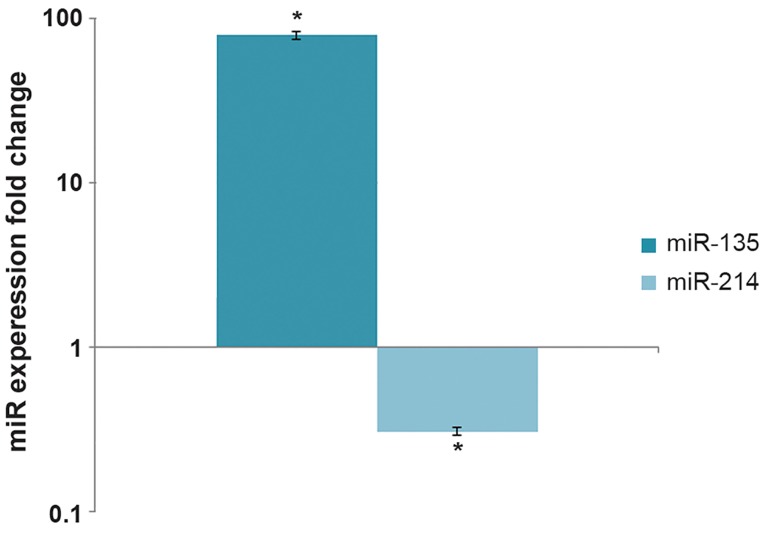Abstract
Objective
MicroRNAs (miRNAs) are a class of small non-coding RNAs that play pivotal roles in many biological processes such as regulating skeletal muscle development where alterations in miRNA expression are reported during myogenesis. In this study, we aimed to investigate the impact of predicted miRNAs and their target genes on the myoblast to myocyte differentiation process.
Materials and Methods
This experimental study was conducted on the C2C12 cell line. Using a bioinformatics approach, miR-214 and miR-135 were selected according to their targets as potential factors in myoblast to myocyte differentiation induced by 3% horse serum. Immunocytochemistry (ICC) was undertaken to confirm the differentiation process and quantitative real-time polymerase chain reaction (PCR) to determine the expression level of miRNAs and their targets.
Results
During myoblast to myocyte differentiation, miR-214 was significantly down- regulated while miRNA-135, Irs2, Akt2 and Insr were overexpressed during the process.
Conclusion
miR-214 and miR-135 are potential regulators of myogenesis and are involved in skeletal muscle development through regulating the IRS/PI3K pathway.
Keywords: Myoblast, Differentiation, miR-214, miR-135
Introduction
Several global approaches have been applied in order to better understand the molecular mechanism of myogenesis. Skeletal muscle is derived from the somites, the embryonic structures in mammalians that produce differentiated muscular tissue after progressive subdivisions (1). Myoblasts (immature muscle cells) exit from cell cycle after a defined proliferation time to then become terminally differentiated myocytes (1, 2). Finding the protein network underlying skeletal muscle differentiation will lead to a better understanding of muscle biology, muscle dysfunction and pathogenesis of various muscular disorders, and may provide new approaches for therapy.
The fate of myogenic precursor cells is first determined by transcription factors Pax3/Pax7, followed by regulation of myogenic differentiation (MyoD) through the expression of myogenic regulatory factors (MRFs) in the skeletal muscle lineage (3). MyoD is thus considered as a marker of terminal commitment to muscle fate. Muscle-specific genes, including myosin heavy chain (MHC) genes, are expressed in the last phase of this multiregulated program, where mononucleated myocytes specifically fuse to each other to form multinucleated myotubes (4). Table 1 provides the most important transcription factors, growth factors and related signaling pathways involved in muscle differentiation.
This myogenic process is controlled by numerous signaling pathways (5). In recent years, the mammalian target of rapamycin (mTOR) has emerged as a key regulator of skeletal myogenesis by controlling multiple stages of myogenic differentiation through distinct mechanisms (6). mTOR is a Ser/Thr kinase that operates as an important regulator of cellular differentiation (7). Several mediators of amino acid signals have been reported to lie upstream of mTOR, including its activator phosphoinositide 3-kinase (PI3K) (8).
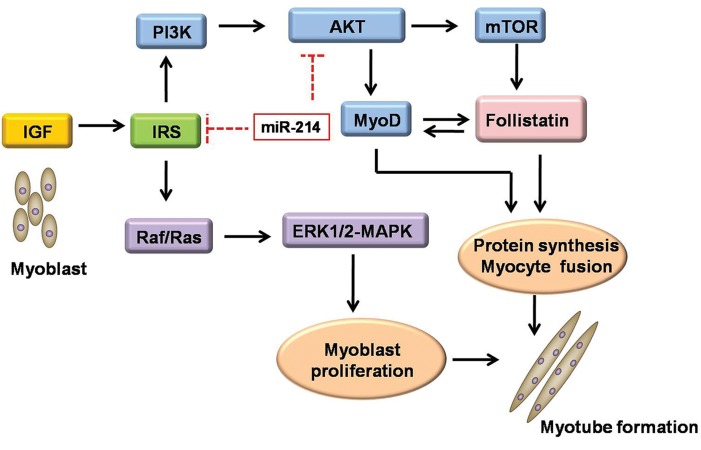
mTOR signaling regulates myoblast differentiation by controlling the myogenic expression of insulin-like growth factor (IGF) at the transcriptional level via a muscle-specific enhancer (9). IGFs are critically involved in skeletal muscle development, adult muscle regeneration and hypertrophy (10). In cultured myoblasts, growth factor deprivation initiates the differentiation process, owing to the induction of IGF (11) and subsequent activation of the IGFreceptor (Fig .1). It then initiates an autocrine signaling cascade through insulin receptor substrates (IRS) 1 and 2 that activate PI3K and in turn a major downstream pathway mediated by and RAC-beta serine/threonine-protein kinase (AKT) (12). The IGF-1/AKT/mTOR pathway is therefore an important regulatory component which controls muscle development (13). Upon binding to its membrane receptor (IGFR1), IGF- 1 activates both extracellular-signal-regulated kinases1/2 (ERK1/2) and PI3K/AKT/mTOR pathways. ERK1/2 is required for myoblast proliferation (14), while the PI3K/Akt/mTOR pathway promotes protein synthesis and is essential for myotube formation through a MyoD/ follistatin pathway, which requires mTOR kinase activity (13, 15). Thus, PI3K/AKT pathway is a vital intracellular signaling mechanism that pivotal for muscle development (16).
Fig.1.
Myogenesis through the IGF/AKT/mTOR pathway. IGF is thought to initiate an autocrine signaling cascade through the IGF-I receptor, and IRS 1 and 2 that activate PI3K and mitogen-activated protein kinases (MAPK). PI3K activates AKT, however, either PI3K or AKT is sufficient for myoblast differentiation and fusion. PI3K and AKT drive differentiation by increasing the transcriptional activity of MyoD and activating the mTOR pathway. mTOR expression and activity increases during differentiation leading to an increase in the activity of its downstream target, follistatin, which prevents myostatin (the most powerful inhibitor of muscle growth) from executing its inhibitory effect on muscle development. AKT; RAC-beta serine/threonine-protein kinase, ERK1/2; Extracellular-signal-regulated kinases1/2, IGF; Insulin-like growth factor, IRS; Insulin receptor substrates, mTOR; Mammalian target of rapamycin, MyoD; Myogenic differentiation, PI3K; Phosphoinositide 3-kinase, Raf; Rapidly accelerated fibrosarcoma and Ras; Rat sarcoma.
The process of myogenesis is extremely complex and requires a specific organization of signaling molecules that regulate the expression of particular genes and miRNAs (17, 18).
Recently, microRNAs (miRNAs), a class of evolutionarily conserved and small non-coding RNAs (19), have emerged as novel and essential regulators of myogenesis (20). Mature miRNAs are 21– 25 nucleotides in length and are partially complementary to one or more mRNA molecules. Their main function is to down-regulate gene expression in a variety of manners including translational repression, mRNA cleavage and de adenylation (13, 21). Regulation of myogenic gene expression by miRNAs has emerged as a new level of control for myogenesis. For example, miRNAs can promote differentiation by repressing negative regulators of transcriptional activity or suppress it by repressing positive regulators. Muscle-specific miRNAs such as miR-1, miR-133, and miR-206 have a central role in myogenesis (22). Other miRNAs have also been implicated in muscle development, including miR-26a (23), miR-27b (24), miR-29 (25), miR- 125b (26), miR-155 (27), miR-128a (28) and miR- 181 (29).
Additional miRNAs have been reported to participate in skeletal myogenesis and include miR-24 (30), miR-378 (31), miR221/222 (32), miR-486 (33), miR-208b/miR-499 (34) and miR-214 (35). Furthermore, several miRNAs have been demonstrated to regulate the PI3K/AKT/mTOR pathway during myogenesis (33). Although an increasing number of miRNAs are found to function in myognenesis, knowledge about the role of individual miRNAs in the molecular network of muscle development remains poorly understood and still mainly unknown. As a new class of regulators of skeletal myogenesis, miRNAs hold the potential to identifying novel biomarkers and developing therapeutic strategies for muscular diseases. The aim of this study was to quantify expression changes of bioinformatically selected miR-214 and miR-135 and their targets, Insulin receptor substrates (Irs2), RAC-beta serine/threonine-protein kinase (Akt2) and insulin receptor (Insr) to better understand the role of miRNAs during the muscle differentiation process.
In our survey, by using the C2C12 myoblast cell line, we identified two miRNA which had significant expression change during C2C12 differentiation process.
Table 1.
Muscle-specific factors during myogenic process
| Proliferation | Differentiation | |
|---|---|---|
| Myogenic factors | PAX3/7, HDAC4, MEF2, IGFI | MRF (MyoD, MRF4, Myf5), MEF2, IGFII, Myogenin (MyoG), MHC, M-CK |
| Signaling pathway | MEK/ERK pathway | mTOR/PI3K/AKT pathway |
Transition from proliferation to differentiation, which is accompanied by the down-regulation of Pax-7 and up-regulation of Myogenin and MRF-4 is dependent on both MyoD and the mTOR/PI3K/AKT pathway. MyoD and Myf5 are both considered markers of terminal com- mitment to muscle fate. Muscle-specific genes such as myosin heavy chain genes (MHC genes) and muscle creatine kinase (M-CK) are expressed in the last phase of this multi-regulated program.
AKT; RAC-beta serine/threonine-protein kinase, ERK; Extracellular-signal-regulated kinases, IGF; Insulin-like growth factor, IRS; Insulin receptor substrates, MEF2; Myocyte enhancer factor2, MEK; Mitogen/extracellular signal-regulated Kinase, MRF; Myogenic regulatory factors, mTOR; Mammalian target of rapamycin, MyoD; Myogenic differentiation, HDAC4; Histone deacetylase 4 and PI3K; Phospho- inositide 3-kinase.
Materials and Methods
Cell culture
463 In our experimental study, C2C12 myoblast cell line was obtained from a cell bank (Stem Cells Technology Research Center, Tehran, Iran). These cells were cultured in growth medium [GM, Dulbecco’s Modified Eagle Medium, (Gibco, UK)] containing 10% fetal bovin serum (Gibco, UK) 24 hours before being induced to differentiate, at 37˚C and 5% CO2.
When cell density reached 70%, cells were digested with 0.25% trypsin (Gibco, UK) and then seeded into culture dishes. When inducing C2C12 myoblasts to differentiate, cell density must reach >90% prior to changing GM to differentiation medium [DM, Dulbecco’s Modified Eagle Medium supplemented with 3% horse serum (Gibco, UK)]. The cells were subsequently incubated with DM for another 72 hours to undergo differentiation. The control cell line (the undifferentiated C2C12 line) was cultured only in growth medium for the same time period. All cell cultures were performed at least in triplicate.
Immunocytochemistry (ICC)
After inducing myogenic differentiation, C2C12 cells cultured in 12-well plates were then washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (Sigma, USA) for 15 minutes. Triton-100 (0.5%, Sigma, USA) was used for permeabilization. The cells were then blocked in 2% goat serum (diluted in PBS, Sigma, USA). After blocking, the cells were incubated with antiPax7 or anti-myosin primary antibody depending on cell type (Sigma, USA) at 37˚C for 1 hour and then with the secondary fluorescent antibodies (Ray Biothech, USA) at 37˚C for 1 hour. The nuclei were stained with 4΄, 6-diamidino-2-phenylindole (DAPI, Invitrogen, USA) for 30 seconds.
Bioinformatics-based microRNA selection
Using Target Scan 6.2 (36), miRWalk (37) and RNAhybrid (38), we generated a list of miR-214 and miR-135 candidate target genes, containing a seed site for these two miRNAs. We merely chose target genes for each miRNA which were predicted with at least two algorithms.
RNA isolation and quantitative real-time polymerase chain reaction (PCR)
Cells were lysed and total RNA was extracted using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. The RNA quality and concentration were estimated using denatured gel electrophoresis and spectrophotometry respectively. About 500 ng of the total RNA was reverse transcribed into cDNA using a reverse transcription kit (Fermentas, USA) with random hexamers for target genes. cDNA synthesis of miRNAs was undertaken using the Reverse Transcription System Kit (Promega, USA) with miR-specific stem-loop primers (Table 2). Quantitative real-time polymerase chain reaction (PCR) was performed in triplicate using a 40 cycle PCR in Rotor-gene Q real-time analyzer (Corbett, Sydney, Australia). Each real-time PCR reaction contained 5 μl of 2×SYBR Premix Dimer EraserTM (TaKaRa, Japan), 3 pmol of forward and reverse primers respectively, 1 μl template of cDNA and dH2O up to the final volume of 10 μl, followed by a melting curve analysis to confirm PCR specificity. The average threshold cycle was used for data analysis by Rotor-gene Q software (Corbett, Sydney, Australia). Gene expression levels were normalized against the expression of β-actin and Snord 47(U47) as internal controls for miRNA expression. The 2 −ΔΔCtmethod was employed to estimate the relative expression level of each gene. All reactions were run in triplicate.
Table 2.
Gene-specific primers designed for real-time PCR assay
| Primer | Sequence |
|---|---|
| Reverse transcription | |
| miR-214-3p-stem-loop | 5ˊGTCGTATGCAGAGCAGGGTCCGAGGTATTCGCACTGCATACGACACTGC 3ˊ |
| miR 135a- stem-loop | 5ˊGTCGTATGCAGAGCAGGGTCCGAGGTATTCGCACTGCATACGACTCACA 3ˊ |
| miR- 214 | Forward : 5ˊGCACAGCAGGCACAGAC3ˊ |
| Forward : 5ˊGCACAGCAGGCACAGAC3ˊ | |
| miR-135a | Forward : 5ˊCGATATGGCTTTTTATTCCTA3ˊ |
| Reverse : 5ˊGAGCAGGGTCCGAGGT 3ˊ | |
| Insr | Forward : 5ˊAACAGATGCCACTAATCCTTC 3ˊ |
| Reverse : 5ˊGCCCTTTGAGACAATAATCC 3ˊ | |
| Irs2 | Forward: 5ˊCAGCCAGGAGACAAGAACTC 3ˊ |
| Reverse: 5ˊCGCTTCACTCTTTCACGAC 3ˊ | |
| Akt2 | Forward : 5ˊTTCGGCAAGGTCATTCTG 3ˊ |
| Reverse: 5ˊTGAGGGCTGTAAGGAAGG 3ˊ | |
Akt2; RAC-beta serine/threonine-protein kinase, Insr; Insulin receptor, Irs2; Insulin receptor substrates and PCR; Polymerase chain reaction.
Comparison of real time-PCR result by a highthroughput method data
We next assessed whether the candidate gene expression levels obtained by real time-PCR are comparable with other methods. To this end, real time-PCR results were compared with microarray data available in Gene Expression Omnibus (GEO, accession #GSE4694) (39).
Statistical analysis
The data were presented as mean ± standard error. To determine statistical significance, Student’s t test was applied. If not indicated otherwise, the criterion for significance was set at P<0.05.
Results
Bioinformatically predicted targets
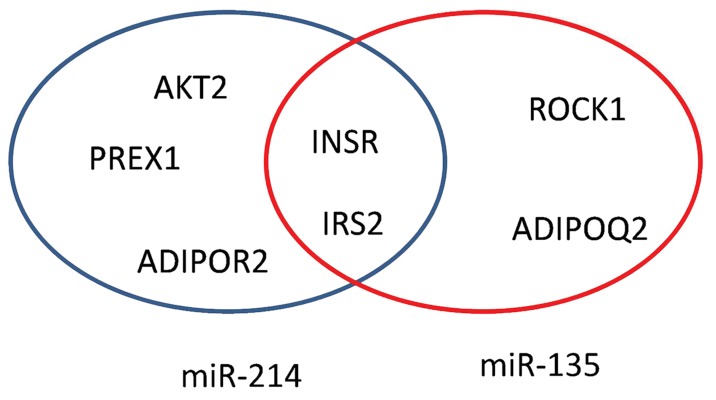
To bioinformatically predict target genes, we initially found that miR-214 and miR-135 targeted several signal molecules regulating the myogenesis process and insulin pathway such as IRS2, AKT2 and INSR (Fig .2).
Fig.2.
Clustering of predicted targets of miRNAs. IRS2 and INSR are mutual predicted targets of both miRNAs.
ADIPOR2; Adiponectin receptor 2, ADIPOQ; Adiponectin, AKT2; RAC-beta serine/threonine-protein kinase, INSR; Insulin receptor, IRS2; Insulin receptor substrate 2, Prex1; Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein and ROCK1; Rho kinase 1.
Characterization of C2C12 differentiation
Differentiation of myoblast cells to myocytes was confirmed by a positive ICC result for the specific skeletal marker, myosin .C2C12 myoblast type was confirmed by a positive ICC result for the precursor cell marker, Pax-7 (Fig .3).
Fig.3.
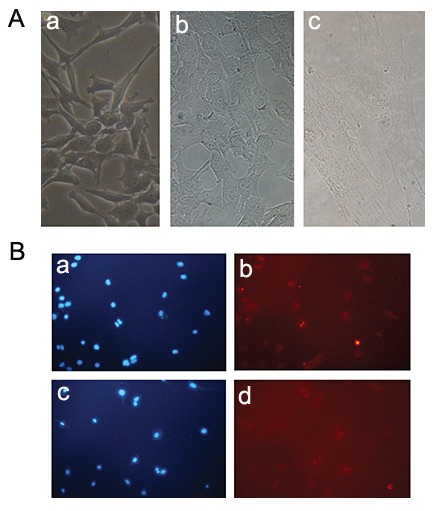
Myoblast to myocyte differentiation. A. Myoblast cells (a) differentiate into myocytes (b, c). Myocyte is indicated in part c and B. C2C12 myoblasts stained with PAX and DAPI as a positive control of precursor cells (a, b). After that, myoblasts were induced to differentiate with DMEM medium containing 3% horse serum for 3 days. The differentiated cells were seeded in a new plate and stained with MHC antibody and DAPI (c, d). DAPI; 4΄,6-diamidino-2-phenylindole, DMEM; Dulbecco’s Minimal Essential Medium and MHC; Myosine heavy chain.
miR-214 and -135 have different expression patterns during myoblast differentiation
Expression profiling of miRNAs showed that miR-214 and miR-135 had significantly altered expression during myoblast differentiation with miR-214 being down-regulated and miR-135 being up-regulated more than 70-fold in differentiated cells (Fig .4).
Fig.4.
Expression pattern of candidate miRNAs during myoblast differentiation. Based on qRT-PCR results, while miR-135 was up-regulated miR-214 was down-regulated during the differentiation process.*; P≤0.05 and qRT-PCR; Quantitative real time polymerase chain reaction.
Changes in expression of predicted targets during C2C12 differentiation
We examined the expression of Irs2, Akt2 and Insr as predicted targets of the two miRNAs studied. Interestingly, expression level of Irs2, Akt2 and Insr, directly targeted by miR-214, were upregulated in differentiated cells (Fig .4).
Comparison of real time -PCR result with microarray data
To extend the results of our quantitative real time-PCR (qRT-PCR) analysis, we analyzed all transcripts in an available microarray dataset. Chen et al. (39) had analyzed three different RNA samples from proliferating and differentiated C2C12 cells individually (6 microarrays in total).
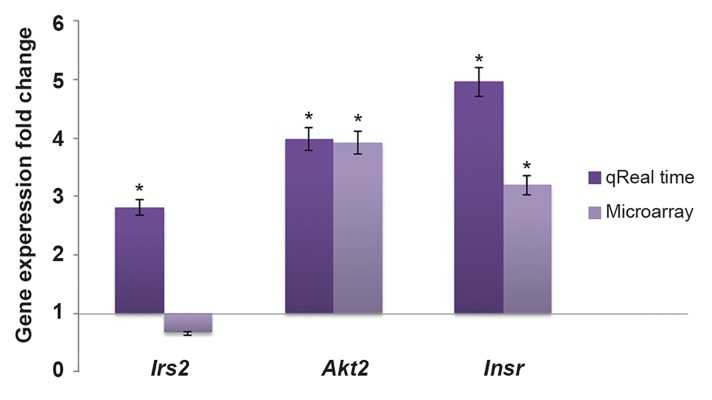
Microarray results for Insr and Akt2 reflected the same up-regulation trend (P value≤0.05) but not for Irs2 (P value=0.473, Fig .5). However, the magnitude of differential expression was different from qRT-PCR results.
Fig.5.
Comparison of the expression levels of predicted targets during myogenesis based on qRT-PCR and microarray analysis. The data were consistent between the two methods except for Irs2 which was not shown to be differentially expressed in the microarray analysis (P=0.473).
Akt2; RAC-beta serine/threonine-protein kinase, Insr; Insulin receptor Irs2; Insulin receptor substrate 2 and *; P≤0.05.
Discussion
MiRNAs play important regulatory roles in many cell processes (40,41) including the multistep differentiation process in mammalian skeletal muscle development (42). The regulation network of myogenic factors and various miRNAs is complex and appears to depend on the cell cycle and fusion stages (43). Currently, differentially expressed miRNAs are thought to be closely related to almost all aspects of muscle development and have been shown to regulate several pathways during myogenesis (43,44).
Moreover, because of important similarities between embryonic muscle development and muscle regeneration in adults, undertaking developmental studies and particularly elucidating the roles of miRNAs in this multi-step process is valuable and may have potential clinical applications (44). In this study, we report that miR-135 was differentially expressed and may thus be involved in skeletal muscle development. Our study, for the first time, also reports that miR-135 expression was up-regulated during myogenic differentiation. miR-135 may participate in the myocyte formation process through targeting unknown components (perhaps inhibitors of muscle growth) of myogenesis in addition to those targets in our prediction (activators). On the contrary, we found that mature miR-214 was already expressed in proliferating C2C12 cells, however, it was significantly downregulated following the induction of differentiation. Furthermore, our qRT-PCR analysis showed that expression level of Irs2, Akt2 and Insr, were up-regulated in differentiated cells. The upregulation of these three predicted target genes indicates that these genes are important in muscle differentiation process.
Using an Affymetrix cDNA microarray dataset (GEO accession #GSE4694) (39), we compared the expression levels of Irs2, Akt2 and Insr in undifferentiated C2C12 cells with differentiated populations. Our data were consistent with those of the microarray (P≤0.05) except for Irs2 (P=0.473). However, qRT-PCR analysis revealed that the levels of Insr, Irs2 and Akt2 transcripts were shown to be more increased in comparison with microarray analysis in terms of fold change. This discrepancy is justifiable because qRT-PCR provides a more accurate representation of changes in the level of specific transcripts than the microarray analysis. This is due to the linearity of qRT-PCR results over a wide concentration range (39,45), as confirmed by serial dilution experiments with different samples (data not shown).
Decreased expression of miR-214 accompanied with overexpression of Irs2, Akt2 and Insr in C2C12 myocytes compared with with primary muscle cells which underwent myogenic differentiation progression.
It has been reported that muscle differentiation is blocked by decreased IRS-1/2 and PI3-K activity (3,46,47). Small RNAi-based gene silencing experiments have shown that insulin signaling pathways are dependent on IRS1/2 and are required for myotube formation and glucose uptake through the activation of AKTs (46,47). In addition, AKT promotes myoblast proliferation in cooperation with mTOR, suggesting that IRS is a key factor in inducing myoblast proliferation and myotube formation by increasing AKTs levels (47). Furthermore, AKT2 is associated with insulin signaling and the AKT/mTOR pathway which lies at the center of the regulatory network controlling muscle development (28). Gene silencing has also revealed specialized roles of AKT2 in myoblast differentiation and glucose metabolism (46).
Interestingly, our study demonstrated that the down-regulation of miR-214 may accelerate myogenesis, because of increasing levels of its targets, Akt2 and Irs2, during myogenic differentiation. Indeed, it might be the result of increasing Irs/Akt activation. As an important regulator of muscle development, a single miRNA can regulate the expression of many mRNA targets. Identifying the regulatory targets of miRNAs in muscle is thus crucial, however, it will be more critical to place them in a biological pathway.
Studies in the past were largely focused on miRNAs regulating a single gene in myogenic signaling pathways (33,35). However, growing evidence suggests that miRNA can also have an effect on signal transduction pathways (13). By targeting a number of members of the same signaling pathway, miRNA can exert more profound effects than regulating one individual gene in a biological process. Thus, miR-214 down-regulation may have positive effects on myogenesis by overexpression of Akt2, Irs2 and Insr in the insulin signaling pathway.
Conclusion
We show that miR-214 and miR-135 are potential regulators of myogenesis through regulating the IRS/AKT/PI3K pathway. Further studies such as luciferase assay and western blot and in vivo experience will be needed to concentrate on the complex regulatory roles of these miRNAs Understanding how miRNA regulate myogenesis will enhance our perception of the muscle development mechanism. Combining bioinformatics, biochemical and genetic approaches together will help us to elucidate the regulatory efficacy of miRNA in myogenesis and will also potentially establish new therapeutic approaches by identifying functional miRNA candidates as potential targets for clinical purposes.
Acknowledgments
This research was funded by Qazvin University of Medical Sciences in cooperation with Stem Cell Technology Center (Bonyakhteh), Tehran, Iran. The authors declare that they have no conflict of interest and also would like to give thanks to Samira Mohammadi Yeganeh and Rezvan Tavakoli for their help during this survey.
References
- 1.Yokoyama S, Asahara H. The myogenic transcriptional network. Cell Mol Life Sci. 2011;68(11):1843–1849. doi: 10.1007/s00018-011-0629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentzinger CF, von Maltzahn J, Rudnicki MA. Extrinsic regulation of satellite cell specification. Stem Cell Res Ther. 2010;1(3):27–27. doi: 10.1186/scrt27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. MicroRNA148a promotes myogenic differentiation by targeting the ROCK1 gene. J Biol Chem. 2012;287(25):21093–21110. doi: 10.1074/jbc.M111.330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4(2) doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16(45):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Ge Y, Chen J. Mammalian target of rapamycin (mTOR) signaling network in skeletal myogenesis. J Biol Chem. 2012;287(52):43928–43935. doi: 10.1074/jbc.R112.406942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OHkinase. Proc Natl Acad Sci USA. 2005;102(40):14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003;163(5):931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton-Davis ER, Shoturma DI, Sweeney HL. Contribution of satellite cells to IGF-I induced hypertrophy of skeletal muscle. Acta Physiol Scand. 1999;167(4):301–305. doi: 10.1046/j.1365-201x.1999.00618.x. [DOI] [PubMed] [Google Scholar]
- 11.Yoon M, Chen J. Distinct amino acid-sensing mTOR pathways regulate skeletalmyogenesis. Mol Biol Cell. 2013;24(23):3754–3763. doi: 10.1091/mbc.E13-06-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry RLS, Rudnicki MA. Molecular mechanisms regulating myogenic determination and differentiation. Front Biosci. 2000;5:D750–767. doi: 10.2741/perry. [DOI] [PubMed] [Google Scholar]
- 13.Jia L, Li YF, Wu GF, Song ZY, Lu HZ, Song CC, et al. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int J Mol Sci. 2013;15(1):296–308. doi: 10.3390/ijms15010296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones NC, Fedorov YV, Rosenthal RS, Olwin BB. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J Cell Physiol. 2001;186(1):104–115. doi: 10.1002/1097-4652(200101)186:1<104::AID-JCP1015>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189(7):1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton D, Philp A, MacKenzie M, Baar K. Prolonged activation of S6K1 does not suppress IRS or PI-3 kinase signaling during muscle cell differentiation. BMC Cell Biol. 2010;11:37–37. doi: 10.1186/1471-2121-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg I, Alexander MS, Kunkel LM. miRNAS in normal and diseased skeletal muscle. J Cell Mol Med. 2009;13(1):2–11. doi: 10.1111/j.1582-4934.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo W, Nie Q, Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genomics. 2013;40(3):107–116. doi: 10.1016/j.jgg.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge Y, Chen J. MicroRNAs in skeletal myogenesis. Cell Cycle. 2011;10(3):441–448. doi: 10.4161/cc.10.3.14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honardoost M, Sarookhani MR, Arefian E, Soleimani M. Insulin resistance associated genes and miRNAs. Appl Biochem Biotechnol. 2014;174(1):63–80. doi: 10.1007/s12010-014-1014-z. [DOI] [PubMed] [Google Scholar]
- 22.Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR206 and miR-133. Dev Biol. 2008;321(2):491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Dey BK, Gagan J, Yan Z, Dutta A. MiR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009;106(32):13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei W, He HB, Zhang WY, Zhang HX, Bai JB, Liu HZ, et al. MiR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668–e668. doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA125b in skeletal myogenesis. J Cell Biol. 2011;192(1):69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seok HY, Tatsuguchi M, Callis TE, He A, Pu WT, Wang DZ. MiR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J Biol Chem. 2011;286(41):35339–35346. doi: 10.1074/jbc.M111.273276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA128a during myogenesis. J Cell Sci. 2013;126(Pt 12):2678–2691. doi: 10.1242/jcs.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8(3):278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 30.Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36(8):2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286(22):19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4(10):e7607–e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Small EM, ORourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. Regulation of PI3-kinase/ Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA. 2010;107(9):4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi K, Lu J, Zhao Y, Wang L, Li J, Qi B, et al. MicroRNA214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone. 2013;55(2):487–494. doi: 10.1016/j.bone.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 37.Dweep H, Gretz N, Sticht C. miRWalk database for miRNAtarget interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 38.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen IH, Huber M, Guan T, Bubeck A, Gerace L. Nuclear envelope transmembrane proteins (NETs) that are upregulated during myogenesis. BMC Cell Biol. 2006;7:38–38. doi: 10.1186/1471-2121-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valsecchi V, Boido M, De Amicis E, Piras A, Vercelli A. Expression of muscle-specific MiRNA 206 in the progression of disease in a murine SMA model. PLoS One. 2015;10(6):e0128560–e0128560. doi: 10.1371/journal.pone.0128560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi L, Zhou B, Li P, Schinckel AP, Liang T, Wang H, et al. MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development. Cell Signal. 2015;27(9):1895–1904. doi: 10.1016/j.cellsig.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Abmayr S, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139(4):641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guller I, Russell AP. MicroRNAs in skeletal muscle: their role and regulation in develop:ent, disease and function. J Physiol. 2010;588(Pt 21):4075–4087. doi: 10.1113/jphysiol.2010.194175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goljanek-Whysall K, Sweetman D, Munsterberg AE. microRNAs in skeletal muscle differentiation and disease. Clin Sci (Lond) 2012;123(11):611–625. doi: 10.1042/CS20110634. [DOI] [PubMed] [Google Scholar]
- 45.Qin LX, Beyer RP, Hudson FN, Linford NJ, Morris DE, Kerr KF. Evaluation of methods for oligonucleotide array data via quantitative real-time PCR. BMC Bioinformatics. 2006;7:23–23. doi: 10.1186/1471-2105-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouzakri K, Zachrisson A, Al-Khalili L, Zhang BB, Koistinen HA, Krook A, et al. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 2006;4(1):89–96. doi: 10.1016/j.cmet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Lim MJ, Choi KJ, Ding Y, Kim JH, Kim BS, Kim YH, et al. RhoA/Rho kinase blocks muscle differentiation via serine phosphorylation of insulin receptor substrate-1 and -2. Mol Endocrinol. 2007;21(9):2282–2293. doi: 10.1210/me.2007-0114. [DOI] [PubMed] [Google Scholar]
- 48.Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, et al. Follistatin complexes Myostatin and antagonizes Myostatin-mediated inhibition of myogenesis. Dev Biol. 2004;270(1):19–30. doi: 10.1016/j.ydbio.2004.01.046. [DOI] [PubMed] [Google Scholar]