Abstract
Findings from the Psychiatric Genomics Consortium genome-wide association study (GWAS) showed that variation at the DRD2 locus is associated with schizophrenia risk. However, the functional significance of rs2514218, the top DRD2 single nucleotide polymorphism in the GWAS, is unknown. Dopamine D2 receptor binding is a common mechanism of action for all antipsychotic drugs, and DRD2 variants were related to antipsychotic response in previous studies. The present study examined whether rs2514218 genotype could predict antipsychotic response, including efficacy and adverse events, in a cohort of patients with first episode of psychosis treated with either risperidone or aripiprazole for 12 weeks. Subjects were genotyped using the Illumina Infinium HumanOmniExpressExome array platform. After standard quality control, data from 100 subjects (49 randomly assigned to treatment with aripiprazole and 51 assigned to risperidone) was available for analysis. Subjects were assessed for psychotic symptomatology and medication-related adverse events weekly for 4 weeks, then biweekly for 8 weeks. Linear mixed model analysis revealed that the homozygotes for the risk (C) allele at rs2514218 had significantly greater reduction in positive symptoms during 12 weeks of treatment compared to the T allele carriers. In the aripiprazole group, C/C homozygotes also reported more akathisia than the T allele carriers, while in the risperidone group, male T allele carriers demonstrated greater prolactin elevations compared to male C/C homozygotes. These findings suggest that the schizophrenia risk variant at the DRD2 locus (or another variant in close proximity) is associated with observable differences in response to treatments which reduce striatal dopamine signaling.
Key words: antipsychotic, pharmacogenetics, DRD2
Introduction
The recent large-scale genome-wide association study (GWAS) from the Psychiatric Genomics Consortium (PGC) represents a watershed moment in schizophrenia research, revealing 108 genomic loci that are reliably associated with schizophrenia risk.1 At the same time, the PGC results raised as many questions as answers, insofar as the functional significance of the risk-associated variants remains largely uncharacterized. One of the loci that may be particularly relevant to schizophrenia treatment is DRD2,2 the gene coding for the dopamine D2 receptor, which is a binding target of all available antipsychotic drugs.3 The top single nucleotide polymorphism (SNP) reported by the PGC at this locus is rs2514218, about 47kb upstream from DRD2; this SNP has not been examined in any pharmacogenetics study to date.
While antipsychotic medications are the primary intervention for schizophrenia and related psychotic disorders,4 many patients frequently discontinue or switch drug regimens due to lack of efficacy and/or treatment-emergent side effects.5 There is currently a lack of clinically relevant, biologically validated predictors of antipsychotic treatment response, so that clinicians often rely on past medication history to determine which antipsychotic to use. However, no such information is available for patients with first-episode psychosis. Pharmacogenetics research focuses on the identification of genetic variants that predict who may optimally benefit from specific drugs6 and can provide a prognostic biomarker that is readily obtainable for any patient regardless of clinical status or history.
Previous research has found that variants in DRD2 may be predictive of antipsychotic drug response.7,8 Therefore, we examined whether rs2514218 was associated with antipsychotic drug response in a cohort of patients with first-episode psychosis. While symptomatology in psychotic disorders is multifaceted (involving positive, negative, and cognitive symptoms), D2 receptor activity of antipsychotic medications is specifically associated with reduction of positive symptoms such as hallucinations and delusions9 but is much less effective in reducing negative symptoms and largely ineffective for cognitive symptoms (see this issue).10 Moreover, a higher degree of D2 blockade is associated with greater risk of specific drug-induced adverse events such as extrapyramidal symptoms (EPS) and akathisia,11 and D2 antagonism in the tuberoinfundibular pathway is thought to underlie prolactin elevation in response to first-generation agents, as well as risperidone (but not aripiprazole).12 Consequently, we aimed to test a primary hypothesis that DRD2 variants will affect change in positive symptoms and secondary hypotheses that the same variant will also be associated with EPS and akathisia, as well as risperidone-induced prolactin elevation. By contrast, we hypothesized that the DRD2 variant would not be associated with change in general psychopathology, global functioning, or negative symptoms.
Methods
Participants
Study subjects were a subset of participants in a randomized, multisite, double-blind, clinical trial for patients with first-episode psychosis (this issue),13 including DSM-IV diagnoses of schizophrenia, schizoaffective disorder, schizophreniform disorder, and psychotic disorder Not Otherwise Specified (NOS). The sample of the clinical trial consisted of 198 subjects, but only a subset participated in the genetic study, which was added subsequent to the enrollment of the first 49 subjects, and was not available at some study sites. The present study had genetic data on 100 patients (75 men and 25 women; mean age = 21.5 years, SD = 5.1, range = 15–39) who provided written informed consent (or assent in the case of minors, with written consent from parent/guardian) for both the trial and genetic analyses. About 24% of subjects (n = 24) were antipsychotic naive; none had more than 2 weeks of lifetime antipsychotic exposure. Subjects were from 6 different sites in the greater New York city area and at sites in San Antonio, TX and Calgary, Alberta, Canada. All study procedures were approved by Institutional Review Boards (IRBs) at each of the participating sites, with the Feinstein Institute for Medical Research IRB as the coordinating center for the clinical trial and genetics studies.
Treatment
Participants were stratified by site, previous antipsychotic exposure (none vs any), and diagnosis (psychotic disorder NOS vs other eligible diagnoses) and were randomly assigned on a 1 to 1 basis to double-masked treatment with either aripiprazole (5–30mg/d) or risperidone (1–6mg/d) for 12 weeks. Study medication was packaged in identically appearing capsules at 3 different dosing levels (level 1: 5mg of aripiprazole or 1mg of risperidone; level 2: 10mg of aripiprazole or 2mg of risperidone; and level 3: 15mg of aripiprazole or 3mg of risperidone). The study allowed for prescription of up to 2 study capsules per day providing a total of 6 possible levels of milligrams of daily study medication. The initial daily dose was 1 study capsule (ie, 5mg of aripiprazole or 1mg of risperidone). Medication doses were advanced according to a titration schedule until response criteria were achieved or dose-limiting side effects occurred. Study psychiatrists had the option of advancing or slowing the titration schedule for clinical needs (eg, side effect management). Limited concomitant medications were permitted to manage side effects: benztropine for EPS; lorazepam or propranolol for akathisia; and lorazepam for agitation or anxiety. As presented in (this issue),13 there was no significant difference in treatment response rate between the 2 treatment arms for the full study cohort (n = 198).
Assessment
Raters blind to treatment condition and DRD2 genotype conducted weekly assessments during the first 4 weeks, then biweekly assessments for the subsequent 8 weeks of the trial. Psychopathology was assessed using the Brief Psychiatric Rating Scale (BPRS),14 Schedule for Assessment of Negative Symptoms (SANS),15 and Clinical Global Impression Scale (CGI-severity).16 As described in the primary trial report (this issue),13 4 of the BPRS items were used to compute the total positive symptoms score: conceptual disorganization, grandiosity, hallucinatory behavior, and unusual thought content. Four global ratings from the SANS were used to represent negative symptoms: affective flattening, alogia, avolition-apathy, and asociality-anhedonia. Interrater reliability, as measured by intraclass correlation coefficients (ICCs), were high for all 4 BPRS items: for conceptual disorganization, ICC = 0.94 (95% CI: 0.76, 0.99); for grandiosity, ICC = 0.92 (95% CI: 0.80, 0.98); for hallucinatory behavior, ICC = 0.93 (0.76, 0.99); and for unusual thought content, ICC = 0.92 (95% CI: 0.82, 0.98) and for the SANS global items affective flattening 0.75 (95% CI: 0.43, 0.93), alogia 0.66 (95% CI: 0.36, 0.89), avolition-apathy 0.69 (95% CI: 0.38, 0.91), and asociality-anhedonia 0.52 (95% CI: 0.17, 0.85). Patients’ diagnoses were confirmed with a Structured Clinical Interview of Axis I DSM-IV Disorders (SCID).
Drug-induced adverse events were measured by Simpson-Angus Scale for EPS17 and Barnes Akathisia Scale (BAS).18 An overall EPS severity score was calculated as the sum of the following Simpson-Angus EPS Scale items: gait, rigidity of major joints, tremor, akinesia, and akathisia. Akathisia was measured by the global item of the BAS. Blood draw was performed at baseline, week 4, week 8, and week 12 to measure prolactin levels.
Genotyping
Peripheral venous blood was drawn for subjects who consented to the genetic study. DNA was extracted from lymphocytes and genotyping was performed using the Illumina Infinium HumanOmniExpressExome array platform. After standard quality control, data from 100 subjects were used in subsequent analysis. For rs2514218, the C allele is the schizophrenia risk allele with a frequency of 81% worldwide.1 Due to the fact that there were only 13 homozygotes for the T allele, patients were dichotomized for primary analyses as homozygous for the common (risk) allele or as carriers of the minor allele (n = 56 for C/C and n = 44 for T allele carriers). Genotype groups (C/C vs T carriers) did not significantly differ on sex, diagnosis, and medication assignment (all P values > .20, see table 1). There were slightly more African American subjects in the C/C homozygotes than in the T carriers, but the difference was not statistically significant (χ2 = 8.89, df = 4, P = .064). To control for the presence of different continental ancestry groups in our cohort, and the consequent risk of population stratification, principal component analysis was conducted in the full Illumina dataset. Following the convention for genomic analysis, the first 5 principal components were used as covariates in subsequent analysis. Genetic analysis was conducted in Golden Helix’s SNP and Variation Suite version 8.3.4 (Golden Helix, Inc.).
Table 1.
Demographic and Clinical Data in 2 Genotype Groups of rs2514218
| C/C Homozygotes (n = 56) | T Carriers (n = 44) | P Values | |
|---|---|---|---|
| Male sex, n (%) | 41 (73.2) | 34 (77.3) | .64 |
| Age (y), mean ± SD | 21.4±5.1 | 21.8±5.0 | .69 |
| Medication (aripiprazole), n (%) | 29 (51.8) | 20 (45.5) | .53 |
| Race, n (%) | .06 | ||
| Asian | 2 (3.6) | 3 (6.8) | |
| African American | 30 (53.6) | 11 (25.0) | |
| Hispanic | 6 (10.7) | 8 (18.2) | |
| Caucasian | 15 (26.8) | 20 (45.5) | |
| Others | 3 (5.4) | 2 (4.5) | |
| Schizophrenia diagnosis, n (%) | 38 (67.8) | 26 (59.1) | .41 |
| BPRS total scores, mean ± SD | 45.4±7.4 | 44.0±8.7 | .37 |
| Duration of untreated psychosis (wk), mean ± SD | 128.8±221.8 | 90.7±110.7 | .30 |
| Medication dose (level)a | 3.3±1.4 | 2.9±1.2 | .12 |
| Treatment responderb, n (%) | 32 (57.1) | 21 (47.7) | .35 |
| Comedications | |||
| Benzodiazepine use, % | 44.6 | 56.8 | .23 |
| Anticholinergics use, % | 21.4 | 25.0 | .67 |
| Beta-blocker use, % | 8.9 | 13.6 | .46 |
Note: BPRS, Brief Psychiatric Rating Scale.
aFor medication dose, levels 1–6 represented 1, 2, 3, 4, 5, and 6mg/d for risperidone and 5, 10, 15, 20, 25, and 30mg/d for aripiprazole, respectively.
bTreatment response: response criteria required (1) a rating of 3 (“mild”) or less on all of the following items of the BPRS: conceptual disorganization, grandiosity, hallucinatory behavior, and unusual thought content and (2) a Clinical Global Impression Scale-Improvement rating of much or very much improved on 2 consecutive rating assessments (this issue).13
Statistical Analysis
Because missing data is inherent in any clinical trial and may be dependent on the observed outcomes, a mixed-model approach to longitudinal data analysis was used (SPSS Mixed Linear Models, IBM), with time as the primary within-subject variable and genotype as the primary between-subject variable. A random intercept in the mixed model was used to account for correlation of measurements over time among subjects with an unstructured covariance matrix. The difference in slopes of the outcomes between the 2 genotype groups was assessed using group-by-time interaction term in the mixed model.
The primary outcome was BPRS positive symptoms. Secondary outcome variables were EPS, akathisia, and prolactin levels. Tertiary outcomes were BPRS total scores, CGI-severity scores, and 4 negative symptoms. Each outcome variable was analyzed separately, while controlling for sex, age, medication assignment, and genomic principal components, using maximum likelihood estimation. Due to differential effects of drugs on EPS, akathisia, and prolactin (see this issue),13 analyses of these variables were conducted in the aripiprazole and risperidone subgroups separately. Final models included genotype group as a fixed factor, time, any significant covariates, and a genotype group-by-time interaction term (if statistically significant).
Results
Table 1 shows descriptive statistics for the 2 genotype groups. There was no significant difference in age, sex, medication assignment, ethnicity, diagnosis, baseline BPRS total score, treatment response, duration of untreated psychosis, medication modal dose, and comedication use.
Positive Symptoms
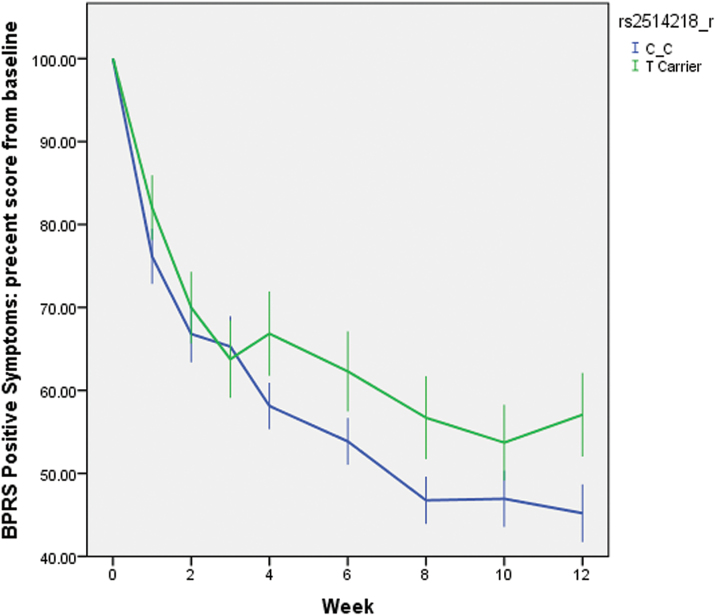
The mixed-model analysis showed that the C/C homozygotes had a greater reduction in positive symptoms during 12 weeks of treatment, compared to the T carriers. For the genotype × time interaction, F(1,111) = 4.13, P = .044, Cohen’s f 2 = 0.036 (effect size measure, equivalent to Cohen’s d = 0.38).19 Unsurprisingly, the main effect of time was also strongly significant in that both genotype groups had large reductions in positive symptoms over 12 weeks, F(1,111) = 178.33, P < .001 . None of the covariates affected the results. Table 2 presents the least square estimates of means and SDs of the outcome variable at each time point, separately for the 2 genotype groups. While none of the single-time point comparisons between the 2 groups in table 2 were statistically significant, the significant interaction term demonstrates that the slopes differed over time. Specifically, the C/C homozygotes were higher in positive symptoms at baseline, but they improved more by week 12. For illustrative purposes, and to represent the data in a clinically meaningful way, we calculated the percent reduction from baseline at each time point. As shown in figure 1, the longitudinal trajectories of the 2 genotype groups separated after week 4, with the curve for the C/C homozygotes being more steep. Using the least square estimates of the percent reduction at each time point in the mixed model, the 2 genotype groups were significantly different at weeks 4, 6, and 12 (P’s = .039, .049, and .030, respectively). Because only 1 subject dropped out between week 3 and 4, it was unlikely that differential drop-out rates might explain the separation of the 2 curves. At the end of 12 weeks, the C/C group had, on average, 10% greater reduction in positive symptoms than the T carriers.
Table 2.
BPRS Positive Symptoms Scores at Each Follow-up (Least Square Estimate, mean ± SE, unadjusted)
| CC Homozygotes | n | T Carriers | n | P Value | |
|---|---|---|---|---|---|
| Week 0 | 14.80±0.46 | 56 | 13.91±0.52 | 44 | .198 |
| Week 1 | 11.05±0.47 | 53 | 11.16±0.54 | 39 | .869 |
| Week 2 | 9.59±0.48 | 49 | 9.54±0.55 | 37 | .950 |
| Week 3 | 9.52±0.48 | 49 | 8.72±0.56 | 35 | .273 |
| Week 4 | 8.51±0.48 | 49 | 9.26±0.56 | 34 | .310 |
| Week 6 | 7.84±0.50 | 44 | 8.41±0.56 | 34 | .447 |
| Week 8 | 6.75±0.49 | 44 | 7.75±0.57 | 32 | .189 |
| Week 10 | 6.76±0.51 | 40 | 7.42±0.60 | 27 | .404 |
| Week 12 | 6.51±0.52 | 38 | 7.64±0.57 | 33 | .143 |
Note: Numbers in the “n” columns are the actual sample size for observed cases at each time point.
Fig. 1.
Percentage of positive symptoms reduction from baseline throughout the 12-week clinical trial, stratified by rs2514218 genotype. N = 100. Error bars are SEs of the mean. The genotype × week interaction was significant in the mixed-models analysis, F(1,111) = 4.13, P = .044. BPRS, Brief Psychiatric Rating Scale.
EPS, Akathisia, and Prolactin Levels
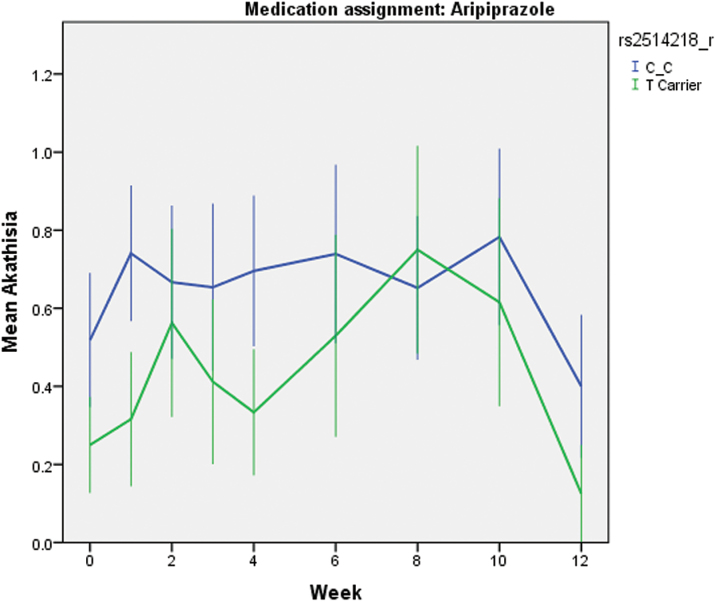
For the BAS global rating of akathisia, mixed-model analysis was conducted in the 2 treatment arms separately, because the full trial results revealed that patients on aripiprazole developed significantly worse akathisia compared to those on risperidone (this issue).13 The genotype × time interaction was not significant in either medication arm, nor was the main effect of time, P’s > .10, which is consistent with the finding in the whole sample (as shown in figure 3, Robinson et al., this issue).13 After removing the nonsignificant interaction term, the main effect of genotype was statistically significant in the aripiprazole group (F(1,46) = 4.19, P = .046, Cohen’s f 2 = 0.014, equivalent to Cohen’s d = 0.24). As shown in figure 2, the C/C homozygotes had worse akathisia than the T carriers, with estimated mean scores of 0.72±0.11 vs 0.34±0.14 (SE). The main effect of genotype was not significant in the risperidone group, P > .05.
Fig. 3.
Mean prolactin levels at baseline and each follow-up visit, stratified by rs2514218 genotype. N = 36. Error bars are SEs of the mean. The genotype × week interaction was significant in the mixed-models analysis, F(3,68) = 2.80, P = .046.
Fig. 2.
Mean akathisia scores at baseline and each follow-up visit, stratified by rs2514218 genotype. N = 49. Error bars are SEs of the mean. The main effect of genotype was significant in the mixed-models analysis, F(1,46) = 4.19, P = .046.
For EPS total scores, neither the genotype × time interaction nor the main effect of genotype was statistically significant in either aripiprazole or risperidone groups (P’s > .10).
Prolactin elevation was a common side effect of risperidone treatment, but aripiprazole is not usually associated with prolactin elevation. Therefore, mixed-model analysis was conducted in risperidone group only. Because prolactin is a reproductive hormone and sex plays an important role, the mixed-model analysis was conducted in males and females separately. In the risperidone arm, the genotype × time interaction was significant, F(3,68) = 2.80, P = .046, Cohen’s f 2 = 0.019 (equivalent to Cohen’s d = 0.28), in male subjects (n = 36). The C/C homozygotes (n = 19) had lower elevation of prolactin at week 8 and week 12, compared to T carriers (n = 17) (figure 3). However, neither interaction nor main effect was significant in females.
BPRS Total Score, CGI, and Negative Symptoms
Mixed-model analysis was conducted for each of these outcome variables. As expected, none of models produced significant genotype × time interaction or genotype main effects. P values for the interaction terms for BPRS total score, CGI-severity, affective flattening, alogia, avolition-apathy, and asociality-anhedonia were .133, .070, .305, .436, .262, and .193, respectively.
Discussion
To our knowledge, no prior study has identified any functional effect of genotype at rs2514218, one of the 108 top hits reported from the PGC GWAS. Within the PGC study, biological assays in blood and postmortem brain tissue identified no detectable relationship between rs2514218 and DRD2 gene expression levels.1 In the present study, we found that the C/C homozygotes improved in positive symptoms more than the T carriers during 12 weeks of treatment. Intriguingly, the C allele is both the schizophrenia risk allele in the PGC GWAS1 and the common allele in the general population. Speculatively, while carriage of the minor T allele may be protective for development of schizophrenia, T carriers who do develop psychotic disorders may be somewhat more likely to have a nonstandard pathophysiology with respect to dopamine signaling. A distinction between 2 subtypes of schizophrenia based on dopamine physiology has been recently proposed.20 Such a dichotomy is also consistent with our own data demonstrating that psychotic patients can be divided into 2 groups based on resting-state functional magnetic resonance imaging patterns of connectivity linking the D2-rich striatum with cortex.21 Patients with psychotic disorders (both first episode and multiepisode) demonstrating the atypical striatal connectivity pattern were replicably less likely to meet criteria for symptomatic response following treatment with antipsychotic medications.
In addition, the C/C homozygotes developed more akathisia during treatment with aripiprazole compared to carriers of the T allele. Only a few prior studies have examined the pharmacogenetics of akathisia as a side effect of antipsychotic medications,22 and none have examined it in the context of aripiprazole treatment, which has been associated with greater liability for akathisia in some studies,23 including our own report (Robinson et al) in this volume. Further studies are needed to replicate and validate this novel finding. In contrast, there was no significant association between the SNP and EPS in either aripiprazole or risperidone groups. Furthermore, rs2514218 was associated with prolactin elevation in males treated with risperidone, in that C/C homozygotes had lower elevation of prolactin compared to the T carriers. This effect seems to be the opposite of what was observed for akathisia and positive symptoms, in which C/C homozygotes were more sensitive to medication effects, as described above. Risperidone is known to cause prolactin elevation and is hypothesized to act by the dopamine tuberoinfundibular pathway.24 It has been theorized that antipsychotic drugs reduce positive symptoms via the mesolimbic pathway and cause EPS and perhaps akathisia via the nigrostriatal pathway.24 Perhaps the effect of the DRD2 risk variant is different across these disparate dopamine pathways, but this needs further research to elucidate. Our side effect findings should be interpreted with caution, because the sample sizes were small for the subgroup analyses. In addition, many patients were prescribed beta-blockers to treat akathisia and received anticholinergics to treat EPS. Although there was no significant difference in comedication use between the 2 genotype groups, the effects of genotypes on akathisia and EPS could have been masked or distorted by administration of these comedications.
Several variants in DRD2 have been found to affect antipsychotic drug response including efficacy and side effects.6,8,25 It is not surprising because dopamine D2 receptor antagonism is a common mechanism of antipsychotic drug action26 and may be “necessary and sufficient” for antipsychotic efficacy.27 Positive symptoms of schizophrenia are likely associated with excessive dopaminergic activity of the mesolimbic pathway,24 therefore, D2 blockade in this region would result in reduction in positive symptoms. Imaging studies have found a threshold relationship between D2 receptor occupancy and antipsychotic drug efficacy.28 However, excessive D2 blockade also leads to EPS such as parkinsonism, dyskinesia, akathisia, dystonia, and other motor side effects.28,29 In the present study, C/C homozygotes experienced greater positive symptom efficacy and greater akathisia, consistent with a hypothesis that the C (risk) allele allows for greater sensitivity to D2 blockade. In vitro studies, as well as neurochemical imaging in vivo studies, would be required to directly test this hypothesis.
Previous studies have found that rs1799732 (−141C Ins/Del), a SNP in the DRD2 promoter region, is associated with antipsychotic efficacy7,8 and drug-induced weight gain.30 Another SNP nearby, rs1799978 (A-241G), was also associated with antipsychotic drug response in a first-episode schizophrenia cohort.7 Both SNPs are carried along the same haplotype with rs2514218 (D’ = 0.93 and 0.99, respectively, in the current dataset), although allele frequencies differ and the overall correlation is low. Still, it is plausible that rs2514218 is tagging the effects of other DRD2 variants in antipsychotic response.
The present study utilized a cohort of patients with first-episode psychosis to study pharmacogenetic markers of drug response. There are many advantages of studying pharmacogenetics in this type of patients, including minimal prior exposure to antipsychotic drugs, less comorbidity and substance abuse, less confounding effects from other psychotropic drugs, and better treatment adherence.31 Nevertheless, there are limitations of the study. The sample size was small, especially for subgroup analysis, so we may not have the statistical power to detect significant genotype-phenotype associations, and findings may be less reliable (due to a greater degree of error variance). Importantly, the 2 drugs used in the clinical trial might have somewhat different mechanisms of action. Aripiprazole is a D2 partial agonist, aiming at normalizing dopaminergic imbalance in schizophrenia and may act as an agonist or antagonist at the D2 receptor, depending on synaptic dopamine levels.32 In contrast, risperidone is more like a typical antipsychotic drug with strong D2 antagonism. These differences in pharmacodynamic processes may contribute noise to a pharmacogenetic study of DRD2. Furthermore, there were potential racial differences between the 2 genotype groups that may influence the findings. There were more African Americans in the C/C group and more Caucasians in the T carriers. Although the difference did not reach statistical significance, P = .064, it is certainly a legitimate concern that racial differences might influence the findings. Therefore, we examined the difference in positive symptom response rates (as defined by 50%, 40%, 30%, and 20% reduction from baseline) and akathisia between African Americans and Caucasians. For response rates, P values for race were all greater than .10. For akathisia, results for race went in the opposite direction and, if anything, weakened the relationship with genotype. In addition, we utilized the genomic principal components to control for potential racial differences and population stratification. Nevertheless, these findings need to be replicated in larger and independent samples to ensure their validity.
In summary, rs2514218, the top SNP from a replicable schizophrenia risk locus, was associated with antipsychotic drug response. These findings provided further evidence to support the important role that DRD2 plays in both schizophrenia risk and antipsychotic drug response. Future studies should examine the pathophysiological mechanisms of DRD2 genetic variation in schizophrenia etiology and treatment.
Funding
This work was supported in part by the National Institutes of Health (K23MH097108 to J.P.Z., R01MH060004 to D.G.R., R21MH099868 to T.L., K23MH100264 to J.A.G., P30MH090590 to J.M.K., and P50MH080173 to A.K.M.) and by NARSAD Young Investigator Grants to J.-P.Z. and J.A.G. from the Brain & Behavior Research Foundation. Medication supplies were donated by Bristol-Myers Squibb and by Janssen Pharmaceuticals.
Acknowledgments
Dr. Zhang has received grant support from the National Institute of Mental Health (NIMH), the Brain & Behavior Research Foundation (BBRF), and Genomind, Inc.. Dr. Robinson has been a consultant to Asubio, Shire, and Otsuka, and he has received grants from Bristol Meyers Squibb, Janssen, and Otsuka. Dr. Gallego has received grant support from the NIMH and the BBRF. Dr. Kane has been a consultant for Alkermes, Amgen, Bristol-Myers Squibb, Eli Lilly, EnVivo Pharmaceuticals (Forum), Forest, Genentech, H. Lundbeck, Intracellular Therapies, Janssen Pharmaceutica, Johnson and Johnson, Merck, Novartis, Otsuka, Pierre Fabre, Reviva, Roche, Sunovion, and Teva, and he has received honoraria for lectures from Bristol-Myers Squibb, Janssen, Genentech, Lundbeck, and Otsuka and is a Shareholder in MedAvante, Inc. and the Vanguard Research Group. Dr. Malhotra is a consultant to Genomind, Inc. and FORUM Pharmaceuticals, and he has received grant support from the NIMH. Dr. Lencz has received grant support from the NIMH, the US-Israel Binational Science Fund, and the BBRF.
References
- 1. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lencz T, Malhotra AK. Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol Psychiatry. 2015;20:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kapur S, Mamo D. Half a century of antipsychotics and still a central role for dopamine D2 receptors. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:1081–1090. [DOI] [PubMed] [Google Scholar]
- 4. Kane JM, Correll CU. Past and present progress in the pharmacologic treatment of schizophrenia. J Clin Psychiatry. 2010;71:1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lieberman JA, Stroup TS, McEvoy JP, et al. ; Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) Investigators. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353:1209–1223. [DOI] [PubMed] [Google Scholar]
- 6. Zhang JP, Malhotra AK. Pharmacogenetics and antipsychotics: therapeutic efficacy and side effects prediction. Expert Opin Drug Metab Toxicol. 2011;7:9–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lencz T, Robinson DG, Xu K, et al. DRD2 promoter region variation as a predictor of sustained response to antipsychotic medication in first-episode schizophrenia patients. Am J Psychiatry. 2006;163:529–531. [DOI] [PubMed] [Google Scholar]
- 8. Zhang JP, Lencz T, Malhotra AK. D2 receptor genetic variation and clinical response to antipsychotic drug treatment: a meta-analysis. Am J Psychiatry. 2010;167:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miyamoto S, Duncan GE, Marx CE, Lieberman JA. Treatments for schizophrenia: a critical review of pharmacology and mechanisms of action of antipsychotic drugs. Mol Psychiatry. 2005;10:79–104. [DOI] [PubMed] [Google Scholar]
- 10. Trampush JW, Lencz T, DeRosse P, et al. Relationship of cognition to clinical response in first-episode schizophrenia spectrum disorders. Schizo Bull, Nov 2015, Doi: 10.1093/schbul/sbv120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kapur S, Remington G, Zipursky RB, Wilson AA, Houle S. The D2 dopamine receptor occupancy of risperidone and its relationship to extrapyramidal symptoms: a PET study. Life Sci. 1995;57:PL103–PL107. [DOI] [PubMed] [Google Scholar]
- 12. Compton MT, Miller AH. Antipsychotic-induced hyperprolactinemia and sexual dysfunction. Psychopharmacol Bull. 2002;36:143–164. [PubMed] [Google Scholar]
- 13. Robinson DG, Gallego JA, John M, et al. A randomized comparison of aripiprazole and risperidone for the acute treatment of first-episode schizophrenia and related disorders: 3 month outcomes. Schizo Bull, Nov 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woerner MG, Mannuzza S, Kane JM. Anchoring the BPRS: an aid to improved reliability. Psychopharmacol Bull. 1988;24:112–117. [PubMed] [Google Scholar]
- 15. Andreason NC. Scale for the Assessment of Negative Symptoms (SANS). Iowa City, IA: University of Iowa; 1983. [Google Scholar]
- 16. Guy W, ed. ECDEU Assessment Manual for Psychopharmacology. US Department of Health, Education, and Welfare publication ADM 76-338. Washington, DC: National Institute of Mental Health; 1976. [Google Scholar]
- 17. Simpson GM, Angus JW. A rating scale for extrapyramidal side effects. Acta Psychiatr Scand Suppl. 1970;212:11–19. [DOI] [PubMed] [Google Scholar]
- 18. Barnes TR. A rating scale for drug-induced akathisia. Br J Psychiatry. 1989;154:672–676. [DOI] [PubMed] [Google Scholar]
- 19. Selya AS, Rose JS, Dierker LC, Hedeker D, Mermelstein RJ. A Practical Guide to Calculating Cohen’s f(2), a Measure of Local Effect Size, from PROC MIXED. Front Psychol. 2012;3:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howes OD, Kapur S. A neurobiological hypothesis for the classification of schizophrenia: type A (hyperdopaminergic) and type B (normodopaminergic). Br J Psychiatry. 2014;205:1–3. [DOI] [PubMed] [Google Scholar]
- 21. Sarpal DK, Argyelan M, Robinson DG, et al. Baseline striatal functional connectivity as a predictor of response to antipsychotic drug treatment. Am J Psychiatry. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lencz T, Malhotra AK. Pharmacogenetics of antipsychotic-induced side effects. Dialogues Clin Neurosci. 2009;11:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rummel-Kluge C, Komossa K, Schwarz S, et al. Second-generation antipsychotic drugs and extrapyramidal side effects: a systematic review and meta-analysis of head-to-head comparisons. Schizophr Bull. 2012;38:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davis KL, Kahn RS, Ko G, Davidson M. Dopamine in schizophrenia: a review and reconceptualization. Am J Psychiatry. 1991;148:1474–1486. [DOI] [PubMed] [Google Scholar]
- 25. Muller DJ, Zai CC, Sicard M, et al. Systematic analysis of dopamine receptor genes (DRD1-DRD5) in antipsychotic-induced weight gain. Pharmacogenomics J. 2012;12:156–164. [DOI] [PubMed] [Google Scholar]
- 26. Miyamoto S, Miyake N, Jarskog LF, Fleischhacker WW, Lieberman JA. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol Psychiatry. 2012;17:1206–1227. [DOI] [PubMed] [Google Scholar]
- 27. Kapur S, Seeman P. Does fast dissociation from the dopamine d(2) receptor explain the action of atypical antipsychotics?: A new hypothesis. Am J Psychiatry. 2001;158:360–369. [DOI] [PubMed] [Google Scholar]
- 28. Remington G, Kapur S. D2 and 5-HT2 receptor effects of antipsychotics: bridging basic and clinical findings using PET. J Clin Psychiatry. 1999;60(suppl 10):15–19. [PubMed] [Google Scholar]
- 29. Remington G, Kapur S. Atypical antipsychotics: are some more atypical than others? Psychopharmacology (Berl). 2000;148:3–15. [DOI] [PubMed] [Google Scholar]
- 30. Lencz T, Robinson DG, Napolitano B, et al. DRD2 promoter region variation predicts antipsychotic-induced weight gain in first episode schizophrenia. Pharmacogenet Genomics. 2010;20:569–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malhotra AK, Zhang JP, Lencz T. Pharmacogenetics in psychiatry: translating research into clinical practice. Mol Psychiatry. 2012;17:760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shapiro DA, Renock S, Arrington E, et al. Aripiprazole, a novel atypical antipsychotic drug with a unique and robust pharmacology. Neuropsychopharmacology. 2003;28:1400–1411. [DOI] [PubMed] [Google Scholar]