Blocking β-catenin/TCF1–mediated transcriptional activation with a specific small molecule or by TCF1 knockdown delays the mouse embryonic stem cell differentiation program and enhances pluripotency.
Abstract
The ability of mouse embryonic stem cells (mESCs) to self-renew or differentiate into various cell lineages is regulated by signaling pathways and a core pluripotency transcriptional network (PTN) comprising Nanog, Oct4, and Sox2. The Wnt/β-catenin pathway promotes pluripotency by alleviating T cell factor TCF3-mediated repression of the PTN. However, it has remained unclear how β-catenin’s function as a transcriptional activator with TCF1 influences mESC fate. Here, we show that TCF1-mediated transcription is up-regulated in differentiating mESCs and that chemical inhibition of β-catenin/TCF1 interaction improves long-term self-renewal and enhances functional pluripotency. Genetic loss of TCF1 inhibited differentiation by delaying exit from pluripotency and conferred a transcriptional profile strikingly reminiscent of self-renewing mESCs with high Nanog expression. Together, our data suggest that β-catenin’s function in regulating mESCs is highly context specific and that its interaction with TCF1 promotes differentiation, further highlighting the need for understanding how its individual protein–protein interactions drive stem cell fate.
Introduction
Pluripotency in mouse embryonic stem cells (mESCs) is controlled by a transcriptional network regulated by three core transcription factors: Nanog, Oct4, and Sox2 (reviewed in Silva and Smith, 2008; Nichols and Smith, 2009; Wray and Hartmann, 2012). Extrinsic signaling molecules including leukemia inhibitory factor (LIF) and Wnts influence the balance between pluripotency and differentiation in a context-dependent manner (Okita and Yamanaka, 2006; Loh et al., 2015). The primary consequence of Wnt stimulus is stabilization of β-catenin, a nuclear effector that activates transcription of target genes together with the lymphoid enhancer factor/T cell factor (TCF) family of transcription factors (Valenta et al., 2012). In addition to its nuclear functions, much of the cellular β-catenin is membrane localized at adherens junctions, where it interacts with E-cadherin and α-catenin (Valenta et al., 2012).
The Wnt/β-catenin pathway is important for early embryonic development of metazoans, particularly in the specification of the body axis and patterning of mesendoderm and neural lineages (Nusse and Varmus, 2012; Oates et al., 2012; Park and Shen, 2012). Exogenous addition of Wnt proteins to mESCs has been shown to activate TCF target genes while promoting self-renewal and inhibiting differentiation (Sato et al., 2004; Ogawa et al., 2006; Singla et al., 2006; Ying et al., 2008; Wagner et al., 2010; ten Berge et al., 2011). By contrast, studies have also suggested that Wnt activity is low in self-renewing embryonic stem cells (ESCs) and is activated during differentiation (Davidson et al., 2012; Marks et al., 2012; Faunes et al., 2013), raising the question of whether TCF-mediated transcription is required for pluripotency. The primary evidence for direct regulation of the pluripotency transcriptional network (PTN) by Wnt/β-catenin pathway comes from studies involving TCF3 (TCF7L1), a transcriptional repressor of Wnt target genes (Cole et al., 2008; Zhang et al., 2013) that promotes differentiation by directly inhibiting the PTN (Wray et al., 2011; Yi et al., 2011). It is thought that β-catenin alleviates TCF3’s repressive function by binding to TCF3 and removing it from the DNA, thereby promoting self-renewal (Wray et al., 2011; Shy et al., 2013). Although β-catenin has been implicated in influencing PTN activity (Merrill, 2012), the precise interactions by which it modulates pluripotency and lineage-specific differentiation in mESCs have remained elusive (Sokol, 2011; Lien and Fuchs, 2014). There is an increasing body of evidence supporting the hypothesis that β-catenin can influence pluripotency independent of its function as a transcriptional activator with TCFs (Takao et al., 2007; Kelly et al., 2011; Lyashenko et al., 2011; Faunes et al., 2013). A host of recent studies have shed some light on how β-catenin/Oct4 protein interactions influence pluripotency (Takao et al., 2007; Kelly et al., 2011; Faunes et al., 2013; Muñoz Descalzo et al., 2013; Zhang et al., 2013).
In this study, we examined the specific mechanisms by which β-catenin/TCF interactions regulate self-renewal and differentiation of mESCs. Transcriptional profiling of sorted cell populations and spontaneously differentiating embryoid bodies revealed that TCF target genes are up-regulated during exit from pluripotency. Furthermore, we found that using a small molecule (inhibitor of β-catenin–responsive transcription [iCRT3]; Gonsalves et al., 2011) to selectively inhibit the interaction between β-catenin and TCF1 confers pluripotent characteristics that are surprisingly reminiscent of the “2i” ground state (Wray et al., 2010), even in the absence of exogenously stabilized β-catenin. Notably, mESCs cultured long term with iCRT3 were uniformly pluripotent and maintained the ability to differentiate into all three germ layers. Finally, transcriptome analysis and biochemical assays revealed that knockdown of TCF1 mimicked the effect of iCRT3 treatment. Overall, our data suggest that β-catenin/TCF1–mediated transcriptional activation promotes differentiation and that blocking it with specific small molecules or by TCF1 knockdown delays the differentiation program, enhancing pluripotent characteristics of mESCs.
Results and discussion
TCF-dependent transcription is up-regulated during mESC differentiation
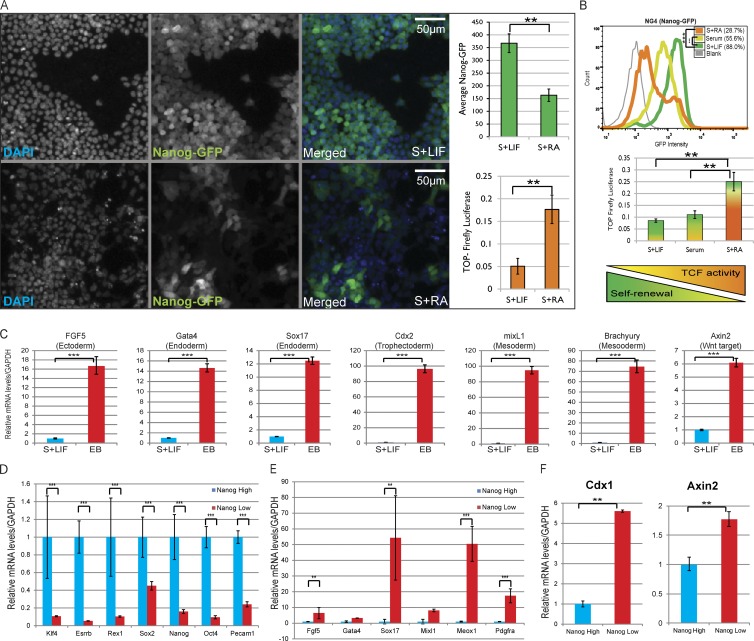
Although several studies have provided insights into the molecular mechanisms, the specific context-dependent function of β-catenin in ESC self-renewal and differentiation remains controversial and poorly understood (Lyashenko et al., 2011; ten Berge et al., 2011; Rudloff and Kemler, 2012; Faunes et al., 2013). Therefore, we decided to further investigate the correlation between β-catenin/TCF–mediated transcriptional activity and the expression of pluripotency markers under serum containing stem conditions (serum plus LIF) or differentiation conditions (LIF withdrawal: serum plus retinoic acid–induced differentiation [serum plus RA]). To accomplish this, we generated a mESC line containing both a β-catenin/TCF–responsive transcriptional reporter (TOP-Firefly Luciferase/TOPFlash) and Nanog-GFP. Although differentiation resulted in the expected down-regulation of Nanog-GFP expression, the TOPFlash reporter showed a concomitant and significant increase (Fig. 1, A and B). In addition, transcriptional profiling of differentiating embryoid bodies (EBs; Desbaillets et al., 2000; Itskovitz-Eldor et al., 2000) revealed an up-regulation of lineage-specific markers, including several TCF target genes such as Brachyury (Arnold et al., 2000), Cdx2 (Zhao et al., 2014), and Axin2 (Jho et al., 2002; Fig. 1 C).
Figure 1.
TCF-dependent transcriptional activation is up-regulated during mESC differentiation. (A) Mean Nanog-GFP levels (green bar graph; mean ± SD of four replicates) in NG4-TOPluc cells maintained in serum plus LIF (S+LIF; top) and serum plus RA (S+RA; bottom). Normalized TOPFlash reporter activity in simultaneously cultured NG4-TOPluc (orange bar graph; mean ± SD of three replicates). Resized original images shown are from a single representative experiment out of four replicates, with number of cells measured ≥1,000 per experiment. (B) Comparison of Nanog-GFP (by flow cytometry, top; representative data of three replicates) and transfected TOPFlash (middle; mean ± SD of three replicates) expression in mESCs cultured for 48 h in S+LIF, serum, or S+RA. Graphical summary reflecting inverse correlation between TCF-mediated transcriptional activity and self-renewal (bottom). (C) qPCR analysis of differentiation markers and TCF target genes in EBs versus the parental ESCs. Mean ± SD of three replicates. (D) Relative mRNA levels of pluripotency markers in sorted NanogHigh and NanogLow mESCs. Mean ± SD of two replicates. (E) Relative mRNA levels of differentiation markers in sorted NanogHigh and NanogLow mESCs. Mean ± SD of two replicates. (F) Relative mRNA levels of TCF target genes Cdx1 (mean ± SD of two replicates) and Axin2 (mean ± SD of three replicates) in sorted NanogHigh (S+LIF) and NanogLow (S+RA) mESCs. **, P < 0.01; ***, P < 0.001.
To rule out the possibility that the TCF activation observed in heterogeneous populations is from self-renewing cells, we measured mRNA levels of pluripotency and differentiation markers and TCF target genes in cells sorted for high Nanog-GFP (NanogHi) to NanogLow cells from both stem (serum plus LIF) and differentiation (serum plus RA) conditions. Quantitative PCR analysis revealed that TCF target genes are specifically up-regulated in NanogLow cells (Fig. S1 A). This activation is more evident when cells are grown in differentiation media. Although pluripotency markers are consistently down-regulated in the NanogLow cell population, Cdx2 was the only TCF target with an appreciable activation in NanogLow cells grown in serum plus LIF. We attribute the lack of significant activation of TCF target genes in NanogLow cells in self-renewing conditions to the fact that the cells are still under selective pressure of LIF, which strongly promotes pluripotency by actively blocking differentiation (Hall et al., 2009). The quantitative PCR (qPCR) plot shown in Fig. S1 (A) clarifies that NanogHigh cells from both conditions generally exhibit higher expression of pluripotency markers than the NanogLow subpopulation. Next, we compared transcriptomes of cells sorted for high Nanog-GFP expression during self-renewal (top 30% NanogHigh from serum plus LIF) with NanogLow cells during differentiation (bottom 30% NanogLow from serum plus RA). The data set analyzed for differential expression and functional enrichment (Table S2) confirmed that although NanogLow cells exhibit lower expression of pluripotency markers (Fig. 1 D), they have significantly higher mRNA levels of differentiation markers (Fig. 1 E) as well as TCF target genes such as Cdx1 (Pilon et al., 2007) and Axin2 (Fig. 1 F). These results concur with previous in vivo observations in which canonical Wnt/β-catenin signaling was not detected in early preimplantation embryos (from which ESCs are derived) but was involved in germ layer specification (Kimura-Yoshida et al., 2005; Ferrer-Vaquer et al., 2010; Nowotschin and Hadjantonakis, 2010).
Small molecule–mediated inhibition of TCF-dependent transcription improves expression of pluripotency markers in cultured mESCs
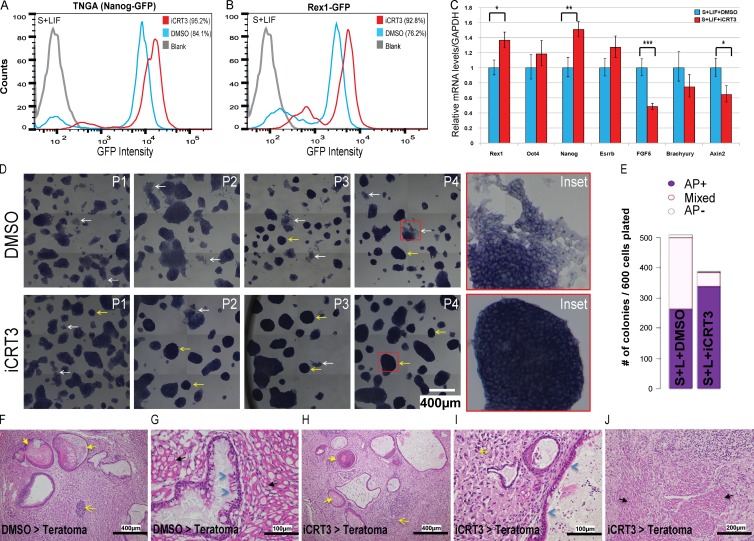
To directly assess the requirement of TCF-mediated transcription for mESC pluripotency and differentiation, we used iCRT3, a small molecule that inhibits the Wnt pathway by blocking binding between β-catenin and activating TCFs (Gonsalves et al., 2011). Surprisingly, TNGA (Nanog reporter; Chambers et al., 2007) and Rex1-GFP (Wray et al., 2011) mESCs maintained for multiple passages in serum plus LIF with iCRT3 exhibited enhanced expression of their respective pluripotency reporters (Fig. 2, A and B). In addition, cells maintained long term with iCRT3 showed enhanced expression of classic pluripotency genes compared with the DMSO control, whereas expression of differentiation markers and TCF target genes was concomitantly reduced (Fig. 2 C). Interestingly, the enhancement of stem-like characteristics in iCRT3-treated cells (as determined by AP staining and colony morphology) progressively increased over passages, and colonies were strikingly reminiscent of the classic 2i morphology by later passages (Ying et al., 2008), despite the absence of exogenous signals to stabilize β-catenin (Wray and Hartmann, 2012; bottom inset in Fig. 2 D). Furthermore, mESCs cultured with iCRT3 for multiple passages remained pluripotent, as judged by their ability to form increased numbers and AP-positive colonies (Fig. 2 E), as well as their ability to successfully generate EBs (Fig. S1 B) with comparable expression of lineage-specific markers (Fig. S1 C). Although mRNA levels of mesodermal markers (mixL1 and Brachyury) and the TCF target gene (Axin2) were lower in EBs generated after iCRT3 treatment (relative to DMSO; Fig. S1 C), the expression levels of these differentiation markers were significantly higher than their self-renewing parental mESCs (not depicted). Furthermore, these EBs could be successfully differentiated into spontaneously contracting mesoderm-derived cardiomyocytes (unpublished data), suggesting that long-term iCRT3 treatment does not inhibit mesodermal differentiation capability of mESCs. Finally, to validate the pluripotency of cells maintained for multiple passages with iCRT3, we performed a teratoma assay to measure their ability to differentiate into all three germ layers in vivo. We found that the iCRT3-treated cells readily differentiated into teratomas upon injection (Fig. S1 D). The disparity in teratoma size may be explained by the observation that stem cell populations with more homogeneous pluripotency (as we see in iCRT3-treated cells) form larger teratomas (Zhang et al., 2008; Gropp et al., 2012). The DMSO-treated cells demonstrated spontaneous differentiation typical to ESCs maintained with serum (Fig. 2 D), which suggests that they have greater heterogeneity and thereby generate teratomas of smaller size (Fig. S1 D). The teratomas generated from cells precultured with iCRT3 demonstrated similar expression levels of lineage markers (Fig. S1 E) and tissue types representing all three germ layers (Fig. 2, F–J; and Fig. S1 F) compared with control teratomas. Together, these results indicate that TCF-dependent transcriptional activation may promote lineage specification and is dispensable for maintenance of mESC pluripotency.
Figure 2.
Long-term inhibition of β-catenin/TCF–mediated transcriptional activity enhances self-renewal of mESCs. (A and B) Flow cytometry of TNGA (Nanog reporter) (A) and Rex1-GFP (B) mESCs grown in serum plus LIF (S+LIF) with iCRT3 (14 d). Two independent experiments were set up with TNGA and Rex1-GFP cell lines. (C) qPCR analysis for pluripotency/differentiation markers and TCF target genes in cells maintained in iCRT3 for four passages. Mean ± SD of three replicates. (D) Representative bright-field images of mESCs maintained in S+LIF over multiple passages of iCRT3 treatment (P1–P4). Note that iCRT3-treated cells demonstrate lower spontaneous differentiation (white arrows and inset), more compact colony morphology, and higher AP expression (yellow arrows and inset), compared with DMSO. Resized original images and additional ×6 digitally magnified insets shown are from a single representative experiment (for NG4) out of three replicates for each of NG4 and Rex1-GFP cell lines. (E) Representative quantification of AP levels of colonies formed from E14Tg2A mESCs after iCRT3 treatment in stem conditions (S+LIF).These results are representative of identical experiments set up for two independent E14Tg2A and CBA cell lines. (F–J) Representative staining of teratomas derived from NG4 cells treated with DMSO (F and G) or iCRT3 (H–J). Detection of tissue derived from all three germ layers is suggestive of teratoma arising from pluripotent cells and is marked by keratin in lumen of cysts lined by squamous keratinized epithelium (ectoderm, yellow arrows), neoplastic neuronal tissue/neuron rosettes (ectoderm, yellow open arrows), cysts lined by single layer of cuboidal or pseudostratified columnar ciliated epithelium (endoderm, blue arrowheads), and skeletal muscles (mesoderm, black arrows). Resized original images provided by IMCB Histopathology Core (A*Star) are from a single representative experiment out of eight pairs of teratomas generated from cells treated with DMSO/iCRT3 for 14 d. Refer to Fig. S1 F for representative immunofluorescence staining of germ layer markers in teratoma generated from iCRT3-treated cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Inhibition of Wnt/β-catenin–dependent TCF target gene activation by iCRT3 allows mESCs to resist induced differentiation
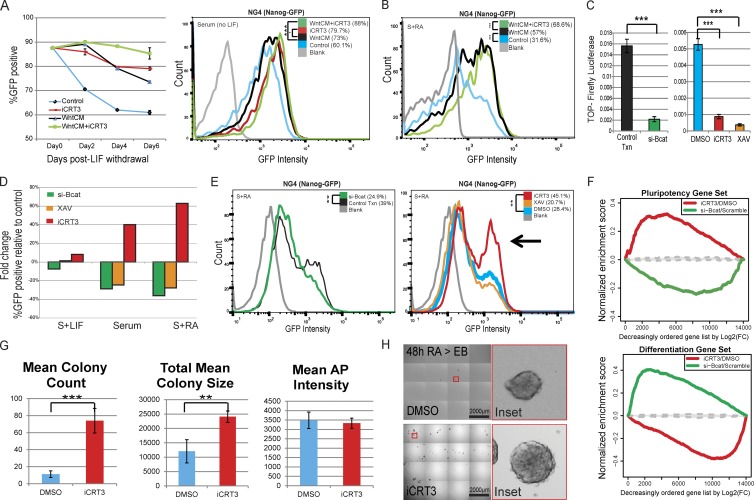
Previous studies have shown that Wnt3a can reduce differentiation of mESCs maintained without LIF (ten Berge et al., 2011). To directly compare how exogenously stabilized β-catenin reduces differentiation relative to merely inhibiting its TCF-dependent transcriptional activity, we passaged mESCs in the absence of LIF with and without Wnt3a-conditioned media (WntCM) and iCRT3. Flow cytometry analysis revealed that iCRT3 treatment resulted in a higher percentage of cells expressing Nanog-GFP than WntCM alone, and the combination of the two (WntCM plus iCRT3) further reduced differentiation, even 6 d after LIF withdrawal (Fig. 3 A). We observed similar results when we activated differentiation with retinoic acid (RA). Although mESCs cultured in Wnt-conditioned media had reduced differentiation relative to control media, the addition of iCRT3 caused a further reduction in differentiation, with a more homogeneous expression of Nanog-GFP (Fig. 3 B).
Figure 3.
iCRT3 treatment allows mESCs to resist induced differentiation. (A) Flow cytometry analysis of Nanog-GFP cells maintained for 6 d (three passages) without LIF to assay the effect of iCRT3 relative to WntCM. (left) In the absence of LIF, WntCM (black) progressively promotes ESC differentiation, as suggested by loss of Nanog-GFP–positive population in contrast with cells maintained with WntCM plus iCRT3 (green) or iCRT3 alone (red). Mean ± SD of three replicates. (right) Histogram plot for Nanog-GFP levels for cells maintained without LIF for 6 d with or without WntCM and iCRT3. Representative data of three replicates. (B) Flow cytometry of mESCs treated with RA (48 h) revealed that cells maintained in WntCM with iCRT3 (green) showed reduced differentiation relative to those grown with WntCM alone (black). Representative data of three replicates. (C) Normalized TOPFlash activity shows a marked reduction in reporter expression upon treatment with si-β-cat, XAV939, or iCRT3. Mean ± SD of three replicates. (D) Bar graph depicting single representative data for fold changes in Nanog-GFP expression upon treatment with XAV939 (orange), si-β-cat (green), or specific inhibition of β-catenin/TCF–dependent transcription using iCRT3 (red) relative to controls (DMSO or Scramble siRNA). (E) Histogram plots depicting changes in Nanog-GFP levels upon loss of β-catenin by si-β-cat or XAV939, and specific inhibition of β-catenin/TCF–dependent transcriptional activity using iCRT3 during RA-induced differentiation. The arrowhead highlights the Nanog-GFP–positive cell population that resists differentiation in presence of iCRT3. Representative data of two replicates. (F) GSEA plots showing how mRNA expression in cells treated with iCRT3 or si-βcat correlates to statistically significant pluripotency and differentiation gene sets (Online supplemental material). Genes were ordered on the x axis by log2 fold changes for treatments of iCRT3 (relative to DMSO) or si-βcat (relative to control transfection) during RA-mediated differentiation. The range of GSEA curves generated by randomly selected gene sets is shown in gray (interquartile range based on 500 size-matched random gene sets). (G) Colony-forming efficiency (CFE) of cells precultured in RA with iCRT3 or DMSO for 48 h. After treatment, cells were plated in limiting dilutions back into serum plus LIF (S+LIF) without any inhibitors for an additional 48 h (please refer to schematic in Fig. S2 C). CFE quantified for mean colony count, colony area, and AP intensities. Mean ± SD of four replicates. (H) Representative resized bright-field images as well as ×10 digitally magnified insets of four replicates showing control mESCs exposed to serum plus RA with DMSO that produced fewer EBs (top) relative to the enhanced efficiency of those grown in serum plus RA with iCRT3 (bottom; please refer to schematic in Fig. S2 C). **, P < 0.01; ***, P < 0.001. FC, fold change; Txn, transfection.
Because we observed increased expression of Nanog-GFP upon inhibition of β-catenin–mediated transcription, we next assayed how the loss of β-catenin protein affects mESC pluripotency in either self-renewing or differentiating conditions. Loss of β-catenin using RNAi (si-βcat) or XAV939 (a tankyrase inhibitor that stabilizes the β-catenin destruction complex; Huang et al., 2009), or specifically interfering with its binding to TCF/lymphoid enhancer factors (iCRT3), resulted in robust down-regulation of the TOPFlash reporter (Fig. 3 C). Corroborating previous studies (Lyashenko et al., 2011; Wray et al., 2011), we did not observe a significant reduction of pluripotency upon loss of β-catenin in self-renewal conditions (serum plus LIF; Fig. 3 D). However, the percentage of Nanog-GFP–positive cells was robustly reduced upon loss of β-catenin protein levels (XAV939 or si-βcat) under differentiation conditions (upon LIF withdrawal or addition of RA; Fig. 3, D and E). In sharp contrast, iCRT3 promoted an increase in the Nanog-GFP population for both differentiation conditions (Fig. 3, D and E, arrow). These results further support the hypothesis that TCF-driven activation of β-catenin target genes may in fact promote differentiation and is dispensable for maintenance of pluripotency, corroborating claims made in previous studies (Lyashenko et al., 2011; Faunes et al., 2013).
Because we observed the most significant variation in pluripotency levels by influencing β-catenin’s role in differentiating cells (Fig. 3, D and E), we profiled the transcriptome of cells treated with iCRT3 or si-βcat during RA-induced differentiation for further insight. Gene set enrichment analysis (GSEA) and comparison of expression profiles revealed that iCRT3-treated cells (relative to DMSO control) were enriched for a “pluripotency gene signature” (genes up-regulated in NanogHigh cells in serum plus LIF; Fig. 3 F and Fig. S2 A). By contrast, the mRNA profile of si-βcat–treated cells (relative to Scrambled siRNA control) had a stronger correlation to a “differentiation gene signature” (genes up-regulated in differentiating NanogLow cells after 48 h in serum plus RA; Fig. 3 F and Fig. S2 A). These unbiased global mRNA profile analyses indicate that although β-catenin plays a role in maintaining pluripotency, blocking its TCF-dependent transcriptional activity lowers expression of differentiation markers such as Brachyury, Cdx2, and Meox1 and confers a gene signature similar to self-renewing mESCs (Fig. S2, A and B).
Next, to determine whether iCRT3 improves functional pluripotency under differentiation cues, we treated mESCs with RA and either iCRT3 or DMSO and tested their ability to form self-renewing colonies when reintroduced to stem conditions or differentiate into EBs in absence of iCRT3 (Fig. S2 C). Measurement of AP-positive colonies revealed that cells that had been cultured in serum plus RA and iCRT3 were able to form colonies that were greater in number (approximately 6.5-fold) and size (approximately twofold) relative to those cultured in serum plus RA and DMSO when reintroduced to self-renewing conditions (serum plus LIF, without iCRT3; Fig. 3 G). Moreover, the cells cultured with DMSO during RA treatment were severely deficient in their ability to form EBs compared with the cells exposed to iCRT3 (Fig. 3 H). Taken together, these observations suggest that although stabilization of β-catenin by Wnt3a improves Nanog-GFP expression, blocking its TCF-mediated transcriptional activator function further enhanced pluripotency, because cells had homogeneous Nanog-GFP expression even when cultured in differentiation conditions. Moreover, although the reduction of β-catenin protein accelerates loss of pluripotency in cells undergoing spontaneous (serum) or induced differentiation (serum plus RA), its function in the maintenance of pluripotency is largely independent of β-catenin/TCF–dependent transcriptional activation.
Inhibition of β-catenin/TCF1 interaction by iCRT3 delays mESC differentiation independent of TCF3
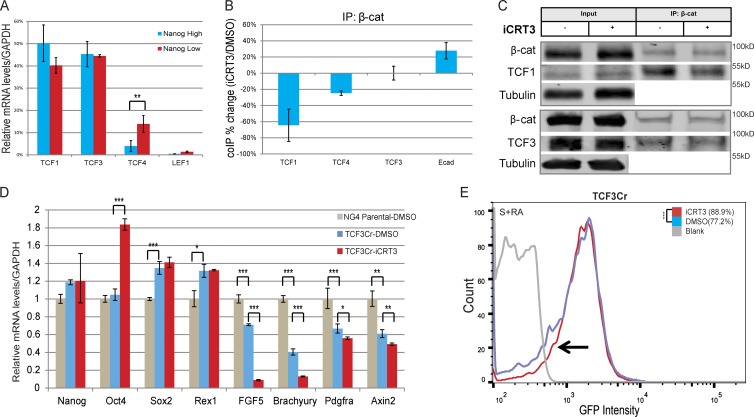
Differential levels of TCF family members have been shown to modulate cell fate choices in a context-dependent manner (Pereira et al., 2006; Mao and Byers, 2011; Lien et al., 2014). Therefore, we used coimmunoprecipitation (CoIP) to measure the influence of iCRT3 on individual β-catenin/TCF interactions, focusing on TCF1 and TCF3, the most significantly expressed TCF members in mESCs (Pereira et al., 2006; Fig. 4 A). As expected, iCRT3 significantly inhibited binding between β-catenin and TCF1 (∼70% reduction relative to DMSO control; Fig. 4, B and C). Surprisingly, we did not observe any significant changes in the interaction between β-catenin and TCF3 (Fig.4, B and C), although the TCF family of proteins shares high sequence similarity in the β-catenin binding domain (Graham et al., 2000). However, in agreement with our previously reported observations in human embryonic kidney 293 and colorectal cancer cells (Gonsalves et al., 2011), iCRT3 reduced β-catenin’s binding to TCF4 (∼25% reduction; Fig. 4 B and Fig. S3 A), which has been shown to be up-regulated during differentiation, although it is expressed at lower levels in self-renewing mESCs (Pereira et al., 2006; Fig. 4 A). Furthermore, we observed an improvement in β-catenin/E-cadherin complex formation (∼30% increase; Fig. 4 B and Fig. S3 A), which has been shown to correlate with ground-state pluripotency (Faunes et al., 2013).
Figure 4.
iCRT3 delays mESC differentiation by inhibiting β-catenin–TCF1 interaction independent of TCF3. (A) Bar graph showing relative mRNA levels for TCF/LEF factors from expression profiles of sorted NanogHigh and NanogLow mESCs, expressed as a percentage of the total pool of TCF/LEF transcripts. Mean ± SD of two replicates. (B) Bar graphs quantifying CoIP analysis from total cell lysate show the altered interactions of β-catenin in the presence of iCRT3 during RA-induced differentiation. Mean ± SD of three replicates. (C) Representative Western blots of three CoIP experiments from total cell lysates of differentiating cells (serum plus RA [S+RA]) maintained with iCRT3 for 48 h show that although β-catenin’s interaction with TCF1 is reduced relative to DMSO control, β-catenin/TCF3 complex formation is unchanged. Tubulin is used as loading control. (D) qPCR analysis of NG4 (parental line) and CRISPR-mediated TCF3-null cells maintained in RA-induced differentiating conditions (48 h) reveal reduced expression of differentiation markers after iCRT3 treatment. Mean ± SD of three replicates. (E) Flow cytometry of TCF3Cr mESCs during RA-induced differentiation reveal an increase in the Nanog-GFP–positive population of iCRT3-treated cells relative to the DMSO control. Representative data of three replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Interaction between β-catenin and TCF3 has been suggested to alleviate TCF3-mediated inhibition of the PTN (Blair et al., 2011; Wray et al., 2011; Shy et al., 2013). To confirm that the effect of iCRT3 is not driven by β-catenin/TCF3 interaction, we knocked out TCF3 (Fig. S3 B) in NG4 cells using the CRISPR/Cas9 system (NG4-TCF3Cr), which resulted in expected reduction of ESC differentiation (Fig. 4 D). When the TCF3-null mESCs were exposed to RA in the presence of iCRT3, flow cytometry revealed an approximately 12% gain in the GFP-positive population of cells relative to the DMSO control (highlighted by the arrow in Fig. 4 E). This was further supported by qPCR analysis, which revealed that iCRT3 enhanced Oct4 expression and reduced levels of differentiation markers in TCF3-null mESCs grown with RA (Fig. 4 D). In summary, these results suggest that selectively inhibiting β-catenin’s ability to activate transcription with TCF1 reduces mESC differentiation without altering β-catenin/TCF3 interaction. These findings also suggest that β-catenin has different affinities for individual TCF proteins (Graham et al., 2000, 2001; Sun and Weis, 2011) and highlights the need for renewed investigations into the structural basis for interactions between β-catenin and its protein partners.
Loss of TCF1 reduces differentiation and enhances expression of pluripotency markers in mESCs
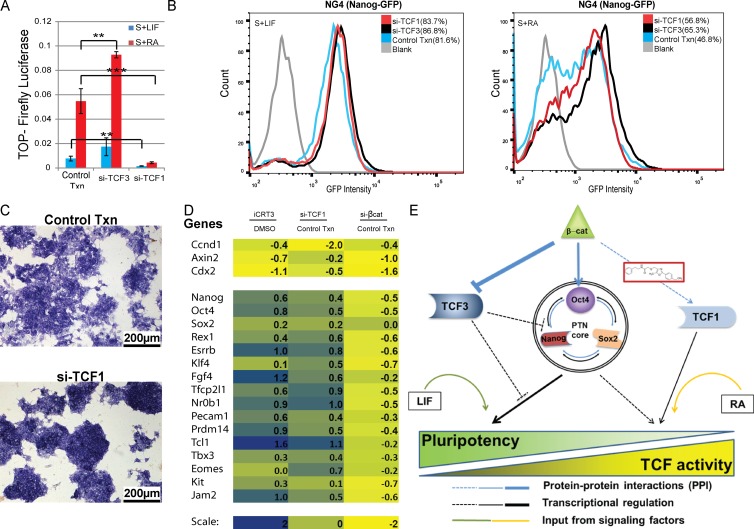
A noteworthy study by Yi et al. (2011) previously showed that simultaneous overexpression of TCF1 and β-catenin proteins up-regulates Nanog-Luciferase expression in addition to the TOPFlash reporter during Wnt stimulation of self-renewal in mESCs. In contrast, Aulicino et al. (2014) reported no significant loss of pluripotency upon loss of TCF1. To specifically test whether TCF1 is required for self-renewal, we performed RNAi-mediated knockdown of TCF1 and assayed levels of pluripotency markers, TCF reporters, and target genes. As expected, similar to iCRT3 treatment (Fig. 3 C), loss of TCF1 significantly reduced TOPFlash reporter activity, whereas si-TCF3 resulted in activation of Wnt signaling (Fig. 5 A). Concomitantly, TCF1 loss resulted in higher Nanog-GFP expression, suggesting reduced differentiation (Fig. 5 B). Furthermore, bright-field microscopy of si-TCF1–treated cells maintained more compact colony morphology and higher AP expression relative to control cells (Fig. 5 C). In addition, mRNA profiling confirmed that knockdown of TCF1 in mESCs increased expression of pluripotency markers, whereas differentiation markers were reduced (Fig. S3 C).
Figure 5.
Loss of TCF1 reduces mESC differentiation. (A) Normalized TOPFlash activity showing a marked reduction in reporter expression upon treatment with si-TCF1. Positive control cells treated with si-TCF3 have relatively higher TOPFlash activity. Mean ± SD of three replicates. (B) Flow cytometry of cells maintained in serum plus LIF (S+LIF; left) and serum + RA (S+RA; right) for 48 h, showing that similar to si-TCF3–treated cells, si-TCF1 tends to enhance Nanog-GFP expression. (C) Bright-field image showing that RNAi-mediated loss of TCF1 delays differentiation of RA-treated cells, as indicated by their higher AP expression and more compact colony morphology. Resized original images shown are from a single representative experiment out of three replicates. (D) Heat map plot showing relative mRNA expression levels of TCF target genes (top) and pluripotency-associated genes (bottom) upon iCRT3 treatment, or siRNA knockdown of TCF1 or β-catenin compared with DMSO or transfection of the Scrambled control, respectively. (E) Model depicting proposed mechanism for how the Wnt pathway regulates pluripotency and differentiation of mESCs. β-Catenin can alleviate the repressive effect of TCF3 on the transcriptional output of the PTN core, and it can also prevent TCF3 from binding to Oct4 and inhibiting transcription of pluripotency-associated genes. Our results suggest that β-catenin’s interaction with TCF1 activates transcription of differentiation promoting target genes. Although inhibition of β-catenin/TCF1 interaction by iCRT3 or loss of TCF1 does not affect β-catenin/TCF3 interaction, it reduces Wnt target gene expression and delays mESC differentiation. iCRT3 treatment also enhances the formation of β-catenin/Oct4 complex, which has been previously correlated to ground-state pluripotency. In addition, extrinsic signaling factors such as LIF and RA promote pluripotency and differentiation, respectively. **, P < 0.01; ***, P < 0.001. Txn, transfection.
Next, we tested wether si-TCF1 had any effect on RA-induced differentiation. qPCR analysis revealed that TCF1 delayed the loss of expression of pluripotency genes such as Rex1, Esrrb, and Oct4 after the addition of RA (Fig. S3 D). In addition, activation of the lineage marker FGF5 was reduced (Fig. S3 D). Comparison of expression profiles of genes known to have a role in self-renewal indicated that both iCRT3 and si-TCF1 treatment resulted in up-regulation of pluripotency-associated genes relative to their respective controls, even under RA-induced differentiation (Fig. 5 D), Alternatively, although si-βcat resulted in loss of TCF target genes similar to iCRT3 and si-TCF1, it also reduced expression of pluripotency markers. (Fig. 5 D). Furthermore, cluster analysis of transcriptome data validated that TCF1-knockdown cells have expression patterns that resemble an enhanced “stem” signature similar to NanogHigh or iCRT3-treated mESCs, suggesting that cells lacking TCF1 are resistant to differentiation (Fig. S3 E).
To test how β-catenin’s interactions with TCF3 and Oct4, which were previously shown to enhance pluripotency (Kelly et al., 2011; Wray et al., 2011; Yi et al., 2011; Zhang et al., 2013), are altered upon TCF1 loss, we performed CoIP from total lysates of control and si-TCF1–treated cells. Western blots revealed that similar to iCRT3 treatment, TCF1 loss did not affect the interaction between β-catenin and TCF3 (Fig. S3 F). However, we observed a significant increase in β-catenin/Oct4 interaction, which has been suggested to be a key TCF-independent mechanism to promote mESC pluripotency (Kelly et al., 2011; Faunes et al., 2013; Muñoz Descalzo et al., 2013). In conclusion, our results indicate that β-catenin/TCF1–mediated transcription promotes differentiation and that blocking this interaction by knocking down TCF1 results in enhanced pluripotency similar to iCRT3-treated cells.
Our findings lead us to suggest the following model (as represented in Fig. 5 E), wherein endogenous β-catenin promotes self-renewal in vitro by both buffering TCF3’s repressive role as well as through its TCF-independent interactions with Oct4, which may fortify regulation of the PTN core. However, β-catenin/TCF1–mediated transcriptional activation coincides with and may promote spontaneous and directed differentiation, whereas its inhibition enhances functional pluripotency of cultured mESCs. In recent years, inhibitor cocktails have emerged as powerful tools to achieve enhanced pluripotency for cultured stem cells without genetic manipulation. However, because stem cells treated with these cocktails exhibit a broad range of changes, including epigenetic modifications and signaling pathway activity, significant future work is required before these inhibitors can be used for patient-specific applications (Zhang et al., 2012; Wu et al., 2013). Notably, this study highlights the usefulness of targeted small molecules such as iCRT3, which can precisely modulate and thereby query the function of specific protein–protein interactions of β-catenin without significantly altering its protein levels. Furthermore, our observations that iCRT3 has similar effects on TCF and pluripotency reporters in various serum-free conditions (unpublished data) provide an important future direction to use analogous approaches for further dissecting the molecular interactions that govern ground-state pluripotency. These observations could have important implications in understanding how context-dependent functions of β-catenin in nuclear transcription and cell adhesion may regulate stem cell characteristics, especially in Wnt-relevant cancers, thereby enabling the development of novel tools to specifically target drug-resistant cancer stem cells.
Materials and methods
Cell lines
Nanog-GFP (NG4) mESCs and subsequent derived lines used in this study are transgenic for a construct coding for Hygromycin-EGFP, inserted in exon 1 of Nanog, resulting in a Nanog-Hygro-GFP fusion gene product (a gift from I. Lemishka, Icahn School of Medicine, New York, NY; Schaniel et al., 2009). For monitoring endogenous β-catenin/TCF–dependent transcription, the Nanog-GFP cells were transduced with a lentiviral expression vector for 7x-TOP-Firefly Luciferase (7FTP plasmid 24308; Addgene; Fuerer and Nusse, 2010) and selected using Puromycin to establish the NG4-TOPluc line. In addition to NG4, TNGA (Nanog reporter) and E14Tg2A (Faunes et al., 2013), Rex1-GFP, and CBA (Ying et al., 2008; Marks et al., 2012) cells were used for long-term culture with iCRT3 (Fig. 2) and were kind gifts from Austin Smith (University of Cambridge, Cambridge, England, UK). For generating the mESC line with CRISPR/Cas9-mediated knockout of TCF3, a 20-bp guide sequence targeting the first exon of TCF3 was identified using the Zhang Laboratory MIT CRISPR Design Tool. Complementary synthetic oligonucleotides with 5′ overhangs (5′-CACCGGTCGTCCCTGGTCAACGAAT-3′ and 5′-AAACATTCGTTGACCAGGGACGACC-3′; Thermo Fisher Scientific) were annealed and ligated into the PX459 vector (48139; Addgene) as described by Cong et al. (2013). The ligation mixture was transformed into One Shot Stbl3 chemically competent cells (Life Technologies), plasmid DNA was extracted (Plasmid Maxi kit; Qiagen) from the clonal colony, and the sequence was verified (GENEWIZ). 100,000 NG4 cells were reverse transfected in 6-well plates with 1 µg plasmid DNA using Lipofectamine 2000 (Life Technologies) and selected with 2 ng/ml Puromycin (Life Technologies) for 48 h. Surviving cells were sorted using a BD MoFlo XDP into 96-well plates, one cell per well, and clones were expanded and screened for absence of TCF3 protein by Western blot analysis to identify null cell lines. The NG4-TCF3Cr9 (TCF3Cr) clonal line was used for further experiments described in Fig. 4.
Culture conditions
mESCs used for this study were maintained in feeder-free conditions in standard ESC Complete media containing DMEM high glucose (Sigma-Aldrich) supplemented with 10% serum (ES Cell FBS; Gibco), LIF (Esgro-LIF; Millipore), Glutamax (Life Technologies), and MEM Nonessential Amino Acid Solution (ATCC; protocols were adapted from Faunes et al., 2013). Healthy cells were maintained with daily media changes and regular passaging every 48–72 h at a proportion of 1:10 on 0.1% gelatin-coated (Millipore) plates after dissociation using TrypLE Express (Life Technologies). For differentiation using RA (Sigma-Aldrich), cells were plated in Complete medium with 0.5 µM RA in the absence of LIF for 48 h. For long-term cultures, cells were plated in limiting dilutions in 6- or 96-well plates for multiple passages (14 d) in stem conditions (serum plus LIF) with DMSO or iCRT3, with daily media changes. AP staining was performed for every passage to monitor the relative pluripotency levels. Small molecules used include 10 µM iCRT3 (ChemDiv) and 1 µM XAV939 (Sigma-Aldrich), which were diluted with DMSO. L-Wnt3a and control L cells were gifts from R.T. Moon (University of Washington, Seattle, WA).
RNAi-mediated knockdowns
0.05 µM (final concentration) of pools for Silencer Select siRNA for respective target genes (Life Technologies) and Scrambled siRNA (Silencer Negative Control No. 1 siRNA; Life Technologies) for control transfection were transfected using 0.3 µl Lipofectamine 2000 (Thermo Fisher Scientific) for 10,000 cells per 100-µl transfection volume in 96-well format. After 72 h of knockdown, cells were further processed as per the experimental design for reporter assays, flow cytometry, or RNA/protein extractions (please refer to the following methods). Transfections for knockdowns in functional analysis experiments and for flow cytometry were performed with scaled-up volumes used for multiwell plates (6-well plates, 2 ml; 12-well plates, 1 ml; or 24-well plates, 0.5 ml). 24 h after transfection, ESCs were treated with 48 or 72 h of RA (0.5 µM) to induce differentiation for experiments in which the effect on RA-mediated differentiation was assayed.
RNA isolation, cDNA synthesis, and qPCR analysis
Nanog-GFP cells were cultured with LIF (serum plus LIF) or RA (serum plus RA) for 48 h (in 10-cm dishes) before flow cytometry–based separation of top 30% NanogHigh and bottom 30% of NanogLow for both media conditions, followed by RNA isolation. Snap-frozen pieces of teratoma were used for RNA isolation to study germ layer marker expression using qPCR. RNA isolation was performed using TRIzol reagent (Life Technologies) from cells cultured in 6-well plates. The aqueous phase containing RNA was collected and cleaned using an RNA cleanup kit (Qiagen) and was eluted in 30–50 µl RNase-free water. RNA was quantified using a Nano-drop and 1–2 µg total RNA was used for cDNA preparation using a high-capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed per the manufacturer’s protocol using Brilliant II SYBR green master mix with low Rox (Agilent) on an Mx3005p qPCR system (Agilent). Relative gene expression was normalized to readings for GAPDH for individual samples and analysis was performed as per the ΔΔCt method (Zhang et al., 2010) using Microsoft Excel. Refer to Table S1 for primers used for qPCR.
RNA sequencing and primary data processing
For RNA sequencing (RNA-seq) from pluripotent and differentiating cells, Nanog-GFP cells were cultured with LIF (serum plus LIF) or RA (serum plus RA) for 48 h (in 10-cm dishes) before flow cytometry–based separation of the top 30% NanogHigh (from LIF) and bottom 30% of NanogLow (from RA), followed by RNA isolation. To understand endogenous β-catenin and TCF1’s role in maintaining pluripotency, we performed expression profiling from Nanog-GFP mESCs which were treated with iCRT3 or DMSO, or transfected with si-βcat, si-TCF1, or Scramble, under RA-induced differentiation conditions (48 h, serum plus RA in 6-well plates). All experiments were performed in duplicate or triplicate (for si-TCF1 and its complementary control transfection) and RNA-seq was performed by LC Sciences and the New York University Genome Technology Center. Raw sequence reads were mapped to the mouse genome (version mm10) with Bowtie (version 0.12.9; Langmead et al., 2009) using the default parameters. Reads between 5.1 million and 27.9 million were aligned per library. RNA-seq data sets were also processed through TopHat (version 2.0.8) with the following settings: -a 10 -g 20, using the mm10 refseq gene annotation as shown by Trapnell et al. (2009). TopHat results were then pipelined to Cufflinks (version 2.1.0) with default settings (Trapnell et al., 2010). Absolute read counts for annotated genomic features were computed using the htseq-count script from the HTSeq (version 0.5.4p2) software suite with the following parameters: –stranded = no–mode = union.
Differential gene expression, GSEA, and statistical analysis
Differential expression analysis for RNA-seq data was performed using DESeq (version 1.14.0)/DESeq2R/Bioconductor with default parameters (Anders and Huber, 2010). We evaluated the enrichment of selected gene sets in RNA-seq differential expression data using the GSEA method (Subramanian et al., 2005) and its R implementation (released by the Broad Institute). For generating differential expression for each experiment, all genes were ordered based on log2 fold change. For all further analysis, we filtered out consistently low abundance transcripts, by only considering transcripts that have a fragments per kilobase of transcript per million mapped reads value of ≥1 in at least one of the 12 RNA-seq experiments we performed. Gene sets were defined as follows. The pluripotency gene set comprised 703 total genes, which were significantly up-regulated in NanogHigh cells when maintained in serum plus LIF, with an adjusted P < 0.01 and log2(fold change) values > 0 (as reported by DESeq), when comparing self-renewing NanogHigh (serum plus LIF) with differentiated NanogLow (serum plus RA) cells. The differentiation gene set comprised 777 total genes, which were up-regulated in NanogLow mESCs after 48 h of RA-induced differentiation when comparing self-renewing NanogHigh (serum plus LIF) with differentiated NanogLow (serum plus RA) cells. All experiments were conducted with biological replicates as described in the respective figure legends, and results where relevant are presented as means ± SD. The highlighted statistical significance represents P values generated using the Student’s t test (using Microsoft Excel or GraphPad) or DESeq (for RNA-seq data).
EB formation, cardiomyocyte differentiation, and teratoma assay
For the EB formation assay, 500 cells were added to each well in 96-well ultra-low attachment plates (Corning). Cells were plated in standard ESC serum medium without LIF or inhibitors. For individual experiments, 36 biological replicates were set up for each condition. EBs were collected from individual wells using a cut tip and were added to a 6-well plate for (Corning) for visualization and imaging after 5 d. To test the differentiation capacity of the mESCs after iCRT3 treatment, the cells were first cultured with DMSO or iCRT3 (10 µM) for 14 d, followed by EB formation (in absence of inhibitors). Representative EBs collected from individual wells were pooled in6-well plates and imaged to show that cells maintained in iCRT3 for 14 d form EBs similar to those formed from DMSO-treated cells (Fig. S1 B). RNA was extracted from EBs for qPCR analysis of differentiation markers. For generation of spontaneously beating cardiomyocytes, after DMSO/iCRT3 treatment, cells were allowed to form EBs (in absence of inhibitors) that differentiate into beating cardiomyocytes (protocol modified from Wang and Yang, 2008). To test how iCRT3 functionally maintained pluripotency during differentiation, mESCs were treated with DMSO or iCRT3 in serum plus RA for 48 h, followed by replating 500 cells per well in a 96-well ultra-low attachment plate for EB formation (representative EBs are shown in Fig. 3 H). For the teratoma assay, mESCs were cultured with DMSO or iCRT3 for 15 d in stem conditions (serum plus LIF). After long-term culture, the cells were dissociated from the culture plates, washed, and then resuspended in cold PBS before injections. Approximately 2.5 × 105 DMSO– and iCRT3–treated cells were injected into opposite abdominal flanks of eight male Balb/C nude mice (aged 6 wk). Teratomas were dissected from the mice 23 d after the injection and were immediately snap frozen for further processing. Hematoxylin and eosin–stained teratoma sections were generated and analyzed by the ICMB Histopathology core (A*Star) for evaluation of germ layer differentiation. Immunostaining of teratoma sections for germ layer markers was performed using rabbit anti–AFP (1:100; Dako/Agilent), rabbit anti–Myosin Heavy Chain (1:200; Abcam), and mouse anti–βIIITubulin (1:200; Thermo Fisher Scientific). Alexa Fluor 488 (ThermoFisher Scientific) for respective primary antibodies and DAPI were used for imaging of the mounted tissue in VectaShield mounting solution.
Immunostaining, colony formation, and alkaline phosphatase staining
For immunostaining, cells were fixed in 4% formaldehyde (Electron Microscopy Sciences) for 20 min, washed three times with 1× PBS, permeabilized, blocked with blocking buffer (0.1% Triton X-100, 1% BSA, and 5% normal goat serum in Dulbecco’s PBS at room temperature for 20 min, and immunostained with rabbit anti–GFP antibody (Invitrogen; 1:500: 4°C overnight), anti–rabbit IgG Alexa 488 goat antibody (Invitrogen; 1:1,000; 2 h at room temperature), and DAPI. After staining, cells were stored in 1× PBS and were then imaged with the ArrayScan (Cellomics Inc.; using a 20× Plan Apo lens with 0.45 NA; Olympus) high-content imaging system fitted with an ORCA-ER 1.00 camera in autofocus mode. Target Activation software (version 4.0 protocol on Cellomics HTS software) was used to measure mean GFP expression (exposure time 0.004 s) of at least 1,000 DAPI-stained (exposure time 0.0025 s) cells per replicate.
For the colony counting assay described in Fig. 2 E, 600 E14Tg2A or CBA cells in serum plus LIF were added to 6-well plates. After 8–12 h, media were refreshed for the next 5 d. The resulting colonies were then fixed and stained for AP expression using an AP leukocyte kit (86R-1KT; Sigma-Aldrich). The stained colonies were mounted in 1× PBS and imaged using a MOTIC A31 inverted microscope using a 4× lens (4×/0.10 working distance 23.5) and a GXCAM (pixel size 2.615 µm; GTVISION) C mounted with a 0.5× lens using TSView7 Image capture software. For each condition, and colonies were manually scored for presence and color (white, stained [AP+], or mixed [colonies containing both white and stained cells]). To test whether iCRT3 treatment delayed RA-induced differentiation, NG4 cells were maintained for 48 h in differentiating conditions (serum plus RA) with DMSO or iCRT3 (6-well plates), followed by counting and replating them in limiting dilutions in stem conditions (serum plus LIF with no inhibitors) for an additional 48 h. For the colony formation assay (Fig. 3 G), dissociated cells were counted using Countess (Life Technologies) and plated at clonal densities in gelatin-coated 96-well plates. Four replicate experiments were performed with 12 wells per condition. To test the colony forming efficiency, media were removed and cells were washed in 1× PBS, followed by 1 min in fixation solution (20 ml nuclease-free water [NFW], 30 ml acetone with 400 µl citrate concentrate [Sigma-Aldrich], volumes scaled as required). After fixation, cells were washed in 1× PBS followed by incubation at 37°C in staining solution (48 ml NFW, 2 ml Naphthol AS-MX phosphate alkaline solution [Sigma-Aldrich], and one complete capsule of Fast Blue RR salt [Sigma-Aldrich]). Staining was monitored for control conditions under a bright-field microscope and was stopped with washes of NFW, and stained cells were stored with 1× PBS at 4°C before imaging. Bright-field images (imaged at autoexposure set up for control well for every experiment) for ESC colonies stained with AP (Fig. 2 D) were acquired using a Nikon TE2000-E microscope using a Coolsnap HQ2 camera with 20× PlanApo (0.75 NA/1.0 mm WD) lenses. Automated quantitative image analysis of AP-stained colonies (Fig. 3 G) was performed using the JOBS module (NIS Elements; Nikon) and TE2000-E microscope (Nikon) using a Coolsnap HQ2 camera with 10× PlanApo (0.45 NA/4.0 mm WD).
Flow cytometry
Cultured cells (two or three replicates in 24-well formats) were washed with 1× PBS and dissociated with Accutase (Millipore). Cells were spun down and resuspended in 1× PBS with 3% filtered FBS and DAPI for dead cell exclusion and collected in 5-ml polystyrene tubes with cell strainer (BD Falcon). Flow cytometry analysis was performed using a BD LSRII machine for at least 10,000 live cell events per sample (DAPI negative) using the UV laser. Doublets were excluded from the live cell analysis using side and forward scatter before measuring GFP expression using the 488-nm laser. The CCE mESC line (gift from I. Aifantis, New York University School of Medicine, New York, NY; Schaniel et al., 2009) was used as a blank for background correction for flow cytometry experiments. Flow cytometry of TNGA and Rex1-GFP lines (Fig. 2, A and B) were performed by Pleckstrin homology as described above, using the E14Tg2A cell line as a blank for background (Faunes et al., 2013). Raw data were reanalyzed and representative histogram plots were generated using FlowJo software.
Luciferase assays
For β-catenin/TCF transcriptional activity reporter assays, 10,000 mESCs were reverse transfected in 96-well plates using Lipofectamine 2000 with 25 ng 14× TOPFlash (Firefly Luciferase) and 25 ng SV-40 Renilla Luciferase (for normalization of cell number and transfection efficiency) reporter constructs. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). For measuring TOP activity in NG4-TOPluc ESCs with virally integrated 7× TOPFlash, the Firefly Luciferase readings were normalized for cell number using Presto Blue Cell Viability reagent (Life Technologies) as per the manufacturer’s protocol.
Biochemistry
Total protein lysates were collected from cells (cultured in 10-cm dishes), which were fixed with 1% PFA for 15 min followed by quenching with 0.1 M glycine for additional 10 min. After quenching, the fixed cells were washed twice with ice-cold 1× PBS and collected using cell lifters. The total protein lysates were collected by resuspending the cells with standard RIPA lysis buffer and were sonicated on ice five times for 10 s, with constant pulses separated by 15-s pauses. Insoluble particles were cleared by centrifugation (10,000 g, 15 min at 4°C). Protein concentrations were determined using the BCA Protein Assay kit (Pierce) and identical volumes at a concentration of 1 µg/µl were used for all CoIP experiments. CoIP was performed for total cell lysates using mouse anti–β-catenin (15B8; Sigma-Aldrich) antibody using the μMACS Protein A/G Kit according to the manufacturer’s instructions (Miltenyi Biotech).
For Western blotting, identical protein quantities were loaded onto precast 4%–15% Tris glycine gradient cells (BioRad) for electrophoretic analysis. Proteins were blotted onto nitrocellulose membranes for immunoblot analysis using a standard protocol. Primary antibodies and dilutions used were mouse anti–β-catenin (15B8, 1:1,000; Sigma-Aldrich), rabbit anti–TCF3 (1:500; a gift from B. Merrill, University of Illinois, Chicago, IL; Yi et al., 2011), rabbit anti–TCF4 (C48H11, 1:500; Cell Signaling), rabbit anti–TCF1 (C63D9, 1:250; Cell Signaling), mouse anti–E-cadherin (1:1,000; BD Transduction 36/E-cadherin), mouse anti–Oct3/4 (sc-5279, 1:250; Santa Cruz Biotechnology, Inc.), and mouse anti–tubulin (T9026, 1:1,000; Sigma-Aldrich). Infrared–conjugated anti–mouse (800 nm) and anti–rabbit (680 nm) antibodies were used for secondary detection. Western blots were imaged and intensities of individual protein bands were quantified using the Odyssey Infrared Imaging System (LiCOR). Mean and standard deviations to represent CoIP experiments reflect two or three biological replicate experiments, as mentioned in the respective figure legends. For analysis of CoIP, raw intensities measured by the LiCOR Odyssey, which represent the quantities of partner proteins, were normalized to the amount of target protein pulled down (for the respective lane) and compared across conditions (SDs for percent change in CoIP from replicate experiments are shown in the respective bar plots). All representative Western blots shown and used for quantification, which corresponded to individual experiments, were run on the same gel. Further protocol details are available upon request.
Online supplemental material
Fig. S1 A supports data presented in Fig. 1 (expression profiles for pluripotency and differentiation markers in sorted NanogHigh and NanoLow subpopulations from serum plus LIF and serum plus RA conditions) and panels B–E additionally show that long-term treatment with iCRT3 (Fig. 2) does not inhibit differentiation capability of mESCs. Expression data in Fig. S2 support the GSEA plot shown in Fig. 3 F and this figure also contains an experimental outline for data presented in Fig. 3 (G and H). Fig. S3 supports data presented in Figs. 4 and 5 and shows that loss of β-catenin/TCF1 interaction by using iCRT3- or siRNA-mediated knockdown of TCF1 results in reduced differentiation and correlates with enhanced β-catenin/Oct4 interaction. Table S1 lists all primers and sequences used for qPCR analysis. Table S2 shows the alphabetically organized identification numbers used for GSEA for RNA-seq data (related to Fig. 3) and functional enrichment analysis for differentially expressed genes in NanogHigh and NanogLow mESCs. Table S3 lists the top 200 significantly differentially expressed (refer to padj values) genes organized by their log2 fold change in the comparison of NanogHigh versus NanogLow and the corresponding mean log2 fold change values for iCRT3, si-TCF1, and si-βcat relative to respective controls, used to plot the clustered heat map in Fig. S4 B. Online supplemental material is available online at http://www.jcb.org/cgi/content/full/jcb.201503017/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201503017.dv.
Supplementary Material
Acknowledgments
We thank A. Smith, B. Merrill, I. Aifantis, C. Basilico, L. Dailey, A. Mansukhani, B. Doble, E. Hernando, and I. Lemishka for cell lines and reagents; and U. Basu-Roy, S. Buckley, P. Cowin, M. Schober, J. Brown, M. Murtha, and members of the RD Laboratory and New York University (NYU) Langone Medical Center Cancer Institute and Stem Cell Program for helpful comments and discussions. We also thank members of the NYU research support groups, especially RNAi Core (Director C. Yun), Cytometry and Cell Sorting Core (Director P. Lopez), Genome Technology Center (Director A. Heguy), and High Performance Computing Facility (Associate Technology Director E. Peskin) for significant help with experimental setup and data processing. Finally, we thank the Institute for Cellular and Molecular Biology Histopathology core (A*Star; Senior Veterinary Pathologist M. Al-Haddawi) for histopathological evaluation of teratoma samples.
This work was supported by the the National Institutes of Health (grant 1R01CA155125-01 to R. DasGupta and NCI-1T32CA160002 to M. Murphy), GIS Institutional Funds (GIS/14-ARB3207), NYSTEM (Shared Facilities for Stem Cell Research grant N09S-012, postdoctoral training fellowship C026880 to S.S. Chatterjee, and C026880 to M. Murphy), NYU RNAi Core (screening subsidy grant to R. DasGupta), Wellcome Trust (to A.M. Arias and P. Hayward), the National Center for Advancing Translational Sciences (grant UL1 TR00038), and National Institutes of Health (to NYU OCS Cores and Shared Resources).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CoIP
- coimmunoprecipitation
- EB
- embryoid body
- ESC
- embryonic stem cell
- GSEA
- gene set enrichment analysis
- iCRT3
- inhibitor of β-catenin–responsive transcription
- LIF
- leukemia inhibitory factor
- mESC
- mouse embryonic stem cell
- NFW
- nuclease-free water
- PTN
- pluripotency transcriptional network
- qPCR
- quantitative PCR
- RA
- retinoic acid
- RNA-seq
- RNA sequencing
- TCF
- T cell factor
- WntCM
- Wnt3a-conditioned media
References
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S.J., Stappert J., Bauer A., Kispert A., Herrmann B.G., and Kemler R.. 2000. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech. Dev. 91:249–258. 10.1016/S0925-4773(99)00309-3 [DOI] [PubMed] [Google Scholar]
- Aulicino F., Theka I., Ombrato L., Lluis F., and Cosma M.P.. 2014. Temporal perturbation of the Wnt signaling pathway in the control of cell reprogramming is modulated by TCF1. Stem Cell Reports. 2:707–720. 10.1016/j.stemcr.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair K., Wray J., and Smith A.. 2011. The liberation of embryonic stem cells. PLoS Genet. 7:e1002019 10.1371/journal.pgen.1002019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., and Smith A.. 2007. Nanog safeguards pluripotency and mediates germline development. Nature. 450:1230–1234. 10.1038/nature06403 [DOI] [PubMed] [Google Scholar]
- Cole M.F., Johnstone S.E., Newman J.J., Kagey M.H., and Young R.A.. 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22:746–755. 10.1101/gad.1642408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson K.C., Adams A.M., Goodson J.M., McDonald C.E., Potter J.C., Berndt J.D., Biechele T.L., Taylor R.J., and Moon R.T.. 2012. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA. 109:4485–4490. 10.1073/pnas.1118777109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbaillets I., Ziegler U., Groscurth P., and Gassmann M.. 2000. Embryoid bodies: an in vitro model of mouse embryogenesis. Exp. Physiol. 85:645–651. 10.1111/j.1469-445X.2000.02104.x [DOI] [PubMed] [Google Scholar]
- Faunes F., Hayward P., Descalzo S.M., Chatterjee S.S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A.K., Dasgupta R., and Arias A.M.. 2013. A membrane-associated β-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 140:1171–1183. 10.1242/dev.085654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A., Piliszek A., Tian G., Aho R.J., Dufort D., and Hadjantonakis A.K.. 2010. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev. Biol. 10:121 10.1186/1471-213X-10-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerer C., and Nusse R.. 2010. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE. 5:e9370 10.1371/journal.pone.0009370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves F.C., Klein K., Carson B.B., Katz S., Ekas L.A., Evans S., Nagourney R., Cardozo T., Brown A.M., and DasGupta R.. 2011. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. USA. 108:5954–5963. 10.1073/pnas.1017496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T.A., Weaver C., Mao F., Kimelman D., and Xu W.. 2000. Crystal structure of a beta-catenin/Tcf complex. Cell. 103:885–896. 10.1016/S0092-8674(00)00192-6 [DOI] [PubMed] [Google Scholar]
- Graham T.A., Ferkey D.M., Mao F., Kimelman D., and Xu W.. 2001. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat. Struct. Biol. 8:1048–1052. 10.1038/nsb718 [DOI] [PubMed] [Google Scholar]
- Gropp M., Shilo V., Vainer G., Gov M., Gil Y., Khaner H., Matzrafi L., Idelson M., Kopolovic J., Zak N.B., and Reubinoff B.E.. 2012. Standardization of the teratoma assay for analysis of pluripotency of human ES cells and biosafety of their differentiated progeny. PLoS One. 7:e45532 10.1371/journal.pone.0045532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R., et al. . 2009. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 5:597–609. 10.1016/j.stem.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S., et al. . 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 461:614–620. 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., and Benvenisty N.. 2000. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 6:88–95. [PMC free article] [PubMed] [Google Scholar]
- Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., and Costantini F.. 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22:1172–1183. 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K.F., Ng D.Y., Jayakumaran G., Wood G.A., Koide H., and Doble B.W.. 2011. β-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell. 8:214–227. 10.1016/j.stem.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida C., Nakano H., Okamura D., Nakao K., Yonemura S., Belo J.A., Aizawa S., Matsui Y., and Matsuo I.. 2005. Canonical Wnt signaling and its antagonist regulate anterior-posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev. Cell. 9:639–650. 10.1016/j.devcel.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C, Pop M, Salzberg S.L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W.H., and Fuchs E.. 2014. Wnt some lose some: transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 28:1517–1532. 10.1101/gad.244772.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W.H., Polak L., Lin M., Lay K., Zheng D., and Fuchs E.. 2014. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat. Cell Biol. 16:179–190. 10.1038/ncb2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Lim B., and Ang L.T.. 2015. Ex uno plures: molecular designs for embryonic pluripotency. Physiol. Rev. 95:245–295. 10.1152/physrev.00001.2014 [DOI] [PubMed] [Google Scholar]
- Lyashenko N., Winter M., Migliorini D., Biechele T., Moon R.T., and Hartmann C.. 2011. Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat. Cell Biol. 13:753–761. 10.1038/ncb2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C.D., and Byers S.W.. 2011. Cell-context dependent TCF/LEF expression and function: alternative tales of repression, de-repression and activation potentials. Crit. Rev. Eukaryot. Gene Expr. 21:207–236. 10.1615/CritRevEukarGeneExpr.v21.i3.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., and Stunnenberg H.G.. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 149:590–604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B.J. 2012. Wnt pathway regulation of embryonic stem cell self-renewal. Cold Spring Harb. Perspect. Biol. 4:a007971 10.1101/cshperspect.a007971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz Descalzo S., Rué P., Faunes F., Hayward P., Jakt L.M., Balayo T., Garcia-Ojalvo J., and Martinez Arias A.. 2013. A competitive protein interaction network buffers Oct4-mediated differentiation to promote pluripotency in embryonic stem cells. Mol. Syst. Biol. 9:694 10.1038/msb.2013.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., and Smith A.. 2009. Naive and primed pluripotent states. Cell Stem Cell. 4:487–492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Nowotschin S., and Hadjantonakis A.K.. 2010. Cellular dynamics in the early mouse embryo: from axis formation to gastrulation. Curr. Opin. Genet. Dev. 20:420–427. 10.1016/j.gde.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., and Varmus H.. 2012. Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J. 31:2670–2684. 10.1038/emboj.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates A.C., Morelli L.G., and Ares S.. 2012. Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development. 139:625–639. 10.1242/dev.063735 [DOI] [PubMed] [Google Scholar]
- Ogawa K., Nishinakamura R., Iwamatsu Y., Shimosato D., and Niwa H.. 2006. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem. Biophys. Res. Commun. 343:159–166. 10.1016/j.bbrc.2006.02.127 [DOI] [PubMed] [Google Scholar]
- Okita K., and Yamanaka S.. 2006. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr. Stem Cell Res. Ther. 1:103–111. 10.2174/157488806775269061 [DOI] [PubMed] [Google Scholar]
- Park M., and Shen K.. 2012. WNTs in synapse formation and neuronal circuitry. EMBO J. 31:2697–2704. 10.1038/emboj.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Yi F., and Merrill B.J.. 2006. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 26:7479–7491. 10.1128/MCB.00368-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon N., Oh K., Sylvestre J.R., Savory J.G., and Lohnes D.. 2007. Wnt signaling is a key mediator of Cdx1 expression in vivo. Development. 134:2315–2323. 10.1242/dev.001206 [DOI] [PubMed] [Google Scholar]
- Rudloff S., and Kemler R.. 2012. Differential requirements for β-catenin during mouse development. Development. 139:3711–3721. 10.1242/dev.085597 [DOI] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., and Brivanlou A.H.. 2004. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10:55–63. 10.1038/nm979 [DOI] [PubMed] [Google Scholar]
- Schaniel C., Ang Y.S., Ratnakumar K., Cormier C., James T., Bernstein E., Lemischka I.R., and Paddison P.J.. 2009. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 27:2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shy B.R., Wu C.I., Khramtsova G.F., Zhang J.Y., Olopade O.I., Goss K.H., and Merrill B.J.. 2013. Regulation of Tcf7l1 DNA binding and protein stability as principal mechanisms of Wnt/β-catenin signaling. Cell Reports. 4:1–9. 10.1016/j.celrep.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., and Smith A.. 2008. Capturing pluripotency. Cell. 132:532–536. 10.1016/j.cell.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla D.K., Schneider D.J., LeWinter M.M., and Sobel B.E.. 2006. wnt3a but not wnt11 supports self-renewal of embryonic stem cells. Biochem. Biophys. Res. Commun. 345:789–795. 10.1016/j.bbrc.2006.04.125 [DOI] [PubMed] [Google Scholar]
- Sokol S.Y. 2011. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development. 138:4341–4350. 10.1242/dev.066209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., and Weis W.I.. 2011. Biochemical and structural characterization of β-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1. J. Mol. Biol. 405:519–530. 10.1016/j.jmb.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao Y., Yokota T., and Koide H.. 2007. β-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 353:699–705. 10.1016/j.bbrc.2006.12.072 [DOI] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., and Nusse R.. 2011. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13:1070–1075. 10.1038/ncb2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L., and Salzberg S.L.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., and Basler K.. 2012. The many faces and functions of β-catenin. EMBO J. 31:2714–2736. 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R.T., Xu X., Yi F., Merrill B.J., and Cooney A.J.. 2010. Canonical Wnt/β-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells. 28:1794–1804. 10.1002/stem.502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Yang P.. 2008. In vitro differentiation of mouse embryonic stem (mES) cells using the hanging drop method. J. Vis. Exp. 17:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J., and Hartmann C.. 2012. WNTing embryonic stem cells. Trends Cell Biol. 22:159–168. 10.1016/j.tcb.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., and Smith A.G.. 2010. The ground state of pluripotency. Biochem. Soc. Trans. 38:1027–1032. 10.1042/BST0381027 [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., and Smith A.. 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13:838–845. 10.1038/ncb2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.L., Pandian G.N., Ding Y.P., Zhang W., Tanaka Y., and Sugiyama H.. 2013. Clinical grade iPS cells: need for versatile small molecules and optimal cell sources. Chem. Biol. 20:1311–1322. 10.1016/j.chembiol.2013.09.016 [DOI] [PubMed] [Google Scholar]
- Yi F., Pereira L., Hoffman J.A., Shy B.R., Yuen C.M., Liu D.R., and Merrill B.J.. 2011. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 13:762–770. 10.1038/ncb2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A.. 2008. The ground state of embryonic stem cell self-renewal. Nature. 453:519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ruschhaupt M., and Biczok. R.. 2010. ddCt method for qRT–PCR data analysis. http://www.bioconductor.org/packages/release/bioc/html/ddCt.html (accessed September 1, 2015).
- Zhang W.Y., de Almeida P.E., and Wu J.C.. 2008. Teratoma formation: A tool for monitoring pluripotency in stem cell research. In StemBook. Harvard Stem Cell Institute, Cambridge, MA. [PubMed] [Google Scholar]
- Zhang X., Peterson K.A., Liu X.S., McMahon A.P., and Ohba S.. 2013. Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells. 31:2667–2679. 10.1002/stem.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li W., Laurent T., and Ding S.. 2012. Small molecules, big roles — the chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci. 125:5609–5620. 10.1242/jcs.096032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Gan Q., Stokes A., Lassiter R.N., Wang Y., Chan J., Han J.X., Pleasure D.E., Epstein J.A., and Zhou C.J.. 2014. β-catenin regulates Pax3 and Cdx2 for caudal neural tube closure and elongation. Development. 141:148–157. 10.1242/dev.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.