Abstract
Background. Viral shedding is often considered to correlate with the infectivity of influenza, but the evidence for this is limited.
Methods. In a detailed study of influenza virus transmission within households in 2008–2012, index case patients with confirmed influenza were identified in outpatient clinics, and we collected nose and throat swab specimens for testing by reverse-transcription polymerase chain reaction from all household members regardless of illness. We used individual-based hazard models to characterize the relationship between viral load (V) and infectivity.
Results. Assuming that infectivity was proportional to viral load V gave the worst fit, because it strongly overestimated the proportion of transmission occurring at symptom onset. Alternative models assuming that infectivity was proportional to a various functions of V provided better fits, although they all overestimated the proportion of transmission occurring >3 days after symptom onset. The best fitting model assumed that infectivity was proportion to Vγ, with estimates of γ = 0.136 and γ = 0.156 for seasonal influenza A(H1N1) and A(H3N2) respectively.
Conclusions. All the models we considered that used viral loads to approximate infectivity of a case imperfectly explained the timing of influenza secondary infections in households. Identification of more accurate correlates of infectivity will be important to inform control policies and disease modeling.
Keywords: influenza, infectiousness, public health, isolation
Characterizing the infectivity of human influenza virus is important for disease control and prevention. For example, determining how long persons are infectious is required to inform recommendations about how long sick individuals should isolate themselves at home to avoid further transmission in the community. Determining which age groups contribute the most to transmission is important to inform policy making, for example on school closure. Although infectivity can be assessed from epidemiological studies documenting transmission in human populations, this approach may be expensive and resource consuming [1–3]. As a consequence, if a good biomarker of infectivity is available, it might be advantageous simply to study the biomarker in a sample of influenza case patients, without having to follow up contacts of those patients.
Viral shedding seems a natural candidate and has indeed been used as a proxy measure of infectivity in many studies [3–11]. One common approach is to use the duration of viral shedding as the infectious period. This leads to estimates of the average infectious period in the range of 4–8 days [3–11]. However, this estimate does not take into account the major variations in viral loads that occur over time and may affect infectivity. If instead infectivity at a given time is proportional to viral load at that time, most transmission would be predicted to occur in the first few days of infection, with a generation time of 2.3 days [5]. However, this assumption has never been tested.
Because viral shedding is commonly used as a proxy measure for infectivity, with important policy implications, it is essential to evaluate the performance of this biomarker in explaining and predicting infectivity. Household transmission studies provide an ideal natural setting to explore this relation, because a substantial fraction of transmission occurs in this setting [12–14], lending generalizability, and because it is feasible to measure exposed household contacts intensively for the period that the index case patient may be infectious. In the current study, analyzing large prospective studies of influenza A virus transmission in Hong Kong households conducted in influenza seasons from 2008 to 2012 [3, 15], we examined the relationship between viral shedding and infectivity.
METHODS
Study Subjects
We have been conducting large community-based studies of the household transmission of influenza virus in Hong Kong [3, 15]. In these studies, outpatients with acute respiratory illness within 2 days after illness onset, who lived in a household with ≥2 other persons, none of whom reported recent illness in the preceding 14 days before the first visit, were recruited and tested with the QuickVue Influenza A + B test (Quidel). Subjects with a positive result on the rapid test were further followed up along with their household contacts, involving 3 home visits over approximately 7 days. Nose and throat swab specimens were collected from all subjects and their household contacts at each visit, regardless of the presence of respiratory symptoms. Daily symptoms for index case patients and their household contacts were recorded in symptom diaries for the duration of follow-up. Subjects recruited from January 2008 to June 2009 were part of a randomized controlled trial of enhanced hand hygiene, with or without surgical face masks, randomly allocated on a household basis [15], and subjects subsequently recruited beginning in the summer of 2009 were part of a comparative study of seasonal and pandemic influenza virus transmission in households, with a simple hand hygiene intervention given to all households [3]. Our analyses only included households in which index case patients had polymerase chain reaction (PCR)–confirmed influenza A virus infection.
Laboratory Methods
Paired nasal and throat swab samples were pooled immediately after collection in viral transport medium and delivered to the laboratory for cryopreservation at −70°C within 24 hours of collection. Swab samples were subsequently tested using quantitative reverse-transcription PCR to detect influenza A virus and quantify virus shedding. Total nucleic acid was extracted using the NucliSens easy MAG extraction system (bioMerieux; Boxtel), according to the manufacturer's instructions, and 12 μL of extracted nucleic acid with a random primer was used to prepare complementary DNA with an Invitrogen Superscript III kit (Invitrogen), as described elsewhere [16]. Influenza A virus was detected with a PCR assay, as described elsewhere [17]. At the end of the assay, PCR products were subjected to a melting-curve analysis to determine the specificity of the assay. The lower limit of detection of the PCR assay was approximately 900 virus gene copies per milliliter.
Ethics Statement
All subjects aged ≥18 years gave written informed consent, and proxy written consent was obtained from parents or legal guardians of children aged ≤17 years , with additional written assent from those aged 8–17 years. The study protocol was approved by the Institutional Review Board of the University of Hong Kong.
Statistical Analysis
PCR-confirmed influenza virus infection was defined as a positive result on testing of ≥1 nasal and throat specimen collected during the follow-up period. Illness onset time for PCR-confirmed influenza virus infection was defined as the first day when the subject reported ≥2 of the following 7 signs or symptoms: runny nose, cough, sore throat, headache, phlegm, myalgia, and fever [7]. Households that included >1 person with symptom onset at recruitment (ie, multiple index case patients) were excluded from the analyses.
To characterize the transmission dynamics in households and the factors affecting infectivity or susceptibility, we used an individual-based hazard model [18] extended to incorporate factors affecting infectivity. The model described the risk of PCR-confirmed infection among household contacts as depending on the time since symptom onset in any other infected persons in each household. The model allowed for infections from outside the household (community infections), or infections via other household contacts rather than the index case (tertiary infections). The community risk of infection was assumed to be time varying and directly proportional to influenza incidence rates in the general community, approximated by local surveillance data [19], with a different constant of proportionality (scaling factor) for each subtype (see Supplementary Appendix). We based our analysis on the times of symptom onset and assumed that the incubation period was 1 day. We also conducted a sensitivity analysis with an incubation period of 2 days.
We consider a number of functional forms to characterize the relationship between infectivity and viral shedding. These included infectivity being proportional to viral load V, or to the logarithm of viral load log V or to a power of these variables (Vγ or [log V]γ). For comparison purposes, we also considered a more standard transmission model [2, 18], denoted the “Epi-only” model, that did not include data on viral shedding and assumed that the infectivity of a case varied with time since symptom onset with a flexible Weibull-shaped function.
Because the viral shedding trajectories were only available for some days at or after symptom onset, we first fitted a log-linear regression model [8, 20], which accounted for censoring due to the lower limit of detection of the PCR assay [21] and allowed for separate intercepts for each individual but a common slope, supported by other studies [5, 7], on the observed viral shedding data. We also explored whether other possible models including curvilinear models or models with random slopes for each individual could better describe the observed viral shedding trajectories (Supplementary Appendix). Then we used the predicted viral shedding trajectories from the fitted model as a proxy for individual trajectory (viral shedding model). After that, we compared these models to determine which model could better describe the infectivity profile since symptom onset. The structures of the Epi-only and viral shedding models are summarized in Table 1.
Table 1.
Structure of the Epi-only Model and the Viral Shedding Models
| Feature | Model Structurea |
|---|---|
| Susceptibility of subject j | All models: based on age and vaccination status of subject j |
| Infectivity of subject i | All models: based on age, being an index case patient, and oseltamivir treatment status of subject i |
| Hazard of infection from community of subject j | All models: based on influenza activity in community and susceptibility of subject j |
| Hazard of infection of subject j from infected subject i | All models: based on infectivity of subject i, susceptibility of subject j and the infectivity profile of subject i |
| Total hazard of infection of subject j | All models: hazard of infection from community of subject j plus sum of hazard of infection of subject j from each infected subject i |
| Infectivity profile | Epi-only model: assumed independent of characteristic of subject i and modeled by a Weibull distribution; viral shedding models: proportional to viral load Vγ (model A, γ estimated; model B, γ = 1) or to logarithm of viral loads (log V)γ (model C, γ estimated; model D, γ = 1), |
a Further details of the models (eg, mathematical formulas) are available in the Supplementary Appendix.
We used the Epi-only model to estimate the proportion of cases attributed to household transmission and explore possible factors affecting susceptibility or infectivity. We considered age, being an index case patient, and oseltamivir treatment as 3 factors that might influence infectivity and age, receipt of influenza vaccination (trivalent inactivated influenza vaccine, which included seasonal A[H1N1], A[H3N2], and B strains) as 2 factors that might influence susceptibility [2, 3, 18, 22–24] and estimated their effects. We also explored whether the presence of specific symptoms were correlated with individual infectivity and whether sex (using age-sex categories to allow for interactions), household interventions (face mask or hand hygiene), smoking, and the presence of chronic conditions were associated with the susceptibility of household contacts to infection.
We conducted our statistical analysis in a Bayesian framework. We constructed a Markov chain Monte Carlo algorithm [25] to fit the transmission model and estimate the parameters. One particular feature of our study design is that there were no household contacts with symptom onset at or before the recruitment day, which we accounted for by using conditional likelihood in the statistical model (Supplementary Appendix). Simulation studies demonstrated that the algorithm could give unbiased parameter estimates (Supplementary Appendix).
The different models of infectivity were compared with each other on the basis of the deviance information criterion (DIC) [26]. DIC differences >5 were considered substantial [27]. We compared the observed distribution of times of infection of secondary cases with the one predicted by the Epi-only and viral shedding models (Supplementary Appendix). The adequacy of model fit was assessed with simulation-based χ2 tests comparing observed and expected distributions of the number of secondary cases in households of different sizes [2]. All statistical analyses were conducted using R (version 3.0.1; R Foundation for Statistical Computing) and MATLAB (version 7.8.0; MathWorks) software.
RESULTS
We compared the characteristics of index case patients with confirmed seasonal influenza A(H1N1) and seasonal influenza A(H3N2) virus infections (abbreviated hereafter as sH1N1 and H3N2, respectively) and their household contacts in 258 households (Table 2). Index case patients with sH1N1 were on average younger than those with H3N2 (P < .001). Other characteristics of household contacts were similar for index case patients in the 2 groups.
Table 2.
Characteristics of Index Case Patients With Seasonal Influenza A(H1N1) or Seasonal Influenza A(H3N2) Virus Infection and Their Household Contacts
| Characteristic | Patients or Contacts, No. (%) |
|
|---|---|---|
| Seasonal A(H1N1) | Seasonal A(H3N2) | |
| Index case patients | 141 | 117 |
| Age, y | ||
| ≤18 | 108 (77) | 65 (56) |
| 18–50 | 27 (19) | 35 (30) |
| >50 | 6 (4) | 17 (15) |
| Male sex | 69 (49) | 62 (53) |
| Prior vaccination | 22 (16) | 26 (22) |
| Oseltamivir treatmenta | 62 (44) | 48 (41) |
| No. of household contacts | ||
| 2 | 48 (34) | 44 (38) |
| 3 | 51 (36) | 52 (44) |
| 4 | 33 (23) | 18 (15) |
| 5 | 6 (4) | 2 (2) |
| 6 | 3 (2) | 1 (1) |
| No. of secondary cases in household | ||
| 0 | 107 (76) | 88 (75) |
| 1 | 30 (21) | 24 (21) |
| 2 | 4 (3) | 4 (3) |
| 3 | 0 (0) | 1 (1) |
| Household contacts | 429 | 332 |
| Age, y | ||
| ≤18 | 81 (19) | 68 (20) |
| 18–50 | 271 (63) | 206 (62) |
| >50 | 77 (18) | 58 (17) |
| Male sex | 158 (37) | 131 (39) |
| Prior vaccination | 50 (12) | 50 (15) |
| No. of infections in age groupsb | ||
| ≤18 y | 11 (14) | 10 (15) |
| 18–50 y | 25 (9) | 19 (9) |
| >50 y | 2 (3) | 6 (10) |
| Asymptomatic infection | 2/38 (5) | 4/35 (11) |
a Only oseltamivir treatment started within 48 hours after onset was classified as oseltamivir treatment.
b Brackets represent secondary infection risk for the corresponding age groups.
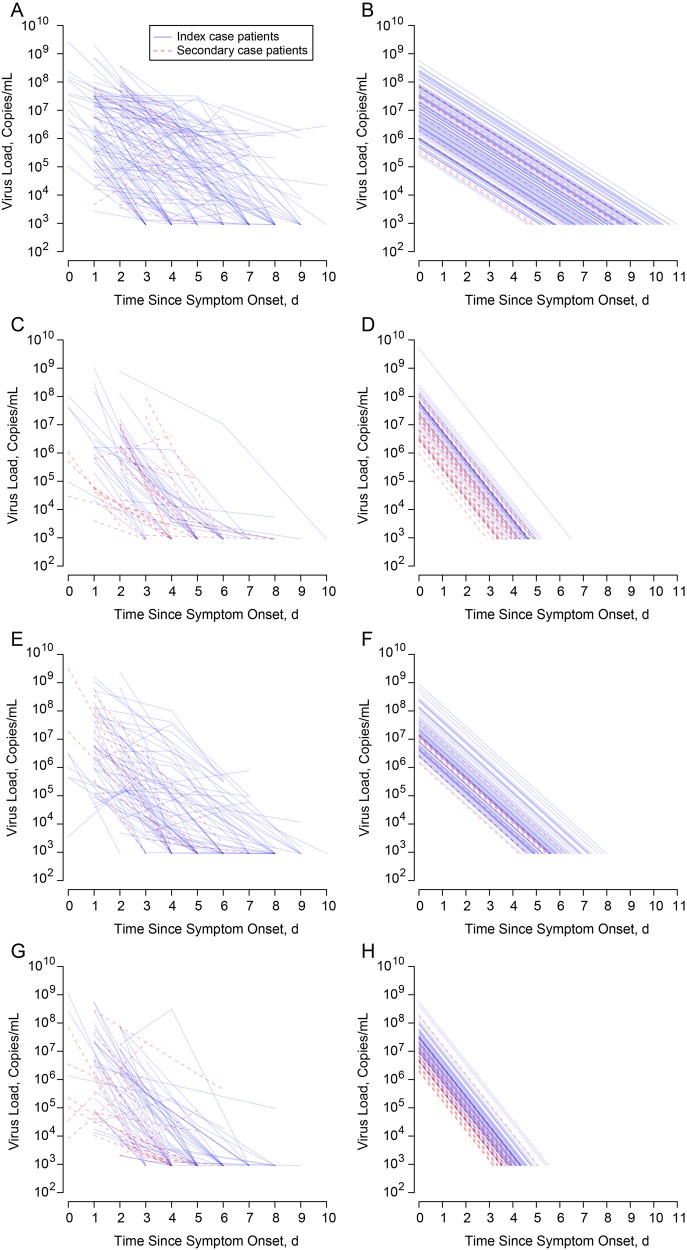
The trajectories of viral shedding, estimated from the fitted log-linear censored models with random intercepts, for subjects with sH1N1 and sH3N2 are shown in Figure 1. We summarized the estimated viral shedding trajectories in Table 3. After adjustment for being an index case patient, no significant differences in the estimated geometric mean viral shedding at symptom onset was found between infected children and adults for all subtypes. The rate of decline of viral shedding was slower for infected children than for infected adults, and the duration of shedding was longer for infected children for both subtypes. We also fitted log-linear censored models with random intercepts and random slopes for infected children, log-curvilinear censored models with random intercepts, and log-linear censored models allowing effect of oseltamivir on slope, but there was no significant improvement in term of model fit compared with the models with random intercepts only (Supplementary Table 1).
Figure 1.
Viral shedding patterns from observed data and predicted from the fitted random effects log-linear censored regression model. A, B, Observed and predicted viral shedding pattern for children with polymerase chain reaction (PCR)–confirmed seasonal influenza A(H1N1) virus infection. C, D, Observed and predicted viral shedding pattern for adults with PCR-confirmed seasonal influenza A(H1N1) virus infection. E, F, Observed and predicted viral shedding pattern for children with PCR-confirmed seasonal influenza A(H3N2) virus infection. G, H, Observed and predicted viral shedding pattern for adults with PCR-confirmed seasonal influenza A(H3N2) virus infection.
Table 3.
Estimates of Viral Shedding Trajectories From Fitted Log-linear Censored Regression Model With Random Effects on Intercepts
| Change in Viral Load (95% CI), Log10 copies/mL |
P Value | ||
|---|---|---|---|
| Childrena | Adultsa | ||
| Seasonal A(H1N1) | |||
| Being an index case patient | 0.11 (−0.73 to 0.95) | 0.63 (−0.29 to 1.55) | .35 |
| Oseltamivir treatmentb | −0.37 (−0.81 to 0.07) | −0.16 (−1.09 to 0.76) | .04 |
| Slope | −0.53 (−0.59 to −0.46) | −1.02 (−1.16 to −0.88) | <.001 |
| Mean duration of shedding, dc | 7.68 (4.98–10.43) | 4.29 (3.25–5.19) | <.001 |
| Intercept random effect SD | 0.98 | 0.87 | … |
| Seasonal A(H3N2) | |||
| Being an index case patient | 0.57 (−0.36 to 1.49) | 0.46 (−0.30 to 1.22) | .74 |
| Oseltamivir treatmentb | −0.33 (−0.93 to 0.28) | 0.26 (−0.50 to 1.02) | .02 |
| Slope | −0.74 (−0.84 to −0.64) | −1.04 (−1.18 to −0.90) | <.001 |
| Mean duration of shedding, dc | 5.6 (4.52–7.64) | 4.06 (3.23–5.11) | <.001 |
| Intercept random effect SD | 0.74 | 0.55 | … |
Abbreviations: CI, confidence interval; SD, standard deviation.
a Unless otherwise specified, values represent change in log10 viral load associated with changes in each covariate, with 95% CIs.
b Only oseltamivir treatment started within 48 hours after onset was classified as oseltamivir treatment.
c Defined as days from symptom onset to viral shedding lower than its detection limit.
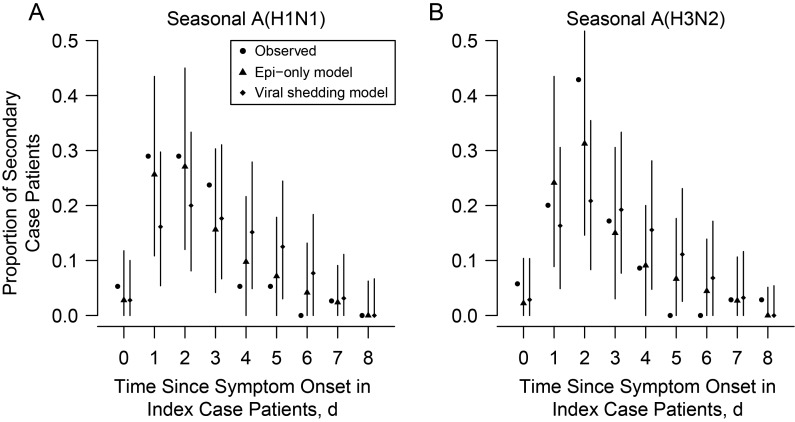
The estimated infectivity profiles from the viral shedding model and the Weibull model are presented in Figure 2. Compared with the Epi-only profile, we found that the assumption that infectivity was proportional to viral shedding V (model B) led to most transmission occurring around the time of symptom onset (>80%), which was not consistent with the data. Models assuming that the infectivity profile was proportional to Vγ (model A), (log V)γ (model C), or log V (model D) gave a better fit, although they tended to overestimate the proportion of transmission occurring after day 3. Indeed, this proportion was 3% (95% credible interval [CrI], 0%–23%) and 1% (0%–15%) for sH1N1 and sH3N2, respectively, in the Epi-only model compared with 29% (21%–41%) and 13% (8%–23%) in model A. Among models A, C, and D, model A with γ = 0.136 (95% CrI, .020–.225) for sH1N1 and γ = 0.156 (.032–.261) for sH3N2 gave a slightly better fit for both sH1N1 (DIC difference, 5.98 between models A and C and 1.80 between models A and D) and sH3N2 (DIC difference, 6.34 between models A and C and 3.32 between models A and D) (Supplementary Table 2). The Epi-only model gave a better fit to the distribution of time of infection than the viral shedding models (Figure 3). Based on the Epi-only model, we found the proportion of cases attributed to household transmission were 97.5% (95% CrI, 89.5%–100%) and 97.1% (88.6%–100%) for sH1N1 and sH3N2, respectively (Supplementary Appendix).
Figure 2.
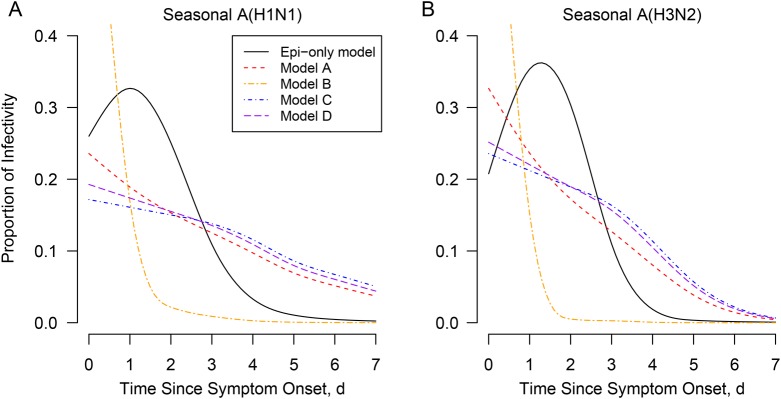
The estimated infectivity profile since symptom onset from the Epi-only and the viral shedding model for seasonal influenza A(H1N1) virus (A) and seasonal influenza A(H3N2) virus (B). It showed proportion of infectivity in an illness episode. In the viral shedding model, infectivity was assumed to be proportional to viral load Vγ (model A, γ estimated; model B, γ = 1) or to logarithm of viral loads (log V)γ (model C, γ estimated; model D, γ = 1).
Figure 3.
Estimated distribution of time of infection in secondary case patients from the Epi-only and viral shedding models for seasonal influenza A(H1N1) virus (A) and seasonal influenza A(H3N2) virus (B). Circles represent the observed proportion of infections that occurred since symptom onset in index case patients; triangles and diamonds, the estimated proportions of secondary infections that occurred since symptom onset in index case patients from the Epi-only and viral shedding model, respectively; vertical lines, 95% credible intervals for the estimated proportions of infectivity. The observed infection times were estimated under the assumption that the incubation period was 1 day.
The results of the Epi-only model are shown in Table 4. Infected children were significantly more infectious than infected adults for sH1N1 (relative infectivity, 2.88; 95% CrI, 1.21–7.68; P = .02). After adjustment for age and being an index case patient, we found that oseltamivir treatment for subjects with sH1N1 infection within 48 hours of illness onset was associated with lower infectivity (relativity infectivity, 0.29; 95% CrI, .01–.89; P = .02). We did not identify any significant association between age and susceptibility or between vaccination and susceptibility. We explored other possible factors that could affect infectivity or susceptibility by including them in the model but did not detect any significant effect (Supplementary Table 3).
Table 4.
Factors Affecting Influenza Susceptibility and Infectivity in the Epi-only Model
| Characteristic | Seasonal A(H1N1) |
Seasonal A(H3N2) |
||
|---|---|---|---|---|
| Risk Ratioa | P Value | Risk Ratioa | P Value | |
| Factors affecting infectivity | ||||
| Age ≤18 vs >18 y (reference) | 2.88 (1.21–7.68) | .02 | 2.05 (0.82–5.65) | .14 |
| Oseltamivir treatmentb | 0.29 (0.01–0.89) | .02 | 1.84 (0.74–5.18) | .19 |
| Being an index case patient | 1.84 (0.76–4.38) | .20 | 0.97 (0.29–3.30) | .96 |
| Factors affecting susceptibility | ||||
| Age | ||||
| ≤18 vs 19–50 y (reference) | 1.67 (0.77–3.33) | .20 | 1.49 (0.65–3.21) | .34 |
| >50 vs 19–50 y (reference) | 0.30 (0.04–1.12) | .07 | 1.05 (0.34–2.54) | .91 |
| Vaccination | 0.33 (0.05–1.25) | .11 | 1.59 (0.67–3.56) | .24 |
a Values represented risk ratios associated with each covariate and the corresponding 95% credible intervals.
b Only oseltamivir treatment started within 48 hours after onset was classified as oseltamivir treatment.
DISCUSSION
We explored the relationship between viral shedding measured by PCR on pooled nose and throat swab specimens and infectivity in influenza virus transmission in households. In particular, we investigated whether infectivity over time could be modeled as a function of viral shedding V, considering different functional forms: Vγ (model A), V (model B), (log V)γ (model C) or log V (model D). We found that the model that assumed infectivity was proportional to viral load V gave the worst fit, because it strongly overestimated the proportion of transmission occurring at symptom onset. Alternative models that assumed infectivity was proportional to a power function Vγ, to log V, or to (log V)γ provided better fits, although they all overestimated the proportion of transmission occurring >3 days after symptom onset. This indicates that the duration of virus shedding detectable by PCR may be longer than the infectious period. It may also indicate that other factors, such as the severity of symptoms [28] or social behavior [29], influence infectivity within households. One caveat of this analysis is that our study design permitted households to be enrolled within 2 days of illness onset in the index case patients, whereas households with multiple index case patients were excluded. Our design therefore selected against households with rapid transmission, and we may have underestimated the amount of transmission that occurs around the time of illness onset.
We found that models relying on geometric mean viral loads overestimated infectivity >3 days following symptom onset. In other studies, we also found that the proportion of the median tissue culture infectious dose after 3 days since symptom onset was lower than that of geometric mean viral loads measured by PCR [7], suggesting the median tissue culture infectious dose may be a more accurate biomarker for infectivity, because this captures infectious virus whereas PCR measures genetic material, a fraction of which may not be from infectious virus.
We found that the duration of viral shedding for infected children was longer than for adults, which was consistent with some previous studies [8, 9] but not others, either because of a possible lack of power [10, 11] or because of inconsistent definitions for the duration of viral shedding [4, 6]. We found that infected children were more infectious than infected adults for sH1N1, which was consistent with previous studies [22, 30, 31]. This might be explained by children having more frequent and intense contacts with other household members on average, compared with adults [32]. Consistent with previous studies, we found that oseltamivir treatment within 2 days since onset was not associated with reduced viral shedding [3, 11, 33, 34]. However, we also found that oseltamivir treatment was associated with a reduction in infectivity for sH1N1 but not for sH3N2. Nonpharmaceutical interventions, including face masks and hand hygiene, were not found to reduce the risk of infection in the household setting. This is consistent with other nonpharmaceutical intervention studies [34–36]. Our estimates of the effectiveness of trivalent influenza vaccine for sH1N1 and sH3N2 were consistent with findings reported elsewhere [18]. We estimated that almost all secondary infections (97%) were acquired from the household, and this findings was also consistent with other studies (see Supplementary Appendix) [37–39]. In particular, our analysis of the homology in virus sequences between infections in index case patients and household contacts confirmed that almost all infections in household contacts were acquired within the household [39].
Our study has a number of limitations. First, although recruiting symptomatic index case patients from outpatient clinics is an effective way to study transmission dynamics in household settings, the generalizability of our results may be limited because index case patients had an illness that warranted medical attention and had a positive result on a rapid test, suggesting they might have higher levels of virus shedding [40]. We accounted for this potential effect in our model, but we did not find a significant difference in infectivity between index and secondary case patients. Second, because index case patients were recruited after symptom onset, information on presymptomatic virus shedding is not available, and presymptomatic infectivity was not considered in our study. It is also possible that we missed the period of highest viral shedding for some patients by sampling at 3-day intervals. Third, the duration of follow-up in our study was from 7–12 days after symptom onset in index case patients; hence, some information on long durations of virus shedding for secondary and tertiary cases may have been missed. However, with an average serial interval of about 3 days [41], the effect of such censoring should be minimal. Fourth, nose and throat swab specimens were pooled in our studies, and while we might expect a high correlation in viral loads in the 2 sites in future iterations of the work, it will be interesting to test a range of sites (including viral load in nose, throat, and exhaled breath) to determine which sites provide the best proxy measure of infectivity. Finally, our study is observational, and although we controlled for age and vaccination in our transmission model we cannot rule out the risk of other unidentified confounders.
In conclusion, we characterized the relationship between viral shedding and infectivity. We found that all the models we considered that used viral loads to approximate infectivity of a case imperfectly explained the timing of influenza secondary infections in households, with the best-fitting models—Vγ, log V, and (log V)γ—still overestimating transmission >3 days after symptom onset. This may be because other factors, such as the severity of symptoms [28] or social behavior [29], may also influence infectivity within households. Identification of more accurate correlates of infectivity will be important to inform control policies and disease modeling.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all the physicians, nurses, and staff members at the participating centers for facilitating recruitment; the dedicated team of healthcare workers who conducted the home visits; and Chan Kit Man, Calvin Cheng, Rita Fung, Ho Yuk Ling, Lam Yiu Pong, Lincoln Lau, Tom Lui, Tong Hok Leung, Sara Poon, and Teresa So for research support.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This project was supported by the National Institute of Allergy and Infectious Diseases (NIAID) (contract HHSN266200700005C), the NIAID Centers for Excellence in Influenza Research and Surveillance (Administrative Data Base contract N01-AI-70005), the Government of the Hong Kong Special Administrative Region (commissioned grant HK-10-04-02 from the Health and Medical Research Fund), the National Institute of General Medical Sciences (grant U54 GM088558 to the Harvard Center for Communicable Disease Dynamics and MIDAS initiative grant 1U01GM110721-01 to S. C.), the Area of Excellence Scheme of the University Grants Committee of Hong Kong (grant AoE/M-12/06), L'Oreal Hong Kong (research scholarship to T. K. T), and the Laboratory of Excellence Integrative Biology of Emerging Infectious Diseases (research funding to S. C.).
Potential conflicts of interest. B. J. C. has received research funding from MedImmune and Sanofi Pasteur and consults for Crucell. D. K. M. I. has received research funding from Hoffmann-La Roche. G. M. L. has consulted for Janssen Pharmaceuticals and received speaker fees from Hongkong and Shanghai Banking Corporation and Credit Lyonnais Securities Asia. J. S. M. P. receives research funding from Crucell and serves as an ad hoc consultant for GlaxoSmithKline and Sanofi. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Cauchemez S, Bhattarai A, Marchbanks TL et al. Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc Natl Acad Sci U S A 2011; 108:2825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cauchemez S, Donnelly CA, Reed C et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med 2009; 361:2619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowling BJ, Chan KH, Fang VJ et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med 2010; 362:2175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattarai A, Villanueva J, Palekar RS et al. Viral shedding duration of pandemic influenza A H1N1 virus during an elementary school outbreak—Pennsylvania, May-June 2009. Clin Infect Dis 2011; 52(suppl 1):S102–8. [DOI] [PubMed] [Google Scholar]
- 5.Carrat F, Vergu E, Ferguson NM et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008; 167:775–85. [DOI] [PubMed] [Google Scholar]
- 6.Fielding JE, Kelly HA, Mercer GN, Glass K. Systematic review of influenza A(H1N1)pdm09 virus shedding: duration is affected by severity, but not age. Influenza Other Respir Viruses 2014; 8:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau LL, Cowling BJ, Fang VJ et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis 2010; 201:1509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau LL, Ip DK, Nishiura H et al. Heterogeneity in viral shedding among individuals with medically attended influenza A virus infection. J Infect Dis 2013; 207:1281–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CC, Wang L, Eng HL et al. Correlation of pandemic (H1N1) 2009 viral load with disease severity and prolonged viral shedding in children. Emerg Infect Dis 2010; 16:1265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb M, Singh PK, Fox J et al. Longitudinal study of influenza molecular viral shedding in Hutterite communities. J Infect Dis 2012; 206:1078–84. [DOI] [PubMed] [Google Scholar]
- 11.Suess T, Remschmidt C, Schink SB et al. Comparison of shedding characteristics of seasonal influenza virus (sub)types and influenza A(H1N1)pdm09; Germany, 2007–2011. PLoS One 2012; 7:e51653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao DL, Halloran ME, Obenchain VJ, Longini IM Jr. FluTE, a publicly available stochastic influenza epidemic simulation model. PLoS Comput Biol 2010; 6:e1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson NM, Cummings DA, Fraser C et al. Strategies for mitigating an influenza pandemic. Nature 2006; 442:448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monto AS. Studies of the community and family: acute respiratory illness and infection. Epidemiol Rev 1994; 16:351–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowling BJ, Chan KH, Fang VJ et al. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Ann Intern Med 2009; 151:437–46. [DOI] [PubMed] [Google Scholar]
- 16.Peiris JS, Tang WH, Chan KH et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis 2003; 9:628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan KH, Peiris JS, Lim W, Nicholls JM, Chiu SS. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. J Clin Virol 2008; 42:65–9. [DOI] [PubMed] [Google Scholar]
- 18.Tsang TK, Cauchemez S, Perera RA et al. Association between antibody titers and protection against influenza virus infection within households. J Infect Dis 2014; 210:684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P, Goldstein E, Ho LM et al. Excess mortality associated with influenza A and B virus in Hong Kong, 1998–2009. J Infect Dis 2012; 206:1862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ip DK, Schutten M, Fang VJ et al. Validation of self-swab for virologic confirmation of influenza virus infections in a community setting. J Infect Dis 2012; 205:631–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaida F, Liu L. Fast implementation for normal mixed effects models with censored response. J Comput Graph Stat 2009; 18:797–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cauchemez S, Carrat F, Viboud C, Valleron AJ, Boelle PY. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat Med 2004; 23:3469–87. [DOI] [PubMed] [Google Scholar]
- 23.Horby P, Mai le Q, Fox A et al. The epidemiology of interpandemic and pandemic influenza in Vietnam, 2007–2010: the Ha Nam household cohort study I. Am J Epidemiol 2012; 175:1062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klick B, Nishiura H, Ng S et al. Transmissibility of seasonal and pandemic influenza in a cohort of households in Hong Kong in 2009. Epidemiology 2011; 22:793–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilks WR, Richardson S, Spiegelhalter D. Markov chain Monte Carlo in practice. London, United Kingdom: Chapman & Hall, 1996. [Google Scholar]
- 26.Spiegelhalter DJ, Best N, Carlin B, Van Der Linde A. Bayesian measures of model complexity and fit. J R Statist Soc B 2002; 64:583–639. [Google Scholar]
- 27.Spiegelhalter DJ, Thomas A, Best NG. WinBUGS version 1.3 user manual. Cambridge, United Kingdom: Medical; Research Council Biostatistics Unit, 2000. [Google Scholar]
- 28.Thai PQ, Mai LQ, Welkers MR et al. Pandemic H1N1 virus transmission and shedding dynamics in index case households of a prospective Vietnamese cohort. J Infect 2014; 68:581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucharski AJ, Kwok KO, Wei VW et al. The contribution of social behaviour to the transmission of influenza A in a human population. PLoS Pathog 2014; 10:e1004206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carcione D, Giele CM, Goggin LS et al. Secondary attack rate of pandemic influenza A(H1N1) 2009 in Western Australian households, 29 May-7 August 2009. Euro Surveill 2011; 16:pii:19765. [PubMed] [Google Scholar]
- 31.Viboud C, Boelle PY, Cauchemez S et al. Risk factors of influenza transmission in households. Br J Gen Pract 2004; 54:684–9. [PMC free article] [PubMed] [Google Scholar]
- 32.Mossong J, Hens N, Jit M et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med 2008; 5:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng S, Cowling BJ, Fang VJ et al. Effects of oseltamivir treatment on duration of clinical illness and viral shedding and household transmission of influenza virus. Clin Infect Dis 2010; 50:707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rainwater-Lovett K, Chun K, Lessler J. Influenza outbreak control practices and the effectiveness of interventions in long-term care facilities: a systematic review. Influenza Other Respir Viruses 2014; 8:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azman AS, Stark JH, Althouse BM et al. Household transmission of influenza A and B in a school-based study of non-pharmaceutical interventions. Epidemics 2013; 5:181–6. [DOI] [PubMed] [Google Scholar]
- 36.Cowling BJ, Fung RO, Cheng CK et al. Preliminary findings of a randomized trial of non-pharmaceutical interventions to prevent influenza transmission in households. PLoS One 2008; 3:e2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gubareva LV, Novikov DV, Hayden FG. Assessment of hemagglutinin sequence heterogeneity during influenza virus transmission in families. J Infect Dis 2002; 186:1575–81. [DOI] [PubMed] [Google Scholar]
- 38.Papenburg J, Baz M, Hamelin ME et al. Household transmission of the 2009 pandemic A/H1N1 influenza virus: elevated laboratory-confirmed secondary attack rates and evidence of asymptomatic infections. Clin Infect Dis 2010; 51:1033–41. [DOI] [PubMed] [Google Scholar]
- 39.Poon LL, Chan KH, Chu DK et al. Viral genetic sequence variations in pandemic H1N1/2009 and seasonal H3N2 influenza viruses within an individual, a household and a community. J Clin Virol 2011; 52:146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng CK, Cowling BJ, Chan KH et al. Factors affecting QuickVue Influenza A + B rapid test performance in the community setting. Diagn Microbiol Infect Dis 2009; 65:35–41. [DOI] [PubMed] [Google Scholar]
- 41.Boelle PY, Ansart S, Cori A, Valleron AJ. Transmission parameters of the A/H1N1 (2009) influenza virus pandemic: a review. Influenza Other Respir Viruses 2011; 5:306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.