Abstract
Select species of the bacterial genus Leptospira are causative agents of leptospirosis, an emerging global zoonosis affecting nearly one million people worldwide annually. We examined two Leptospira pathogens, L. interrogans serovar Lai str. 56601 and L. borgpetersenii serovar Hardjo-bovis str. L550, as well as the free-living leptospiral saprophyte, L. biflexa serovar Patoc str. ‘Patoc 1 (Ames)’. The transport proteins of these leptospires were identified and compared using bioinformatics to gain an appreciation for which proteins may be related to pathogenesis and saprophytism. L. biflexa possesses a disproportionately high number of secondary carriers for metabolite uptake and environmental adaptability as well as an increased number of inorganic cation transporters providing ionic homeostasis and effective osmoregulation in a rapidly changing environment. L. interrogans and L. borgpetersenii possess far fewer transporters, but those that they have are remarkably similar, with near-equivalent representation in most transporter families. These two Leptospira pathogens also possess intact sphingomyelinases, holins, and virulence-related outer membrane porins. These virulence-related factors, in conjunction with decreased transporter substrate versatility, indicate that pathogenicity was accompanied by progressively narrowing ecological niches and the emergence of a limited set of proteins responsible for host invasion. The variability of host tropism and mortality rates by infectious leptospires suggests that small differences in individual sets of proteins play important physiological and pathological roles.
Keywords: Spirochaetes, Leptospira, leptospirosis, genome analyses, molecular transport, pathogenesis
Introduction
Leptospirosis is an emerging zoonotic disease that affects about 900,000 people annually worldwide. It is caused by members of the bacterial genus Leptospira within the Spirochaete phylum [1]. The disease poses a tremendous public health risk in tropical environments, especially as it is transmitted through contaminated water, infected tissues, and the urine of mammalian hosts [1]. Once infected, patients can potentially experience a variety of symptoms ranging from fever, myalgia, and fatigue to refractory shock, jaundice, renal failure, and pulmonary hemorrhage [1]. At-risk populations for this disease are primarily, but not exclusively, found in tropical climates in developed and underdeveloped countries. Cases throughout the United States have also been reported, e.g. in Hawaii, Maryland, Louisiana, etc. [2–4]. Factors that increase risk include conditions of slum living, recreational water activities, and flooding [1]. Nonetheless, leptospirosis is found globally, and various animals such as rats, bats, and marsupials as well as domesticated animals such as dogs and cows can serve as reservoirs for its transmissions to humans [5–7]. Human to human transmission is rare, but it is believed that globalization and ecotourism contribute significantly to the emergence of this zoonosis [8].
The causative agents of leptospirosis, Leptospira spp., are spiral-shaped, thin, aerobic, Gram-negative bacteria of highly divergent spirochetes, whose primary carbon sources are long-chain fatty acids [9]. Despite its threat to global public health, the Leptospira genus is not entirely filled with pathogenic species [10]. It is divided into saprophytes (e.g. L. biflexa, L. wolbachii, L. meyeri), pathogens (e.g. L. interrogans, L. borgpetersenii, L. kmetyi), and intermediate pathogens (e.g. L. licerasiae, L. wolfii, L. broomii) [10]. Among these characteristically distinct Leptospira, practice among scientists has been to distinguish leptospires from each other by means of serotyping and antigenic similarity instead of genetic similarity [10]. Consequently, serotyping has resulted in the identification of over 230 different serovars among Leptospira with serovars crossing species lines [10]. The limited ability of sequencing technology at the advent of the identification and characterization of members of Leptospira was a driving force for this convention, but rapid sequencing technology such as qPCR has begun to enable scientists and clinicians to rapidly and effectively identify infecting leptospires and their phylogenetic relationships [11].
L. interrogans and L. borgpetersenii are two of many species that have been identified as pathogenic mediators of leptospirosis [9]. Their most common reservoir animals are rats and cows, respectively, although these two spirochetes are not exclusive to these two animals as noted above [12, 13]. Working closely with these and other aforementioned animals, or being in proximity to them or their urine, gives increased likelihood to contracting the pathogen [14]. It should be noted, however, that L. interrogans is highly capable of survival (but not growth) outside of the host whereas L. borgpetersenii is not [1, 15]. L. biflexa, on the other hand, is a free-living saprophyte isolated from stream water, the antigenic properties of which have been used as a basis for antigenic testing of pathogenic leptospires [16].
In the present study, the publicly available genomes of the following representative organisms were examined: L. interrogans serovar Lai str. 56601, Leptospira borgpetersenii serovar Hardjo-bovis str. L550, and Leptospira biflexa serovar Patoc str. ‘Patoc 1 (Ames)’. Table 1 presents the nomenclature for the organisms used in this study as well as additional information about them. L. interrogans and L. borgpetersenii allow comparison and identification of hallmarks of pathogenic leptospires, and L. biflexa enables comparison for identification of transport proteins and mechanisms unlikely to be related to pathogenicity.
Table 1.
Overview of three Leptospira species and their basic traits.
| Species Name | Leptospira interrogans serovar Lai str. 56601 | Leptospira borgpetersenii serovar Hardjo- bovis str. L550 | Leptospira biflexa serovar Patoc str. ‘Patoc 1 (Ames)’ |
|---|---|---|---|
| Chromosome I RefSeq | NC_004342.2 | NC_008508.1 | NC_010842.1 |
|
|
|||
| Chromosome I Size (Mb) | 4.339 | 3.614 | 3.604 |
|
|
|||
| Chromosome II RefSeq | NC_004343.2 | NC_008509.1 | NC_010845.1 |
|
|
|||
| Chromosome II Size (Mb) | 0.359 | 0.317 | 0.278 |
|
|
|||
| Plasmid p74 RefSeq | N/A | N/A | NC_010846.1 |
|
|
|||
| Plasmid p74 Size (Mb) | N/A | N/A | 0.074 |
|
|
|||
| Total Genome Size (Mb) | 4.698 | 3.931 | 3.956 |
|
|
|||
| Total Protein # | 3,683 | 2,945 | 3,600 |
|
|
|||
| Transporters | 254 | 222 | 294 |
|
|
|||
| Transporters as % of Proteins | 6.90 | 7.54 | 8.17 |
|
|
|||
| Pathogenic? | Yes | Yes | No |
In general, the mechanisms of pathogenesis in leptospirosis are relatively poorly understood. However, there are several suggested mechanisms [9]. The coiled shape of Leptospira is relevant to its corkscrew-like motility through viscous media, which provides an efficient mechanism of dissemination after entry into various organs such as the lungs, liver, kidneys, eyes, and brain [9]. Genes associated with motility and chemotaxis are known to play a role in virulence [17]. Among proposed factors that may facilitate virulence during migration through host tissues are hemolytic sphingomyelinases and phospholipases [17]. Additional components of leptospiral virulence promote adhesion and invasion of host cells, although intracellular pathogenicity has not been demonstrated [17].
Chronically infected animals (particularly rats, bats, and marsupials) are usually asymptomatic but show high levels of leptospiral excretion through the urine, supporting the hypothesis that renal colonization is important for Leptospira in reservoir selection and pathogenesis [9, 18]. Kidney histological studies further support this hypothesis as kidneys show interstitial nephritis during infection, but no such damage in chronic carriers [19]. Kidney nephritis, along with damage to connective tissues, evident from hemorrhagic manifestations in lungs, supports virulence mechanisms involving invasion and damage to connective tissues [20]. Leptospiral lipopolysaccharide (LPS), known to be less toxic than the typical LPSs of other Gram-negative bacteria, more strongly activates Toll-like receptor 2 (TLR2) than TLR4, conventionally achieved by Gram-negative LPS in macrophages [21].
Ultimately, hemolytic sphingomyelinase and phospholipase activities, together with the identified motility and chemotaxis factors of Leptospira, damage host tissue and activate the inflammatory response of the host immune system to potentially cause significant damage, eventually resulting in death of the host [1]. Identification of transporters relevant to pathogenesis might reveal the presence of pore-forming toxins, transporters facilitating basic nutrient uptake, and the protein secretion systems necessary to release proteinaceous virulence factors. Furthermore, Leptospira pathogenic species are known to maintain poor viability in acidic urine relative to alkaline urine, which suggests a preference of sodium cations (Na+) in some transport systems that utilize the proton motive force (pmf) or the sodium motive force (smf) [9].
Pathogenic species of Leptospira must encode the proteins that mediate virulence. Both L. interrogans and L. borgpetersenii might be expected to show similar pathogenesis-related transporters, but this postulate had not been examined. L. biflexa would be expected to have few, or incomplete sets of these proteins [22]. Studies on L. biflexa, L. interrogans, and L. borgpetersenii have suggested that L. biflexa is most closely related to the common Leptospira ancestor, and that pathogenicity was an acquired feature [22]. Consequently, the saprophytic and free-living nature of L. biflexa suggests that its genome enables it to live with high versatility in a range of environments [15, 22]. The suggested flexibility of L. interrogans to live within a host and also in the external environment combined with the greater dependence of L. borgpetersenii for survival in the host in spite of its relatively small genome, due to insertion sequence (IS)-mediated reduction [15], suggests that increased pathogenicity and host tropism favor decreased versatility and reduced genetic diversity. The encoded transport proteins should reflect these characteristics.
Materials and Methods
The spirochete genomes analyzed were the most complete and up to date versions for each organism at the time these studies were initiated. The FASTA formatted protein coding sequences of Leptospira interrogans serovar Lai str. 56601, Leptospira biflexa serovar Patoc str. ‘Patoc 1 (Ames)’, and Leptospira borgpetersenii serovar Hardjo-bovis str. L550 were used [15, 22, 23]. Each protein sequence from the respective proteomes was queried and blasted against the Transporter Classification Database (TCDB; www.tcdb.org [24]) using the program GBlast [25]. GBlast retrieves the TC top hit sequence, TC number, protein size in number of amino acyl residues, the number of predicted TMSs using the HMMTop 2.0 Program, the E-value for the query and hit proteins, regions of sequence similarity, and regions of TMS overlap. The low complexity filter was not used as it is normally of value only for larger datasets including proteins with multiple repeat elements. GBlast can be used with any e-value cutoff desired. The Web-based Hydropathy, Amphipathicity and Topology (WHAT) program [26, 27] was used with a window size of 19 residues and an angle of 100 degrees (as is appropriate for alpha helices) to display the hydropathy and amphipathicity plots for individual proteins in conjunction with TOPCONS for consensus prediction (TOPCONS, www.topcons.net) in order to resolve the differences in the numbers of TMSs between the proteins retrieved and their TCDB homologues. The plots generated by WHAT allow the user to judge if a program such as HMMTOP has missed a TMS or has predicted a TMS inappropriately. A cut-off E-value of 0.001 was used with the GBlast program to eliminate most false positives and proteins with unreliable degrees of sequence divergence, but all hits recorded were rigorously tested to determine homology.
Proteins with no predicted TMSs were eliminated so that only integral membrane proteins, primarily multispanning membrane proteins, were retrieved. Proteins with only an N-terminal signal sequence are numerous because these proteins include almost all secreted proteins that are exported via the general secretory (Sec) pathway or twin arginine translocase (TAT) [28, 29]. The topological prediction programs often miss these TMSs, recording the proteins to have zero TMSs. Consequently, the number of zero or one TMS proteins retrieved were not reliable, and were therefore not always recorded. They were generally excluded from our analyses. While this approach yields fewer transport proteins, the representation of transport systems should remain unaffected since virtually all transport systems include integral multi-spanning membrane proteins. Such a method favors representation of transporter types and reveals their substrate diversity. TMSs detected by GBlast are those of the α-helix-type transporters and not the β-sheet-type porins. Transporters known to have β-sheet porins found primarily in TC subclasses 1.B and partly in 1.C, were further analyzed for β-strands using PRED-TMBB with all three decoding methods [30].
Transport proteins thus obtained from query Leptospira sequences were tabulated, and unusual characteristics were identified based in part on topologies that differed from corresponding family members in TCDB as well as E-values obtained with GBlast. Unusual properties can result from events such as genetic deletion or fusion, sometimes resulting in the gain or loss of extra domains or the generation of multifunctional proteins. Such results can be reflective of the protein sequence, but they can be artifacts due to sequencing errors or incorrect initiation codon assignment. In the latter cases, but not the former, the protein sequences were either corrected when possible or eliminated from our study.
Candidate proteins were examined in greater detail to estimate their probable substrate specificities on the basis of their predicted structures and numbers and degrees of sequence similarity with entries of known function in TCDB. Homology was examined using the Protocol 1 and Protocol 2 programs as well as GSAT [25]. Transport proteins were also classified into families and subfamilies of homologous transporters according to the TC classification system [24, 31, 32]. Regions of sequence similarity were examined to ensure that homology was in the transmembrane region(s) and not merely in hydrophilic domains [33]. The substrate specificities of particular homologues identified in the sequenced genomes were initially predicted based on homology to functionally characterized proteins. Assignment to a family or subfamily within the TC system often allows prediction of substrate type with confidence [34–36]. For each of the families and well-characterized proteins in TCDB (over 10,000), relevant references are provided. Not all of these can be provided in this paper, so only those relevant to Leptospira will be included.
Results
Three Leptospira genomes were analyzed for the occurrence of transport proteins using the Transporter Classification Database (TCDB) [24] and the GBlast program [25]. The results are summarized according to TC subclass in Table S1. Examining the total number of transport proteins present in these three genomes, we see that L. borgpetersenii has the fewest at 222. The most found in any genome is 294 for L. biflexa, 72 more than in L. borgpetersenii.
Transport Protein Subclasses
TC subclass 1.A in TCDB includes all α-type channels except for holins, which are found in the 1.E subclass. 15, 12, and 18 of these α-type channel proteins were identified in L. interrogans, L. borgpetersenii, and L. biflexa, respectively. L. biflexa possesses the greatest number of unique families in subclass 1.A, many of which are cationic channels including two mercuric ion channels, suggesting greater versatility in the saprophyte than in the pathogens. Surprisingly, L. interrogans possesses five proteins belonging to the 1.A.30 Outer Membrane Transporter Energizer (Mot-Exb) Superfamily compared to the seven and six found in both L. biflexa and L. borgpetersenii, respectively.
TC subclass 1.B includes outer membrane β-type porins. 43 were identified in L. interrogans, 32 in L. borgpetersenii, and 40 in L. biflexa. The distribution of these porins does not suggest a major contribution to pathogenicity. Since these proteins localize to the outer membrane via β-strands instead of α-helixes, those containing zero or one predicted α-helical TMSs were included in our study.
TC subclass 1.C includes pore-forming toxins. L. interrogans encodes ten putative toxins showing sequence similarity to established toxins belonging to four families, whereas L. borgpetersenii contains eight and L. biflexa contains seven. The 1.C.67 family of SphH Hemolysins is notably absent in L. biflexa but present in L. interrogans and L. borgpetersenii with four and two members, respectively. Although hemolysins have not been unequivocally shown to be essential for leptospiral pathogenesis, their presence in pathogens is likely to be of significance [17]. Hemolysins have been shown to strongly induce proinflammatory cytokines [21]. The three other families represented in subclass 1.C contain similar representation in the three leptospires examined. It should be noted that toxins with zero or one predicted transmembrane α-helices were included in this study as many secreted toxins can exist in both soluble and membrane integrated forms, and many are known to be pore-forming β-type toxins (see TCDB).
TC subclass 1.E consists of Holins. Both L. interrogans and L. borgpetersenii encode a protein that hits the Mycobacterial 4 TMS Phage Holin (MP4 Holin) (TC#1.E.40.3.6). Holins have a variety of proposed functions in prokaryotes and may play a role in cell lysis and biofilm formation [37]. The presence of these holins in L. interrogans and L. borgpetersenii and their aforementioned functions may promote pathogenicity in these leptospires.
The largest number of transporters for all three species is found in TC subclass 2.A, secondary carriers. L. interrogans encodes 79, L. borgpetersenii 67, and L. biflexa 105. Given the substantially greater number of secondary carriers found in L. biflexa, the relative presence (or lack thereof) of a variety of transporters may help to distinguish between the two Leptospira pathogens and free living bacteria. L. biflexa appears to have much greater metabolic flexibility than its two pathogenic cousins.
TC subclass 3.A are pyrophosphate hydrolysis-driven primary active transporters, usually multi-component systems. With 42, 37, and 53 integral membrane transport proteins of this subclass found in L. interrogans, L. borgpetersenii, and L. biflexa, respectively, these proteins make up a significant proportion of the total transport proteins found in these organisms. The variety and wealth of transporters found in this subclass clearly suggests that they play an important role in spirochetes. The 3.D TC subclass of ion-pumping electron carriers are represented in L. interrogans, L. borgpetersenii, and L. biflexa with 19, 22, and 23 proteins, respectively.
TC class 4 includes group translocators that are believed to modify their substrates in processes coupled to transport. The TC 4.B subclass includes members of the nicotinamide ribonucleoside (NR) group translocating uptake permease (PnuC) family. Only L. biflexa encodes such a protein.
TC subclass 4.C includes fatty-acyl-coenzyme A ligases that activate fatty acids for lipid biosynthesis and may function in transport via group translocation. Each of the three species contains one member of this family. GBLAST revealed more fatty-acyl-coenzyme A ligases in all three leptospires, but the absence of transmembrane segments in addition to an unproven transport function in leptospires warranted their exclusion from our analyses.
All three Leptospira species have proteins belonging to TC subclass 4.D, probable group translocating glycosyl transferases. L. borgpetersenii and L. biflexa encode four, whereas L. interrogans encodes three. Proteins in this family have demonstrated exopolysaccharide synthesis activities thought to be coupled to polysaccharide secretion [38]. As exopolysaccharides can contribute to biofilm formation, all three leptospires likely benefit from the presence of these proteins for both free-living and host colonization purposes.
Subclass 5.A includes electron-carriers that transfer electron pairs from one side of the membrane to the other, thereby influencing cellular energetics. L. interrogans was found to have two, whereas L. borgpetersenii and L. biflexa have three. Among these are disulfide bond oxidoreductases and prokaryotic molybdopterin-containing oxidoreductases. These proteins might play a role in establishing the proton motive force, but they probably do not contribute to pathogenicity.
Subclass 5.B in TCDB consists of one electron transmembrane carriers. None of the leptospires was found to contain integral membrane carriers in this subclass. However, all three contain multiple copies of cytochrome c peroxidases (TC#5.B.3.1.1) for extracellular reduction of Fe2O3 (unpublished results).
Subclass 8.A represents auxiliary proteins with one in L. interrogans and L. borgpetersenii, and two in L. biflexa. All three encode a stomatin-like protein that may help with localization and insertion of proteins into the outer membrane.
Subclass 9.A in TCDB contains known transport proteins whose biochemical mechanisms of transport are unknown. All three leptospires have the same two subclass 9.A protein homologs (TC#9.A.8.1.4 and 9.A.40.2.2).
TC Subclass 9.B includes a variety of proteins that are putatively classified as transporters. Further study of a given 9.B protein might either confirm its involvement in transport, or warrant its removal from the TC classification system if a transport function is disproven. L. interrogans has 36, L. borgpetersenii has 32, and L. biflexa has 35 of these proteins.
Transporter Superfamilies and Families
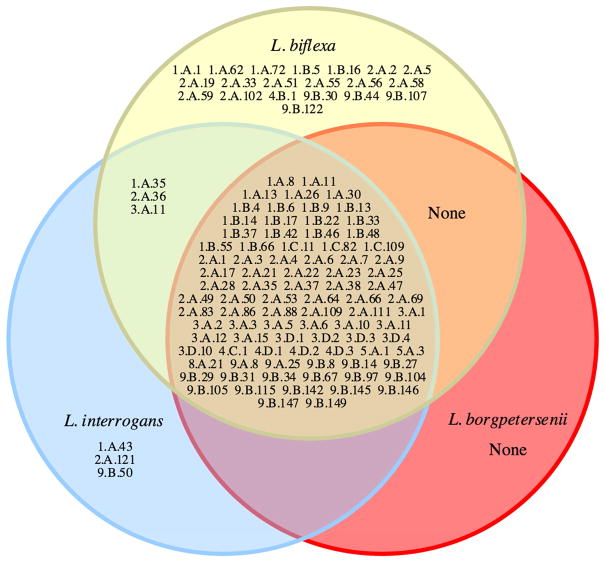
Figure 1 details protein families found in some, but not all three leptospiral species. 20 families are shown to be unique to L. biflexa, dwarfing the three families each unique to L. interrogans and L. borgpetersenii. This is reflective of the large differences in the numbers of total transporters (Table S1) and the disproportionately high number of 2.A carriers (Figure 1). Analyses of these families may reveal key features of free-living saprophytes.
Figure 1.
Representation of transporter families unique to L. interrogans, L. borgpetersenii, L. biflexa, both L. biflexa and L. interrogans, both L. biflexa and L. borgpetersenii, and both L. interrogans and L. borgpetersenii. Families found in all three species are listed in the central area.
Unique families of transporters found in L. biflexa, presented in Figure 1, reveals ten families in the 2.A subclass. Families of unique transporters with notable substrates are TC#1.A.72, 2.A.51, 2.A.56, 2.A.59, 2.A.102, and 4.B.1 that transport mercury, chromate, dicarboxylates, arsenic, sulfites, and nicotinamide mononucleotide as well as related compounds, respectively. All of these substrates are transported only by L. biflexa, suggesting increased versatility over L. borgpetersenii and L. interrogans.
The only families of transporters unique to L. interrogans (Figure 1) include TC#1.A.43 and TC#2.A.121, members of which transport fluoride and sulfate, respectively. L. borgpetersenii does not appear to have unique transporter families. Families belonging to both pathogens are TC#1.A.23, 1.C.67, 1.E.40, 2.A.114, 2.A.115, 9.B.1, and 9.B.125, encoding small mechanosensitive ion channels, hemolysins, holins, peptide transporters, multidrug exporters, proteases, and unknown substrates, respectively. The families unique to L. interrogans may confer increased environmental versatility over L. borgpetersenii, but the shared transport families may play roles in pathogenesis.
Interesting Facets of Channel Proteins
A limited number of channel protein families is represented in the three Leptospira examined. Most channel proteins are involved in ionic and water homeostasis, but some also serve functions in stress responses. These will be described below.
Only a single member of the Voltage-gated Ion Channel Superfamily (TC#1.A.1) was identified, and this protein was found only in L. biflexa. It proved to be a 6 TMS cyclic nucleotide-dependent channel, almost certainly a potassium channel like those characterized in cyanobacteria and other spirochetes [39].
A single member of the MIP Family (TC#1.A.8) of aquaporins and glycerol facilitators was found in each of the three leptospires. These three proteins are probably aquaporins capable of transporting three-carbon compounds such as glycerol and dihydroxyacetone [40]. The high scores, all matching the same TCDB entry, suggest that these three proteins are orthologous.
Ammonium channels are prevalent in leptospires but in variable numbers; thus, L. interrogans has two dissimilar paralogs, and L. biflexa has three, but L. borgpetersenii has only one. Interestingly, it appears that one of these proteins in each organism hits the homolog from Azospirillum brasilense (TC#1.A.11.1.4) with excellent comparable scores. These three proteins are undoubtedly orthologs. It is worth noting that the A. brasilense protein is subject to multiple mechanisms of regulation, which may be applicable to the spirochete proteins as well [41].
The three Leptospira species examined possess either one or two homologs of Epithelial Chloride Channels (TC#1.A.13), characterized only in animals. Although bacterial homologs have been identified, in none of them is the function known. Our results reveal that these spirochete proteins exhibit the same topology as the mammalian proteins, suggesting a similar function. These proteins may prove to exhibit chloride channel activities comparable to those found in eukaryotes.
The three leptospires display either zero or one small mechanosensitive ion channel (MscS; TC#1.A.23), and interestingly, L. biflexa is the one that lacks such a protein. All three organisms lack an MscL channel (TC#1.A.22). Proteins of both of these families are known to function in osmotic adaptation [42].
All three spirochetes possess a member of the MgtE family (TC#1.A.26) of magnesium uptake channels. These three proteins hit the same TC entry with the same high score, clearly indicating orthology.
The three Leptospira species examined possess multiple paralogs of the H+ or Na+-translocating MotAB/ExbBD/TolQR channel-forming constituents (TC#1.A.30). While MotAB proteins function to energize motility [43, 44], ExbBD channels energize transport across the outer membrane [45], and TolQR channels are believed to energize assembly of the outer membrane, promoting stability of this structure [46]. All three spirochetes possess two MotAB energizers, which presumably function in motility, possibly one utilizing the proton motive force and the other utilizing the sodium motive force [47]. On the other hand, the occurrence of ExbBD/TolQR energizers is variable in these three species with three in L. interrogans, four in L. borgpetersenii, and five in L. biflexa. These results suggest that L. interrogans, like E. coli, possesses at least the equivalent of one ExbBD complex and one TolQR complex [48–50] However, the other two leptospires have an increased number of these H+ or Na+ channel proteins. The functions of these proteins will be interesting targets of future investigations.
Remaining families of channel proteins are present only in select Leptospira species. The CorA Metal Ion Transporter Family (TC#1.A.35) is only represented in L. interrogans and L. biflexa. Only L. interrogans possesses a member of the Camphor Resistance Family (TC#1.A.43). These proteins have recently been shown to be fluoride export channels which protect the bacterium against the toxic effects of fluoride [51, 52]. For both the Homotrimeric Cation Channel Family (TC#1.A.62) [53] and the Mercury (Mer) Superfamily (TC#1.A.72) [54, 55] only L. biflexa has constituent channels. While the former proteins have not been characterized in bacteria, the latter function in the uptake of mercuric ions for the purpose of reduction to metallic mercury by a cytoplasmic mercuric reductase, a detoxification reaction [56].
Interesting Facets of β-type porins
β-type porins represent a significant portion of the channel proteins found in the Leptospira examined. The leptospiral outer membrane is of particular interest as it contains cell surface antigens that can be used for vaccine production, and they can also serve as potential drug targets [57]. Furthermore, the outer membrane composition of Leptospira is unlike other spirochetes, possessing more β-type porins and lipopolysaccharide in addition to surface-exposed lipoproteins [58]. Members of sixteen different families of outer membrane porins were identified in at least one of the three Leptospira species examined, and interestingly, fourteen of these families are represented in all three species. Just two of the families (POP; 1.B.5 and SAP; 1.B.16) were found only in L. biflexa, not in the two pathogenic species. While the POP Family is concerned with anion transport, the SAP Family mediates urea and short-chain amide transport.
Some of the families represented in all three organisms have only a single protein per organism, and these may be orthologs of each other as all three proteins hit the same TC entry (see for example 1.B.4, 1.B.6, 1.B.9, and 1.B.13). However, striking differences occur in some of the other families, for example, the Outer Membrane Receptor (OMR) Family (TC#1.B.14), where each organism exhibits different sets of these pore-forming receptors. This fact can be explained by the different specificities of these receptors as illustrated in Table S1. Similar observations were made for the Outer Membrane Factor (OMF) Family (TC#1.B.17), where again, the different specificities of these porins provide an explanation. It seems likely that the complement of OMRs and OMFs reflect the specific environments in which these organisms are found.
The remaining families in this subclass consist of macromolecular transporters for protein secretion (TC#1.B.22; TC#1.B.48), outer membrane protein insertion (TC#1.B.33), lipid export (TC#1.B.42), outer membrane lipid insertion (TC#1.B.46), and polysaccharide export for protection and biofilm formation (TC#1.B.55). All leptospires possess members of these families, which represent core components of the Leptospira outer membrane proteome.
Interesting Facets of Secondary Carriers (TC Subclass 2.A)
In most organisms, the Major Facilitator Superfamily (MFS; TC#2.A.1) is the largest superfamily of secondary carriers. However, in the pathogenic leptospires, the MFS is poorly represented. L. interrogans has eight MFS members and L. borgpetersenii has six, with zero members of the related GPH Family (TC#2.A.2) for both. By contrast, L. biflexa has fifteen MFS porters and two GPH porters. Thus, its MFS representation is two to three times that of the pathogens. Of particular note is the presence of multiple multidrug efflux pumps of MFS subfamily 21, nitrate/nitrite transporters, and several MFS porters of unknown function. Also, while the pathogens possess only a single member of the APC amino acid transporter family, L. biflexa has three such members, each derived from a separate subfamily.
Divalent cation transporters can mediate either uptake or efflux of these essential but potentially toxic substances. While the CDF Family (TC#2.A.4) catalyzes heavy metal export and has equal numbers of its members in all three leptospires, members of the ZIP Family (TC#2.A.5) and the NRAMP Family (TC#2.A.55) catalyze heavy metal uptake and are found only in L. biflexa. Interestingly, the Chromate Resistance (CHR) Family (TC#2.A.51) and the Arsenical Resistance-3 (ACR3) Family (TC#2.A.59) are also restricted to L. biflexa.
The RND Superfamily is by far the largest superfamily of secondary carriers present in these spirochetes with fourteen members in L. interrogans and L. biflexa, and nine in L. borgpetersenii. These proteins are divided about equally between heavy metal efflux pumps and multidrug resistance pumps. Only the SecD and SecF proteins, present in single copy in all three organisms, fall outside of these two groups. These two proteins function together as a single RND pump to facilitate proton-driven protein secretion via the General Secretory Pathway (Sec; TC#3.A.5) [59]. Finally, the two pathogens, but not L. biflexa, possess a single member of the poorly characterized putative Hydrophobe/Amphiphile Efflux-3 (HAE3) subfamily (TC#2.A.6.7) within the RND superfamily.
The Drug/Metabolite Transporter (DMT) Superfamily is the third largest superfamily in these spirochetes. L. interrogans has four such members, L. borgpetersenii has five, and L. biflexa has nine. Most of the top hits in TCDB have not been functionally characterized, so specific substrates cannot be assigned. However, all known members of this superfamily function in the transport of small metabolites and drugs.
Interestingly, all three leptospires have a single homolog of the Sweet family of putative sugar transporters (TC#2.A.123). The homologs identified have a 3 TMS subunit structure. Several 7 TMS Sweet family members have been shown to transport sugars such as glucose and fructose [60]. Presently, two 3D structures of a 3 TMS Sweet glucose transporter from L biflexa have been solved [61]. Based on these structures, transport mediated by this protein appears to be that of a secondary carrier [61].
Table S1 reveals the presence of secondary carriers belonging to many other families, and almost all of these are well-represented in all three leptospires. These families will not be further discussed here.
Interesting Facets of Primary Active Transporters
TC subclass 3.A contains the largest superfamily of transporters found in all three Leptospira species, the ABC Superfamily (TC#3.A.1). While L. interrogans and L. borgpetersenii both possess 20 of these proteins, L. biflexa possesses 31. The ABC Superfamily is represented in all domains of life and is known to transport a wide variety of substrates for both uptake and export. Of note, L. biflexa is the only leptospire to possess ABC transporters for putrescine/spermidine (polyamines), zinc (Zn2+), iron siderophores, and fatty acyl-CoA. However, all three organisms possess good representation of oligopeptide transporters, suggesting that these substances are important for the nutrition of these organisms. All three organisms have ABC uptake systems for sulfate and lipids.
ABC efflux systems are present in numbers that are similar to those of the uptake systems in all three spirochetes. The primary substrates for these exporters are 1) lipids and lipoproteins, 2) proteins and peptides, 3) exopolysaccharides, and 4) multiple drugs. Most of these transporters, except for those specific for lipids, are found in similar numbers in the three spirochetes examined. Only a few ABC export systems are specific to L. biflexa. One of these is a putative organoanion (fatty acid?) exporter (TC#3.A.1.203.8), and the others undoubtedly exhibit specificity for specific proteins (TC#3.A.1.109, 3.A.1.110, and 3.A.1.111).
All three leptospires possess orthologous sets of the integral membrane components of the ATP synthases in the F-ATPase Superfamily (TC#3.A.2) for subunits a, b, and c. The reversibility of the enzyme for both the establishment of the proton motive force and ATP synthesis is a key characteristic of this system. Additionally, all three Leptospira have an H+- or Na+-translocating pyrophosphatase (TC#3.A.10). While the TC hit for L. biflexa (TC#3.A.10.1.1) is different from those for L. interrogans and L. borgpetersenii (TC#3.A.10.1.6), sequence comparisons of these entries showed that these proteins are probably orthologous.
Of note is the variance of P-type ATPases (TC#3.A.3) in Leptospira species. L. interrogans possesses five of these transporters with substrate specificities for magnesium (Mg2+), copper (Cu2+), and potassium (K+). L. borgpetersenii, however, possesses only two, one specific for copper (Cu2+) and the other for calcium (Ca2+). While the Mg2+ and K+ systems catalyze uptake, the Cu2+ and Ca2+ systems probably catalyze efflux. L. biflexa has six including a putative Na+/K+ ATPase, two copper (Cu2+) transporters, a calcium (Ca2+) transporter, and a heavy metal (Co2+, Zn2+, Cd2+) transporter. The diversity of these transporters presumably reflects the types of stress that these organisms encounter. Thus, most prokaryotic P-type ATPases function in stress relief [62, 63].
All three leptospires have proteins with sequence similarity to the three integral membrane components of the General Secretory Pathway (Sec) Family (TC#3.A.5), which transports most secreted proteins across the inner cytoplasmic membrane. The presence of SecDF (TC#2.A.6.4) in all three species reveals the genus-wide presence of the integral membrane constituents of the general secretory pathway. In addition, we found constituents of the outer membrane protein secreting Main Terminal Branch (MTB) Family (TC#3.A.15). These proteins proved to be distantly related to the MTB constituents previously tabulated in TCDB, and consequently we have entered all constituents of this system from L. interrogans into TCDB under TC# 3.A.15.4.1. MTB family members are believed to export hundreds of proteins across the outer membranes of Gram-negative bacteria, initially secreted across the cytoplasmic membrane by the Sec system [64].
Not surprisingly, all motile leptospires possess the flagellar (Type III) secretion complex (TC#3.A.6). The constituents recorded in Table S1 include six integral membrane constituents of these systems in all three leptospires whose near-identical E-values for all components suggest orthology for the three complete systems in this genus. The striking structural and functional similarity between Type III secretion systems and flagellar export systems gives rise to the possibility that these systems export virulence factors in addition to flagellar subunits as has been demonstrated for these systems in other bacteria [65, 66]. The flagellar apparatus has been shown to secrete virulence-associated proteins [67, 68].
As expected, all leptospires have the Septal DNA Translocase (TC#3.A.12), involved in DNA transfer across the completed septa of newly dividing cells. However, while these organisms lack a type IV protein secretion system involved in conjugation, they do possess components of Bacterial Competence-related DNA Transformation (DNA-T) systems (TC#3.A.11). Interestingly, while L. interrogans and L. biflexa appear to have all constituents of these systems, only two were found in L. borgpetersenii. Possibly this last organism has lost some of the constituents of these systems and therefore has lost competence. Surprisingly, nothing seems to have been published on competence for DNA uptake in Leptospira species.
None of the spirochetes examined appears to have a Na+-transporting carboxylic acid decarboxylase of the NaT-DC Family (TC#3.B.1). These organisms do have decarboxylases, but they lack the integral membrane protein, which is required for Na+ extrusion. We therefore conclude that this mechanism for generating a sodium motive force is lacking in these organisms, in agreement with the conclusion that these ion pumps are largely restricted to anaerobes [69].
Constituents of most, but not all, of the primary proton pumping electron transfer complexes present in mitochondria and many aerobic bacteria were found in the leptospires. These include the proton-translocating NADH dehydrogenase (TC#3.D.1), proton-translocating transhydrogenase (TC#3.D.2), and proton-translocating cytochrome oxidase (TC#3.D.4), but not the proton-translocating quinol:cytochrome c reductase (TC#3.D.3). Additionally, leptospires possess prokaryotic succinate dehydrogenase (TC#3.D.10). These results are consistent with the conclusion that leptospires use electron transfer as a primary mechanism for generating a proton motive force, subsequently used for ATP synthesis. As expected, these aerobic bacteria possess members of the disulfide bond oxidoreductase D (DsbD) and Molybdopterin-containing Oxidoreductase (PMO) Families (TC#5.A.1 and 5.A.3, respectively), but surprisingly not the single electron transferring DsbB complex. In this regard, it is important to note that members of the DsbD family can serve any of a variety of biological functions (see TCDB).
Possible Group Translocators (TC Class 4)
None of the leptospires possess a phosphoenolpyruvate-dependent sugar transporting phosphotransferase system (PTS) although proteins of such systems have been found in other spirochetes [70]. However, L. biflexa appears to have a nicotinamide ribonucleoside uptake permease (TC#4.B), thought to function by a group translocation mechanism [71]. Each spirochete also has a membrane-associated acyl-CoA ligase (TC#4.C) that could function in transport [72]. Finally, each leptospire possesses three or four polysaccharide synthase/exporters (TC#4.D), all of which give low scores to the proteins in TCDB. These putative enzyme/porters may catalyze vectorial glycosyl polymerization [38, 73].
Poorly Characterized Transporters (TC Class 9)
TC subclass 9.A represents known transport systems that function by an unknown mechanism of action. Three such systems are found in all three spirochetes, and no other member of this subclass was identified. The first of these families is the FeoB family of ferrous iron uptake transporters prominent in iron acquisition (TC#9.A.8) [74]. Members of the second family, (HlyC/CorC; TC#9.A.40) may function as divalent cation channels. TC subclass 9.B includes putative transporters, where even transport function is not established. These proteins are listed in Table S1 but will not be discussed.
Transporter Substrates
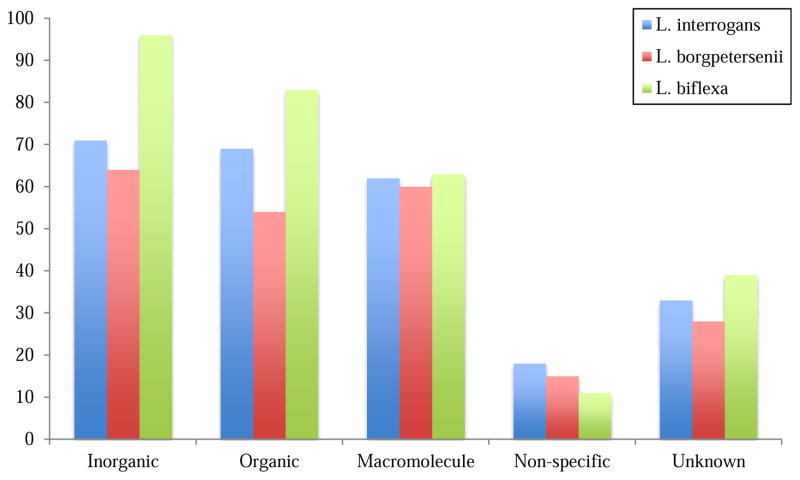
The substrates of transporters found in the three leptospires likely reflect the physiological characteristics of these organisms. In Figure 3, the distribution of substrates by category is presented.
Figure 3.
Distribution of transporters based on substrate category in L. interrogans, L. borgpetersenii, and L. biflexa. Table 1. Overview of three Leptospira species and their basic traits.
All three spirochetes have very similar percentages of the various substrate categories and subcategories. The most obvious difference between the three species is the relatively larger size of certain categories and subcategories in L. biflexa. Whereas L. interrogans and L. borgpetersenii have 71 and 64 inorganic substrate transporters, respectively, L. biflexa has 96, suggesting that this organism must be capable of maintaining intracellular ionic homeostasis under a much greater range of environmental conditions than for the two pathogens. Similarly, L. biflexa has a greater number of transporters for each category of substrate except nonselective transporters. The same can be seen in most subcategories where L. biflexa has more transporters than the other two spirochetes.
Correlating with its greater capacity for maintaining ionic homeostasis, L. biflexa has 73 proteins involved in cation transport whereas L. interrogans and L. borgpetersenii have 53 and 48, respectively. The prevalence of these transporters correlates with the disproportionately high number of L. biflexa secondary carriers that can utilize protons (H+) or sodium (Na+) for symport or antiport. This fact also indicates a reliance on transport that is energized by the proton (or sodium) motive force over other energy-coupling mechanisms such as those driven by ATP hydrolysis. In addition to ionic homeostasis, cation symport and antiport facilitate osmotic regulation [42] and heavy metal resistance [75]. Iron also serves as a vital di- and trivalent cation in Leptospira spp., required for growth and taken up into the cell via a bevy of iron uptake systems [76]. Other inorganic monovalent and divalent cationic substrates pumped by these three organisms include potassium (K+), calcium (Ca2+), magnesium (Mg2+), and various cations of metals including copper, iron, zinc, cobalt, cadmium, mercury, and manganese.
Transporters specific for inorganic anions number 16 for L. interrogans, 13 for L. borgpetersenii, and 20 for L. biflexa. Anions, compared to cations, represent a much smaller proportion of the inorganic substrates transported by these leptospires. The latter play roles in redox processes, establishing, for example, the pmf or smf for energization. Anion transporters are found primarily in TC subclass 2.A, taking up or exporting bicarbonate, phosphate, arsenate, arsenite, telluride, chromate, chloride, and fluoride. Sulfate uptake, on the other hand, is mediated primarily by homologs of the CysPTWA ATP-dependent ABC system.
The three Leptospira species have only a small percentage of their transporters dedicated to organoanion (non-carboxylic organic anion) transport. L. biflexa has four of these transporters, whereas L. interrogans has three and L. borgpetersenii has one. Fatty acids and other carboxylic acids, bile acids and their conjugates, taurine, and carnitine are the main substrates in this subclass of carbon sources. These spirochetes also exhibit good representation of carboxylate transporters with seven in L. biflexa, four in L. borgpetersenii, and six in L. interrogans. Pyruvate, malate, succinate, acetate, hydroxybenzoate, citrate, and fumarate are all probably transported. These leptospires have outer membrane porins dedicated to fatty acid and hydrophobic compound uptake (TC#1.B.9.3.3). Fatty acid group translocation across the inner membrane may be catalyzed by transporters in the FAT Family (TC#4.C.1) [77–79].
Sugars and polyols taken up include glycerol, glycerol-3-phosphate, monosaccharides, and disaccharides. L. biflexa possesses eleven such proteins, whereas L. interrogans possesses eight and L. borgpetersenii seven. As Leptospira utilize fatty acids as primary carbon sources, this subclass of carbon sources may play roles in osmoregulation, alternative metabolic pathways involving differentially expressed genes, and membrane construction [77–79].
L. biflexa has disproportionately high numbers of proteins involved in the transport of amines, amides, and polyamines (nine proteins compared to three each in L. interrogans and L. borgpetersenii). Among such transported substrates are putrescine, spermidine, ethanolamine, choline, and quaternary ammonium compounds. Transport of polyamines is associated with cellular growth and proliferation and may alleviate stress resulting from elevated external pH [80–82].
All three leptospires have transporters for amino acids and their conjugates, primarily members of TC subclass 2.A. Representatives can be found in the APC (TC#2.A.3), DMT (TC#2.A.7), AGCS (TC#2.A.25), and ABC (TC#3.A.1) families. There appears to be substantial diversity in the types of amino acids transported. Of additional note, the three Leptospira species possess at least seven homologs of a putative alanyl teichoic acid synthesis protein DltB (TC#2.A.50). These homologs possess domains that strongly match with a DltB domain (e-48), and another weaker match for an O-acyltransferase (e-24) (unpublished results). The strong match warrants inclusion of these proteins in this study as potential transporters of activated d-alanine, an amino acid conjugate.
The three spirochetes included in this study possess several proteins that function in the transport of peptides and their conjugates. These peptides can be di-/tripeptides, oligopeptides, and peptidoglycan fragments. These transporters additionally transport antibacterial agents, various nitrogen sources, and precursors of cell wall biosynthesis. They primarily belong to the POT/PTR and ABC families.
Proteins can be secreted by leptospires using multiple systems. The General Secretory Pathway (TC#3.A.5) and the outer membrane secreting Main Terminal Branch (MTB) (TC#3.A.15) probably provide the primary pathways for protein secretion across the two membranes of the cell envelope. However, flagellar proteins and possibly some virulence proteins are secreted by the Type III Secretory Pathway (TC#3.A.6) [67]. Finally, the Outer Membrane Insertion BAM complex (TC#1.B.33) probably inserts most outer membrane proteins into this structure.
A key feature of Leptospira is its outer membrane, composed of lipids, porins, lipoproteins, and lipopolysaccharide (LPS); this last constituent consists primarily of lipid A and O-antigen. The Outer Membrane Lipopolysaccharide Export Porin (LPS-EP) Family (TC#1.B.42), the Outer Membrane LolAB Lipoprotein Insertion Apparatus Family (TC#1.B.46), the Multidrug/Oligosaccharidyl-lipid/Polysaccharide Flippase Family (TC#2.A.66), and members of the ABC Superfamily (TC#3.A.1) are the primary systems dedicated to the export of lipids and LPS precursors to the outer membrane.
All three leptospires in this study possess proteins for the transport of drugs with fifteen in L. borgpetersenii, and nineteen in both L. interrogans and L. biflexa. Multidrug resistance pumps are known to be prevalent in free living organisms which need to defend themselves against toxic substances produced by other microbes [83]. Further characterization of the members of the Drug/Metabolite Transporter Superfamily (TC#2.A.7) should provide a more accurate representation of the substrates transported by members of this superfamily. Drug exporters function to protect the cell from endogenously produced antibiotics, to remove exogenous and harmful substances produced by other microbes, and to export drug-like secondary metabolites such as siderophores, lipids, signalling peptides and periplasmic redox cofactors [83].
The siderophores category of substrates (labeled for convenience but includes vitamins and cofactors) in these three organisms is comprised of thirteen transporters in L. interrogans and twelve in L. biflexa, with only seven in L. borgpetersenii. All three leptospires transport Vitamin B12 (cobalamin). Siderophore transporters, also of the ABC-type, are found in all three organisms, consistent with a need for iron in these aerobes.
Nonselective channels include α-type channels, β-barrel porins, and pore-forming toxins. The three leptospires examined have transporters in these subclasses with eighteen in L. interrogans, fifteen in L. borgpetersenii, and eleven in L. biflexa. These nonselective transporters can play receptor roles in the outer membrane, induce toxin-like effects in other bacteria, and regulate cellular osmolarity (see TCDB). The greater numbers of these proteins in these two pathogens suggests that they play important roles in virulence.
All three leptospires have a substantial proportion of transporters that are poorly defined, about 13%. This represents a diverse subset of all leptospiral proteins with no clearly demarcated function [15, 22, 23]. Nonetheless, these proteins, which may transport unknown substrates via ill-defined mechanisms of action, are likely to serve important, possibly genus-specific roles in metabolism and pathogenicity.
DISCUSSION
Comparative Leptospiral Proteome Analyses
Members of the genus Leptospira cause leptospirosis, a zoonotic disease with global prevalence, affecting nearly one million people annual with mortality ranging 5–25%. Current treatment of its wide variety of symptoms relies heavily on symptom management and antibiotic administration. Antibiotics currently in use to treat leptospirosis include penicillin, doxycycline, and cephalosporins, the efficacies of which remains mixed and questionable [84–88]. Prophylaxis usually involves vaccines of typically heat-attenuated leptospires with limited results. Chemoprophylactic treatment involves extended administration of doxycycline, a procedure that has been shown to reduce the incidence of the disease [89–91]. Variation in the susceptibility of different strains of Leptospira to these treatments has been reported, but instances of asymptomatic carriers in humans complicate the issue [92, 93]. The causes of the variable clinical manifestations of leptospirosis are poorly understood, and the entire Leptospira genus is poorly characterized.
Comparative analyses of transport proteins provide clues as to the metabolic, pathological, and drug resistance properties of these spirochetes. By comparing and contrasting two known pathogenic members, L. interrogans and L. borgpetersenii, with a free-living saprophyte, L. biflexa [15, 22, 23, 94], we have generated data that will help define the metabolic capabilities of these organisms, allow identification of transporters common to these leptospires, and distinguish transport systems required for pathogenic versus saprophytic life. While our analyses makes the primary distinction between pathogens and free-living organisms, it is still important to consider L. interrogans and L. borgpetersenii as organisms with dissimilar abilities to survive outside of a host [1, 15]. The genetic differences between L. interrogans and L. borgpetersenii, while relatively small compared to L. biflexa, are vast compared to virulence-attenuated strains of L. interrogans [95].
Bioinformatics is a powerful tool for analyzing and assessing biological data, and this field enables supplementation of findings in the wet lab. The results provided by GBlast facilitated rudimentary compilation of potential transport proteins within an organism’s genome. However, since sequence similarity is the primary method used to determine a match of an unknown query to a known entry in TCDB, functionally distinct proteins with variations in sequence may result in false positives for a given protein being declared a transporter, depicting sequence convergence where divergence would otherwise be expected. A few examples of this occurred in our study, most notably with query entries matching with members of the Pore-forming RTX Toxin Family (TC#1.C.11). For most of these proteins, the three leptospires examined were shown to have methyl-accepting chemotaxis protein domains, indicating that these toxin homologs may have other functions in these three organisms. All three organisms possess proteins that demonstrate strong sequence similarity to an activated D-alanine derivative exporter, belonging to the Glycerol Uptake Porter family (TC#2.A.50). Activated D-alanine transporters have been found in Gram-positive bacteria, and it is possible that these Leptospira proteins are membrane-bound o-acyltransferases.
Leptospires exhibit profound changes in their transcriptomes in response to a changing external environment [96, 97]. The findings in our study reflect the total potential transporters available to respective Leptospira species. Other investigators have studied the transcriptional profiles of specific pathovars under various environmental conditions [97–99]. Future efforts will attempt to integrate transporter proteomic studies with transcriptomic studies and the bioinformatic analyses explored here.
Distinguishing Transporters of Three Leptospires
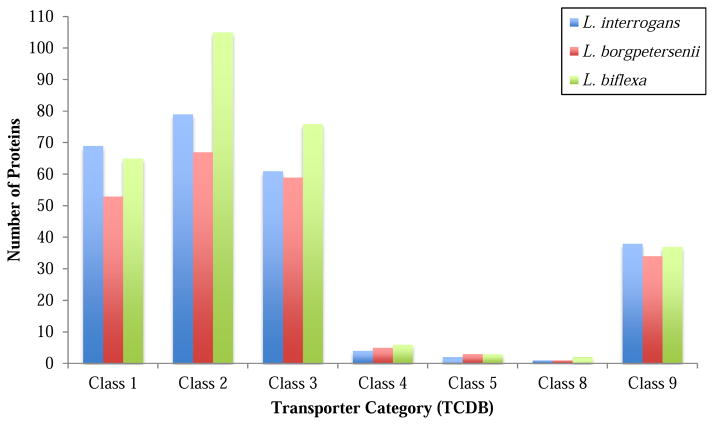
All identified transporters are compiled with their characteristics in Table S1, revealing many of the conclusions drawn in our study. We show that L. borgpetersenii possesses 222 integral membrane transport proteins, L. interrogans possesses 254, and L. biflexa possesses 294, showing that the two pathogens have fewer transporters than the free-living L. biflexa. This difference arises because of decreases in the secondary carriers, primary active transporters, and channel proteins in the former two organisms as seen in figure 2. This fact reflects decreased transport capabilities and therefore metabolic diversity and potential for homeostatic control of the pathogens relative in the free-living saprophyte.
Figure 2.
Distribution of transporters based on TC classes in L. interrogans, L. borgpetersenii, and L. biflexa.
Figure 3 shows the very significant difference in the inorganic cation transporters of L. biflexa (73) compared to L. interrogans (53) and L. borgpetersenii (48). Transporters associated with these substrates include pmf and/or smf generators, osmotic and ionic homeostatic stress response regulators, and heavy metal resistance proteins. These cation transporters confer upon L. biflexa the ability to survive in external environments through effective osmotic regulation, metabolic versatility, and by competing with other environmental microbes.
L. biflexa has increased numbers of transporters for carbon sources and amino acids relative to L. interrogans and L. borgpetersenii. Among these substrates are carboxylates, sugars, polyols, non-carboxylic organoanions, amines, amino acids & their conjugates, and peptides & their conjugates. With just one exception, L. biflexa has more transporters in each of these subcategories of substrates. This undoubtedly reflects its superior metabolic versatility, conferring the ability of this free-living organism to grow under a wide range of environmental conditions. Pathogenic species of Leptospira are known to lack proteins related to carbohydrate, nitrogen, and amino acid metabolism that correlate with their protracted growth in artificial media [100]. Many of the aforementioned transporters, as seen in Table S1, belong to the 2.A TC subclass, the carriers of which are known to demonstrate lower affinities, but greater efficiencies at lower energy cost, than ABC transporters.
The relatively high affinity ABC transporters are well represented in all three leptospires, but L. biflexa has significantly more (31) of these transport proteins than L. interrogans (20) or L. borgpetersenii (20). Uptake systems for peptides and sulfate are present, but L. biflexa possesses a system for putrescine/spermidine uptake, as well as ones for siderophore, zinc (Zn2+), and vitamin acquisition. High affinity acquisition of putrescine, critical in cell survival, demonstrates a crucial component of L. biflexa saprophytism [101]. Similarly, uptake systems for iron siderophores and zinc (Zn2+) serve to accumulate them to high concentrations within the cell so pathogens have better access within a host. Macromolecular ABC export systems are similarly well represented in all three species, transporting proteins and polysaccharides. These spirochetes also exhibit differential abilities to export drugs including antibiotics. L. biflexa, however, uniquely has homologs of exporters for fatty acyl CoA and putative adhesin proteins. Fatty acyl CoA export may function to acylate the outer membrane and adhesin proteins, likely to play a role in biofilm formation.
Interspecies differences in transporter classes and substrate categories are limited in Leptospira. Individual transport proteins serve as likely contributors to pathogenesis in L. interrogans and L. borgpetersenii. Both pathogens encode members of the SphH Hemolysin Family (TC#1.C.67) that are notably absent in L. biflexa [102–104]. Sph2 (Uniprot # P59116; Table S1), is a sphingomyelinase with all active site residues essential for catalysis in vitro. Sphingomyelinase C (Uniprot # Q04XS2) of L. borgpetersenii is closest in sequence to Sph2. Both pathogens, but not L. biflexa, possess proteins with strong sequence similarity to a member of the Mycobacterial 4 TMS Phage Holin Family (TC#1.E.40). The proposed roles of prokaryotic holins in cell lysis and biofilm formation indicate the potential role these proteins may play in pathogenesis [105]. Both L. interrogans and L. borgpetersenii possess a member of the Putative Peptide Transporter Carbon Starvation CstA Family (TC#2.A.114). Mutation of a homolog in C. jejuni displayed decreased host-pathogen interactions [106]. All three leptospires possess proteins exhibiting sequence similarity to a member (TC#1.B.6.1.20) of the OmpA-OmpF Porin family. The L. interrogans protein queried has been shown to be Loa22, a protein essential for leptospiral virulence [107]. The relative dissimilarity of the L. biflexa homolog could render it avirulent.
Transporter Hallmarks of Leptospira
Leptospira is a branch of a divergent phylum and represents a genetically isolated group of bacteria [22]. Transporters identified in the leptospires in this study potentially serve novel roles for these Gram-negative aerobes. All three leptospires possess a significant number of α-type channels, most of which transport inorganic ions and small metabolites. The largest representative of these channels are the MotAB/ExbBD/TolQR channel-forming constituents (TC#1.A.30) with roles in motility, energized outer membrane transport, and outer membrane stability, respectively. The presence of two MotAB energizers may permit one system to utilize the proton motive force utilization and the other the sodium motive force [47]. L. interrogans is slightly deficient in ExbBD/TolQR energizers relative to L. borgpetersenii and L. biflexa, which raises questions about the role of these energizers in Leptospira [48–50].
Of strong clinical relevance is the leptospiral outer membrane proteome (surfaceome), constituted largely by a variety of β-type porins. It contains cell surface antigens for potential vaccine production and drug targets [57]. As mentioned above, all three leptospires possess Loa22 (or a homolog), a surface-exposed porin, necessary for leptospiral virulence. Aside from this virulence factor, leptospires possess outer membrane transporters for the transport of small molecules, as well as larger molecules including siderophores, proteins and membrane constituents. The leptospiral outer membrane plays a role in transport of substrates from the extracellular space to the periplasm.
The largest TC subclass identified in L. interrogans, L. borgpetersenii, and L. biflexa is 2.A, carrier proteins catalyzing uniport, antiport, and symport. The diversity of substrates transported is due to members of about 40 families of carriers. This distribution of transporters suggests an important role of secondary carriers in nutrient acquisition over primary active transporters such as ABC systems. The distribution of secondary carriers and primary active transporters reveals the prioritization of the acquisition and export of various molecules. Metabolic flexibility should dictate utilization of low-affinity secondary carriers, whereas a specific metabolic need might necessitate other higher-affinity systems.
P-type ATPase (TC#3.A.3) distribution varies between all three leptospires, but all three possess an exporter of copper (Cu2+), indicating a critical need for strict intracellular copper regulation. Only L. interrogans possesses ATPases for the uptake of Mg2+ and K+, suggesting these ions play critical roles for this organism to survive in the external environment and/or in the host [108]. L. biflexa possesses ATPases for Ca2+ export, heavy metal resistance (Co2+, Zn2+, Cd2+), and Na+/K+. These proteins, unique to L. biflexa, likely highlight its effective osmoregulation and capacity for membrane potential maintenance.
All three leptospires possess multiple systems for protein secretion. The primary pathways for protein secretion across the two membranes of the cell envelope are probably provided by the General Secretory Pathway (TC#3.A.5) and the outer membrane secreting Main Terminal Branch (TC#3.A.15). Flagellar proteins and potential virulence proteins are secreted by the Type III Secretory Pathway (TC#3.A.6) [67]. This particular pathway is likely critical for virulence, as inhibition of flagellar motility in leptospires has been shown to render them avirulent [65].
Common in all three leptospires are primary proton pumping electron transfer complexes inherently present in mitochondria and many aerobic bacteria. These systems include the proton-translocating NADH dehydrogenase (TC#3.D.1), proton-translocating transhydrogenase (TC#3.D.2), and proton-translocating cytochrome oxidase (TC#3.D.4). The presence of these proton-translocating systems is consistent with the conclusion that electron transfer is used as a primary mechanism to generate a proton motive force, subsequently used for ATP synthesis.
As leptospires are genetically divergent compared to most well studied bacteria, they are expected to share a strong core of proteins and possess unique systems for pathogenesis and free-living [15, 22, 23]. A significant portion of the identified transporter proteome in these leptospires are incompletely characterized proteins from TC subclasses 9.A and 9.B. Further identification and characterization of these proteins, in addition to the remaining encoded non-transport proteins, should provide a more complete understanding of leptospiral pathogenesis and saprophytism.
Key attributes of Leptospira are aligned with their motile and chemotactic abilities. The embedded flagella permitting cork-screw like motility favors these organisms in host dissemination and environmental survival [109]. Transport proteins identified in this study, including a flagellar export system, chemotaxis proteins, and flagellar motor energizers, play roles in survival and virulence. Chemotaxis toward specific molecules like glucose may facilitate tissue tropism of Leptospira pathogens [109]. L. biflexa can persist over long periods of time in distilled water by forming biofilms, and aggregation has been suggested to allow environmental survival and host colonization [79, 110]. Transport proteins that excrete exopolysaccharides, signaling molecules, and adhesion proteins also promote biofilm formation for persistence [111–113].
Transport proteins represent a subset (about 10%) of the entire proteome of an organism. However, intracellular processes are dependent on what materials are available in the cell. By providing an overview of the molecules transported and how they are imported and exported, conclusions about the potential metabolism and physiology of an organism can be drawn. In the case of Leptospira, the overall transportome (transporters of the proteome), reveals key characteristics of saprophytism and pathogenesis. L. biflexa demonstrates high flexibility and versatility in its transportome with a relatively large subset of secondary carriers and transporter families not found in the pathogens. In addition, L. biflexa possesses high-affinity transporters for critical cofactor import and increased numbers of uptake systems for carbon and nitrogen sources. Meanwhile, the pathogens possess remarkably similar transport protein profiles, suggesting the host tropism and environmental survival for the two relies on similar transporters. Both possess factors that may be associated with, or facilitate, pathogenesis, absent in L. biflexa, such as pore-forming toxins, holins, and virulence-related outer membrane porins. The increased versatility of L. biflexa as a free-living organism likely reflects the inverse as the decreased versatility in the pathogens correlates to the realization of progressively narrower ecological niches. As leptospirosis manifests itself in a variety of symptoms, small differences within individual proteins and the leptospiral proteome may play roles in determining virulence and mortality in humans [114]. The findings reported here on these leptospiral transporters should improve our understanding of the pathology of leptospirosis and allow more specific experimentation with L. biflexa as a model system for the Leptospira genus.
Supplementary Material
Table S1. TC classification and functional prediction of transport-related proteins found in L. interrogans, L. borgpetersenii, and L. biflexa. Sequences were retrieved using GBlast with E-values larger than e-12 are highlighted to indicate low substrate-specificity confidence.
Table S2. Counts and percentages of transporters in L. interrogans, L. borgpetersenii, and L. biflexa based on substrate specificity and TC classification.
Genomes of three Leptospira species were analyzed for encoded transport proteins.
Pathogenic Leptospira encode fewer transporters overall for metabolites and cations.
These pathogens exhibit decreased metabolic flexibility and stress adaptability.
The pathogens encode more sphingomyelinases, holins, and virulence-associated porins.
Pathogenesis may have evolved by host adaptation with gain of virulence proteins.
Acknowledgments
This work was supported by NIH grants GM077402 and GM109895.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–71. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 2.Duplessis CA, Sklar MJ, Maves RC, Spichler A, Hale B, Johnson M, et al. Hemoptysis associated with leptospirosis acquired in Hawaii, USA. Emerging infectious diseases. 2011;17:2375–7. doi: 10.3201/eid1712.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinetz JM, Glass GE, Flexner CE, Mueller P, Kaslow DC. Sporadic urban leptospirosis. Ann Intern Med. 1996;125:794–8. doi: 10.7326/0003-4819-125-10-199611150-00002. [DOI] [PubMed] [Google Scholar]
- 4.Toliver HL, Krane NK. Leptospirosis in New Orleans. The American journal of the medical sciences. 2014;347:159–63. doi: 10.1097/MAJ.0b013e3182787068. [DOI] [PubMed] [Google Scholar]
- 5.dos Paixao MS, Alves-Martin MF, da Tenorio MS, Starke-Buzetti WA, Alves ML, da Silva DT, et al. Serology, isolation, and molecular detection of Leptospira spp. from the tissues and blood of rats captured in a wild animal preservation centre in Brazil. Preventive veterinary medicine. 2014;115:69–73. doi: 10.1016/j.prevetmed.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Vashi NA, Reddy P, Wayne DB, Sabin B. Bat-associated leptospirosis. Journal of general internal medicine. 2010;25:162–4. doi: 10.1007/s11606-009-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayral FC, Bicout DJ, Pereira H, Artois M, Kodjo A. Distribution of Leptospira Serogroups in Cattle Herds and Dogs in France. The American journal of tropical medicine and hygiene. 2014 doi: 10.4269/ajtmh.13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandara M, Ananda M, Wickramage K, Berger E, Agampodi S. Globalization of leptospirosis through travel and migration. Globalization and health. 2014;10:61. doi: 10.1186/s12992-014-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adler B, de la Pena Moctezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010;140:287–96. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Cerqueira GM, Picardeau M. A century of Leptospira strain typing. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2009;9:760–8. doi: 10.1016/j.meegid.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez S, Geymonat JP, Hernandez E, Marques JM, Schelotto F, Varela G. Usefulness of real-time PCR assay targeting lipL32 gene for diagnosis of human leptospirosis in Uruguay. Journal of infection in developing countries. 2013;7:941–5. doi: 10.3855/jidc.4110. [DOI] [PubMed] [Google Scholar]
- 12.Loffler SG, Pavan ME, Vanasco B, Samartino L, Suarez O, Auteri C, et al. Genotypes of pathogenic Leptospira spp isolated from rodents in Argentina. Memorias do Instituto Oswaldo Cruz. 2014;109:163–7. doi: 10.1590/0074-0276140295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang F, Collins-Emerson JM, Cullum A, Heuer C, Wilson PR, Benschop J. Shedding and Seroprevalence of Pathogenic Leptospira spp. in Sheep and Cattle at a New Zealand Abattoir. Zoonoses and public health. 2014 doi: 10.1111/zph.12146. [DOI] [PubMed] [Google Scholar]
- 14.Munoz-Zanzi C, Mason MR, Encina C, Astroza A, Romero A. Leptospira contamination in household and environmental water in rural communities in southern Chile. International journal of environmental research and public health. 2014;11:6666–80. doi: 10.3390/ijerph110706666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A. 2006;103:14560–5. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Victoria B, Ahmed A, Zuerner RL, Ahmed N, Bulach DM, Quinteiro J, et al. Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PloS one. 2008;3:e2752. doi: 10.1371/journal.pone.0002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adler B. Pathogenesis of leptospirosis: cellular and molecular aspects. Vet Microbiol. 2014;172:353–8. doi: 10.1016/j.vetmic.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Hsu SH, Lo YY, Tung JY, Ko YC, Sun YJ, Hung CC, et al. Leptospiral outer membrane lipoprotein LipL32 binding on toll-like receptor 2 of renal cells as determined with an atomic force microscope. Biochemistry. 2010;49:5408–17. doi: 10.1021/bi100058w. [DOI] [PubMed] [Google Scholar]
- 19.Ferrer MF, Scharrig E, Alberdi L, Cedola M, Pretre G, Drut R, et al. Decayaccelerating factor 1 deficiency exacerbates leptospiral-induced murine chronic nephritis and renal fibrosis. PloS one. 2014;9:e102860. doi: 10.1371/journal.pone.0102860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson B, Choy HA, Pinne M, Rotondi ML, Miller MC, Demoll E, et al. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PloS one. 2007;2:e1188. doi: 10.1371/journal.pone.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Wu Y, Ojcius DM, Yang XF, Zhang C, Ding S, et al. Leptospiral hemolysins induce proinflammatory cytokines through Toll-like receptor 2-and 4- mediated JNK and NF-kappaB signaling pathways. PLoS One. 2012;7:e42266. doi: 10.1371/journal.pone.0042266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, et al. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One. 2008;3:e1607. doi: 10.1371/journal.pone.0001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–93. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 24.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic acids research. 2014;42:D251–8. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy VS, Saier MH., Jr BioV Suite--a collection of programs for the study of transport protein evolution. The FEBS journal. 2012;279:2036–46. doi: 10.1111/j.1742-4658.2012.08590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusnady GE, Simon I. Topology of membrane proteins. J Chem Inf Comput Sci. 2001;41:364–8. doi: 10.1021/ci0001280. [DOI] [PubMed] [Google Scholar]
- 27.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–2. [PubMed] [Google Scholar]
- 28.Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargent F, Berks BC, Palmer T. Pathfinders and trailblazers: a prokaryotic targeting system for transport of folded proteins. FEMS Microbiol Lett. 2006;254:198–207. doi: 10.1111/j.1574-6968.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- 30.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic acids research. 2004;32:W400–4. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–8. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youm J, Saier MH., Jr Comparative analyses of transport proteins encoded within the genomes of Mycobacterium tuberculosis and Mycobacterium leprae. Biochim Biophys Acta. 2012;1818:776–97. doi: 10.1016/j.bbamem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 35.Felce J, Saier MH., Jr Carbonic anhydrases fused to anion transporters of the SulP family: evidence for a novel type of bicarbonate transporter. J Mol Microbiol Biotechnol. 2004;8:169–76. doi: 10.1159/000085789. [DOI] [PubMed] [Google Scholar]
- 36.Saier MH., Jr Vectorial metabolism and the evolution of transport systems. J Bacteriol. 2000;182:5029–35. doi: 10.1128/jb.182.18.5029-5035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saier MH, Jr, Reddy BL. Holins in Bacteria, Eukaryotes, and Archaea: Multifunctional Xenologues with Potential Biotechnological and Biomedical Applications. J Bacteriol. 2015;197:7–17. doi: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis JK. Combining polysaccharide biosynthesis and transport in a single enzyme: dual-function cell wall glycan synthases. Front Plant Sci. 2012;3:138. doi: 10.3389/fpls.2012.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brams M, Kusch J, Spurny R, Benndorf K, Ulens C. Family of prokaryote cyclic nucleotide-modulated ion channels. Proc Natl Acad Sci U S A. 2014;111:7855–60. doi: 10.1073/pnas.1401917111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bienert GP, Desguin B, Chaumont F, Hols P. Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. The Biochemical journal. 2013;454:559–70. doi: 10.1042/BJ20130388. [DOI] [PubMed] [Google Scholar]
- 41.Huergo LF, Merrick M, Pedrosa FO, Chubatsu LS, Araujo LM, Souza EM. Ternary complex formation between AmtB, GlnZ and the nitrogenase regulatory enzyme DraG reveals a novel facet of nitrogen regulation in bacteria. Mol Microbiol. 2007;66:1523–35. doi: 10.1111/j.1365-2958.2007.06016.x. [DOI] [PubMed] [Google Scholar]
- 42.Pivetti CD, Yen MR, Miller S, Busch W, Tseng YH, Booth IR, et al. Two families of mechanosensitive channel proteins. Microbiol Mol Biol Rev. 2003;67:66–85. doi: 10.1128/MMBR.67.1.66-85.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo CJ, Sowa Y, Pilizota T, Berry RM. Mechanism and kinetics of a sodium-driven bacterial flagellar motor. Proc Natl Acad Sci U S A. 2013;110:E2544–51. doi: 10.1073/pnas.1301664110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci U S A. 2011;108:2498–503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annual review of microbiology. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett. 1999;177:191–7. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- 47.Ito M, Terahara N, Fujinami S, Krulwich TA. Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. Journal of molecular biology. 2005;352:396–408. doi: 10.1016/j.jmb.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang F, Saier MH., Jr Transport proteins promoting Escherichia coli pathogenesis. Microbial pathogenesis. 2014;71–72:41–55. doi: 10.1016/j.micpath.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Held KG, Postle K. ExbB and ExbD do not function independently in TonBdependent energy transduction. J Bacteriol. 2002;184:5170–3. doi: 10.1128/JB.184.18.5170-5173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goemaere EL, Devert A, Lloubes R, Cascales E. Movements of the TolR Cterminal domain depend on TolQR ionizable key residues and regulate activity of the Tol complex. J Biol Chem. 2007;282:17749–57. doi: 10.1074/jbc.M701002200. [DOI] [PubMed] [Google Scholar]
- 51.Stockbridge RB, Robertson JL, Kolmakova-Partensky L, Miller C. A family of fluoride-specific ion channels with dual-topology architecture. eLife. 2013;2:e01084. doi: 10.7554/eLife.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S, Smith KD, Davis JH, Gordon PB, Breaker RR, Strobel SA. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc Natl Acad Sci U S A. 2013;110:19018–23. doi: 10.1073/pnas.1310439110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverio AL, Saier MH., Jr Bioinformatic characterization of the trimeric intracellular cation-specific channel protein family. J Membr Biol. 2011;241:77–101. doi: 10.1007/s00232-011-9364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi A, Tamang DG, Saier MH. Mercury transport in bacteria. Water Air and Soil Pollution. 2007;182:219–34. [Google Scholar]
- 55.Mok T, Chen JS, Shlykov MA, Saier MH. Bioinformatic Analyses of Bacterial Mercury Ion (Hg2+) Transporters. Water Air and Soil Pollution. 2012;223:4443–57. [Google Scholar]
- 56.Miller SM. Bacterial detoxification of Hg(II) and organomercurials. Essays in biochemistry. 1999;34:17–30. doi: 10.1042/bse0340017. [DOI] [PubMed] [Google Scholar]
- 57.Raja V, Natarajaseenivasan K. Pathogenic, diagnostic and vaccine potential of leptospiral outer membrane proteins (OMPs) Critical reviews in microbiology. 2013 doi: 10.3109/1040841X.2013.787387. [DOI] [PubMed] [Google Scholar]
- 58.Haake DA, Matsunaga J. Leptospira: a spirochaete with a hybrid outer membrane. Mol Microbiol. 2010;77:805–14. doi: 10.1111/j.1365-2958.2010.07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–8. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–32. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Y, Tao Y, Cheung LS, Fan C, Chen LQ, Xu S, et al. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature. 2014 doi: 10.1038/nature13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan H, Babayan V, Blyumin E, Gandhi C, Hak K, Harake D, et al. The p-type ATPase superfamily. J Mol Microbiol Biotechnol. 2010;19:5–104. doi: 10.1159/000319588. [DOI] [PubMed] [Google Scholar]
- 63.Thever MD, Saier MH., Jr Bioinformatic characterization of p-type ATPases encoded within the fully sequenced genomes of 26 eukaryotes. J Membr Biol. 2009;229:115–30. doi: 10.1007/s00232-009-9176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nivaskumar M, Francetic O. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta. 2014;1843:1568–77. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 65.Lambert A, Picardeau M, Haake DA, Sermswan RW, Srikram A, Adler B, et al. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infect Immun. 2012;80:2019–25. doi: 10.1128/IAI.00131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen L, Paulsen IT, Tchieu J, Hueck CJ, Saier MH., Jr Phylogenetic analyses of the constituents of Type III protein secretion systems. Journal of molecular microbiology and biotechnology. 2000;2:125–44. [PubMed] [Google Scholar]
- 67.Saier MH., Jr Protein secretion and membrane insertion systems in gramnegative bacteria. J Membr Biol. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 68.Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci U S A. 1999;96:6456–61. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Granjon T, Maniti O, Auchli Y, Dahinden P, Buchet R, Marcillat O, et al. Structurefunction relations in oxaloacetate decarboxylase complex. Fluorescence and infrared approaches to monitor oxomalonate and Na(+) binding effect. PLoS One. 2010;5:e10935. doi: 10.1371/journal.pone.0010935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saier MH, Jr, Newman MJ, Rephaeli AW. Properties of a phosphoenolpyruvate: mannitol phosphotransferase system in Spirochaeta aurantia. J Biol Chem. 1977;252:8890–8. [PubMed] [Google Scholar]
- 71.Foster JW, Park YK, Penfound T, Fenger T, Spector MP. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J Bacteriol. 1990;172:4187–96. doi: 10.1128/jb.172.8.4187-4196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Black PN, DiRusso CC. Vectorial acylation: linking fatty acid transport and activation to metabolic trafficking. Novartis Found Symp. 2007;286:127–38. doi: 10.1002/9780470985571.ch11. discussion 38–41, 62–3, 96–203. [DOI] [PubMed] [Google Scholar]
- 73.Hubbard C, McNamara JT, Azumaya C, Patel MS, Zimmer J. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 2012;418:21–31. doi: 10.1016/j.jmb.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 74.Louvel H, Saint Girons I, Picardeau M. Isolation and characterization of FecAand FeoB-mediated iron acquisition systems of the spirochete Leptospira biflexa by random insertional mutagenesis. J Bacteriol. 2005;187:3249–54. doi: 10.1128/JB.187.9.3249-3254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annual review of microbiology. 1996;50:753–89. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 76.Louvel H, Bommezzadri S, Zidane N, Boursaux-Eude C, Creno S, Magnier A, et al. Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp. J Bacteriol. 2006;188:7893–904. doi: 10.1128/JB.00711-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nevoigt E, Stahl U. Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS microbiology reviews. 1997;21:231–41. doi: 10.1111/j.1574-6976.1997.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 78.Patarakul K, Lo M, Adler B. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC microbiology. 2010;10:31. doi: 10.1186/1471-2180-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brihuega B, Samartino L, Auteri C, Venzano A, Caimi K. In vivo cell aggregations of a recent swine biofilm-forming isolate of Leptospira interrogans strain from Argentina. Revista Argentina de microbiologia. 2012;44:138–43. [PubMed] [Google Scholar]
- 80.Igarashi K. Physiological functions of polyamines and regulation of polyamine content in cells. Yakugaku zasshi : Journal of the Pharmaceutical Society of Japan. 2006;126:455–71. doi: 10.1248/yakushi.126.455. [DOI] [PubMed] [Google Scholar]
- 81.Grillo MA, Colombatto S. Polyamine transport in cells. Biochemical Society transactions. 1994;22:894–8. doi: 10.1042/bst0220894. [DOI] [PubMed] [Google Scholar]
- 82.Tomitori H, Kashiwagi K, Igarashi K. Structure and function of polyamineamino acid antiporters CadB and PotE in Escherichia coli. Amino acids. 2012;42:733–40. doi: 10.1007/s00726-011-0989-9. [DOI] [PubMed] [Google Scholar]
- 83.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12:205–13. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 84.Brett-Major DM, Coldren R. Antibiotics for leptospirosis. Cochrane Database Syst Rev. 2012;2:CD008264. doi: 10.1002/14651858.CD008264.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guerrier G, D’Ortenzio E. The Jarisch-Herxheimer reaction in leptospirosis: a systematic review. PLoS One. 2013;8:e59266. doi: 10.1371/journal.pone.0059266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei YF, Chiu CT, Lai YF, Lai CH, Lin HH. Successful treatment of septic shock and respiratory failure due to leptospirosis and scrub typhus coinfection with penicillin, levofloxacin, and activated protein C. J Microbiol Immunol Infect. 2012;45:251–4. doi: 10.1016/j.jmii.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 87.Griffith ME, Moon JE, Johnson EN, Clark KP, Hawley JS, Hospenthal DR, et al. Efficacy of fluoroquinolones against Leptospira interrogans in a hamster model. Antimicrob Agents Chemother. 2007;51:2615–7. doi: 10.1128/AAC.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffith ME, Hospenthal DR, Murray CK. Antimicrobial therapy of leptospirosis. Curr Opin Infect Dis. 2006;19:533–7. doi: 10.1097/QCO.0b013e3280106818. [DOI] [PubMed] [Google Scholar]
- 89.Guidugli F, Castro AA, Atallah AN. Antibiotics for preventing leptospirosis. Cochrane Database Syst Rev. 2000:CD001305. doi: 10.1002/14651858.CD001305. [DOI] [PubMed] [Google Scholar]
- 90.Brett-Major DM, Lipnick RJ. Antibiotic prophylaxis for leptospirosis. Cochrane Database Syst Rev. 2009:CD007342. doi: 10.1002/14651858.CD007342.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ricaldi JN, Vinetz JM. Leptospirosis in the tropics and in travelers. Curr Infect Dis Rep. 2006;8:51–8. doi: 10.1007/s11908-006-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ressner RA, Griffith ME, Beckius ML, Pimentel G, Miller RS, Mende K, et al. Antimicrobial susceptibilities of geographically diverse clinical human isolates of Leptospira. Antimicrob Agents Chemother. 2008;52:2750–4. doi: 10.1128/AAC.00044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, Vinetz JM. Asymptomatic renal colonization of humans in the peruvian Amazon by Leptospira. PLoS Negl Trop Dis. 2010;4:e612. doi: 10.1371/journal.pntd.0000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johnson RC. Leptospira. In: Baron S, editor. Medical Microbiology. 4. Galveston (TX): 1996. [Google Scholar]
- 95.Zhong Y, Chang X, Cao XJ, Zhang Y, Zheng H, Zhu Y, et al. Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res. 2011;21:1210–29. doi: 10.1038/cr.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xue F, Dong H, Wu J, Wu Z, Hu W, Sun A, et al. Transcriptional responses of Leptospira interrogans to host innate immunity: significant changes in metabolism, oxygen tolerance, and outer membrane. PLoS Negl Trop Dis. 2010;4:e857. doi: 10.1371/journal.pntd.0000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caimano MJ, Sivasankaran SK, Allard A, Hurley D, Hokamp K, Grassmann AA, et al. A model system for studying the transcriptomic and physiological changes associated with mammalian host-adaptation by Leptospira interrogans serovar Copenhageni. PLoS Pathog. 2014;10:e1004004. doi: 10.1371/journal.ppat.1004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu DL, Bao L. Research on the relevance between the virulent genes differential expression and pathogenecity of Leptospira with microarray. Sichuan Da Xue Xue Bao Yi Xue Ban. 2015;46:11–5. [PubMed] [Google Scholar]
- 99.Eshghi A, Becam J, Lambert A, Sismeiro O, Dillies MA, Jagla B, et al. A putative regulatory genetic locus modulates virulence in the pathogen Leptospira interrogans. Infect Immun. 2014;82:2542–52. doi: 10.1128/IAI.01803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ricaldi JN, Fouts DE, Selengut JD, Harkins DM, Patra KP, Moreno A, et al. Whole genome analysis of Leptospira licerasiae provides insight into leptospiral evolution and pathogenicity. PLoS Negl Trop Dis. 2012;6:e1853. doi: 10.1371/journal.pntd.0001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–90. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 102.Lee SH, Kim S, Park SC, Kim MJ. Cytotoxic activities of Leptospira interrogans hemolysin SphH as a pore-forming protein on mammalian cells. Infect Immun. 2002;70:315–22. doi: 10.1128/IAI.70.1.315-322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Narayanavari SA, Sritharan M, Haake DA, Matsunaga J. Multiple leptospiral sphingomyelinases (or are there?) Microbiology. 2012;158:1137–46. doi: 10.1099/mic.0.057737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang YX, Geng Y, Yang JW, Guo XK, Zhao GP. Cytotoxic activity and probable apoptotic effect of Sph2, a sphigomyelinase hemolysin from Leptospira interrogans strain Lai. BMB Rep. 2008;41:119–25. doi: 10.5483/bmbrep.2008.41.2.119. [DOI] [PubMed] [Google Scholar]
- 105.Saier MH, Jr, Reddy BL. Holins in Bacteria, Eukaryotes and Archaea: Multifunctional Xenologues with Potential Biotechnological and Biomedical Applications. J Bacteriol. 2014 doi: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rasmussen JJ, Vegge CS, Frokiaer H, Howlett RM, Krogfelt KA, Kelly DJ, et al. Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J Med Microbiol. 2013;62:1135–43. doi: 10.1099/jmm.0.059345-0. [DOI] [PubMed] [Google Scholar]
- 107.Ristow P, Bourhy P, da Cruz McBride FW, Figueira CP, Huerre M, Ave P, et al. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 2007;3:e97. doi: 10.1371/journal.ppat.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsunaga J, Coutinho ML. Positive regulation of Leptospira interrogans kdp expression by KdpE as Demonstrated with a novel beta-galactosidase reporter in Leptospira biflexa. Appl Environ Microbiol. 2012;78:5699–707. doi: 10.1128/AEM.00713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Islam MS, Takabe K, Kudo S, Nakamura S. Analysis of the chemotactic behaviour of Leptospira using microscopic agar-drop assay. FEMS Microbiol Lett. 2014;356:39–44. doi: 10.1111/1574-6968.12495. [DOI] [PubMed] [Google Scholar]
- 110.Barragan VA, Mejia ME, Travez A, Zapata S, Hartskeerl RA, Haake DA, et al. Interactions of leptospira with environmental bacteria from surface water. Curr Microbiol. 2011;62:1802–6. doi: 10.1007/s00284-011-9931-3. [DOI] [PubMed] [Google Scholar]
- 111.Balsanelli E, de Baura VA, de Pedrosa FO, de Souza EM, Monteiro RA. Exopolysaccharide biosynthesis enables mature biofilm formation on abiotic surfaces by Herbaspirillum seropedicae. PloS one. 2014;9:e110392. doi: 10.1371/journal.pone.0110392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chagnot C, Zorgani MA, Astruc T, Desvaux M. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Frontiers in microbiology. 2013;4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miyashiro T, Oehlert D, Ray VA, Visick KL, Ruby EG. The putative oligosaccharide translocase SypK connects biofilm formation with quorum signaling in Vibrio fischeri. MicrobiologyOpen. 2014;3:836–48. doi: 10.1002/mbo3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spichler A, Athanazio D, Seguro AC, Vinetz JM. Outpatient follow-up of patients hospitalized for acute leptospirosis. Int J Infect Dis. 2011;15:e486–90. doi: 10.1016/j.ijid.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. TC classification and functional prediction of transport-related proteins found in L. interrogans, L. borgpetersenii, and L. biflexa. Sequences were retrieved using GBlast with E-values larger than e-12 are highlighted to indicate low substrate-specificity confidence.
Table S2. Counts and percentages of transporters in L. interrogans, L. borgpetersenii, and L. biflexa based on substrate specificity and TC classification.