Abstract
Background. We aimed to characterize the natural history of hepatitis C virus (HCV) reinfection and spontaneous clearance following reinfection (reclearance), including predictors of HCV reclearance.
Methods. Data were synthesized from the 9 prospective cohorts of the International Collaboration of Incident Human Immunodeficiency Virus and HCV in Injecting Cohorts study, which evaluated HCV infection outcomes among people who inject drugs. Participants with primary HCV infection were classified as having achieved viral suppression if they had negative results of at least 1 subsequent HCV RNA test. Those with positive results of an HCV RNA test following viral suppression were investigated for reinfection. Viral sequence analysis was used to identify reinfection (defined as detection of heterologous virus with no subsequent detection of the original viral strain).
Results. Among 591 participants with acute primary HCV infection, 118 were investigated for reinfection. Twenty-eight participants were reinfected (12.3 cases/100 person-years; 95% confidence interval [CI], 8.5–17.8). Peak HCV RNA level was lower during reinfection than primary infection (P = .011). The proportion of individuals with reclearance 6 months after reinfection was 52% (95% CI, 33%–73%). After adjustment for study site, females with the IFNL4 (formerly IFNL3 and IL28B) rs12979860 CC genotype detected were more likely to have reclearance (hazard ratio, 4.16; 95% CI, 1.24–13.94; P = .021).
Conclusions. Sex and IFNL4 genotype are associated with spontaneous clearance after reinfection.
Keywords: hepatitis C, re-infection, viral resolution, cohort study, IFNL4, sex, injecting drug use
Spontaneous clearance of primary hepatitis C virus (HCV) occurs in 25% of individuals [1]. However, reinfection following spontaneous clearance suggests that natural immunity is short-lived or has limited breadth or magnitude, with implications for vaccine development [2].
Studies of HCV reinfection in people who inject drugs have produced contradictory results [2], with variations in reinfection rates and proportions of reinfections clearing spontaneously [3–12]. These discrepancies may be attributed to methodological limitations, including variations in frequency of follow-up testing and data capture [2, 13], and to classification of viral recurrence as reinfection without confirmation that viremic episodes are genetically distinct [12].
The International Collaboration of Incident HIV and HCV in Injecting Cohorts (InC3) study, which pooled data from 9 prospective cohorts in Australia, Canada, the Netherlands, and the United States [14], mainly comprising from people who inject drugs, enables assessment of HCV reinfection in well-characterized HCV-infected participants. The aims of our study were to characterize the natural history of spontaneous clearance following primary infection (primary clearance), HCV reinfection, and spontaneous clearance following reinfection (reclearance) in the InC3 study; assess differences in peak HCV RNA levels during reinfection, compared with primary infection; assess predictors of HCV reclearance; and assess differences in the time to primary clearance and reclearance.
PARTICIPANTS AND METHODS
Study Population and Design
The InC3 study has been described previously [14]. All cohorts followed up participants at regular intervals, using standardized methods (Table 1). Participants were recruited and followed up between 1985 and 2010. For the current study, only individuals with documented acute primary HCV infection and results of ≥2 subsequent HCV RNA tests were included.
Table 1.
Incidence Rate of Reinfection, by Cohort
| Variable | Participants Evaluated, No.a | Reinfections, No. | Test Interval, mos | Test Interval Acute Primary HCV Infection, mosb | Person-Years | Rate, Cases/100 Person-Years (95% CI) |
|---|---|---|---|---|---|---|
| Overall | 128 | 28 | … | … | 227.8 | 12.3 (8.5–17.8) |
| Cohort (location) | ||||||
| ACS (the Netherlands) | 13 | 4 | 6 | 6 | 67.0 | 6.0 (2.2–15.9) |
| ATAHC (Australia) | 25 | 2 | …c | 3 | 31.0 | 6.5 (1.6–25.8) |
| BAHSTION (United States) | 12 | 1 | …d | 1e | 11.3 | 8.9 (1.2–62.9) |
| BBAASH (United States) | 27 | 15 | 1 | 1 | 41.2 | 36.4 (21.9–60.4) |
| HEPCO (Canada) | 7 | 0 | 6 | 1f | 19.7 | 0.0 |
| HITS-c (Australia) | 3 | 0 | 6 | 0.5–1g | 2.7 | 0.0 |
| HITS-p (Australia) | 18 | 2 | 6 | 0.5–1g | 23.5 | 8.5 (2.1–34.1) |
| N2 (Australia) | 4 | 2 | 3 | 3 | 7.4 | 27.1 (6.8–108.5) |
| UFO (United States) | 19 | 2 | 3 | 1 | 24.0 | 8.3 (2.1–33.3) |
Abbreviations: ACS, Amsterdam Cohort Studies; ATAHC, Australian Trial in Acute Hepatitis C study; BAHSTION, Boston Acute HCV Study: Transmission, Immunity, and Outcomes Network; BBAASH, Baltimore Before and After Acute Study of Hepatitis; CI, confidence interval; HCV, hepatitis C virus; HEPCO, St. Luc Cohort; HITS-c, Hepatitis C Incidence and Transmission Study–Community; HITS-p, Hepatitis C Incidence and Transmission Study–Prison; N2, Networks 2; UFO, UFO study.
a Includes participants with persistent spontaneous clearance and reinfection. Participants with intercalation are excluded because they are assumed not to have spontaneously cleared infection. Participants with indeterminate reoccurring viremia are excluded because the reinfection outcome is unknown.
b Many cohort sites perform tests more frequently after identification of acute primary HCV infection.
c Eligibility is based on recent HCV seroconversion or confirmed acute HCV infection.
d Eligibility of seronegative participants or those with acute HCV infection is based on recent exposure to HCV or suspected or acute HCV infection.
e For 6 months, after which the interval between tests is reduced.
f For 24 weeks, after which the normal test schedule resumes.
g For 3 months among participants with early acute infection (ie, HCV RNA is still present), after which the normal test schedule resumes.
Primary HCV infection is defined as an individual's first HCV infection. Documented acute primary HCV infected is defined as either (1) HCV seroconversion with an HCV antibody–negative test result followed by an HCV antibody– or HCV RNA–positive test result within 2 years or (2) evidence of symptomatic infection (defined by jaundice or an alanine transaminase level of >400 U/L, a positive HCV RNA or HCV antibody test result, and recent high-risk exposure). All participants provided written informed consent, and cohort protocols were approved by local institutional human research review committees.
Laboratory Testing
Choice of HCV RNA testing and HCV sequencing methods and regions varied between but not within cohorts. Qualitative and quantitative HCV RNA, HCV genotype and serotype, and interferon lambda 4 (IFNL4) rs12979860 genotype (formerly known as interferon lambda 3 and interleukin 28B) assays have been described previously [15]. Regions of the virus sequenced to confirm reinfection are listed in Supplementary Table 1. Polymerase chain reaction amplification conditions, primers, and sequencing methods have been described previously [16–20]. Cohort sites used distance [6, 7, 11] or phylogenetic methods [5] to distinguish heterologous from homologous virus within viral subtypes.
Estimated Date of Primary HCV Infection
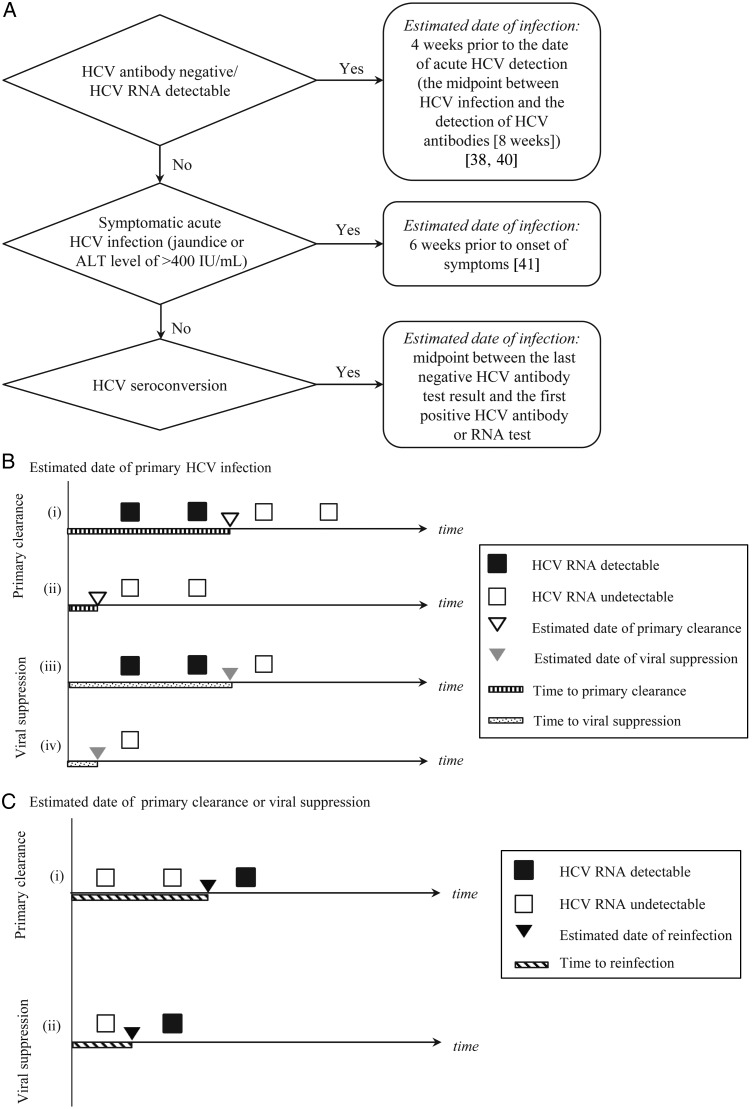
The estimated date of primary HCV infection was determined using the flow chart in Figure 1A.
Figure 1.
Timing and classification of hepatitis C virus (HCV) primary infection, viral suppression, spontaneous clearance, and reinfection. A, Flowchart for determining the estimated date of primary HCV infection. B, Estimated dates of and times to primary clearance and viral suppression. All timelines represent participants with confirmed primary HCV infection followed by either viral suppression or primary clearance. The timelines begin at the estimated date of primary infection. After primary infection, HCV RNA test results are depicted on the timeline by squares. Black squares represent HCV RNA–positive test results, and white squares represent HCV RNA–negative test results. Primary clearance is distinguished from viral suppression by the number of HCV RNA–negative test results (white squares). The top timelines (i and ii) depict primary clearance (as indicated by the 2 consecutive HCV RNA–negative test results), and the bottom timelines (iii and iv) depict viral suppression (defined as 1 HCV RNA–negative test result). In timelines ii and iv, HCV RNA was undetectable at the time of primary infection detection, as indicated by the white initial squares (HCV RNA tests) in the timelines. The estimated date of primary clearance is illustrated using a white triangle, and the estimated date of viral suppression is illustrated using a gray triangle. These dates are both estimated as follows: if HCV RNA is detectable at the time of detection of primary infection (i and iii), the estimated date of primary clearance or viral suppression is the midpoint between the HCV RNA–positive test result prior to primary clearance or viral suppression and the first HCV RNA–negative test result.
Antiviral Treatment for HCV Infection
The natural history of HCV reinfection and reclearance may differ following antiviral treatment, compared with spontaneous clearance of primary HCV infection. Therefore, this analysis was limited to studying HCV reinfection and reclearance in the absence of a history of antiviral treatment. Individuals who were treated >26 weeks after the estimated date of primary infection were censored from the treatment date. Individuals treated for HCV infection were excluded if the estimated duration of primary infection was <26 weeks, to reduce misclassification bias due to uncertainty around subsequent spontaneous clearance without treatment (n = 37).
Primary HCV Infection Outcomes
After acute primary HCV infection, subjects with 1 subsequent HCV RNA–negative test result and those with 2 consecutive HCV RNA tests (≥28 days apart) with negative results were classified as having viral suppression and primary clearance, respectively. Those with viral suppression at the final follow-up visit (n = 34) were excluded because the final outcome could not be determined. Those with detectable HCV RNA following viral suppression or primary clearance were classified as having reoccurring viremia. Among those with reoccurring viremia, viral genotype/subtype and results of viral sequence analysis were used to distinguish reinfection (defined as detection of heterologous virus with no subsequent detection of the original viral strain), intercalation (defined as detection of homologous virus), and indeterminate cases (defined as cases for which viral sequencing data were unavailable or detection of heterologous virus with subsequent detection of the original viral strain). If primary clearance occurred prior to detection of seroconversion, such that no HCV RNA could be isolated from the primary infection, serotyping was performed to classify primary infection.
Estimated Dates and Times of Primary Clearance, Viral Suppression, and Reinfection
Methods for determining the estimated dates of primary clearance, viral suppression, and reinfection are illustrated in Figure 1B–D. In participants with evidence of primary clearance prior to reinfection, the time to reinfection was calculated as the time from the date of primary clearance to the date of reinfection. In participants with viral suppression prior to reinfection, the time to reinfection was calculated as the time from the date of viral suppression to the date of reinfection (Figure 1C). The time to reappearance of viremia in intercalation and indeterminate intermittent viremia was calculated similarly to the time to reinfection.
Figure 1.
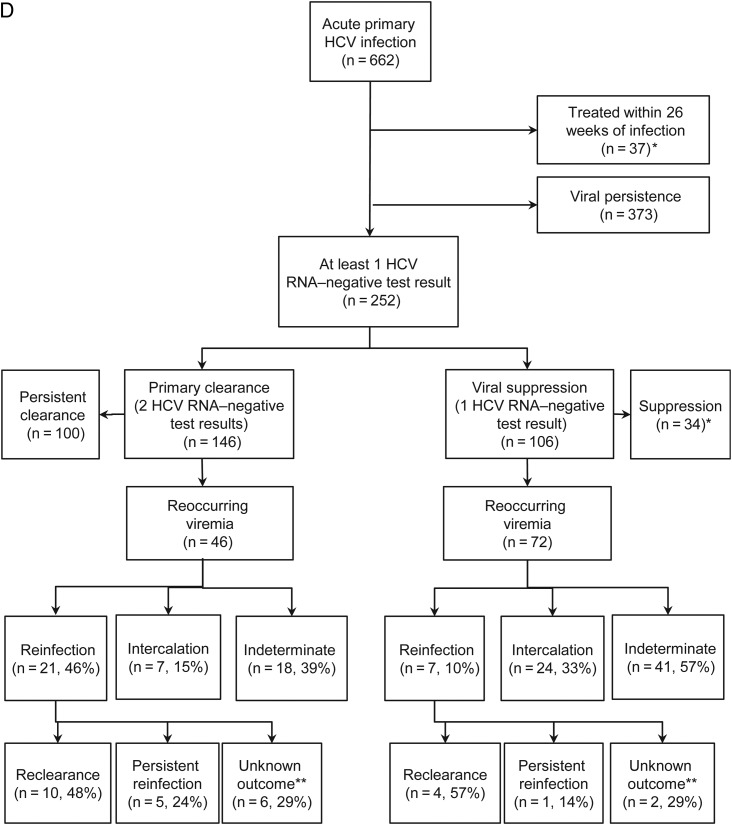
If HCV RNA is undetectable at the time of detection of primary infection (ii and iv), the estimated date of primary clearance or viral suppression is the midpoint between the estimated date of primary infection (the beginning of the illustrated timeline) and the first HCV RNA–negative test result. In all 4 cases, the time to primary clearance or viral suppression is the time from the estimated date of infection until the estimated date of primary clearance or viral suppression. C, Estimated dates of and times to reinfection. Both timelines represent participants with confirmed primary HCV infection followed by either viral suppression or primary clearance and confirmed reinfection. The timelines begin at the estimated date of primary clearance or viral suppression. HCV RNA test results are depicted on the timeline by squares. Black squares represent HCV RNA–positive test results, and white squares represent HCV RNA–negative test results. Primary clearance is distinguished from viral suppression by the number of HCV RNA–negative test results (white squares). The top timeline (i) depicts primary clearance (as indicated by the 2 consecutive HCV RNA–negative test results), and the bottom timeline (ii) depicts viral suppression (define as 1 HCV RNA–negative test result). The estimated date of reinfection is the midpoint between the last HCV RNA–negative test result and the first HCV RNA–positive test result. The time to reinfection is defined as the time from the estimated date of primary clearance or viral suppression until the estimated date of reinfection. D, Flowchart of reinfection classification. *Primary HCV infection outcome unknown; **Reinfection outcome unknown: includes five cases with insufficient follow-up to determine outcome and three cases with change in genotype after reinfection.
HCV Reinfection Outcomes
Reclearance was defined as 2 consecutive HCV RNA–negative tests (performed ≥28 days apart) with negative results following reinfection. The date of reclearance was calculated similarly to the date of primary clearance, and the time to reclearance was calculated as the time from the date of reinfection to the date of reclearance. For those without clearance, follow-up time was calculated from the date of reinfection until the date of the last therapy-naive positive HCV RNA test result. Persistent reinfection was defined as continuous viremia with the confirmed reinfecting virus for >6 months.
Classification of Peak HCV RNA Load During Primary Infection and Reinfection
Peak HCV RNA load was defined as the maximum quantitative RNA value measured within 3 months of the date of infection for both primary infection and reinfection (Supplementary Materials).
Statistical Analyses
Wilcoxon signed rank tests were used to evaluate the median within-participant difference between peak log HCV RNA load in reinfection, compared with primary infection. It was hypothesized that the peak log RNA load would be lower during reinfection, compared with primary infection [6]. Participants with at least 1 quantitative HCV RNA test in the first 3 months of primary HCV infection and reinfection were included.
Predictors of HCV reclearance were assessed using Cox proportional hazards regression. Models included shared frailty terms for study site to capture unobserved heterogeneity between sites that may have contributed to the time to reclearance. Potential interactions between study site—categorized as the Baltimore Before and After Acute Study of Hepatitis (BBAASH; the study contributing largest number of reinfection events) versus other sites—and hypothesized predictors were evaluated. Hypothesized predictors were determined a priori on the basis of established predictors of primary clearance, including age [21], sex [1, 15, 22, 23], IFNL4 genotype (SNP rs12979860; CC vs CT/TT) [24–26], the combined effect of sex and IFNL4 genotype (female rs12979860 CC vs others) [15, 23], HCV genotype during reinfection (genotype 1 vs non–genotype 1) [15, 27], and reinfection with the same versus a different HCV genotype from that in the primary infection. It was hypothesized that participants reinfected with the same HCV genotype would have a greater propensity toward reclearance [28, 29]. The combined effect of sex and IFNL4 genotype was investigated by comparing females with the rs12979860 CC genotype and all other participants because, although this study combined the largest number of HCV reinfections studied to date, there were not sufficient numbers of reinfections to investigate the interaction between sex and IFNL4 genotype. The effect of human immunodeficiency virus (HIV) infection [30] was not assessed owing to small numbers of HIV-infected participants. The effects of jaundice and elevated alanine aminotransferase levels were not assessed because most of the participating studies did not collect this information at the time of HCV reinfection.
Differences in times to primary clearance and reclearance in participants with reclearance were assessed using gap-time unrestricted proportional hazards regression, which is appropriate for analysis of predictors of time-to-event outcomes with multiple events [31]. The hypothesis was that time to primary clearance would be longer than the time to reclearance [6].
For all investigations, sensitivity analyses were performed to assess the effect of excluding participants infected with HIV at reinfection (n = 3), excluding reinfections defined on the basis of serotyping (n = 2), excluding reinfections with viral suppression rather than primary clearance prior to reinfection (n = 7), and stratifying by study site (BBAASH vs others). For the analysis of differences between peak HCV RNA loads in primary HCV infection and reinfection, sensitivity analysis of the effect of defining the peak RNA load as the peak within 1 month of infection, rather than 3 months, were conducted (Supplementary Materials). For the analysis of predictors of reinfection, sensitivity analyses excluding participants with <2 HCV RNA tests after reinfection were also performed.
Characteristics at the time of primary HCV infection and reappearance of viremia were analyzed by primary infection outcome, using Kruskal–Wallis, χ2, and Fisher exact tests, as appropriate. Proportions of reinfections resulting in spontaneous clearance and persistent infection 6 months after reinfection were estimated using Kaplan–Meier survivor functions. Finally, classifications of reoccurring viremia after primary clearance versus reoccurring viremia after viral suppression were compared using χ2 tests. Statistically significant differences were those with P values of <.05; P values are 2-sided. All analyses were performed using Stata, version 11.0 (College Station, Texas). Participant timeline figures were prepared in R [32, 33].
RESULTS
Participant Characteristics
Of 662 participants with acute primary HCV infection, 591 had a defined infection outcome (Figure 1D and Table 2). At the time of primary infection, the median age was 26 years, and 36% of participants were female. Most participants (96%) had injected drugs. A small minority (7%) were infected with HIV.
Table 2.
Characteristics, Exposures, and Risk Behaviors of 591 Participants With Acute Primary Hepatitis C Virus (HCV) Infection
| Characteristic at Time of Incident Primary HCV Infection | Persistent HCV, Participants, No. (%) (n=373) | Cleared or Intermittent HCV Infection |
||||
|---|---|---|---|---|---|---|
| Overall, Participants, No. (%) (n=218) | Reinfection | Intercalation | Indeterminate Intermittent Viremia | Persistent Cleared | ||
| Overall | … | … | 28/218 (13) | 31/218 (14) | 59/218 (27) | 100/218 (46) |
| Site | ||||||
| ACS (the Netherlands) | 18 (5) | 24 (11) | 4/24 (17) | 1/24 (4) | 10/24 (42) | 9/24 (38) |
| ATAHC (Australia) | 84 (23) | 29 (13) | 2/29 (7) | 1/29 (3) | 3/29 (10) | 23/29 (79) |
| BAHSTION (United States) | 21 (6) | 21 (10) | 1/21 (5) | 3/21 (14) | 6/21 (29) | 11/21 (52) |
| BBAASH (United States) | 52 (14) | 59 (27) | 15/59 (25) | 20/59 (34) | 12/59 (20) | 12/59 (20) |
| HEPCO (Canada) | 48 (13) | 18 (8) | 0/18 (0) | 0/18 (0) | 11/18 (61) | 7/18 (39) |
| HITS-c (Australia) | 6 (2) | 3 (1) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 3/3 (100) |
| HITS-p (Australia) | 63 (17) | 20 (9) | 2/20 (10) | 1/20 (5) | 1/20 (5) | 16/20 (80) |
| N2 (Australia) | 10 (3) | 7 (3) | 2/7 (29) | 3/7 (43) | 0/7 (0) | 2/7 (29) |
| UFO (United States) | 71 (19) | 37 (17) | 2/37 (5) | 2/37 (5) | 16/37 (43) | 17/37 (46) |
| Age, y, median (IQR)a | 27 (23–34) | 26 (22–30) | 24 (20–30) | 25 (24–28) | 26 (21–30) | 26 (23–32) |
| Sex | ||||||
| Male | 259 (69)b | 118 (54) | 14/118 (12) | 16/118 (14) | 38/118 (32) | 50/118 (42) |
| Female | 113 (30) | 100 (46) | 14/100 (14) | 15/100 (15) | 21/100 (21) | 50/100 (50) |
| Data missing | 1 (0) | 0 (0) | 0/0 (0) | 0/0 (0) | 0/0 (0) | 0/0 (0) |
| Ethnicity/race | ||||||
| European origin | 305 (82) | 177 (81) | 26/177 (15) | 25/177 (14) | 48/177 (27) | 78/177 (44) |
| Other | 41 (11) | 23 (11) | 2/23 (9) | 4/23 (17) | 6/23 (26) | 11/23 (48) |
| Data missing | 27 (7) | 18 (8) | 0/18 (0) | 2/18 (11) | 5/18 (28) | 11/18 (61) |
| History of IDU | 358 (96) | 210 (96) | 28/210 (13) | 30/210 (14) | 59/210 (28) | 93/210 (44) |
| HIV infectiona | ||||||
| No | 328 (88) | 199 (91) | 25/199 (13) | 28/199 (14) | 55/199 (28) | 91/199 (46) |
| Yes | 30 (8) | 12 (6) | 3/12 (25) | 1/12 (8) | 2/12 (17) | 6/12 (50) |
| Data missing | 15 (4) | 7 (3) | 0/7 (0) | 2/7 (29) | 2/7 (29) | 3/7 (43) |
| Peak HCV RNA load, log IU/mL, median (IQR)c | 5.5 (4.5–6.4) | 5.8 (4.0–7.0) | 6.7 (5.3–7.0) | 5.7 (5.1–6.8) | 5.3 (2.9–6.5) | 5.9 (3.2–7.0) |
| HCV genotypea | ||||||
| 1 | 174 (47)d | 114 (52) | 18/114 (16) | 24/114 (21) | 27/114 (24) | 45/114 (39) |
| 2 | 22 (6) | 10 (5) | 1/10 (10) | 2/10 (20) | 2/10 (20) | 5/10 (50) |
| 3 | 130 (35) | 46 (21) | 8/46 (17) | 5/46 (11) | 9/46 (20) | 24/46 (52) |
| 4 | 2 (1) | 4 (2) | 1/4 (25) | 0/4 (0) | 3/4 (75) | 0/4 (0) |
| 6 | 4 (1) | 0 (0) | 0/0 (0) | 0/0 (0) | 0/0 (0) | 0/0 (0) |
| Mixed | 10 (3) | 2 (1) | 0/2 (0) | 0/2 (0) | 0/2 (0) | 2/2 (100) |
| Unknown | 31 (8) | 42 (19) | 0/42 (0) | 0/42 (0) | 18/42 (43) | 24/42 (57) |
| Recent IDUe | ||||||
| No | 57 (15) | 19 (9) | 2/19 (11) | 2/19 (11) | 1/19 (5)f | 14/19 (74) |
| Yes | 238 (64) | 113 (52) | 9/113 (8) | 5/113 (4) | 40/113 (35) | 59/113 (52) |
| Data missing | 5 (1) | 6 (3) | 1/6 (17) | 1/6 (17) | 0/6 (0) | 4/6 (67) |
| Data not collected at cohort site | 73 (20) | 80 (37) | 16/80 (20) | 23/80 (29) | 18/80 (23) | 23/80 (29) |
| Recent IDU frequencye | ||||||
| No recent injecting | 41 (11) | 16 (7) | 1/16 (6) | 2/16 (13) | 1/16 (6) | 12/16 (75) |
| Daily or more | 127 (34) | 48 (22) | 7/48 (15) | 3/48 (6) | 13/48 (27) | 25/48 (52) |
| Less than daily but at least weekly | 72 (19) | 48 (22) | 3/48 (6) | 1/48 (2) | 19/48 (40) | 25/48 (52) |
| Less than weekly | 55 (15) | 19 (9) | 0/19 (0) | 0/19 (0) | 8/19 (42) | 11/19 (58) |
| Data missing | 5 (1) | 7 (3) | 1/7 (14) | 2/7 (29) | 0/7 (0) | 4/7 (57) |
| Data not collected at cohort site | 73 (20) | 80 (37) | 16/80 (20) | 23/80 (29) | 18/80 (23) | 23/80 (29) |
| Primary drug injected recentlye,g | ||||||
| Heroin/other opioids | 122 (33) | 72 (33) | 5/72 (7) | 3/72 (4) | 26/72 (36) | 38/72 (53) |
| Psychostimulants | 88 (24) | 34 (16) | 3/34 (9) | 1/34 (3) | 14/34 (41) | 16/34 (47) |
| Other | 6 (2) | 0 (0) | 0/0 (0) | 0/0 (0) | 0/0 (0) | 0/0 (0) |
| Data missing | 84 (23) | 32 (15) | 4/32 (13) | 4/32 (13) | 1/32 (3) | 23/32 (72) |
| Data not collected at cohort site | 73 (20) | 80 (37) | 16/80 (20) | 23/80 (29) | 18/80 (23) | 23/80 (29) |
| Recent receptive needle sharinge | ||||||
| No | 162 (43) | 77 (35) | 7/77 (9) | 1/77 (1) | 26/77 (34) | 43/77 (56) |
| Yes | 75 (20) | 30 (14) | 3/30 (10) | 3/30 (10) | 11/30 (37) | 13/30 (43) |
| Data missing | 63 (17) | 31 (14) | 2/31 (6) | 4/31 (13) | 4/31 (13) | 21/31 (68) |
| Data not collected at cohort site | 73 (20) | 80 (37) | 16/80 (20) | 23/80 (29) | 18/80 (23) | 23/80 (29) |
Data are proportion (%) of participants, unless otherwise indicated. Data exclude 34 participants who did not have intermittent viremia or spontaneous clearance but had a negative result of their final HCV RNA test (ie, viral suppression) and 37 participants treated within 26 weeks of infection.
Abbreviations: ACS, Amsterdam Cohort Studies; ATAHC, Australian Trial in Acute Hepatitis C; BAHSTION, Boston Acute HCV Study: Transmission, Immunity, and Outcomes Network; BBAASH, Baltimore Before and After Acute Study of Hepatitis; HCV, hepatitis C virus; HEPCO, St. Luc Cohort; HITS-c, Hepatitis C Incidence and Transmission Study–Community; HITS-p, Hepatitis C Incidence and Transmission Study–Prison; HIV, human immunodeficiency virus; IDU, injection drug use; IQR, interquartile range; N2, Networks 2; UFO, UFO study.
a At the time of primary HCV infection.
b Statistically significant differences from the group with persistent spontaneous clearance, the group with reinfection, and the group with intercalation.
c In the first 3 months of primary HCV infection.
d Statistically significant difference in the genotype distribution (genotype 1 vs other genotypes) from the group with intercalation.
e Reported at the interview prior to primary HCV infection diagnosis, recent indicates last 1–6 months prior to interview.
f Statistically significant differences from the group with persistent infection and the group persistent spontaneous clearance.
g Heroin/other opioids includes heroin, other opioids, and speedball; psychostimulants include amphetamines (including methamphetamines) and cocaine.
Primary HCV Infection Characteristics and Outcomes
Among those with known HCV genotypes during primary infection (518 [88%]), the most common genotypes were 1 (in 288 participants [56%]) and 3 (in 176 [34%]; Table 2). Following primary infection, 252 participants had viral suppression (at least 1 negative HCV RNA test result), of whom 146 (58%) had primary clearance (at least 2 consecutive HCV RNA tests performed ≥28 days apart that had negative results). Overall, 118 (47%) had reoccurring viremia, 72 after viral suppression and 46 after primary clearance (Figure 1D). Among those with reoccurring viremia, viral sequence analysis was used to distinguish reinfection (in 28 participants), intercalation (in 31), and indeterminate cases (in 55, viral sequencing data were not available; and in 4, heterologous virus with subsequent detection of the original viral strain was observed). Reinfection was more common after primary clearance (46% of cases) than viral suppression (10% of cases; P < .001; Figure 1D).
Study Retention and Frequency of HCV Testing
In the 28 reinfected participants, the median length of follow-up after the date of primary HCV infection was 4.6 years (interquartile range [IQR], 3.2–7.3 years) years. The median number of HCV RNA tests in this time was 17 (IQR, 9–35), and the median interval between tests was 48 days (IQR, 33–122 days). The median interval between tests prior to primary clearance (49 days; IQR, 32–140 days) was similar to the median interval during the risk period for reinfection and until reclearance or the end of follow-up (53 days; IQR, 33–138 days). In participants without reinfection, the median length of follow-up was 1.5 years (IQR, 0.7–2.8 years), the median number of HCV RNA tests was 5 (IQR, 3–9), and the median interval between tests was 84 days (IQR, 38–140 days). HCV RNA assay lower limits are provided in Supplementary Table 3.
HCV Reinfection
Twenty-eight participants had at least 1 reinfection (Figure 1D), and the incidence rate was 12.3 cases/100 person-years (95% CI, 8.5–17.8 cases/100 person-years); data were calculated by including participants with persistent primary clearance and reinfection, and Table 1 shows incidence rates stratified by study site. Fifteen reinfections involved a viral genotype different from that detected during primary infection, 3 involved a different viral subtype, and 10 involved the same genotype and subtype (Table 3).
Table 3.
Viral Genotype Detected During Primary Hepatitis C Virus (HCV) Infection and Reinfection Among 28 Participants With Reinfection
| Characteristic at Time of HCV Infection | Primary HCV Infection (n = 28) | HCV Reinfection (n = 28) |
|---|---|---|
| HCV genotype | ||
| 1 | 18 (64) | 17 (61) |
| 2 | 1 (4) | 4 (14) |
| 3 | 8 (29) | 6 (21) |
| 4 | 1 (4) | 1 (4) |
| HCV genotype/subtype in primary infection vs reinfection | ||
| Different genotype | … | 15 (54) |
| Different subtype | … | 3 (11) |
| Same genotype | … | 10 (36) |
Peak HCV RNA Load in Reinfection, Compared With Primary Infection
Fifteen reinfections had quantitative HCV RNA measurements available within the first 3 months of both primary infection and reinfection. The median peak HCV RNA load in reinfection (3.4 log IU/mL; IQR, 2.6–6.5 log IU/mL) was lower than that in primary infection (6.7 log IU/mL [IQR, 5.3–7.0 log IU/mL]; median difference, 1.46 log IU/mL [IQR, 0.34–4.18 log IU/mL]; P = .011). Timelines for quantitative HCV RNA measurements for 3 participants with reinfection, illustrating a range of trajectories, are included in Supplementary Figure 1.
Reinfection Outcomes
For 23 of the 28 reinfection cases, follow-up was sufficient (defined as the occurrence of at least 2 subsequent study visits) to classify the outcome. Of the 9 participants with reinfection without reclearance and with sufficient follow-up, the median estimated duration of reinfection at the end of follow-up was 66.9 months (range, 10.1–226.6 months). All participants had HCV genotype data from >1 time point following reinfection. The Kaplan–Meier estimate of the reclearance proportion 6 months after reinfection was 52% (95% CI, 33%–73%), after excluding the 3 individuals with changes in genotype or subtype.
Time to Reclearance
The median time to reclearance after reinfection was 3.0 months (IQR, 2.0–4.4 months). In the same participants, the median time to primary clearance was 5.5 months (IQR, 2.6–11.2 months).There was a tendency toward a shorter time to reclearance, compared with the time to primary clearance; this did not reach statistical significance (hazard ratio [HR], 1.86; 95% CI, .70–4.91; P = .211).
Predictors of HCV Reclearance
In shared frailty (for cohort site) but otherwise unadjusted Cox proportional hazards analysis of participants with reinfection, female participants with IFNL4 rs12979860 CC genotype were 4-fold more likely to reclear at any given time, compared with other participants (HR, 4.16; 95% CI, 1.24–13.94; P = .021; Table 4). There were no other statistically significant factors associated with reclearance.
Table 4.
Cox Proportional Hazards Regression Analysis of Predictors of Reclearance of Hepatitis C Virus (HCV), Adjusted for Cohort Site by Using a Shared Frailty Model
| Predictor | Reclearances, No. |
Reclearance Rate, Cases/100 Person-Years | HR (95% CI) | P Value | Shared Frailty for Cohort |
|
|---|---|---|---|---|---|---|
| θ | P Value | |||||
| Overall | 14 | 56.6 | … | … | ||
| Age at reinfection, y | ||||||
| ≤25 | 7 | 45.1 | 1.00 | … | ||
| >25 | 5 | 60.9 | 1.72 (.45–6.53) | .426 | 0.72 | .119 |
| Sexa | ||||||
| Male | 5 | 36.0 | 1.00 | … | ||
| Female | 9 | 82.9 | 2.45 (.72–8.29) | .150 | 0.74 | .125 |
| IFNL4 rs12979860 | ||||||
| CC | 10 | 69.8 | 2.00 (.56–7.18) | .285 | 1.18 | .053 |
| CT/TT | 4 | 38.3 | 1.00 | … | ||
| Combined effect of sex and IFNL4b | ||||||
| Female and CC | 7 | 126.9 | 4.16 (1.24–13.94) | .021 | 1.08 | .063 |
| Other | 7 | 36.4 | 1.00 | … | ||
| Reinfection HCV genotype | ||||||
| 1 | 9 | 53.2 | 1.00 | … | ||
| Other | 5 | 63.9 | 2.79 (.62–12.51) | .181 | 1.26 | .041 |
| Reinfected with different genotype from that in primary infection | ||||||
| Yes | 6 | 53.1 | 4.69 (.90–24.36) | .066 | 2.22 | .017 |
| No | 8 | 59.5 | 1.00 | |||
Abbreviations: CI, confidence interval; HR, hazard ratio.
a Data are missing for 2 participants with reclearance. No data were missing data for participants without reclearance.
b Schoenfeld residuals were used to evaluate the proportional hazards assumption. P = .990 by the test of proportional hazards.
Multiple HCV Reinfections
Of the 14 participants with reclearance, 5 had further reinfections. Three participants had 2 reinfections, and 2 participants had 3 reinfections. Overall, 35 reinfections were observed (Figure 2).
Figure 2.
Timeline of reinfection events from the estimated date of primary hepatitis C virus (HCV) infection. Timelines illustrate results of HCV RNA tests and HCV genotyping for the 28 participants with reinfection. Each box represents 1 HCV RNA test. Empty boxes are tests in which HCV RNA was undetectable, whereas filled boxes (black or colored) indicate that HCV RNA was detectable. HCV genotyping results are indicated using color and box labels. Within individuals, distinct strains within a single viral genotype and subtype that were confirmed by viral sequencing are illustrated using different shades of the same color. Reinfection events (R) are defined by the appearance of a new viral genotype, viral subtype, or distinct strain confirmed by viral sequencing following a period during which HCV RNA was undetectable. HCV from the primary infections of participants UFOVT0134 and AUS206 was serotyped.
HCV Intercalation
Compared with reinfection, intercalation was more likely to occur after a shorter period of HCV RNA undetectability (P < .001; Table 5). Similarly, intercalation tended to be observed earlier following primary infection (timing of reappearance of viremia after the estimated date of primary HCV infection in intercalation, P = .002; Table 5). However, considerable variation was observed.
Table 5.
Time From Estimated Date of Primary Infection Until Reappearance of Viremia, Number of Hepatitis C Virus (HCV) RNA–Negative Test Results, and Duration of Preceding Period of HCV RNA Undetectability
| No. | Median | IQR | Range | |
|---|---|---|---|---|
| Time to reappearance of viremia, da | ||||
| Intercalation | 31 | 250 | 174–433 | 113–2219 |
| Reinfection | 28 | 546 | 312–984 | 93–3747 |
| Indeterminate cases without sufficient viral sequencing | 55 | 231 | 140–399 | 46–3167 |
| Indeterminate cases with viral sequencingb | 4 | 173 | 168–645 | 166–1114 |
| HCV RNA–negative test results, no.c | ||||
| Intercalation | 31 | 1 | 1–2 | 1–6 |
| Reinfection | 28 | 3 | 2–7 | 1–15 |
| Indeterminate cases without sufficient viral sequencing | 55 | 1 | 1–2 | 1–18 |
| Indeterminate cases with viral sequencingb | 4 | 1 | 1–6 | 1–10 |
| Duration of HCV undetectability, dd | ||||
| Intercalation | 7 | 88 | 32–221 | 31–1293 |
| Reinfection | 21 | 210 | 131–412 | 28–1031 |
| Indeterminate cases without sufficient viral sequencing | 22 | 119 | 63–282 | 28–2707 |
| Indeterminate cases with viral sequencingb | 1 | 603 | … | … |
Abbreviation: IQR, interquartile range.
a Statistically significant differences between the reinfection group and the intercalation group (P = .002) and between the reinfection group and indeterminate group without sufficient viral sequencing (P < .001).
b Classified as indeterminate on the basis of detection of heterologous virus with subsequent detection of the original viral strain.
c Preceding reappearance of viremia; statistically significant differences between the reinfection group and the intercalation group (P = .001) and between the reinfection group and indeterminate group without sufficient viral sequencing (P < .001).
d Among cases with at least 2 HCV RNA–negative test results at least 28 days apart, calculated as the period from the first negative test result until the last negative test result.
Indeterminate Intermittent Viremia
Of the 59 cases of indeterminate intermittent viremia, 4 were classified on the basis of sequencing of heterologous virus with subsequent detection of the original viral strain, and the remaining 55 were classified on the basis of insufficient viral sequencing. The duration of the period of HCV RNA undetectability preceding reappearance of viremia and the timing of reappearance of viremia in the latter 55 cases were similar to that in intercalation (Table 5).
Sensitivity Analyses
Results were not sensitive to any of the factors tested (Supplementary Tables 3–5).
DISCUSSION
This study characterizes the natural history of viral suppression, primary clearance, HCV reinfection and reclearance in the largest sample of participants (mostly people who inject drugs) with well-defined primary HCV infection and reinfection reported to date. The peak HCV RNA load at the time of HCV reinfection was lower than in primary infection, providing further evidence of protective immunity in humans. Six months after reinfection, the clearance proportion was 52% (95% CI, 33%–73%). The combined effect of female sex and rs12979860 CC IFNL4 genotype was predictive of reclearance following reinfection.
Reclearance was predicted by a combined effect of sex and IFNL4 genotype. To the best of our knowledge, this is the first study to investigate predictors of reclearance. The propensity toward reclearance was 4 times greater among females with the rs12979860 CC IFNL4 genotype. This is particularly notable because, by definition, all participants with reinfection have already cleared 1 HCV infection and therefore would be expected to have a greater tendency toward spontaneous clearance a priori. In the context of primary clearance, similar findings have been reported with respect to female sex [1, 22] and IFNL4 genotype [24, 25] predicting clearance independently and in combination [15, 23], including within the InC3 study population [15]. The IFNL4 gene region encodes the interferon λ3 protein and is involved in viral control, although the precise mechanism remains unknown. It is possible that female sex influences HCV clearance through a mechanism related to general sex-based differences in immunity [34, 35]; however, the pathways by which these differences affect HCV control require elucidation. Further research is required to assess whether the combined effect of IFNL4 genotype and sex on primary clearance and reclearance is simply the product of the 2 independent effects or whether there is a synergistic effect [15, 23]. The importance of sex and IFNL4 genotype in both primary HCV infection and reinfection suggests that these factors have a crucial role in long-term protection from persistent HCV infection. Although IFNL4 genotype and sex are fixed genetic traits, the fact that spontaneously clearing infections are controlled better with subsequent exposures suggests the existence of an adaptive component. A better understanding of the mechanisms behind the immune response in females with the IFNL4 rs12979860 CC genotype has the potential to provide insights for vaccine development.
The findings presented here suggest partial protective immunity following primary clearance of HCV. The peak HCV RNA level was lower during reinfection, compared with that during primary infection. Reclearance following reinfection was observed in half of the participants, with the time to reclearance tending to be shorter following reinfection, compared with that for primary clearance. These findings are consistent with previous findings by Osburn et al and are not sensitive to stratification or adjustment by study site (BBAASH vs others) [6]. Mathematical modeling studies have shown that the interval between tests influences the proportions of reinfections resulting in reclearance and persistence [13]. In this study, heterogeneity in test intervals between study sites limited the interpretation of these proportions. Nonetheless, the identification of persistent reinfections indicates that, while primary HCV infection appears to confer protection against persistent HCV reinfection in some cases, there are limits to this protection. Chimpanzee studies indicate inadequate cross-strain protection in some cases [28]; however, this study did not find a higher probability of reclearance in participants reinfected with the same genotype as that found during their primary infection. Further studies to characterize the viral genomes of the primary and reinfection strains and to resolve the detailed characteristics of the immune responses against these viruses, including both neutralizing antibodies and HCV-specific T cells, are warranted.
The identification of diverse outcomes of HCV reinfection illustrates the complexity of the natural history of HCV. Among the 23 participants with follow-up after HCV reinfection, approximately one third experienced persistent reinfection, and one third experienced multiple reinfections after resolution of their first reinfection. The remaining third was composed of participants with reclearance but without further reinfections (possibly partly due to shorter follow-up) and of participants with changes in viral genotype or subtype following reinfection. This report adds to the few cases of multiple consecutive reinfection that have been reported previously [5, 6, 11], highlighting the ongoing risk of reinfection among those who continue to be exposed to HCV and emphasizing the need for education about reinfection risk in these groups and for delivery of HCV antiviral treatment to those who become reinfected.
In contrast to reinfection, intercalation was usually observed within the first 2 years of primary HCV infection, consistent with previous reports of fluctuations in HCV RNA loads in early HCV infection [36, 37]. However, intercalation cases were also observed later in HCV infection and after lengthier periods of HCV RNA undetectability, as has previously been reported [6, 12, 38, 39]. This highlights the importance of viral sequencing for classification of HCV reinfection. Intercalation may occur as a result of transient control of viral replication by the host immune response, but further research is required to develop a more detailed understanding of such events.
Despite bringing together the largest number of well-defined HCV reinfection events following spontaneous clearance or viral suppression reported to date, the number of events is low for detailed statistical analysis. Therefore, our analysis of predictors of HCV reclearance could not be adjusted for the effect of potential confounders. Participants with identified reinfections were typically followed longer and more frequently than other participants, and a large proportion of reoccurring viremia events could not be classified as reinfections or intercalations. These factors suggest that the true reinfection rate in the InC3 population is likely to be higher than the observed rate. While standard methods were used to classify outcomes of infection, there were differences between cohort sites in terms of methods of recruitment, test intervals, HCV RNA monitoring methods, and the region of HCV sequenced to assess reinfection. In some of the participating cohorts, data on HCV-related risk behaviors were not collected; therefore, risk behaviors could not be assessed as predictors of HCV reinfection or reclearance. The analysis of time to HCV reclearance versus primary clearance only included participants with reclearance during the study period. While the participants with persistent reinfection were all followed for at least 10 months, indicating that future reclearance would be unlikely, late clearance can occur, so there is a small risk of bias from excluding right-censored data (Supplementary Materials) [39].
This is the first study to investigate predictors of HCV reclearance. Similar to primary clearance, there appears to be a combined effect of sex and IFNL4 genotype on reclearance of HCV, suggesting that these factors together have considerable impact on long-term protection from persistent HCV infection. This study also highlights the complexity of acute HCV infection and reinfection and the factors that contribute to viral clearance. These findings suggest that HCV reinfection is associated with lower levels of viremia and a possibly shorter time to spontaneous reclearance, supporting a role for immunologic memory in conferring partial protection against persistent infection. Nonetheless, there is considerable heterogeneity in reinfection outcomes, and participants with ongoing exposure to HCV risk developing persistent reinfection.
STUDY GROUP MEMBERS
Steering committee: Kimberly Page (chair, UFO), Julie Bruneau (HEPCO), Andrea L. Cox (BBAASH), Gregory J. Dore (ATAHC), Jason Grebely (ATAHC), Margaret Hellard (N2), Georg Lauer (BAHSTION), Arthur Y. Kim (BAHSTION), Andrew R. Lloyd (HITS-p), Lisa Maher (HITS-c), Barbara H. McGovern (BAHSTION), Maria Prins (ACS), and Naglaa H. Shoukry (HEPCO). Coordinating center: Meghan Morris (study coordinator), Judy Hahn (coinvestigator), and Thomas M. Rice (data manager). Site data managers: Maryam Alavi (ATAHC), Rachel Bouchard (HEPCO), Jennifer Evans (UFO), Bart Grady (ACS), Jasneet Aneja (BAHSTION), Rachel Sacks-Davis (Networks 2), Suzy Teutsch (HITS-p), Bethany White (HITS-c), Brittany Wells (BBAASH), and Geng Zang (HEPCO). InC3 researchers: Tanya Applegate, Gail Matthews, and Barbara Yeung (ATAHC); Bart Grady and Thijs van de Laar (ACS); Jasneet Aneja and Leslie Erin Prince (BAHSTION); Elise Roy and Geng Zang (HEPCO); Anna Bates, Jarliene Enriquez, Sammy Chow, and Ju Park (HITS-c); Luke McCredie and Suzy Teutsch (HITS-p); Campbell Aitken, Scott Bowden, Peter Higgs, and Lilly Tracy (N2); and Alya Briceno (UFO).
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (NIH; grants U19 AI088791 [to A. L. C.] and U19 AI066345 [to A. Y. K., T. M. A. and B. H. M.]); the National Institute on Drug Abuse, NIH (grants R01 DA031056 [to the InC3 Study], R01 DA033541 [to A. Y. K.], R01 DA016017 [to K. P. and M. M.], and R01 DA15999-01 [to G. J. D., J. G., A. R. L., and M. H.]); the Victorian Operational Infrastructure Support Program (to the Burnet Institute); the National Health and Medical Research Council (postgraduate scholarship to R. S.-D., practitioner research fellowships to G. J. D. and A. R. L., senior research fellowships to M. H. and L. M., career development fellowship to J. G., and project grants 630483 [to the Hepatitis C Virus Incidence and Transmission Study–Community {HITS-c}] and 331312 [to the Networks 2 {N2} project]); Fonds de la Recherche du Québec–Santé (research career awards to J. B. and N. H. S.); an Australian postgraduate PhD award (to B. H.); the Canadian Institutes of Health Research (grants MOP-103138 and MOP-106468 to J. B. and N. H. S.); the Netherlands National Institute for Public Health and the Environment (to the Amsterdam Cohort Study); the University of New South Wales Hepatitis C Vaccine Initiative (to the HITS-c); the Australian Centre for HIV and Hepatitis Virology Research (to the N2 project); and the Centre for Research Excellence into Injecting Drug Use (to M. H., G. J. D., L. M., J. G., and R. S. D.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Collaborators: on behalf of the InC3 study group, Kimberly Page, Julie Bruneau, Andrea L. Cox, Gregory J. Dore, Jason Grebely, Margaret Hellard, Georg Lauer, Arthur Y. Kim, Andrew R. Lloyd, Lisa Maher, Barbara H. McGovern, Maria Prins, Naglaa H. Shoukry, Meghan Morris, Judy Hahn, Thomas M. Rice, Maryam Alavi, Rachel Bouchard, Jennifer Evans, Bart Grady, Jasneet Aneja, Rachel Sacks-Davis, Suzy Teutsch, Bethany White, Brittany Wells, Geng Zang, Tanya Applegate, Gail Matthews, Barbara Yeung, Bart Grady, Thijs van de Laar, Jasneet Aneja, Leslie Erin Prince, Elise Roy, Geng Zang, Anna Bates, Jarliene Enriquez, Sammy Chow, Ju Park, Luke McCredie, Suzy Teutsch, Campbell Aitken, Scott Bowden, Peter Higgs, Lilly Tracy, and Alya Briceno
References
- 1.Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat 2006; 13:34–41. [DOI] [PubMed] [Google Scholar]
- 2.Grebely J, Prins M, Hellard M et al. Towards a hepatitis C virus vaccine: insights from studies of hepatitis C virus reinfection in injection drug users. Lancet Infect Dis 2012; 12:408–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken CK, Lewis J, Tracy SL et al. High incidence of hepatitis C virus reinfection in a cohort of injecting drug users. Hepatology 2008; 48:1746–52. [DOI] [PubMed] [Google Scholar]
- 4.Micallef JM, Macdonald V, Jauncey M et al. High incidence of hepatitis C virus reinfection within a cohort of injecting drug users. J Viral Hepat 2007; 14:413–8. [DOI] [PubMed] [Google Scholar]
- 5.van de Laar TJW, Molenkamp R, van den Berg C et al. Frequent HCV reinfection and superinfection in a cohort of injecting drug users in Amsterdam. J Hepatol 2009; 51:667–74. [DOI] [PubMed] [Google Scholar]
- 6.Osburn WO, Fisher BE, Dowd KA et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology 2010; 138:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham ST, Bull RA, Bennett JM et al. Frequent multiple hepatitis C virus infections among injection drug users in a prison setting. Hepatology 2010; 52:1564–72. [DOI] [PubMed] [Google Scholar]
- 8.Grebely J, Conway B, Raffa JD, Lai C, Krajden M, Tyndall MW. Hepatitis C virus reinfection in injection drug users. Hepatology 2006; 44:1139–45. [DOI] [PubMed] [Google Scholar]
- 9.Currie SL, Ryan JC, Tracy D et al. A prospective study to examine persistent HCV reinfection in injection drug users who have previously cleared the virus. [erratum appears in Drug Alcohol Depend. 2008 Jul;96(1–2):192] Drug Alcohol Depend 2008; 93:148–54. [DOI] [PubMed] [Google Scholar]
- 10.Mehta SH, Cox A, Hoover DR et al. Protection against persistence of hepatitis C. Lancet 2002; 359:1478–83. [DOI] [PubMed] [Google Scholar]
- 11.Sacks-Davis R, Aitken C, Higgs P et al. High Rates of Hepatitis C Virus Reinfection and Spontaneous Clearance of Reinfection in People who Inject Drugs: a Prospective Cohort Study. PLoS One 2013; 8:e80216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page K, Osburn W, Evans J et al. Frequent Longitudinal Sampling of Hepatitis C Virus Infection in Injection Drug Users Reveals Intermittently Detectable Viremia and Reinfection. Clin Infect Dis 2013; 56:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickerman P, Grebely J, Dore GJ et al. The more you look the more you find - Effects of hepatitis C virus testing interval on re-infection incidence and clearance: Implications for future vaccine study design. J Infect Dis 2012; 205:1342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grebely J, Morris MD, Rice TM et al. Cohort Profile: The International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) Study. Int J Epidemiol 2012; 42:1649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grebely J, Page K, Sacks-Davis R et al. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology 2014; 59:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dev AT, McCaw R, Sundararajan V, Bowden S, Sievert W. Southeast Asian patients with chronic hepatitis C: the impact of novel genotypes and race on treatment outcome. Hepatology 2002; 36:1259–65. [DOI] [PubMed] [Google Scholar]
- 17.Ray S, Arthur R, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout Egypt. J Infect Dis 2000; 182:698–707. [DOI] [PubMed] [Google Scholar]
- 18.Van de Laar T, Langendam M, Bruisten S et al. Changes in risk behavior and dynamics of hepatitis C virus infections among young drug users in Amsterdam, the Netherlands. J Med Virol 2005; 77:509–18. [DOI] [PubMed] [Google Scholar]
- 19.Tu E, Bull R, Greening G et al. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin Infect Dis 2008; 46:413–20. [DOI] [PubMed] [Google Scholar]
- 20.Kuntzen T, Timm J, Berical A et al. Viral Sequence Evolution in Acute Hepatitis C Virus Infection. J Virol 2007; 81:11658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Rosenberg PS, Brown DL et al. Correlates of spontaneous clearance of hepatitis C virus among people with hemophilia. Blood 2006; 107:892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page K, Hahn JA, Evans J et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis 2009; 200:1216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Berg CHBS, Grady BPX, Schinkel J et al. Female Sex and IL28B, a Synergism for Spontaneous Viral Clearance in Hepatitis C Virus (HCV) Seroconverters from a Community-Based Cohort. PLoS One 2011; 6:e27555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas DL, Thio CL, Martin MP et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461:798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tillmann HL, Thompson AJ, Patel K et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology 2010; 139:1586–92, 92 e1. [DOI] [PubMed] [Google Scholar]
- 26.Grebely J, Petoumenos K, Hellard M et al. Potential role for IL28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology 2010; 52:1216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson EC, Fleming VM, Main J et al. Predicting spontaneous clearance of acute hepatitis C virus in a large cohort of HIV-1-infected men. Gut 2011; 60:837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prince AM, Brotman B, Lee D-H et al. Protection against Chronic Hepatitis C Virus Infection after Rechallenge with Homologous, but Not Heterologous, Genotypes in a Chimpanzee Model. J Infect Dis 2005; 192:1701–9. [DOI] [PubMed] [Google Scholar]
- 29.Lanford R, Guerra B, Chavez D et al. Cross-genotype immunity to hepatitis C virus. J Virol 2004; 78:1575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas DL, Astemborski J, Rai RM et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA 2000; 284:450–6. [DOI] [PubMed] [Google Scholar]
- 31.Kelly PJ, Lim LL-Y. Survival analysis for recurrent event data: an application to childhood infectious diseases. Stat Med 2000; 19:13–33. [DOI] [PubMed] [Google Scholar]
- 32.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer, 2009. [Google Scholar]
- 33.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2011. [Google Scholar]
- 34.Bouman A, Heineman M, Faas M. Sex hormones and the immune response in humans. Hum Reprod Update 2005; 11:411–23. [DOI] [PubMed] [Google Scholar]
- 35.Klein S, Jedlicka A, Pekosz A. The Xs and Y of immune responses to viral vaccines. Lancet Infectious Diseases 2010; 10:338–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hajarizadeh B, Grebely J, Applegate T et al. Dynamics of HCV RNA levels during acute hepatitis C virus infection. J Med Virol 2014; 86:1722–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glynn SA, Wright DJ, Kleinman SH et al. Dynamics of viremia in early hepatitis C virus infection. Transfusion 2005; 45:994–1002. [DOI] [PubMed] [Google Scholar]
- 38.Cox AL, Netski DM, Mosbruger T et al. Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin Infect Dis 2005; 40:951–8. [DOI] [PubMed] [Google Scholar]
- 39.Mosley JW, Operskalski EA, Tobler LH et al. The course of hepatitis C viraemia in transfusion recipients prior to availability of antiviral therapy. J Viral Hepat 2008; 15:120–8. [DOI] [PubMed] [Google Scholar]
- 40.Page-Shafer K, Pappalardo BL, Tobler LH et al. Testing strategy to identify cases of acute hepatitis C virus (HCV) infection and to project HCV incidence rates. J Clin Microbiol 2008; 46:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofer H, Watkins-Riedel T, Janata O et al. Spontaneous viral clearance in patients with acute hepatitis C can be predicted by repeated measurements of serum viral load. Hepatology 2003; 37:60–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.