to require neither ESCRT-II nor ALIX. ALIX binds to CHMP4 through its Bro1 domain; however, several other Bro1-domain-containing proteins exist in animals. One of these is HD-PTP, which is required for neuron pruning and whose function requires an intact CHMP4-binding Bro1 domain. In this study, it was possible to partially separate the roles of the ESCRTs in MVB biogenesis from their specialized role in neuron pruning on the basis of the differential requirement for ESCRT-II. By doing so, an essential non-MVB role for ESCRTs was demonstrated. Imaging of ESCRT localization shows concentrations of ESCRT-III at sites in dendrites that go on to become sites of severing. At present, it is unclear what upstream signal recruits and activates the ESCRTs at scission sites. It is also unclear whether Ptc or Nrg is involved in negatively regulating this aspect of ESCRT function, which if so would represent a double-negative feedback loop. What is clear is that membrane scission of dendrites and axons represents another important addition to the list of genuinely new functions for the ESCRTs (Loncle et al, 2015).
ESCRTs and the nucleus
In 2001, two back-to-back papers from Hollenberg and co-workers identified CHMP1, respectively, as both a cytoplasmic vesicle trafficking protein (Howard et al, 2001) and a nuclear matrix protein (Stauffer et al, 2001). Given its ambiguous nature, it was named chromatin modifying protein/charged multivesicular body protein-1. In the subsequent decade, the cytosolic membrane-remodeling functions of CHMP1 and many other CHMPs were thoroughly explored. While no evidence has emerged for a putative chromatin remodeling function for CHMP1, other functions of CHMP1 in the nucleus have come to the fore. Evidence for nuclear roles for the ESCRTs began to emerge in 2012, with virological studies leading the way. Epstein–Barr virus (EBV) assembles in the nucleus and is too large for export through the nuclear pore complex (NPC). Instead, EBV exits the nucleus by budding through the nuclear envelope. ESCRTs were shown to be required for the EBV budding through the nuclear membrane (Lee et al, 2012). The BFRF1 protein of EBV recruits ALIX, which in turn recruits CHMP4B.
It took until 2014 for physiological (as opposed to pathophysiological) functions to emerge for nuclear ESCRTs. In budding yeast, the nuclear envelope does not break down during the cell cycle. NPCs are long-lived and their quality control is important. ESCRT-III subunits and Vps4 were identified as factors important for NPC integrity in an epistatic screen in yeast (Webster et al, 2014). None of the upstream ESCRTs are involved in the pathway. Rather, the nuclear inner membrane protein and NPC quality control factor Heh2 appear to directly recruit ESCRT-III. The molecular mechanism for the detection and removal of flawed NPCs remains uncertain. One attractive possibility is that Vps4 might extract subunits of misassembled NPCs. Indeed, Vps4 is a disassembly machine that completely unfolds its substrates (Yang et al, 2015), like other AAA+ ATPases involved in disaggregation and degradation. In this scenario, ESCRT-III would serve as an adaptor for Vps4 rather than as a membrane-remodeling machine.
Finally, 2015 brought compelling evidence that nuclear ESCRTs have a physiological role in membrane remodeling. In animals, the nuclear envelope breaks down and must be reformed in every round of cell division. The mechanism by which nuclear envelope fragments are resealed has been unknown. Resealing entails the same changes in membrane topology that have become familiar from other ESCRT reactions. Resealing occurs during telophase. Armed with the hypothesis that ESCRTs might be involved, two groups imaged ESCRT-III localization in telophase (Olmos et al, 2015; Vietri et al, 2015). As with NPC quality control, upstream ESCRT proteins do not appear to be required. Rather, UFD1, a subunit of the p97 complex, seems able to directly recruit ESCRT-III to sites of sealing (Olmos et al, 2015). The “orphan” ESCRT-III protein CHMP7 is required for the process, giving it a job to do for the first time (Vietri et al, 2015). Nuclear envelope formation requires the severing of spindle microtubules that would otherwise physically obstruct membrane sealing (Fig4B). A similar problem needs to be solved in cytokinesis, where the central spindle must be severed before the membrane neck surrounding it can be sealed. The two ESCRT-III subunits CHMP1B and IST1 bind with high affinity to the microtubule-severing enzyme spastin (Yang et al, 2008; Yu et al, 2008; Connell et al, 2009). In cytokinesis, CHMP1B recruits the microtubule-severing enzyme spastin such that microtubule and membrane neck severing are coordinated at the same site by an elegant mechanism (Yang et al, 2008). In the case of nuclear envelope reformation, CHMP7 recruits IST1 to the same end (Vietri et al, 2015). The parallels to the mechanism of membrane abscission in cytokinesis, and the consistency with the biochemistry and biophysics of these proteins, are truly satisfying. The reconciliation of the apparently separate functions of ESCRTs in membrane remodeling and in the nucleus is equally satisfying.
ESCRTs and viral replication compartments
Plus-stranded RNA viruses such as the tombusviruses and bromoviruses of plants replicate in compartments that are protected by a limiting membrane. Tomato bushy stunt virus (TBSV), which can also replicate in yeast, uses its p33 protein to recruit ESCRT-I and Bro1 (yeast ALIX), and in turn, ESCRT-III (Barajas et al, 2009). The ESCRTs are involved in the budding of the viral replication complex into a membrane-delimited vesicular compartment. The bromovirus brome mosaic virus (BMV) also uses ESCRT-III to build itself a protected replication compartment (Diaz et al, 2015). The 1a protein of BMV recruits the ESCRT-III protein Snf7 directly (Diaz et al, 2015), without the apparent need for upstream factors. In contrast to the situation with retroviral budding from the plasma membrane, these budded replication compartment are not actually severed from the limiting membrane. Since they use much of the same machinery, what prevents membrane scission here is an interesting question.
ESCRTs and autophagy
The term autophagy refers to any of several cellular self-consumptive processes, macroautophagy, microautophagy, and chaperone-mediated autophagy. These processes are essential both for cell survival during starvation and for the clearance of a wide variety of unnecessary or harmful materials from the cell. In microautophagy, cytosolic materials are directly taken up into the lysosome. In the endosomal version of this, material is taken up in intralumenal vesicles of MVBs, which subsequently fuse with the lysosome. Since the ESCRTs are so central to ILV formation and MVB biogenesis, it perhaps should not be surprising that they are also required for endosomal microautophagy (Sahu et al, 2011). The chaperone Hsc70 appears to act as an adaptor to select certain cytosolic protein and recruit them to the late endosomal limiting membrane for internalization by ESCRTs (Sahu et al, 2011). This role for the ESCRTs could be considered a new use of their classical function in MVB biogenesis.
The term “autophagy” is often used as a synonym for “macroautophagy”, the uptake of bulk cytosol or large subcellular structures by a double-membrane phagophore that grows and engulfs its substrates. “Macroautophagy” will be used here since this article also concerns microautophagy. It has been known for some time that the ESCRTs are required for macroautophagy (Lee et al, 2007; Rusten et al, 2007). It is becoming clear that the ESCRT pathway is upregulated in advance of and in coordination with the larger macroautophagic response to starvation (Jones et al, 2012; Mueller et al, 2015). What has been much less certain is whether the ESCRTs have a direct role in the remodeling of the phagophore membrane as it grows and closes around its cargo. In principle, the macroautophagic phenotypes of ESCRT mutants, as reported through 2014, could be accounted for by their essential but indirect roles in the MVB pathway (Filimonenko et al, 2007). This has remained the prevailing model in the field, in part because it is essentially impossible to distinguish closed autophagosomes from nearly closed ones by electron microscopy. In mammals, incorporation of the SNARE protein syntaxin-17 into autophagosomes is important for their fusion with lysosomes, and syntaxin-17 incorporation seems to depend upon their closure (Itakura et al, 2012). Thus, failure to close late in autophagosome biogenesis would presumably appear as a defect in fusion with the lysosome.
The prevailing view notwithstanding, the topology of the closure of the phagophore double membrane should make it, in theory, an ideal substrate for sealing by the ESCRTs. Now, incisive and exciting imaging studies of chmp1 Arabidopsis show large and abundant unclosed phagophores (Spitzer et al, 2015). Ultimately, autophagic cargo does reach its destination in chmp1 cells, but the process is substantially delayed. If these images are the smoking gun, the eyewitness account of the crime has yet to be produced: direct imaging of ESCRTs localized at shrinking rim of the autophagic cup at the moment before closure. This unique and transient event may be extraordinarily difficult to capture. In the absence of this final piece of evidence, the preponderance of the data now favor a direct role in phagophore closure, at least for ESCRT-III, and at least for plastid autophagy in plants. Given the conservation of the ESCRT system and its function, though, it would be surprising if this role were not more widespread.
Concluding remarks
The last few years has seen in quiet revolution in ESCRT cell biology. The new functions discovered or confirmed outnumber the classical function several times over. It is now hard to think of any example of “reverse” topology membrane budding and scission in eukaryotes (or in some Archaea) that does not involve the ESCRTs. Many of the most interesting discoveries are being made by cell and developmental biologists who are new to the ESCRT field—more evidence that the ESCRTs have gone mainstream in cell biology. It is hard to know how much farther the field can go in finding new functions. The frontier questions now may not be so much what do the ESCRTs do or why, but rather how they do what they do.
Acknowledgments
I thank A. Johnson for making the figures and the members of my laboratory for many stimulating discussions. Work on the ESCRTs in the Hurley laboratory is supported by National Institutes of Health grant R01AI112442.
Conflict of interest
The author declares that he has no conflict of interest.
References
- Andrews NW, Almeida PE, Corrotte M. Damage control: cellular mechanisms of plasma membrane repair. Trends Cell Biol. 2014;24:734–742. doi: 10.1016/j.tcb.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Barajas D, Jiang Y, Nagy PD. A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog. 2009;5:e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the ESCRT machinery at membrane-bound HIV-1 gag clusters. Proc Natl Acad Sci USA. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Choudhuri K, Llodra J, Roth EW, Tsai J, Gordo S, Wucherpfennig KW, Kam LC, Stokes DL, Dustin ML. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Zhang JT, Ollwerther A, Wang XF, Ahlquist P. Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 2015;11:e1004742. doi: 10.1371/journal.ppat.1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Gan X, Gould SJ. Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol Biol Cell. 2011;22:817–830. doi: 10.1091/mbc.E10-07-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Goliand I, Nachmias D, Gershony O, Elia N. Inhibition of ESCRT-II-CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol Biol Cell. 2014;25:3740–3748. doi: 10.1091/mbc.E14-08-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschuh K, Feenstra J, Koss M, Ferretti E, Risolino M, Zewdu R, Sahai MA, Benazet JD, Peng XP, Depew MJ, Quintana L, Sharpe J, Wang BL, Alcorn H, Rivi R, Butcher S, Manak JR, Vaccari T, Weinstein H, Anderson KV, et al. ESCRT-II/Vps25 constrains digit number by endosome-mediated selective modulation of FGF-SHH signaling. Cell Rep. 2014;9:674–687. doi: 10.1016/j.celrep.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5:a016766. doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TL, Stauffer DR, Degnin CR, Hollenberg SM. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT complexes: It’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Odorizzi G. Get on the exosome bus with ALIX. Nat Cell Biol. 2012;14:654–655. doi: 10.1038/ncb2530. [DOI] [PubMed] [Google Scholar]
- Im YJ, Wollert T, Boura E, Hurley JH. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev Cell. 2009;17:234–243. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issman-Zecharya N, Schuldiner O. The PI3K class III complex promotes axon pruning by downregulating a ptc-derived signal via endosome-lysosomal degradation. Dev Cell. 2014;31:461–473. doi: 10.1016/j.devcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- Jones CB, Ott EM, Keener JM, Curtiss M, Sandrin V, Babst M. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 2012;13:468–482. doi: 10.1111/j.1600-0854.2011.01314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-P, Liu P-T, Kung H-N, Su M-T, Chua H-H, Chang Y-H, Chang C-W, Tsai C-H, Liu F-T, Chen M-R. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr virus. PLoS Pathog. 2012;8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Agromayor M, Martin-Serrano J, Williams DW. An ESCRT module is required for neuron pruning. Sci Rep. 2015;5:8461. doi: 10.1038/srep08461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Payne JA, Aboian M, Smith W, Katzmann DJ, Piper RC. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev Cell. 2015;33:328–342. doi: 10.1016/j.devcel.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Matusek T, Wendler F, Poles S, Pizette S, D’Angelo G, Fuerthauer M, Therond PP. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature. 2014;516:99–103. doi: 10.1038/nature13847. [DOI] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi S, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Hamilton IB, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M, Schmidt O, Angelova M, Faserl K, Weys S, Kremser L, Pfaffenwimmer T, Dalik T, Kraft C, Trajanoski Z, Lindner H, Teis D. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife. 2015;4:e07736. doi: 10.7554/eLife.07736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Mehlum A, Stang E, Stenmark H. Hrs recruits clathrin to early endosomes. EMBO J. 2001;20:5008–5021. doi: 10.1093/emboj/20.17.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Sachse M, Urbe S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17:507–513. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Ashour J, Maruyama T, Altenburg AF, Cragnolini JJ, Bilate A, Avalos AM, Kundrat L, Garcia-Sastre A, Ploegh HL. Type I interferon imposes a TSG101/ISG15 checkpoint at the Golgi for glycoprotein trafficking during influenza virus infection. Cell Host Microbe. 2013;14:510–521. doi: 10.1016/j.chom.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer LL, Sreetama SC, Sharma N, Medikayala S, Brown KJ, Defour A, Jaiswal JK. Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat Commun. 2014;5:5646. doi: 10.1038/ncomms6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Hayden MS, Lee KY, Trombetta ES, Pypaert M, Nara A, Yoshimori T, Wilm B, Erdjument-Bromage H, Tempst P, Hogan BL, Mellman I, Ghosh S. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J Cell Biol. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jorgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Muller S, Hanisch FG, Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Spitzer C, Li FQ, Buono R, Roschzttardtz H, Chung TJ, Zhang M, Osteryoung KW, Vierstra RD, Otegui MS. The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell. 2015;27:391–402. doi: 10.1105/tpc.114.135939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating Notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. Spastin and ESCRT-III coordinates mitotic spindle disassembly and nuclear envelope resealing. Nature. 2015;522:231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Krempler A, Qi Y, Park K, Henry MD, Triplett AA, Riedlinger G, Rucker Iii EB, Hennighausen L. Tsg101 is essential for cell growth, proliferation, and cell survival of embryonic and adult tissues. MolCell Biol. 2003;23:150–162. doi: 10.1128/MCB.23.1.150-162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman JG, Leung KF, Field MC, Dacks JB. The cell biology of the endocytic system from an evolutionary perspective. Cold Spring Harb Perspect Biol. 2014;6:a016998. doi: 10.1101/cshperspect.a016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Stjepanovic G, Shen QT, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol. 2015;22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Quiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Wong JJL, Lim K-L, Liou Y-C, Wang H, Yu F. Endocytic pathways downregulate the L1-type cell adhesion molecule neuroglian to promote dendrite pruning in Drosophila. Dev Cell. 2014;30:463–478. doi: 10.1016/j.devcel.2014.06.014. [DOI] [PubMed] [Google Scholar]