Abstract
Objective
Cytochrome P450 is one of the major drug metabolizing enzyme families and its role in metabolism of cancer drugs cannot be less emphasized. The association be- tween single nucleotide polymorphisms (SNPs) in CYP1A1 and pathogenesis of chronic myeloid leukemia (CML) has been investigated in several studies, but the results observed vary based on varied risk factors. The objective of this study was to investigate the risk factors associated with the CYP1A1*2C [rs1048943: A>G] polymorphism in CML patients and its role in therapeutic response to imatinib mesylate (IM) affecting clinico-pathological parameters, in the Indian population.
Materials and Methods
In this case-control study, CYP1A1*2C was analysed in CML patients. After obtaining approval from the Ethics Committee of oncology hospital, we collected blood samples from 132 CML patients and 140 matched controls. Genom- ic DNA was extracted and all the samples were analysed for the presence of the CYP1A1*2C polymorphism using allele-specific polymerase chain reaction, and we examined the relationship of genotypes with risk factors such as gender, age, phase of the disease and other clinical parameters.
Results
We observed a significant difference in the frequency distribution of CYP1A1*2C genotypes AA (38 vs. 16%, P=0.0001), AG (57 vs. 78%, P=0.0002) and GG (5 vs. 6%, P=0.6635) between patients and controls. In terms of response to IM therapy, significant variation was observed in the frequencies of AA vs AG in major (33 vs 67%) and poor (62 vs 31%) hematological responders, and AA vs AG in major (34 vs. 65%) and poor (78 vs. 22%) cytogenetic responders. However, the patients with the GG homozygous genotype did not show any significant therapeutic outcome.
Conclusion
The higher frequency of AG in controls indicates that AG may play a protec- tive role against developing CML. We also found that patients with the AG genotype showed favorable treatment response towards imatinib therapy, indicating that this polymorphism could serve as a good therapeutic marker in predicting response to such therapy.
Keywords: Cytochrome P-450 Enzyme System, CYP1A1, Polymorphism, Chronic Myeloid Leukemia, Imatinib
Introduction
Chronic myeloid leukemia (CML) is an acquired hematopoietic stem cell disease, characterized by an increased production of immature granulocytes (blasts) which accumulate in the bone marrow and interfere with the normal blood cell production, accounting for 30% of adult leukemias (1). The symptoms of CML include bone marrow hypercellularity, anaemia, splenomegaly and leucocytosis (1). CML progresses slowly in three phases of chronic phase (CP), accelerated phase (AP) and blast phase (BP) which are differentiated by the number of blast cells in the blood and the bone marrow, and the severity of the symptoms. In 95% of CML cases, chromosomal translocation resulting in the formation of the Philadelphia (Ph) chromosome is observed (2, 3), which in turn leads to the formation of the BCR-ABL fusion gene. This reciprocal translocation, creating an elongated chromosome 9 der(9) and a truncated chromosome 22 (Ph chromosome), is the hallmark of CML and the main drugs which target this oncogenic fusion gene (BCR-ABL) are imatinib and sunitinib. The protein encoded by the fusion gene exhibits enhanced tyrosine kinase activity and plays a key role in the initiation and maintenance of CML by activating various intracellular signalling pathways resulting in uncontrolled proliferation, decreased apoptosis and survival of leukemic stem cells (4).
Since the ABL gene expresses a membraneassociated tyrosine kinase protein, the BCR-ABL transcript is also translated into a tyrosine kinase. Although the activity of this enzyme is typically controlled by other molecules, the mutant tyrosine kinase encoded by the BCR-ABL transcript results in a protein that is "always on" or constitutionally expressed, resulting in unregulated cell division (i.e. cancer). Though the BCR region also codes for serine/threonine kinases, the tyrosine kinase activity is more pertinent for drug therapy. The tyrosine kinase inhibitor imatinib mesylate (IM, Gleevec®), the CML drug of choice, inhibits the BCR-ABL tyrosine kinase function in Ph (Ph+) cells, causing functional alterations in genes involved in cell cycle control and cell adhesion, and ultimately results in the death of Ph+ cells via apoptosis . It is most effective in treating patients in the CP phase and controls the disease in about 75% of these patients (5, 6). However, in some cases, resistance develops after an initial favourable response and in some CML-CP patients it is not effective at all (7). Though the exact mechanism underlying the drug resistance is not yet clear, it has been suggested that additional acquired chromosomal aberrations and mutations in BCR-ABL kinase domain may be responsible for this resistance (8, 9). Patients in the advanced phases of AP and BP may not respond to the drug due to the acquisition of various molecular abnormalities in undifferentiated leukemic cells (10).
The genetic variability single nucleotide polymorphisms (SNPs) in genes encoding phase I and phase II drug metabolizing enzymes which detoxify the xenobiotics [xenobiotic metabolizing enzymes (XMEs)], have been linked with the variation in susceptibility of different individuals toward leukemia (11, 12) as well as with therapeutic response of individuals toward drugs (13). Our previous work on SNPs in genes encoding drug metabolizing enzymes and drug transporters showed that these polymorphisms affect drug response in breast cancer (14-16), and head and neck cancer (17).
Cytochrome P450, family 1, Subfamily A, polypeptide 1 (CYP1A1), a polymorphic gene which codes for the important phase-I XME aryl hydrocarbon hydroxylase, is involved in drug metabolism and activation of a number of exogenous procarcinogens (18) into highly reactive electrophilic carcinogenic molecules. These electrophiles can bind to DNA and form adducts, leading to mutations in tumor suppressor genes and proto-oncogenes, thus initiating carcinogenesis if not repaired by the DNA repair system. Hence, this gene may play an important role in both the etiology of cancers and as a determinant of cancer therapy response (19, 20).
Three single nucleotide polymorphisms of CYP1A1 have been studied in relation to various cancers, namely T6235C (*2A), A4889G (*2C) and C4887A (*4). CYP1A1*2C SNP A4889G (rs1048943; exon 7) (21, 22) leads to the substitution of Isoleucine (Ile) by Valine (Val) at position 462 in the protein, which results in a two-fold higher catalytic activity and mutagenicity due to its greater hydrophobicity. The association of the CYP1A1*2C polymorphism with increased susceptibility to acute lymphoid leukemia (ALL) has been reported (23). Contradictory reports are present in the literature about the association of CYP1A1*2C in ALL (24) and AML (25) patients, however, not much is known about its relation with CML except a Turkish cohort study reporting that SNPs in this gene are associated with the risk of CML (26). More importantly, to our knowledge, no reports are available on the association of this polymorphism with the CML risk in the Indian population. So, we hypothesized that this SNP in CYP1A1 may act synergistically with the BCR-ABL fusion oncogene in causing CML. This means that the individuals carrying SNPs in the CYP1A1 gene are at higher risk of developing CML than those who do not carry the SNP. The CYP1A1*2C polymorphism has also been reported to vary in frequency depending on the ethnicity. For instance, this polymorphism was less prevalent in Iran and was thus suggested to play no role in CML development in Iranian patients (27), however, it was found to be involved in the Turkish population (26). CYP1A1 may also play a role in imatinib metabolism (28). In view of such conflicting results, we have conducted a case-control study in the Indian population to examine the association of CYP1A1*2C polymorphism with CML risk and whether this polymorphism affects the therapeutic response of the patients to Imatinib.
Materials and Methods
In this case-control study, the association of CYP1A1*2C with CML was analyzed. Approval for the study was obtained from the Ethics Committee of the Hospital (Bhagwan Mahavir Medical Research CentreHyderabad, Oncology hospital) and informed consent was obtained from all participants by explaining the importance of the study.
Study population and their stratification
132 patients diagnosed with CML and 140 healthy age and sex matched controls formed our study group. The cohort of patients consisted of 76 (58%) male and 56 (42%) female patients with a mean age of 37.5 years. In addition, 102 (77%) patients were in the chronic phase, 20 (15%) patients were in the accelerated phase and 10 (8%) patients were in the blast crisis phase. CML patients were diagnosed at the Department of Medical Oncology, Nizam’s Institute of Medical Sciences (NIMS), Hyderabad, India. The control group consisted of individuals without the history of cancer and other diseases like genetic disorders, allergy, asthma, etc.
Inclusion and exclusion criteria
In this study, fully diagnosed CML patients receiving Imatinib treatment (n=129) and few CML patients prior to Imatinib treatment (n=3) were included as cases. Age range of the CML patients was 14-60 years in females and 15-64 years in males. The diagnosis of CML was based on the standard clinico-hematological criteria by detecting the Ph chromosome (karyotyping) and/or the BCR-ABL fusion gene by reverse transcriptionpolymerase chain reaction (RT-PCR). The exclusion criteria included the patients suffering from any other disease such as chronic myelomonocytic leukemia and other myeloproliferative disorders.
Clinico-pathologic data
Patient’s data including occupational history, complete clinical examination and routine laboratory tests such as complete blood picture [white blood cell (WBC) count and platelet count], liver and kidney functions were obtained. In addition, clinical data including phase of the disease and response to imatinib therapy were collected from the medical records with the permission of the attending medical oncologist. Patients were stratified based on response according to the European LeukemiaNet (ELN) criteria. Response status of patients to imatinib therapy (hematological, cytogenetic) was classified on the basis of WBC count, percentage of Ph+ cells and the duration of response to imatinib therapy (29, 30). We stratified the patients in our study based on these parameters to examine the efficacy of their treatment regimen. The effect of imatinib treatment was assessed after 3, 6 and 12 months. An ideal treatment regimen leads to a major/complete hematological response (MHR) and at least a minor cytogenetic response (mCyR) within 3 months, a partial CyR (PCyR) within 6 months and a complete CyR (CCyR) within 12 months
New method of stratification of chronic myeloid leukemia patients
We established one more criterion of stratification of the CML patients, with reference to imatinib therapy, based on the genotypes of the drug metabolizing enzyme gene CYP1A1 and its role in response to the treatment. For this reason, we carried out genomic studies for the gene CYP1A1 using samples from both the patient and control groups.
DNA isolation
Venous blood samples (5 ml) from patients diagnosed with CML and treated with imatinib therapy, and from control individuals were collected in ethylenediamine tetraacetic acid (EDTA, BD, India) coated vacutainers. Genomic DNA was isolated by a standard salting out method (31) and then stored at -20˚C until use for mutational analysis.
Genotyping and the detection of CYP1A1*2C
To detect the CYP1A1*2C polymorphism, an allelespecific PCR was performed using two sets of primers. Primers were purchased from Bioserve (Hyderabad, India), dNTPs and Taq polymerase from Labpro (India). Two allele-specific forward primers (F1P- 1A1A; 5ˊ-GAAGTGTATCGGTGAGACCA-3ˊ and F2P-1A1G; 5ˊ-GAAGTGTATCGGTGAGACCG- 3ˊ), were used for PCR amplification together with a reverse primer (RP-1A1.1 5ˊ-GTAGACAGAGTCTAGGCCTCA-3ˊ). Two amplification reactions were needed for each individual analysed, one with primers 1A1.1 (common reverse primer)/1A1A (forward primer-1, specific for Ile allele) and the other with primers 1A1.1 (common reverse primer)/1A1G (forward primer-2 specific, for Val allele) which recognize the Val462 allele. The PCR reactions were performed in a final volume of 25 μl containing 50-150 ng of genomic DNA, 5 μl of 10X PCR buffer containing 20 mM MgCl2 (Bioserve, India), 0.5 μl of 10 pmol of each primer, 0.5 μl of dNTPs mix (200 μM), 1 U/μl of taq DNA polymerase and deionized water. The PCR cycling conditions were an initial denaturation at 94˚C for 3 minutes, followed by 30 cycles of denaturation at 94˚C for 30 seconds, annealing at 65˚C for 45 seconds and extension at 72˚C for 1 minute along with a final extension at 72˚C for 10 minutes in a VeritiTM 96 well thermal cycler (Applied Biosystems, USA).
Statistical analysis
The relationship between the CYP1A1*2C polymorphism and the development of CML with respect to the clinical characteristics was examined by using odds ratio (OR) and 95% confidence intervals (CI) derived from the logistic regression analysis using MedCalc version 7.4.1.0 (MedCalc software, Mariakerke, Belgium). P values of less than 0.05 were regarded as statistically significant.
Results
The mean age of CML patients was 36.7 years in females and 38.3 years in males. The CML patients were divided into 4 groups according to their age at diagnosis (i.e., <20, 20-30, 31-40 and 41-60 years). Incidence of CML was found to be highest in the age group 21-30 (39%), followed by 41-60 years (34%). In contrast, the incidence was found to be low in the age group 31-40 years (22%) and lowest in those less than 20 years (5%). This indicates that the onset of CML is generally after 20 years of age.
CYP1A1*2C genotyping analysis
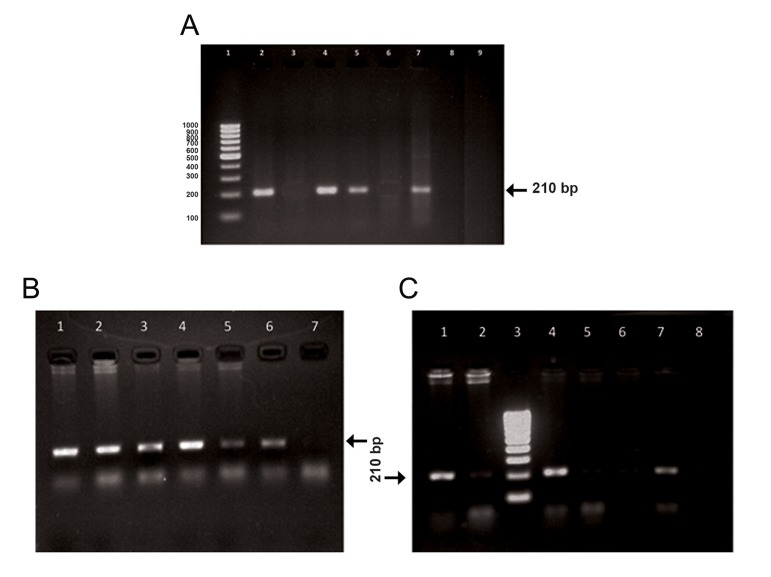
CYP1A1*2C gene polymorphism was genotyped by identifying both the wild type Ile allele (210 bp DNA fragment with the 1A1A1/1A1.1 primers) and the mutant Val allele (210 DNA fragment with 1A1G/1A1.1 primers) (Fig .1).
Fig.1.
Gel electrophoresis of CYP1A1*2C PCR products using allele-specific primers. A. Lane 1 represents the 100 bp DNA marker. Lanes 2, 3 correspond to the Ile/Ile homozygous wild type individuals, lanes 4, 5 correspond to the Ile/Val heterozygous individuals and lanes 6, 7 show Val/Val homozygous mutant genotype. Lanes 8, 9 correspond to the negative controls, B. Lanes 1, 2 correspond to the Ile/Val heterozygous mutant genotype, lane 3 represents the positive control, lanes 4, 5 correspond to the Ile/Val heterozygous mutant genotype, lanes 6, 7 correspond to Ile/Ile homozygous wild type genotype and C. Lanes 1, 2 correspond to the Ile/Val heterozygous mutant genotype, lane 3 represents the 100 bp DNA marker, lanes 4, 5 correspond to the Ile/Val heterozygous mutant genotype, lanes 6, 7 correspond to the Ile/Val heterozygous mutant genotype and lane 8 represents the negative control.
PCR; Polymerase chain reaction.
The distribution of CYP1A1*2C polymorphism genotypes in CML patients and in control individuals is shown in table 1. The frequencies of CYP1A1 Ile/Ile (A4889A), Ile/Val (A4889G), and Val/Val (G4889G) genotypes were 38, 57, and 5% in CML patients and 16, 78, and 6% in controls respectively. We observed differences in the distribution of CYP1A1 Ile/Ile wild type and Ile/Val heterozygous genotypes between patients and controls in the study population. Our analysis of the frequency distribution of genotypes between patients and controls showed that the AG genotype was lower in CML patients (95% CI=0.2176 to 0.6296, OR=0.3701, P=0.0002) when compared with the controls, suggesting that this genotype may play a protective role in reducing CML risk in our cohort. Further, the distribution of homozygous wild type Ile/Ile genotype was found to be higher in the CML patients (38%) than that in the controls (16%) (95% CI=1.84 to 5.8133, OR=3.2705, P=0.0001). We did not find any significant difference in the distribution of GG genotype between patients (5%) and controls (6%) (95% CI=0.265 to 2.33, OR=0.7857, P=0.6635) (Table 1). The frequencies of the major (A, Ile) and minor (G, Val) alleles are shown in table 2.
Table 1.
Distribution of CYP1A1*2C polymorphism genotypes in CML patients and controls
| Genotype | Patients (n=132) n (%) | Controls (n=140) n (%) | OR | 95% CI | P value |
|---|---|---|---|---|---|
| Ile/Ile (AA)- Wild | 50 (38) | 22 (16) | 3.2705 | 1.84 - 5.81 | 0.0001 |
| Ile/Val (AG)- Hetero | 76 (57) | 110 (78) | 0.3701 | 0.218-0.63 | 0.0002 |
| Val/Val (GG)- Homo mutant | 6 (5) | 8 (6) | 0.7857 | 0.265-2.33 | 0.6635 |
CML; Chronic myeloid leukemia, n; Number of subjects, OR; Odds ratio and CI; Confidence interval.
Demographic characteristics of the study population
The mean age at the onset of CML was found to be 37.5 years and ranged between 14 and 60 years. Male predominance has been observed in the present study with a sex ratio of 1.3:1 indicating that the male population are at higher risk for CML development. The results of our analysis of clinical characteristics in CML patients are presented in table 3.
Table 2.
Allele frequencies of CYP1A1*2C polymorphism in CML patients and controls
| Allele | Patients allele frequency | Controls allele frequency |
|---|---|---|
| Ile | 0.67 | 0.55 |
| Val | 0.33 | 0.45 |
CML; Chronic myeloid leukemia.
Table 3.
Association of CYP1A1*2C polymorphism in CML patients with demographic and clinical parameters
| Characteristics | Total CML cases n=132 (%) | Wild type AA (Ile/Ile) n=51 (38%) | Heterozygous AG (Ile/Val) n=75 (58%) | Homo mutant GG (Val/Val) n=6 (4%) | P value |
|---|---|---|---|---|---|
| Gender | |||||
| Males | 76 (58) | 32 (42) | 39 (51) | 5 (6.6) | 0.23 |
| Females | 56 (42) | 19 (34) | 36 (64) | 1 (1.8) | 0.23 |
| Age of diagnosis (Y) | |||||
| <20 | 7 (5) | 3 (43) | 3 (43) | 1 (14) | 0.62 |
| 20-30 | 51 (39) | 19 (37) | 32 (63) | - | 0.54 |
| 31-40 | 29 (22) | 12 (41) | 16 (55) | 1 (3.4) | 0.77 |
| >40 (up to 60) | 45 (34) | 17 (38) | 24 (53) | 4 (8.8) | 0.87 |
| CML phase | |||||
| Chronic | 114 (86) | 46 (40) | 62 (54) | 6 (5) | 0.24 |
| Accelerated | 13 (10) | 3 (23) | 10 (77) | - | 0.18 |
| Blast crisis | 5 (4) | 2 (40) | 3 (60) | - | 0.98 |
| WBC count (cells/ cubic.mm of blood) | |||||
| <20 000 | 15 (11) | 10 (67) | 5 (33) | - | 0.03* |
| >20 000 | 117 (89) | 41 (35) | 70 (60) | 6 (5) | 0.03* |
| Platelet count | |||||
| Normal count (1.5-4.0 lakhs cells/cubic.mm of blood) | 71 (54) | 28 (40) | 38 (54) | 5 (7) | 0.64 |
| Thrombocytopenia (<1.5 lakhs/cubic.mm) | 3 (2.3) | 2 (67) | 1 (33) | - | 0.37 |
| Thrombocytosis (>4 lakhs/cubic.mm) | 58 (44) | 21 (36) | 36 (62) | 1 (1.7) | 0.45 |
| Spleen size | |||||
| Splenomegaly present | 104 (79) | 36 (35) | 64 (62) | 4 (3.8) | 0.04* |
| Splenomegaly absent | 28 (21) | 15 (53.5) | 11 (39) | 2 (7) | 0.04* |
WBC; White blood corpuscles, CML; Chronic myeloid leukemia and *; Indicates statistically significant (P<0.05).
Treatment response of chronic myeloid leukemia patients carrying the CYP1A*2C polymorphism
Association of the CYP1A1*2C polymorphism with respect to therapy responses, such as hematological and cytogenetic responses to imatinib treatment, are presented in table 4.
Influence of the CYP1A1*2C polymorphism on chronic myeloid leukemia with respect to hematological response
MHR was observed to be higher in patients carrying the AG genotype (67%) than non-carriers (P=0.03), whereas, patients without the polymorphism (AA) (62%) exhibited a poorer hematological response than the patients with genotype AG (31%) (P=0.04), indicating that the patients carrying the polymorphism (AG) showed positive therapeutic response compared with wild type (AA) patients. Patients with the homozygous genotype (GG) (8%) exhibited poor hematological response (Table 4).
Table 4.
Association of CYP1A1*2C polymorphism with therapeutic response in CML patients
| Imatinib treatment response | Total n=95 n (%) | Total n=37 n (%) AA | Total n=57 n (%) AG | Total n=1 n (%) GG | P value |
|---|---|---|---|---|---|
| i. Hematological response | |||||
| Major (MHR) | 70 (74) | 23 (33) | 47 (67) | - | 0.03* |
| Minor (mHR) | 12 (13) | 6 (50) | 6 (50) | - | 0.42 |
| Poor (PHR) | 13 (14) | 8 (62) | 4 (31) | 1 (8) | 0.04* |
| ii.Cytogenetic response | |||||
| Major (MCyR) | 71 (75) | 24 (34) | 46 (65) | 1 (1) | 0.08 |
| Minor (mCyR) | 15 (16) | 6 (40) | 9 (60) | - | 0.90 |
| Poor (pCyR) | 9 (9) | 7 (78) | 2 (22) | - | 0.02* |
AA; Wild genotype, AG; Heterozygous genoype, GG; Homozygous variant genotype, MHR; Major hematological response, mHR; Minor hematological response, PHR; Poor hematological response, MCyR; Major cytogenetic response, mCyR; Minor cytogenetic response, pCyR; Poor cytogenetic response and *; Indicates statistically significant (P<0.05).
Influence of CYP1A1*2C polymorphism on chronic myeloid leukemia with respect to cytogenetic response
We observed that most of the major cytogenetic responders (65%) carried a high frequency of Ile/ Val variant genotype, whereas most poor cytogenetic responders (78%) carried a high frequency of wild genotype (AA) (P=0.02). This indicates that this polymorphism may be associated with positive cytogenetic response to imatinib therapy. Patients with homozygous variant genotype (GG) (1%) exhibited major cytogenetic response (Table 4).
Discussion
Polymorphisms in genes encoding the drug metabolizing enzyme CYP1A1 contribute to the variability in susceptibility to various cancers (24, 26, 32). A few studies have reported a significant association of CYP1A1 polymorphism with solid tumors (32), acute lymphoblastic leukemia (24) and CML (26). However, to our knowledge, not much information is available on the role of CYP1A1 in relation to CML patients undergoing imatinib treatment. Generally, 95% of CML patients display genetic abnormality in the form of chromosomal translocation, i.e., Ph+ [t(9;22)], which causes the formation of the fusion oncogene BCR-ABL. The fusion protein formed from this fusion gene possesses enhanced tyrosine kinase activity which causes leukemogenesis (2).
Presence of the A4889G SNP in CYP1A1 results in increased catalytic activity which leads to enhanced DNA adduct formation. These DNA adducts are responsible for causing mutations in the tumor suppressor genes and oncogenes, and thus trigger the uncontrolled hematopoietic cell proliferation, reduced differentiation and decreased apoptosis of malignant hematopoietic blast cells. Thus, individuals exhibiting an ability to activate procarcinogens due to sequence changes in CYP1A1 may be at higher risk for various cancers including CML (26).
We found that although CML occurs at any age, a higher incidence was found in adults of age above 20 with most diagnosed to be in the chronic phase. CML patients are generally treated with the drug imatinib mesylate (Gleevec®, Novartis) with chronic phase CML patients responding well to this therapy. Hence it may be possible that SNPs in the CYP1A1 may affect the response of the CML patients to imatinib therapy.
Our findings suggest that AG genotype may play a protective role in reducing CML risk in our cohort. We had also analysed the influence of CYP1A1*2C polymorphism with respect to various demographic and clinical characteristics of CML patients such as gender, age, CML phase, WBC count, platelet count and spleen size. With respect to gender we found that the males are more susceptible to developing CML than females. This may be attributed to the higher percentage of protective AG genotype in females and higher frequency of AA genotype in males of our study cohort. The frequency of AG genotype was higher than the AA wild genotype. This indicates that this SNP is not associated with the age of onset of CML.
We also found that the AG genotype was associated with good therapeutic response towards IM, which was measured in terms of hematological and cytogenetic responses, hence a good predictor of imatinib therapy.
Consistent with our findings, AML patients carrying the CYP1A1*2C polymorphism were reported to show a better prognosis compared with those carrying the wild-type genotype exhibiting a longer survival rate (33). Similarly, breast cancer patients carrying CYP1A1*2C polymorphism were shown to develop less aggressive tumors (34). Based on these reports, it is worth mentioning that the association of cancer with a particular polymorphism in one population may not be of the same significance in another population due to the variation in population demographics and other influencing factors such as the environment. Such results were also documented previously with respect to other genes including MTHFR (11).
Taspinar et al. (26) reported a higher distribution of CYP1A1 Ile/Val heterozygous genotype in the Turkish CML patients (P<0.001) as compared with the controls, suggesting that carriers of CYP1A1 Ile/Val (AG) genotype had an increased risk of developing CML. On the contrary, no association between the CYP1A1*2C polymorphism and CML risk was reported in the Iranian population (27). In addition, CYP1A1 Val/Val homozygous variant genotype was associated with four-fold risk to ALL in Indian children (24). Some reports also suggested the association of CYP1A1*2C polymorphism with increased risk of solid tumors such as head and neck cancer in patients with Indian descent (17, 32). The above differences reported in the literature may be due to the differences in tumor type, ethnicity, environmental exposures, life style and habits.
Analysis of SNPs with respect to therapeutic response is expected to aid in designing novel, alternative therapeutic strategies to treat CML which may be efficient alone or may be useful in combination with existing therapies. With the advent of new techniques, obtaining the complete genomic sequence of every individual is becoming a possibility with which the SNPs in the genome could be identified and subsequently the risk of developing cancer may be predicted. In addition, the identified SNPs may not only serve as genetic markers in determining the susceptibility of an individual to a disease, it can also predict response to a therapy. Such advances are expected to pave the way for the development of personalized medicine.
Conclusion
We showed that the AG genotype of CYP1A1*2C polymorphism may play a protective role against CML. Our analysis on influence of this polymorphism on therapy of CML patients of Indian descent suggests that this genotype serves as a good predictor to assess response to imatinib therapy.
Acknowledgments
We thank Dr. Sailaja Kagita and Mr. Kirmani Natukula for their technical support and JNIAS for the facilities provided. No financial support was received for this study. The authors declare no conflict of interest.
References
- 1.Guilhot F, Apperley J, Kim DW, Bullorsky EO, Baccarani M, Roboz GJ, et al. Dasatinib induces significant hematologic and cytogenetic responses in patients with imatinibresistant or -intolerant chronic myeloid leukemia in accelerated phase. Blood. 2007;109(10):4143–4150. doi: 10.1182/blood-2006-09-046839. [DOI] [PubMed] [Google Scholar]
- 2.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 3.Kalidas M, Kantarjian H, Talpaz M. Chronic myelogenous leukemia. JAMA. 2001;286(8):895–898. doi: 10.1001/jama.286.8.895. [DOI] [PubMed] [Google Scholar]
- 4.Melo JV, Barnes DJ. Chronic myeloid leukemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 5.Deininger MW. Milestones and monitoring in patients with CML treated with imatinib. Hematology Am Soc Hematol Educ Program. 2008:419–426. doi: 10.1182/asheducation-2008.1.419. [DOI] [PubMed] [Google Scholar]
- 6.Druker BJ, Guilhot F, OBrien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 7.Hochhaus A, Hughes T. Clinical resistance to imatinib: mechanisms and implications. Hematol Oncol Clin North Am. 2004;18(3):641-656, ix. doi: 10.1016/j.hoc.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphiapositive patients: by the GIMEMA working party on chronic myeloid leukemia. Clin Cancer Res. 2006;12(24):7374–7379. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 9.Khorashad JS, de Lavallade H, Apperley JF, Milojkovic D, Reid AG, Bua M, et al. Finding of kinase domain mutations in patients with chronic phase myeloid leukemia responding to imatinib may identify those at high risk of disease progression. J Clin Oncol. 2008;26(29):4806–4813. doi: 10.1200/JCO.2008.16.9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 11.Reddy H, Jamil K. Polymorphisms in the GST (M1 and T1) gene and their possible association with susceptibility to childhood acute lymphocytic leukemia (ALL) in the Indian population. Afr J Biotechnol. 2006;5(16):1454–1456. doi: 10.1080/10428190600562773. [DOI] [PubMed] [Google Scholar]
- 12.Jamil K, Reddy H. Can polymorphisms in genes relate to overall survival in leukemias? Leuk Lymphoma. 2007;48(6):1070–1071. doi: 10.1080/10428190701287353. [DOI] [PubMed] [Google Scholar]
- 13.Hemminki K, Shields PG. Skilled use of DNA polymorphisms as a tool for polygenic cancers. Carcinogenesis. 2002;23(3):379–380. doi: 10.1093/carcin/23.3.379. [DOI] [PubMed] [Google Scholar]
- 14.Kumar CK, Reddy M, Jamil K, Vamsy M. Genotyping of tamoxifen metabolizing enzyme (CYP2D6*4) and its clinical impact in breast cancer patients. Int J Genet Mol Biol. 2010;2(1):006–013. [Google Scholar]
- 15.Suman G, Jamil K, Suseela K, Vamsy MCh. Novel mutations of CYP3A4 in fine needle aspiration cytology samples of breast cancer patients and its clinical correlations. Cancer Biomark. 2009;5(1):33–40. doi: 10.3233/CBM-2009-0569. [DOI] [PubMed] [Google Scholar]
- 16.Khan S, Jamil K, Das GP, Vamsy MCh, Murthy S. Polymorphic sites (1236 and 3435) in multi drug resistance gene 1 influencing drug response in breast cancer patients. Int J Pharmacol. 2007;3(6):453–460. [Google Scholar]
- 17.Sabitha K, Reddy MV, Jamil K. Smoking related risk involved in individuals carrying genetic variants of CYP1A1 gene in head and neck cancer. Cancer Epidemiol. 2010;34(5):587–592. doi: 10.1016/j.canep.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Induski JA, Lutz W. Metabolic genotype in relation to individual susceptibility to environmental carcinogens. Int Arch Occup Environ Health. 2000;73(2):71–85. doi: 10.1007/pl00007942. [DOI] [PubMed] [Google Scholar]
- 19.Oyama T, Kagawa N, Kunugita N, Kitagawa K, Ogawa M, Yamaguchi T, et al. Expression of cytochrome P450 in tumor tissues and its association with cancer development. Front Biosci. 2004;9:1967–1976. doi: 10.2741/1378. [DOI] [PubMed] [Google Scholar]
- 20.Rooseboom M, Commandeur JN, Vermeulen NP. Enzymecatalyzed activation of anticancer prodrugs. Pharmacol Rev. 2004;56(1):53–102. doi: 10.1124/pr.56.1.3. [DOI] [PubMed] [Google Scholar]
- 21.Balta G, Yuksek N, Ozyurek E, Ertem U, Hicsonmez G, Altay C, et al. Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes in childhood acute leukemia. Am J Hematol. 2003;73(3):154–160. doi: 10.1002/ajh.10339. [DOI] [PubMed] [Google Scholar]
- 22.Canalle R, Burim RV, Tone LG, Takahashi CS. Genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukemia. Environ Mol Mutagen. 2004;43(2):100–109. doi: 10.1002/em.20003. [DOI] [PubMed] [Google Scholar]
- 23.Gallegos-Arreola MP, Batista-González CM, Delgado-Lamas JL, Figuera LE, Puebla-Perez AM, Arnaud-Lopez L, et al. Cytochrome P4501A1 polymorphism is associated with susceptibility to acute lymphoblastic leukaemia in adult Mexican patients. Blood Cells Mol Dis. 2004;33(33):326–329. doi: 10.1016/j.bcmd.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Joseph T, Kusumakumary P, Chacko P, Abraham A, Radhakrishna Pillai M. Genetic polymorphism of CYP1A1, CYP2D6, GSTM1 and GSTT1 and susceptibility to acute lymphoblastic leukaemia in Indian children. Pediatr Blood Cancer. 2004;43(5):560–567. doi: 10.1002/pbc.20074. [DOI] [PubMed] [Google Scholar]
- 25.Pelloso LA, Da Silva ID, De Souza NC, Yamamoto M, Chauffaille Mde L. Increased risk of acute myeloid leukemia in patients with CYP1A1 polymorphisms. J Cancer Ther. 2013;4(5):971–977. [Google Scholar]
- 26.Taspinar M, Aydos SE, Comez O, Elhan AH, Karabulut HG, Sunguroglu A. CYP1A1, GST gene polymorphisms and risk of chronic myeloid leukemia. Swiss Med Wkly. 2008;138(1-2):12–17. doi: 10.4414/smw.2008.12036. [DOI] [PubMed] [Google Scholar]
- 27.Razmkhah F, Pazhakh V, Zaker F, Atashrazm F, Sheikhi M. Frequency of CYP1A1*2C polymorphism in patients with leukemia in the Iranian population. Lab Med. 2011;42(4):220–223. [Google Scholar]
- 28.Rochat B, Zoete V, Grosdidier A, von Grunigen S, Marull M, Michielin O. In vitro biotransformation of Imatinib by the tumor expressed CYP1A1 and CYP1B1. Biopharm Drug Dispos. 2008;29(2):103–118. doi: 10.1002/bdd.598. [DOI] [PubMed] [Google Scholar]
- 29.Druker BJ, Lee SJ. Chronic leukemias. In: Devita VT, Hellman S, Rosenberg SA, editors. Cancer, principles and practice of Oncology. 7th ed. Philadelphia: Lippincott; 2005. pp. 2124–2131. [Google Scholar]
- 30.Baccarani M, Cortes J, Pane F, Niederwieser D, Saglio G, Apperley J, et al. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27(35):6041–6051. doi: 10.1200/JCO.2009.25.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahiri DK, Nurnberger Jl Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood RFLP studies. Nucleic Acids Res. 1991;19(19):5444–5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh AP, Shah PP, Ruwali M, Mathur N, Pant MC, Parmar D. Polymorphism in cytochrome P4501A1 is significantly associated with head and neck cancer risk. Cancer Invest. 2009;27(8):869–876. doi: 10.1080/07357900902849657. [DOI] [PubMed] [Google Scholar]
- 33.Pelloso LA, Da Silva ID, De Souza NC, Yamamoto M, Botelho CA, Chauffaille Mde L. CYP1A1 polymorphisms modify overall survival in acute myeloid leukemia patients. Leuk Lymphoma. 2007;48(6):1211–1215. doi: 10.1080/10428190701332431. [DOI] [PubMed] [Google Scholar]
- 34.Cardoso-Filho C, Sarian LO, de Oliveira CB, da Silveira Bossi L, Lourenço GJ, Lima CS, et al. Clinical effects of A4889G and T6235C polymorphisms in cytochrome P-450 CYP1A1 for breast cancer patients treated with tamoxifen: implications for tumor aggressiveness and patient survival. Cancer Chemother Pharmacol. 2013;72(3):529–535. doi: 10.1007/s00280-013-2221-y. [DOI] [PubMed] [Google Scholar]