Abstract
In the growing field of personalised medicine, the analysis of numerous potential targets is becoming a challenge in terms of work load, tissue availability, as well as costs. The molecular analysis of non-small cell lung cancer (NSCLC) has shifted from the analysis of the epidermal growth factor receptor (EGFR) mutation status to the analysis of different gene regions, including resistance mutations or translocations. Massive parallel sequencing (MPS) allows rapid comprehensive mutation testing in routine molecular pathological diagnostics even on small formalin-fixed, paraffin-embedded (FFPE) biopsies. In this study, we compared and evaluated currently used MPS platforms for their application in routine pathological diagnostics. We initiated a first round-robin testing of 30 cases diagnosed with NSCLC and a known EGFR gene mutation status. In this study, three pathology institutes from Germany received FFPE tumour sections that had been individually processed. Fragment libraries were prepared by targeted multiplex PCR using institution-specific gene panels. Sequencing was carried out using three MPS systems: MiSeq™, GS Junior and PGM Ion Torrent™. In two institutes, data analysis was performed with the platform-specific software and the Integrative Genomics Viewer. In one institute, data analysis was carried out using an in-house software system. Of 30 samples, 26 were analysed by all institutes. Concerning the EGFR mutation status, concordance was found in 26 out of 26 samples. The analysis of a few samples failed due to poor DNA quality in alternating institutes. We found 100% concordance when comparing the results of the EGFR mutation status. A total of 38 additional mutations were identified in the 26 samples. In two samples, minor variants were found which could not be confirmed by qPCR. Other characteristic variants were identified as fixation artefacts by reanalyzing the respective sample by Sanger sequencing. Overall, the results of this study demonstrated good concordance in the detection of mutations using different MPS platforms. The failure with samples can be traced back to different DNA extraction systems and DNA quality. Unknown or ambiguous variations (transitions) need verification with another method, such as qPCR or Sanger sequencing.
Keywords: lung cancer, mutation testing, massive parallel sequencing, fixation artefacts
Introduction
In the growing field of personalised medicine, the increasing number of molecular targets for individualised therapies requires the analysis of numerous, potential genetic alterations, which is becoming a challenge in terms of workload, tissue availability, as well as costs (1). For non-small cell lung cancer (NSCLC), molecular analysis has shifted from the analysis of the epidermal growth factor receptor (EGFR) mutation status to the analysis of additional gene target regions, including resistance mutations and gene fusion events (2).
Taking these developments into account, massive parallel sequencing (MPS) has come into focus, as it allows rapid, comprehensive and cost-effective mutation testing for routine molecular pathological diagnostics, even on small formalin-fixed, paraffin-embedded (FFPE) biopsies (3–6). However, the implementation of MPS platforms into routine diagnostics raises questions about feasibility, sensitivity and specificity, as the results of mutation testing are the basis for therapeutic decision making (1,7). The ever-increasing pace of MPS adoption presents enormous challenges, in terms of data processing, storage, management and interpretation, as well as sequencing quality control, which impede the translation of research into clinical practice (8,9).
Additionally, the preanalytical steps are important to consider: the manual macrodissection of selected tumour areas has become a standard procedure in molecular pathology and is a powerful tool to reduce false negative results resulting from wild-type contamination (10). Selecting the right tumour area influences not only the result of the analysis, but also the allele frequency, the value of which is pivotal when reporting diagnostic findings (11). Automated DNA extraction systems are helpful in a routine laboratory with respect to expenditure of time, sample tracking and reproducible sample quality. In addition, an accurate and reliable DNA quantification system is necessary for good and constant MPS performance (12).
In the present study, we compared three different MPS platforms: PGM Ion Torrent™ from Life Technologies™, MiSeq™ from Illumina® and GS Junior from Roche. We used lung cancer samples, obtained from the clinical setting, with a known EGFR and KRAS mutation status. Samples included large tumour resections, as well as small fine needle biopsies. In our comparison, three different multiplex primer panels, tailored to the needs of the respective sequencing platforms were used in the participating institutes, mirroring the individual approaches that may be used for routine testing.
Materials and methods
Samples
A total of 30 tumour samples was collected from 2010 to 2013. All samples were lung adenocarcinomas and each institute contributed 10 samples. Tumours were diagnosed by experienced pathologists and the tumour content was determined by the visual inspection of hematoxylin and eosin (H&E)-stained corresponding sections. The mutation status of the samples was determined previously in routine molecular diagnostics in each institute using conventional methods.
DNA isolation
All tissue specimens were fixed in neutral-buffered formalin prior to paraffin embedding (FFPE samples). Tumour areas were marked by a pathologist on an H&E-stained slide and DNA was extracted from corresponding unstained 10-µm-thick slides by manual macrodissection. Following treatment with proteinase K, the DNA was isolated by either automated or manual extraction: BioRobot M48 (institute A), the QIAamp DNA FFPE Tissue kit (institute B), QIASymphony SP (institute C) (all from Qiagen, Hilden, Germany) or the Maxwell 16 Research system (institute C; Promega, Madison, WI, USA) following the manufacturer's instructions.
DNA quality and quantity
The quality and quantity of the isolated DNA samples were assessed by agarose gel electrophoresis and measured fluorimetrically using the Qubit® HS DNA assay (Life Technologies, Darmstadt, Germany) in institute A. The quantity of the isolated DNA was measured spectrophotometrically using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) in institute B. In institute C, the DNA content was measured fluorimetrically using the Qubit HS DNA assay (Life Technologies) and using a qPCR-based method (RNaseP Detection system; Life Technologies).
Massive parallel sequencing
Illumina® MiSeq™ platform
MiSeq (Illumina, San Diego, CA, USA) was used in institute A. The custom-made lung cancer panel consisted of 102 amplicons for the detection of hotspot mutations in 14 lung cancer-related genes. A full list of the covered amplicons is provided in Table I. Isolated DNA (20 ng) was amplified with 2 customised Ion AmpliSeq™ Primer Pools for 15 sec at 99°C and 4 min at 60°C for 29 cycles, with an initial denaturating step at 99°C for 2 min. PCR products from the same patient were pooled following treatment with FuPa reagent. Following purification with Agencourt AMPure XP (Beckman Coulter, Brea, CA, USA), the PCR products were incubated with NEXTflex™ DNA Adenylation Mix (Bioo Scientific Corp., Austin, TX, USA). Adapters were supplied by NEXTflex™ DNA Barcodes (Bioo Scientific Corp.). After the bead-mediated size selection, NEXTflex™ PCR Master Mix (Bioo Scientific Corp.) was used for the final PCR amplification at 98°C for 15 sec and 60°C for 1 min for 10 cycles, with an initial denaturating step at 98°C for 2 min. Library products were quantified using a Qubit® 2.0 Fluorometer (Qubit® dsDNA HS kit; Life Technologies), diluted and pooled in equal amounts. A total of 6–8 pM was spiked with 5% PhiX DNA and sequenced using the MiSeq™ reagent kit V2 (300 cycles) (both from Illumina). Data were exported as FASTQ files.
Table I.
Overview of the institute-specific gene panels.
| Chromosome | From (hg19) | To (hg19) | Gene name | Exon |
|---|---|---|---|---|
| Custom panel Heidelberg | ||||
| chr1 | 27056234 | 27056365 | ARID1A | 2 |
| chr1 | 27057662 | 27057775 | ARID1A | 3 |
| chr1 | 27057875 | 27058001 | ARID1A | 3 |
| chr1 | 27092899 | 27093023 | ARID1A | 10 |
| chr1 | 27094337 | 27094460 | ARID1A | 11 |
| chr1 | 27099336 | 27099464 | ARID1A | 14 |
| chr1 | 27100275 | 27100411 | ARID1A | 17 |
| chr1 | 27105906 | 27106030 | ARID1A | 20 |
| chr1 | 27106449 | 27106570 | ARID1A | 20 |
| chr1 | 27106750 | 27106883 | ARID1A | 20 |
| chr1 | 115256484 | 115256587 | NRAS | 3 |
| chr1 | 115258676 | 115258805 | NRAS | 2 |
| chr1 | 150549826 | 150549952 | MCL-1 | 3 |
| chr1 | 150551531 | 150551670 | MCL-1 | 1 |
| chr2 | 178098765 | 178098890 | NFE2L2 | 2 |
| chr3 | 41266029 | 41266147 | CTNNB1 | 3 |
| chr3 | 41266893 | 41267010 | CTNNB1 | 5 |
| chr3 | 41275089 | 41275211 | CTNNB1 | 9 |
| chr3 | 178916892 | 178917000 | PIK3CA | 2 |
| chr3 | 178921523 | 178921633 | PIK3CA | 5 |
| chr3 | 178928050 | 178928160 | PIK3CA | 8 |
| chr3 | 178936022 | 178936106 | PIK3CA | 10 |
| chr3 | 178938830 | 178938960 | PIK3CA | 14 |
| chr3 | 178952038 | 178952157 | PIK3CA | 21 |
| chr3 | 181430178 | 181430283 | SOX2 | 1 |
| chr3 | 181430516 | 181430649 | SOX2 | 1 |
| chr4 | 1803550 | 1803636 | FGFR3 | 7 |
| chr4 | 1808277 | 1808409 | FGFR3 | 16 |
| chr4 | 55131108 | 55131222 | PDGFRA | 5 |
| chr4 | 55139749 | 55139881 | PDGFRA | 10 |
| chr4 | 55140692 | 55140818 | PDGFRA | 11 |
| chr4 | 55141036 | 55141156 | PDGFRA | 12 |
| chr4 | 55152001 | 55152128 | PDGFRA | 18 |
| chr4 | 55156632 | 55156764 | PDGFRA | 22 |
| chr4 | 55592107 | 55592203 | KIT | 9 |
| chr4 | 55593595 | 55593684 | KIT | 11 |
| chr4 | 153245407 | 153245522 | FBXW7 | 11 |
| chr4 | 153247237 | 153247369 | FBXW7 | 10 |
| chr4 | 153249405 | 153249530 | FBXW7 | 9 |
| chr5 | 1264501 | 1264634 | TERT | 11 |
| chr5 | 1293392 | 1293528 | TERT | 2 |
| chr6 | 66115100 | 66115214 | EYS | 7 |
| chr6 | 66204680 | 66204810 | EYS | 5 |
| chr7 | 55241602 | 55241732 | EGFR | 18 |
| chr7 | 55242411 | 55242544 | EGFR | 19 |
| chr7 | 55248974 | 55249100 | EGFR | 20 |
| chr7 | 55259416 | 55259546 | EGFR | 21 |
| chr7 | 92300724 | 92300853 | CDK6 | 5 |
| chr7 | 92403995 | 92404124 | CDK6 | 3 |
| chr7 | 116411944 | 116412066 | MET | 14 |
| chr7 | 116417426 | 116417508 | MET | 16 |
| chr7 | 140453110 | 140453232 | BRAF | 15 |
| chr7 | 140481387 | 140481511 | BRAF | 11 |
| chr8 | 38275705 | 38275835 | FGFR1 | 10 |
| chr8 | 38282107 | 38282241 | FGFR1 | 7 |
| chr8 | 128751156 | 128751293 | MYC | 2 |
| chr8 | 128752956 | 128753086 | MYC | 3 |
| chr9 | 5069993 | 5070100 | JAK2 | 12 |
| chr9 | 5073678 | 5073788 | JAK2 | 14 |
| chr9 | 5126715 | 5126797 | JAK2 | 25 |
| chr9 | 21970912 | 21971032 | CDKNA2 | 2 |
| chr9 | 21971086 | 21971218 | CDKNA2 | 2 |
| chr9 | 21974672 | 21974792 | CDKNA2 | 1 |
| chr9 | 139401722 | 139401834 | NOTCH1 | 22 |
| chr9 | 139404170 | 139404306 | NOTCH1 | 18 |
| chr9 | 139412260 | 139412400 | NOTCH1 | 8 |
| chr9 | 139413034 | 139413159 | NOTCH1 | 6 |
| chr10 | 89624207 | 89624322 | PTEN | 1 |
| chr10 | 89685258 | 89685374 | PTEN | 3 |
| chr10 | 89692864 | 89692987 | PTEN | 5 |
| chr10 | 89711806 | 89711936 | PTEN | 6 |
| chr10 | 89717622 | 89717747 | PTEN | 7 |
| chr10 | 89720778 | 89720902 | PTEN | 8 |
| chr10 | 123256020 | 123256129 | FGFR2 | 13 |
| chr10 | 123279495 | 123279622 | FGFR2 | 7 |
| chr11 | 533800 | 533929 | HRAS | 3 |
| chr11 | 534220 | 534349 | HRAS | 2 |
| chr11 | 69456096 | 69456216 | CCND1 | 1 |
| chr11 | 69458624 | 69458747 | CCND1 | 3 |
| chr11 | 119103162 | 119103275 | CBL | 2 |
| chr11 | 119148912 | 119149006 | CBL | 8 |
| chr11 | 119149215 | 119149290 | CBL | 9 |
| chr12 | 25380249 | 25380348 | KRAS | 3 |
| chr12 | 25398183 | 25398310 | KRAS | 2 |
| chr12 | 69210596 | 69210679 | MDM2 | 4 |
| chr12 | 69233038 | 69233165 | MDM2 | 11 |
| chr13 | 48881433 | 48881526 | RB1 | 2 |
| chr13 | 48916793 | 48916902 | RB1 | 3 |
| chr13 | 48923124 | 48923208 | RB1 | 6 |
| chr13 | 48951050 | 48951160 | RB1 | 13 |
| chr13 | 48954320 | 48954437 | RB1 | 16 |
| chr13 | 48955427 | 48955539 | RB1 | 17 |
| chr13 | 49027105 | 49027191 | RB1 | 18 |
| chr13 | 49033834 | 49033935 | RB1 | 20 |
| chr13 | 49037844 | 49037955 | RB1 | 21 |
| chr13 | 49039144 | 49039221 | RB1 | 22 |
| chr13 | 49039304 | 49039410 | RB1 | 23 |
| chr14 | 36987081 | 36987213 | NKX-2.1 | 2 |
| chr14 | 36988227 | 36988351 | NKX-2.1 | 1 |
| chr14 | 105246470 | 105246589 | AKT1 | 3 |
| chr17 | 7573886 | 7574019 | TP53 | 10 |
| chr17 | 7576836 | 7576950 | TP53 | 9 |
| chr17 | 7577028 | 7577157 | TP53 | 8 |
| chr17 | 7577492 | 7577629 | TP53 | 7 |
| chr17 | 7578180 | 7578289 | TP53 | 6 |
| chr17 | 7578425 | 7578555 | TP53 | 5 |
| chr17 | 7579278 | 7579397 | TP53 | 4 |
| chr17 | 7579454 | 7579566 | TP53 | 4 |
| chr17 | 37880169 | 37880287 | ERBB2 | 19 |
| chr17 | 37880958 | 37881089 | ERBB2 | 20 |
| chr18 | 48581196 | 48581323 | SMAD4 | 5 |
| chr18 | 48584702 | 48584826 | SMAD4 | 7 |
| chr18 | 48591813 | 48591934 | SMAD4 | 9 |
| chr18 | 48604680 | 48604811 | SMAD4 | 12 |
| chr19 | 1206977 | 1207113 | STK11 | 1 |
| chr19 | 1218379 | 1218488 | STK11 | 2 |
| chr19 | 1220390 | 1220504 | STK11 | 4 |
| chr19 | 1220594 | 1220684 | STK11 | 5 |
| chr19 | 1221205 | 1221340 | STK11 | 6 |
| chr19 | 1223020 | 1223155 | STK11 | 8 |
| chr19 | 10599879 | 10600011 | KEAP1 | 5 |
| chr19 | 10600372 | 10600496 | KEAP1 | 4 |
| chr19 | 10602263 | 10602390 | KEAP1 | 3 |
| chr19 | 10602579 | 10602708 | KEAP1 | 3 |
| chr19 | 10602796 | 10602912 | KEAP1 | 3 |
| chr19 | 10610088 | 10610218 | KEAP1 | 2 |
| chr19 | 10610289 | 10610416 | KEAP1 | 2 |
| chr19 | 10610465 | 10610599 | KEAP1 | 2 |
| chr19 | 11094812 | 11094945 | SMARCA4 | 2 |
| chr19 | 11136088 | 11136220 | SMARCA4 | 22 |
| chr19 | 11138426 | 11138556 | SMARCA4 | 23 |
| chr19 | 11141448 | 11141561 | SMARCA4 | 25 |
| chr19 | 11144042 | 11144179 | SMARCA4 | 26 |
| chr19 | 30308024 | 30308156 | CCNE1 | 5 |
| chr19 | 30313134 | 30313262 | CCNE1 | 10 |
| chrX | 47028755 | 47028888 | RBM10 | 3 |
| chrX | 47034396 | 47034523 | RBM10 | 5 |
| chrX | 63411268 | 63411399 | FAM123B/AMER1 | 1 |
| chrX | 63412836 | 63412964 | FAM123B/AMER1 | 1 |
| Custom panel Cologne | ||||
| chr1 | 115256352 | 115256453 | NRAS | 3 |
| chr1 | 115256453 | 115256550 | NRAS | 3 |
| chr1 | 115256550 | 115256672 | NRAS | 3 |
| chr1 | 115258676 | 115258798 | NRAS | 2 |
| chr1 | 162688829 | 162688951 | DDR2 | 3 |
| chr1 | 162722872 | 162722995 | DDR2 | 4 |
| chr1 | 162724359 | 162724466 | DDR2 | 5 |
| chr1 | 162724466 | 162724586 | DDR2 | 5 |
| chr1 | 162724586 | 162724687 | DDR2 | 5 |
| chr1 | 162724850 | 162724967 | DDR2 | 6 |
| chr1 | 162724967 | 162725094 | DDR2 | 6 |
| chr1 | 162725447 | 162725572 | DDR2 | 7 |
| chr1 | 162729566 | 162729694 | DDR2 | 8 |
| chr1 | 162729681 | 162729782 | DDR2 | 8 |
| chr1 | 162730973 | 162731107 | DDR2 | 9 |
| chr1 | 162731107 | 162731197 | DDR2 | 9 |
| chr1 | 162731197 | 162731276 | DDR2 | 9 |
| chr1 | 162735765 | 162735879 | DDR2 | 10 |
| chr1 | 162736904 | 162737029 | DDR2 | 11 |
| chr1 | 162737029 | 162737154 | DDR2 | 11 |
| chr1 | 162740090 | 162740201 | DDR2 | 12 |
| chr1 | 162740201 | 162740327 | DDR2 | 12 |
| chr1 | 162741756 | 162741887 | DDR2 | 13 |
| chr1 | 162741887 | 162742002 | DDR2 | 13 |
| chr1 | 162742002 | 162742088 | DDR2 | 13 |
| chr1 | 162743204 | 162743301 | DDR2 | 14 |
| chr1 | 162743301 | 162743421 | DDR2 | 14 |
| chr1 | 162745384 | 162745513 | DDR2 | 15 |
| chr1 | 162745513 | 162745634 | DDR2 | 15 |
| chr1 | 162745915 | 162746038 | DDR2 | 16 |
| chr1 | 162746038 | 162746162 | DDR2 | 16 |
| chr1 | 162748317 | 162748432 | DDR2 | 17 |
| chr1 | 162748432 | 162748519 | DDR2 | 17 |
| chr1 | 162749866 | 162749977 | DDR2 | 18 |
| chr1 | 162749977 | 162750066 | DDR2 | 18 |
| chr2 | 29432650 | 29432776 | ALK | 25 |
| chr2 | 29436843 | 29436974 | ALK | 24 |
| chr2 | 29443565 | 29443688 | ALK | 23 |
| chr2 | 29443688 | 29443772 | ALK | 23 |
| chr2 | 29445200 | 29445332 | ALK | 22 |
| chr2 | 29445369 | 29445489 | ALK | 21 |
| chr3 | 41266072 | 41266193 | CTNNB1 | 3 |
| chr3 | 178935940 | 178936023 | PIK3CA | 9 |
| chr3 | 178936023 | 178936105 | PIK3CA | 9 |
| chr3 | 178936092 | 178936180 | PIK3CA | 9 |
| chr3 | 178951824 | 178951942 | PIK3CA | 20 |
| chr3 | 178951942 | 178952063 | PIK3CA | 20 |
| chr3 | 178952063 | 178952155 | PIK3CA | 20 |
| chr7 | 55241596 | 55241679 | EGFR | 18 |
| chr7 | 55241679 | 55241800 | EGFR | 18 |
| chr7 | 55242411 | 55242539 | EGFR | 19 |
| chr7 | 55248984 | 55249117 | EGFR | 20 |
| chr7 | 55249117 | 55249200 | EGFR | 20 |
| chr7 | 55259367 | 55259486 | EGFR | 21 |
| chr7 | 55259484 | 55259567 | EGFR | 21 |
| chr7 | 116411701 | 116411801 | cMET | intron 13/14 |
| chr7 | 116411801 | 116411909 | cMET | 14 |
| chr7 | 116411894 | 116411998 | cMET | intron 13/14 |
| chr7 | 116411998 | 116412072 | cMET | 14 |
| chr7 | 140453023 | 140453099 | BRAF | 15 |
| chr7 | 140453099 | 140453224 | BRAF | 15 |
| chr7 | 140481297 | 140481387 | BRAF | 11 |
| chr7 | 140481387 | 140481511 | BRAF | 11 |
| chr10 | 89624207 | 89624322 | PTEN | 1 |
| chr10 | 89653745 | 89653817 | PTEN | 2 |
| chr10 | 89653816 | 89653930 | PTEN | 2 |
| chr10 | 89685258 | 89685374 | PTEN | 3 |
| chr10 | 89690819 | 89690917 | PTEN | 4 |
| chr10 | 89692713 | 89692819 | PTEN | 5 |
| chr10 | 89692819 | 89692920 | PTEN | 5 |
| chr10 | 89692920 | 89693032 | PTEN | 5 |
| chr10 | 89711802 | 89711928 | PTEN | 6 |
| chr10 | 89711917 | 89712018 | PTEN | 6 |
| chr10 | 89717580 | 89717695 | PTEN | 7 |
| chr10 | 89717694 | 89717792 | PTEN | 7 |
| chr10 | 89720692 | 89720768 | PTEN | 8 |
| chr10 | 89720769 | 89720842 | PTEN | 8 |
| chr10 | 89724948 | 89725061 | PTEN | 9 |
| chr10 | 89725058 | 89725147 | PTEN | 9 |
| chr10 | 89725207 | 89725320 | PTEN | 9 |
| chr12 | 25380167 | 25380240 | KRAS | 3 |
| chr12 | 25380240 | 25380357 | KRAS | 3 |
| chr12 | 25398183 | 25398304 | KRAS | 2 |
| chr12 | 25398304 | 25398379 | KRAS | 2 |
| chr14 | 105246406 | 105246502 | AKT1 | 4 |
| chr14 | 105246500 | 105246583 | AKT1 | 4 |
| chr15 | 66727356 | 66727487 | MAP2K1 | 2 |
| chr15 | 66727487 | 66727602 | MAP2K1 | 2 |
| chr17 | 7577017 | 7577142 | TP53 | 8 |
| chr17 | 7577140 | 7577233 | TP53 | 8 |
| chr17 | 7577392 | 7577509 | TP53 | 7 |
| chr17 | 7577508 | 7577611 | TP53 | 7 |
| chr17 | 7578141 | 7578234 | TP53 | 6 |
| chr17 | 7578234 | 7578362 | TP53 | 6 |
| chr17 | 7578310 | 7578425 | TP53 | 5 |
| chr17 | 7578425 | 7578555 | TP53 | 5 |
| chr17 | 7579278 | 7579385 | TP53 | 4 |
| chr17 | 7579385 | 7579502 | TP53 | 4 |
| chr17 | 7579502 | 7579590 | TP53 | 4 |
| chr17 | 37880155 | 37880283 | HER2 | 19 |
| chr17 | 37880960 | 37881074 | HER2 | 20 |
| chr17 | 37881074 | 37881206 | HER2 | 20 |
GS Junior platform
GS Junior (Roche, Basel, Switzerland) was used in institute B. Genomic DNA (10–250 ng) was used for the amplification of EGFR exons 18–21 in a single multiplex reaction using the EGFR 18–21 MASTR assay and the 454 MID kit 1–8 (both from Multiplicom N.V., Niel, Belgium) according to the manufacturer's instructions. Libraries were purified, quantified, diluted to a final concentration of 1×106 molecules, multiplexed, clonally amplified by emulsion PCR and sequenced on the GS Junior (Roche) following the manufacturer's instructions. Amplicon libraries were sequenced in two runs on 454 GS Junior with 15 samples each.
PGM Ion Torrent platform
PGM Ion Torrent (Life Technologies) was used in institute C. For library preparation, the multiplex PCR-based Ion Torrent™ AmpliSeq™ technology (Life Technologies) with a custom-made lung cancer panel was used. The panel consisted of 139 primer pairs for the detection of hotspot mutations in 41 lung cancer-related genes. A full list of the covered amplicons is provided in Table I. Amplicon library preparation was performed with the Ion AmpliSeq™ Library kit v2.0 using approximately 10 ng of DNA as advised by the manufacturer. The PCR cycling conditions were as follows: initial denaturation: 99°C for 2 min, cycling: 21 cycles of 99°C, 15 sec and 60°C, 4 min. PCR products were partially digested using FuPa reagent as instructed, followed by the ligation of barcoded sequencing adapters (Ion Xpress Barcode Adapters 1–16 kit; Life Technologies). The final library was purified using Agencourt AMPure XP magnetic beads (Beckman Coulter) and quantified using qPCR (Ion Library Quantitation kit) on a StepOne qPCR machine (both from Life Technologies). The individual libraries were diluted to a final concentration of 100 pM and eight to ten libraries were pooled and processed to library amplification on Ion Spheres using an Ion PGM™ Template OT2 200 kit. Unenriched libraries were quality-controlled using Ion Sphere quality control measurement on a Qubit instrument. Following library enrichment (Ion OneTouch ES), the library was processed for sequencing using the Ion Torrent 200 bp sequencing v2 chemistry and the barcoded libraries were loaded onto a single 318 chip.
Data analysis
Illumina MiSeq platform
The FASTQ files were aligned against reference NCBI build 37 (hg19) and annotated using a modified version of a previously described method (13). The resulting BAM files were visualized using the Integrative Genomics Viewer (IGV; http://www.broadinstitute.org/igv/). Called variants were then imported into a FileMaker (FileMaker GmbH, Germany) database for further analysis, annotation and reporting. A 5% cut-off for variant calls was used and the results were only interpreted if the coverage was >100x.
GS Junior platform
Alignment against reference NCBI build 37 (hg19) and variant calling was carried out using AVA software (Roche). Thresholds for variant calling were set to a minimum allele frequency of 5% with a coverage of at least 100x. All variants were visually inspected using the AVA software (Roche). Annotation of variants was done according to the HGVS nomenclature.
PGM Ion Torrent platform
Raw data processing, sequence generation and alignment to the reference hg19 genome were conducted using the Torrent Suite software (version 4.0; Life Technologies). Variants were identified using the variant caller plug-in package. For hotspot mutations, a minimum allele frequency of 3% was set and for novel mutations, at least a 5% allele frequency was set as the cut-off level (with coverage >100x). Annotation of variants was performed with the CLC genomics workbench (version 6.5) followed by the visual inspection of putative mutations using the IGV browser.
Results
DNA concentration
DNA extraction from the 30 NSCLC samples was carried out with three different DNA extraction systems and the DNA concentration was measured using individual methods as described above. Table II summarises the resulting DNA concentrations. While the DNA concentration ranges measured with the Qubit 2.0 fluorometer in institutes A and C were comparable, the values measured using the NanoDrop 2000c spectrophotometer in institute B were generally higher due to the different principles of measurement. We observed a 1.4- to 856-fold and a 3.9- to 156-fold difference in the concentrations of institute B compared with the concentration values in institutes A and C, respectively with average differences of 133- and 30-fold. Particulary in samples with concentrations below 10 ng/µl, the measurements showed high deviations (Table II). Although only minimal amounts of DNA were measured in some samples from institutes A and C, the maximum volume possible was used for the massive parallel analysis for comparative purposes.
Table II.
DNA concentration.
| Sample no. | Institute A (ng/µl) | Institute B (ng/µl) | Institute C (ng/µl) |
|---|---|---|---|
| 1 | 31 | 362.9 | 20.8 |
| 2 | 2.9 | 7.84 | 0.85 |
| 3 | 3.32 | 109.16 | 7.81 |
| 4 | 0.1 | 4.03 | 0.41 |
| 5 | 12.8 | 186.49 | 11.7 |
| 6 | 7.5 | 14.92 | 1.15 |
| 7 | 16.6 | 374.76 | 4.55 |
| 8 | 2.44 | 24.24 | 1.48 |
| 9 | 26.6 | 504.4 | 44.8 |
| 10 | 0.1 | 3.61 | <0.5 |
| 11 | 10.3 | 26.1 | 3.42 |
| 12 | 5.7 | 266.83 | 2.36 |
| 13 | 8.06 | 11.58 | 2.94 |
| 14 | 4.56 | 21.62 | 1.18 |
| 15 | 2.06 | 28.72 | 4.94 |
| 16 | 2.7 | 15.64 | 1.25 |
| 17 | 1.29 | 25.68 | 3.58 |
| 18 | 3.78 | 17.32 | 1.99 |
| 19 | 0.1 | 19.24 | 4.3 |
| 20 | 8.8 | 204.08 | 1.3 |
| 21 | 0.1 | 1.3 | 0.1 |
| 22 | 18.4 | 470.92 | 12.2 |
| 23 | 0.83 | 204.61 | 6.08 |
| 24 | 0.97 | 52.5 | 5.34 |
| 25 | 0.16 | 103.01 | 2.85 |
| 26 | 0.3 | 103.01 | 8.52 |
| 27 | 0.24 | 60.02 | 1.31 |
| 28 | 0.1 | 85.57 | 1.06 |
| 29 | 0.1 | 47.74 | 0.7 |
| 30 | 0.1 | 56.77 | 4.2 |
DNA extraction from 30 non-small cell lung cancer (NSCLC) samples was carried out with three different DNA extraction systems from Qiagen: BioRobot M48, QIA Symphony SP as well as manual extraction. After the extraction, concentration was measured with the Qubit 2.0 fluorometer in institutes A and C, or with the NanoDrop 2000c spectrophotometer in institute B.
Platform comparison summary
The median amplicon sizes for all platforms ranged from 125–345 bp, allowing the amplification of target sequences from degraded DNA obtained from FFPE material (Table III). The number of analysed amplicons ranged from 4 up to 137. Depending on the platform used, the number of samples analysed in one single run varied from 8 up to 48. The maximum number of median reads per sample was approximately 500.000 on the PGM followed by approximately 350.000 on the MiSeq and 5007 reads on the GS Junior. In general, the read coverage for each amplicon was considered to be sufficient for each sample with median values of between 1290 and 7409.
Table III.
Sequencing statistics.
| MiSeq™ | PGM Ion Torrent™ | GS Junior | |
|---|---|---|---|
| No. of Amplicons | 102 | 137 | 4 |
| Median amplicon size | 150 bp | 125 bp | 345 bp |
| Samples/run | 48 | 8–10 | 15 |
| Median reads/sample | ~350.000 | ~ 500.000 | 5007 |
| Median coverage/amplicon | 7409x | 2500x | 1290x |
Overview of the different massive parallel sequencing (MPS) platforms. bp, base pairs.
Influence of macrodissection
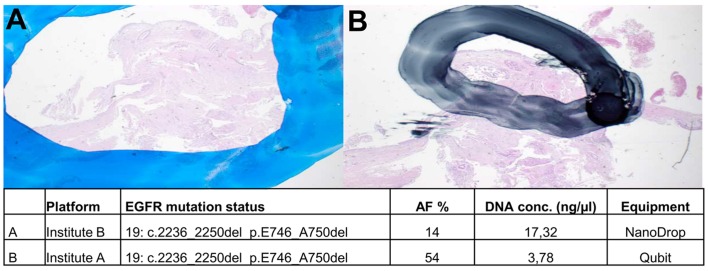
Manual macrodissection of marked regions on unstained sections was performed to enrich for tumour cells in the extraction. Depending on the strictness of separating tumour cells from normal cells, the resulting allele frequencies for mutant vs. wild-type alleles can vary. This is of particular importance when analysing samples with low tumour cell content or when allele frequencies are expected to be low. Depending on the size of the marked area, the proportion of tumour and normal cells and therewith the allele frequencies could differ in the same sample. This is exemplified in Fig. 1; the area used for DNA extraction was larger in institute B than in institute A. Thus, the corresponding allele frequencies for the EGFR mutation of this sample were determined to be 14 and 54%, respectively.
Figure 1.
Macrodissection. Tumor cells on H&E-stained slides were marked by experienced pathologists. Manual macrodissection of marked regions in (A) resulted in an AF of 14% whereas manual macrodissection of tissue in (B) resulted in 54% AF. H&E, hematoxylin and eosin; AF, allele frequency.
Detection of EGFR mutations
Concerning the expected EGFR mutation status, we found concordance in 26 out of 26 samples (Table IV). In all samples, the EGFR mutation status was correctly identified by all participants using a 5% threshold for allele frequencies and at least a coverage rate of 100 (Table IV). The EGFR mutation status of our sample cohort was comprised of 12 single point mutations, 9 complex exon 19 deletions/insertions and 11 wild-type samples. In three cases, two EGFR mutations were present (Table IV, nos. 1, 20 and 21).
Table IV.
EGFR mutation status.
| Case | Expected result | A | B | C | Tumor cell content | A | B | C | A AF% | B AF% | C AF% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | p.G719A | √ | √ | √ | 50 | 13936 | 4917 | 3001 | 20 | 15 | 24 |
| 1 | p.V834L | √ | √ | √ | 50 | 9112 | 4917 | 4829 | 17 | 18 | 22 |
| 2 | p.L838R | √ | √ | √ | 80 | 1430 | 10143 | 5885 | 17 | 17 | 17 |
| 3 | p.E746_A750del | √ | √ | √ | 60 | 10584 | 3379 | 9216 | 79 | 45 | 44 |
| 4 | p.E746_A750del | √ | √ | √ | 10 | 1102 | 512 | 5116 | 23 | 18 | 22 |
| 5 | wt | √ | √ | √ | 90 | wt | wt | wt | |||
| 6 | wt | √ | √ | √ | 70 | wt | wt | wt | |||
| 7 | wt | √ | √ | √ | 60 | wt | wt | wt | |||
| 8 | wt | √ | √ | √ | 30 | wt | wt | wt | |||
| 9 | wt | √ | √ | √ | 30 | wt | wt | wt | |||
| 10 | – | n.a. | n.a. | n.a. | 80 | n.a. | n.a. | n.a. | |||
| 11 | p.E746_A750del | √ | √ | √ | 60 | 9562 | 5020 | 1947 | 67 | 60 | 49 |
| 12 | p.L858R | √ + p.T790M | √ | √ | 50 | 29429/34779 | 8291 | 2820 | 28/1.03 | 21 | 12 |
| 13 | p.E746_A750del | √ | √ | √ | 40 | 9936 | 11132 | 2820 | 31 | 29 | 25 |
| 14 | p.L858R | √ | √ | √ | 30 | 35355 | 6911 | 5693 | 36 | 33 | 13 |
| 15 | p.L858R | √ | √ | √ | 50 | 14143 | 1381 | 3407 | 31 | 41 | 31 |
| 16 | p.E746_A750del | √ | √ | √ | 70 | 11546 | 1472 | 1975 | 34 | 51 | 33 |
| 17 | p.L858R | n.a. | n.a. | √ | 70 | n.a. | n.a. | 3336 | n.a. | n.a. | 20 |
| 18 | p.E746_A750del | √ | √ | √ | n.d. | 4179 | 406 | 1521 | 54 | 14 | 10 |
| 19 | p.L747_A751delinsP | √ | n.a. | √ | 70 | 7445 | n.a. | 1585 | 75 | n.a. | 54 |
| 20 | p.L747_P753delinsS | √ | √ | √ | 80 | 8010 | 5816 | 4221 | 75 | 59 | 49 |
| 20 | p.A755D | √ | √ | √ | 80 | 7297 | 5816 | 4221 | 74 | 59 | 59 |
| 21 | p.E709A | √ | √ | √ | 80 | 716 | 3273 | 4662 | 23 | 24 | 22 |
| 21 | p.G719S | √ | √ | √ | 80 | 2102 | 3273 | 4640 | 9 | 24 | 20 |
| 22 | p.E746_A750del | √ | √ | √ | 50 | 8391 | 33615 | 1968 | 62 | 56 | 50 |
| 23 | p.L858R | √ + p.T790M | √ | √ | 30 | 20413/10246 | 11389 | 1994 | 27/1.42 | 20 | 18 |
| 24 | p.L858R | √ | √ | √ | 30 | 9794 | 16509 | 1714 | 34 | 26 | 30 |
| 25 | wt | n.a. | √ | √ | 60 | wt | wt | wt | |||
| 26 | wt | √ | √ | √ | 60 | wt | wt | wt | |||
| 27 | wt | √ | √ | √ | 70 | wt | wt | wt | |||
| 28 | wt | √ | √ | √ | 60 | wt | wt | wt | |||
| 29 | wt | √ | √ | √ | 70 | wt | wt | wt | |||
| 30 | wt | √ | √ | √ | n.d. | wt | wt | wt |
Concerning the epidermal growth factor receptor (EGFR) mutation status, we found concordance in 26/26 samples. The mutation status was analysed previously with conventional methods. Institute A found two resistance mutations in samples 12 and 23. AF%, allele frequency; hook, concordant EGFR result; n.a., not analysable, n.d., not determined; wt, wild-type.
In only one case (no. 10), parallel sequencing was unsuccessful due to either failed PCR amplification or insufficient coverage. This case, which could not be analysed by conventional methods previously, was included intentionally to test the limits of parallel sequencing. In three cases (nos. 17, 19 and 25) with limited tumour material, parallel sequencing failed depending on the DNA extraction method. Institute A, using the BioRobot M48, did not get any sequencing results for samples 17 and 25, which was due to high salt concentrations that inhibited the multiplex PCR. Samples 17 and 19 could not be analysed by institute B due to the high degradation of samples and failed amplification.
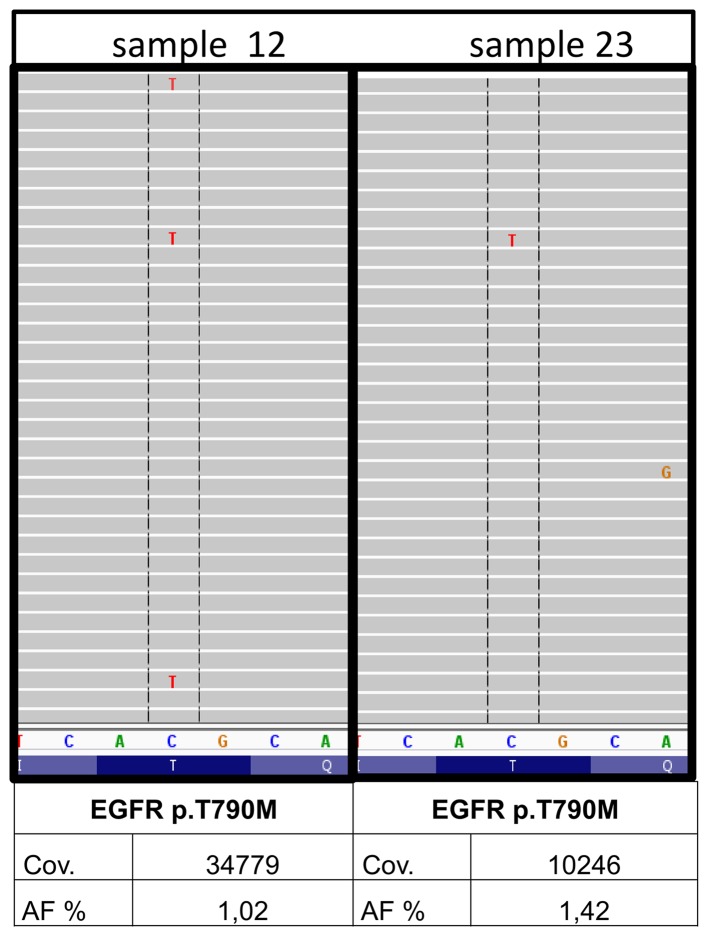
In 2 out of the 30 samples, minor p.T790M clones of the EGFR gene were detected (nos. 12 and 23) by institute A. The underlying mutation was found with 1.03 and 1.42% allele frequency with a coverage of 34779 and 10246, respectively and balanced forward and reverse reads (Fig. 3). A qPCR system (therascreen® EGFR RGQ PCR kit; Qiagen) with a detection limit of 1% allele frequency was used for the verification of originally extracted DNA samples (BioRobot M48; Qiagen), newly extracted DNA samples (Maxwell 16 Research system; Promega) from both samples as well as the corresponding DNA samples from institutes B and C. The minor variants could not be confirmed in any of the DNA samples. Thus, the EGFR p.T790M found in the first analysis most likely constitutes a fixation artefact.
Figure 3.
Minor variants. Minor variants could be detected in two out of 30 samples in institute A (nos. 12 and 23). The resistance mutation p.T790M in EGFR was found with 1.03 and 1.42% AF with a coverage of 34779 and 10246. AF, allele frequency; cov, coverage.
Additional mutations and fixation artefacts
Besides the EGFR mutations, additional variants were identified by institutes A and C using more comprehensive primer sets (Table V). Concordance was found in 15 additional variants, whereas 16 variants could not be confirmed due to the missing inclusion of the respective primers in the individual panels. Seven samples (nos. 1, 4, 8, 13, 20, 24 and 30) showed no additional mutations, which was confirmed by both institutes.
Table V.
Additional variations.
| Case | Gene | Nucleotide change | AA change | AF A (%) | AF C (%) |
|---|---|---|---|---|---|
| 1 | – | – | – | – | – |
| 2 | TP53 | c.469G>T | p.V157F | 80 | 79 |
| 3 | TP53 | c.637C>T | p.R213* | 79 | 34 |
| 4 | – | – | – | – | – |
| 5 | NKX2.1 | c.515A>C | p.Q172P | n.i. | 23 |
| RB1 | c.2267delA | p.Y756fs | n.i. | 91 | |
| TP53 | c.733G>T | p.G245C | 87 | 91 | |
| 6 | TP53 | c.641A>G | p.H214R | 33 | 23 |
| 7 | TP53 | c.830G>T | p.C277F | 23 | 44 |
| 8 | – | – | – | – | – |
| 9 | KRAS | c.35G>A | p.G12D | 2 | 5 |
| 10 | n.a. | n.a. | |||
| 11 | TP53 | c.1073C>T | p.P295S | 1 | 5 |
| JAK3 | c.2164G>A | p.V722I | n.i. | 37 | |
| 12 | TP53 | c.610G>T | p.E204* | 7 | 25 |
| 13 | – | – | – | – | – |
| 14 | ATM | c.2572T>C | p.F858L | n.i. | 66 |
| 15 | TP53 | c.913A>T | p.K305 | 26 | 20 |
| KIT | c.1621A>C | p.M541L | n.i. | 57 | |
| 16 | SMO | c.979G>A | p.A327T | n.i. | 45 |
| 17 | – | – | – | n.a. | – |
| 18 | TP53 | c.530C>G | p.P177R | 26 | 8 |
| 19 | TP53 | c.725G>A | p.C242Y | 81 | 34 |
| TP53 | c.555C>G | p.S185R | 73 | n.i. | |
| KIT | c.1621A>C | p.M541L | n.i. | 78 | |
| PIK3CA | c.1633G>A | p.E545K | 44 | 4 | |
| 20 | – | – | – | – | – |
| 21 | PIK3CA | c.1624G>A | p.E542K | 18 | 17 |
| 22 | CTNNB1 | c.98C>G | p.S33C | 33 | 31 |
| 23 | NOTCH1 | c.3604C>T | p.P1202S | n.i. | 5 |
| RBM10 | c.79delG | p.G27fs | n.i. | 17 | |
| 24 | – | – | – | – | – |
| 25 | SMARCA4 | c.3634G>A | p.E1212K | n.i./n.a. | 5 |
| KRAS | c.35G>A | p.G12D | n.a. | 10 | |
| 26 | KRAS | c.35G>A | p.G12D | 26 | 29 |
| 27 | KEAP1 | c.1426G>T | p.G476W | n.i. | 45 |
| MAP2K1 | c.171G>T | p.K57N | 45 | n.i. | |
| 28 | CDK6 | c.584G>T | p.S195I | n.i. | 13 |
| CDKN2A | c.253C>T | p.Q85 | n.i. | 6 | |
| 29 | HRAS | c.59C>T | p.T20I | n.i. | 5 |
| BRAF | c.1406G>A | p.G469E | FA | – | |
| NRAS | c.178G>A | p.G60R | FA | – | |
| PIK3CA | c.1633G>A | p.E545K | FA | – | |
| 30 | – | – | – | – | – |
Besides the epidermal growth factor receptor (EGFR) mutations, additional mutations could be identified with the extended primer sets used in institutes A and C. Concordance was found in 15 additional variations whereas 16 variants could not be confirmed by the other institute due to missing primer panel inclusion. Fixation artefacts were observed in sample 29. AA, amino acid; AF, allele frequency; FA, fixation artefact; n.a., not analysable; n.i., not included in primer panel; -, no variant found.
Concordant results were found in the genes CTNNB1 (no. 22), PIK3CA (nos. 19 and 21) and most frequently in TP53 (nos. 2, 3, 5, 6, 7, 11, 12, 15, 18 and 19). In two samples (nos. 9 and 26), a recurrent KRAS p.G12D mutation was identified. Notably, in sample 9 this KRAS mutation with a low allele frequency of 2.36 and 5%, respectively, was identified by both institutes, thereby confirming the true nature of this mutation (Table V).
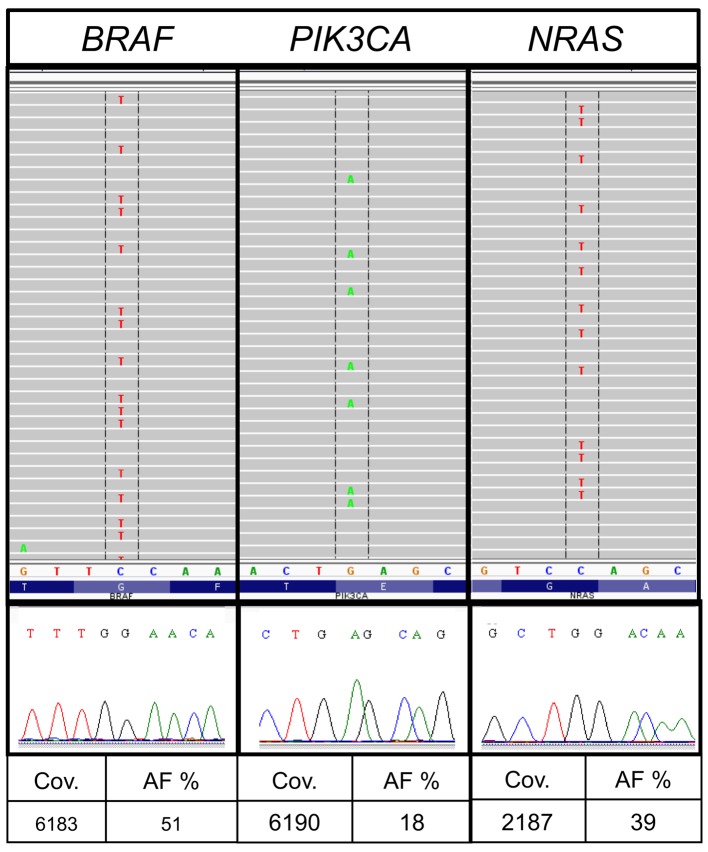
Divergent results were discovered in sample no. 29. The average number of reported variants for each sample was 172 for all allele frequencies and 23 for allele frequencies above 5% in institute A. Sample no. 29 showed a markedly higher number of variants (157) following bioinformatic analysis institute A. The sample from institute A had a very low DNA concentration (Table II) and the variants were predominantly G>A or T>C substitutions. The results included besides other variants different hotspot mutations such as BRAF c.1406G>A, p.G469E [allele frequency (AF), 51%; coverage (cov), 6813], PIK3CA c.1633G>A, p.E545K (AF, 18%; cov, 6190) and NRAS c.178G>A, p.G60R (AF, 39%; cov, 2187) (Table V and Fig. For verification, the respective regions were reanalysed with Sanger sequencing as previously described (14). The mutations could not be confirmed and were categorized as fixation artefacts.
Discussion
In routine pathological diagnostics mostly FFPE material is available for molecular characterisation. With decreasing sample sizes and increasing numbers of molecular analyses, a targeted sequencing approach using MPS systems seems to be required. Since it is well known that DNA extracted from FFPE is degraded, with a maximum size of about 350 bp (15), approaches such as whole genome, transcriptome or exome sequencing are, besides being labour-intensive and expensive, not suitable for routine diagnostics. Targeted sequencing with the focus on hotspot regions is suitable for analysing FFPE material, in a cost-effective and technically feasible way. Comparing the benchtop systems available for parallel sequencing, they show all method-specific advantages and disadvantages. The 454 GS Junior has a low throughput, but generates at the same time long runs (16,17). The Ion Torrent PGM™ is a cost-saving and fast system, but has a limited accuracy in homopolymeric regions, which also applies to the 454 GS Junior (1,16). The MiSeq has a very high throughput and low error rates, but the runtime is long (17) and it needs a higher number of samples per run to be cost efficient.
In this study, in comparing 30 lung cancer samples with three different MPS platforms, we observed good concordance in the detection of mutations using different DNA extraction methods, quantification systems and individually designed primer panels. All institutes analysed 26 out of 26 samples accurately concerning the EGFR status.
Independently of the downstream methods used, the crucial step in mutation analyses from tumour material is macrodissection and therewith the selection of the right areas. A tumour burden of 40% is recommended for Sanger sequencing (18). As MPS is more sensitive than Sanger sequencing, the amount of tumour cells required may be lower (19,20). Samples with low tumour cell content are at risk of being reported as false-negative. In contrast to our results (21) found no correlation between H&E-based morphologic assessment of tumour burden and the actual mutant allele frequency. In our cohort, the absolute allele frequencies for certain variants showed differences between the three laboratories, depending mainly on the selection of the macrodissected area. Restricted marking of tumour cells increases the detection thresholds, which may be critical for variants with low allele frequencies. Unfortunately at the same time there is an enhanced risk of 'mispicking' during the manual dissecting process. The important role of manual macrodissection is also emphasized by Ausch et al because the combination of the content of tumour cells and the allele frequency leads to the diagnostic study (22). We recommend a careful pathologic review of each individual case because the minimum percentage of tumour cells for doubtless results has not yet been defined (23). From our results, we suggest a tumour cell burden of at least 10%, which can also be reached in small biopsies.
Through the development of minimally invasive techniques biopsy sizes are decreasing. This is in contrast to the ever increasing demands of immunohistochemistry stainings and molecular analyses. Minimally invasive biopsies often deliver insufficient amounts of tissue material for subsequent analyses. We included one extra small tissue sample (no. 10) on purpose, which was originally difficult to analyse by conventional methods, to explore how the different MPS systems would cope with such a sample. None of the institutes were able to extract sufficient DNA for a reliable molecular analysis using next-generation sequencing (NGS) technologies.
In institute A, two further samples could not be analysed due to the high salt concentrations in BioRobot M48 extracts (12). The multiplex PCR for the library generation was inhibited and samples failed completely. Institute B could not analyse two samples as well due to strong DNA degradation. This can be attributed to the manual extraction method chosen byin institute B as it has been reported that automated nucleic acid extraction ensures a standardisation of sample processing and decreases time and variability in the clinical laboratory (24,25). Additionally, it is well known that manual extraction delivers less DNA than automated extraction (26). In this study, a comparison of the total DNA amounts is not possible due to the different systems used for measuring of DNA concentration. In institute C, using the automated QIASymphony SP system, only one sample failed. This extraction system was previously shown to generate DNA extracts with higher quality and concentration [Heydt et al (12)].
In FFPE material, non-reproducible sequence artefacts caused by DNA deamination induced by the sample fixation are frequently detected by all sequence analysis methods. The characteristic nucleotide transitions G>A and T>C had been found by several groups (27–29). Sequence artefacts arising from FFPE DNA are especially problematic when only limited amounts of template DNA are used for PCR amplification [Wong et al (29)]. In one of our samples, we detected mutations in hotspot regions with the typical C>T and G>A exchange which could not be validated by Sanger sequencing although they had sufficient allele frequency and coverage in MPS (Fig. 2).
Figure 2.
Fixation artefacts. In our cohort, sample 29 showed a high number of variants after the bioinformatic analysis in institute A. Hotspot mutations in BRAF, NRAS and PIK3CA were selected for validation by Sanger sequencing. The mutations could not be confirmed and were therefore assessed to be fixation artefacts. AF, allele frequency; cov, coverage.
Since the fixation artefacts are amplified during all PCR-based methods and appear as false-positive variants, it is advisable to reduce the DNA amplification steps during mutational analyses. Hybrid selection methods like Nanostring® or SureSelect (Agilent Technologies) work without a preamplification step. Also, an approach from Udar et al where the two DNA strands were processed individually minimises fixation artefacts (30). Two independent libraries were combined and sequenced on the MiSeq (Illumina) instrument. Variant frequencies were calculated using information from both strands and are narrowed down.
Notably, the KRAS mutation (c.35G>A, p.G12D) in sample nine, which could also be attributed to a fixation artefact, was identified by two institutes with allele frequencies of 2.36 and 5% confirming the true nature of this mutation (Table V). Most of the artefacts appear once but not in duplicates so one solution to detect C>T (and G>A) sequence artefacts when using FFPE-DNA is to prepare analysis in duplicates. Verification of such low allele frequencies with an alternative method is a challenge, because most methods (Sanger sequencing, high resolution melting) have a higher detection limit than MPS.
The majority of patients with lung cancer receiving EGFR-tyrosine kinase inhibitor (TKI) therapy acquire resistance after a median of 10–16 months (31). Intense study in these NSCLCs has identified two major mechanisms of developing resistance to first generation TKIs: secondary resistance mutations within the same gene and 'oncogene kinase switch' systems with an overlap into another pathway (32). Also, new sensitive detection methods like MPS have identified a proportion of TKI-naive tumours that carry the secondary resistance mutation p.T790M in the EGFR gene; these resistant clones may be selected after exposure to TKI inhibitors (32–35). In institute A, two samples (nos. 12 and 23) with minor clones for the EGFR resistance mutation p.T790M were found (Table IV). Due to the low allele frequency, validation with Sanger sequencing seemed to be impossible. We therefore used a qPCR approach with a detection limit of 1%. Neither the DNA extracts from institutes B and C, nor the newly prepared or the primary DNA extracts from institute A, showed the resistance mutation (data not shown). Therefore, for the analysis of DNA from FFPE tissues, a general detection limit of 5% seems to balance sensitivity vs. reproducibility.
Acknowledgments
We thank Professor Wolfgang Hartmann (Institute of Pathology, University Hospital Muenster) for performing the pathological review of clinical material.
References
- 1.Endris V, Penzel R, Warth A, Muckenhuber A, Schirmacher P, Stenzinger A, Weichert W. Molecular diagnostic profiling of lung cancer specimens with a semiconductor-based massive parallel sequencing approach: feasibility, costs, and performance compared with conventional sequencing. J Mol Diagn. 2013;15:765–775. doi: 10.1016/j.jmoldx.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Clinical Lung Cancer Genome Project (CLCGP) Network Genomic Medicine (NGM) A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulahannan D, Kovac MB, Mulholland PJ, Cazier JB, Tomlinson I. Technical and implementation issues in using next-generation sequencing of cancers in clinical practice. Br J Cancer. 2013;109:827–835. doi: 10.1038/bjc.2013.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, Al-Kateb H, Nguyen TT, Duncavage EJ, Cottrell CE, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121:631–639. doi: 10.1002/cncr.29089. [DOI] [PubMed] [Google Scholar]
- 5.Tops BB, Normanno N, Kurth H, Amato E, Mafficini A, Rieber N, Le Corre D, Rachiglio AM, Reiman A, Sheils O, et al. Development of a semi-conductor sequencing-based panel for genotyping of colon and lung cancer by the Onconetwork consortium. BMC Cancer. 2015;15:26. doi: 10.1186/s12885-015-1015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JY, Kim SH, Lee YS, Lee SY, Hwang JA, Kim JY, Yoon SJ, Lee GK. Comparison of targeted next-generation sequencing with conventional sequencing for predicting the responsiveness to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy in never-smokers with lung adenocarcinoma. Lung Cancer. 2014;85:161–167. doi: 10.1016/j.lungcan.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 7.de Koning TJ, Jongbloed JD, Sikkema-Raddatz B, Sinke RJ. Targeted next-generation sequencing panels for monogenetic disorders in clinical diagnostics: the opportunities and challenges. Expert Rev Mol Diagn. 2014;15:61–70. doi: 10.1586/14737159.2015.976555. [DOI] [PubMed] [Google Scholar]
- 8.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: A practical perspective. Clin Biochem Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 9.Sikkema-Raddatz B, Johansson LF, de Boer EN, Almomani R, Boven LG, van den Berg MP, van Spaendonck-Zwarts KY, van Tintelen JP, Sijmons RH, Jongbloed JD, Sinke RJ. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–1042. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]
- 10.Snow AN, Stence AA, Pruessner JA, Bossler AD, Ma D. A simple and cost-effective method of DNA extraction from small formalin-fixed paraffin-embedded tissue for molecular oncologic testing. BMC Clin Pathol. 2014;14:30. doi: 10.1186/1472-6890-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchetti I, Iervasi G, Mazzanti CM, Lessi F, Tomei S, Naccarato AG, Aretini P, Alberti B, Di Coscio G, Bevilacqua G. Detection of the BRAF(V600E) mutation in fine needle aspiration cytology of thyroid papillary microcarcinoma cells selected by manual macrodissection: an easy tool to improve the preoperative diagnosis. Thyroid. 2012;22:292–298. doi: 10.1089/thy.2011.0107. [DOI] [PubMed] [Google Scholar]
- 12.Heydt C, Fassunke J, Künstlinger H, Ihle MA, König K, Heukamp LC, Schildhaus HU, Odenthal M, Büttner R, Merkelbach-Bruse S. Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics. PLoS One. 2014;9:e104566. doi: 10.1371/journal.pone.0104566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ihle MA, Fassunke J, König K, Grünewald I, Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Büttner R, Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JH, Gouda-Vossos A, Dzamko N, Halliday G, Huang Y. DNA extraction from fresh-frozen and formalin-fixed, paraffin-embedded human brain tissue. Neurosci Bull. 2013;29:649–654. doi: 10.1007/s12264-013-1379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey KG, Herrera-Galeano JE, Redden CL, Luu TV, Servetas SL, Mateczun AJ, Mokashi VP, Bishop-Lilly KA. Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood. BMC Genomics. 2014;15:96. doi: 10.1186/1471-2164-15-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 18.Warth A, Penzel R, Brandt R, Sers C, Fischer JR, Thomas M, Herth FJ, Dietel M, Schirmacher P, Bläker H. Optimized algorithm for Sanger sequencing-based EGFR mutation analyses in NSCLC biopsies. Virchows Arch. 2012;460:407–414. doi: 10.1007/s00428-012-1219-x. [DOI] [PubMed] [Google Scholar]
- 19.Moskalev EA, Stöhr R, Rieker R, Hebele S, Fuchs F, Sirbu H, Mastitsky SE, Boltze C, König H, Agaimy A, et al. Increased detection rates of EGFR and KRAS mutations in NSCLC specimens with low tumour cell content by 454 deep sequencing. Virchows Arch. 2013;462:409–419. doi: 10.1007/s00428-013-1376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hlinkova K, Babal P, Berzinec P, Majer I, Mikle-Barathova Z, Piackova B, Ilencikova D. Evaluation of 2-year experience with EGFR mutation analysis of small diagnostic samples. Diagn Mol Pathol. 2013;22:70–75. doi: 10.1097/PDM.0b013e31827e6984. [DOI] [PubMed] [Google Scholar]
- 21.Portier BP, Kanagal-Shamanna R, Luthra R, Singh R, Routbort MJ, Handal B, Reddy N, Barkoh BA, Zuo Z, Medeiros LJ, et al. Quantitative assessment of mutant allele burden in solid tumors by semiconductor-based next-generation sequencing. Am J Clin Pathol. 2014;141:559–572. doi: 10.1309/AJCP1JUGQMW7ZNTL. [DOI] [PubMed] [Google Scholar]
- 22.Ausch C, Buxhofer-Ausch V, Oberkanins C, Holzer B, Minai-Pour M, Jahn S, Dandachi N, Zeillinger R, Kriegshäuser G. Sensitive detection of KRAS mutations in archived formalin-fixed paraffin-embedded tissue using mutant-enriched PCR and reverse-hybridization. J Mol Diagn. 2009;11:508–513. doi: 10.2353/jmoldx.2009.090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirker R, Herth FJ, Kerr KM, Filipits M, Taron M, Gandara D, Hirsch FR, Grunenwald D, Popper H, Smit E, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010;5:1706–1713. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- 24.Dundas N, Leos NK, Mitui M, Revell P, Rogers BB. Comparison of automated nucleic acid extraction methods with manual extraction. J Mol Diagn. 2008;10:311–316. doi: 10.2353/jmoldx.2008.070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esona MD, McDonald S, Kamili S, Kerin T, Gautam R, Bowen MD. Comparative evaluation of commercially available manual and automated nucleic acid extraction methods for rotavirus RNA detection in stools. J Virol Methods. 2013;194:242–249. doi: 10.1016/j.jviromet.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Eijk R, Stevens L, Morreau H, van Wezel T. Assessment of a fully automated high-throughput DNA extraction method from formalin-fixed, paraffin-embedded tissue for KRAS, and BRAF somatic mutation analysis. Exp Mol Pathol. 2013;94:121–125. doi: 10.1016/j.yexmp.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: Causes and strategies for minimization. Clin Chem. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 28.Marchetti A, Felicioni L, Buttitta F. Assessing EGFR mutations. N Engl J Med. 2006;354:526–528. doi: 10.1056/NEJMc052564. [DOI] [PubMed] [Google Scholar]
- 29.Wong SQ, Li J, Tan AY, Vedururu R, Pang JM, Do H, Ellul J, Doig K, Bell A, MacArthur GA, et al. CANCER 2015 Cohort: Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics. 2014;7:23. doi: 10.1186/1755-8794-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udar N, Haigis R, Gros T, Kerry N, Barnes B, Pokholok D, Ross M, Lucio-Eterovic AK, Zhang Q, Zenali M, Jaeger E. A novel technique that distinguishes low-level somatic DNA variants from FFPE-induced artifacts in solid tumors by next-generation sequencing (NGS) International Association for the Study of Lung Cancer. 2013 [Google Scholar]
- 31.Oxnard GR, Arcila ME, Sima CS, Riely GJ, Chmielecki J, Kris MG, Pao W, Ladanyi M, Miller VA. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10:281–289. doi: 10.3816/CLC.2009.n.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 34.Rosell R, Molina MA, Costa C, Simonetti S, Gimenez-Capitan A, Bertran-Alamillo J, Mayo C, Moran T, Mendez P, Cardenal F, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17:1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- 35.Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, Ho BC, Chang GC, Shih JY, Yu SL, Yang PC. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]