3D structural analysis of a yeast [PSI+] prion model by correlative fluorescence and electron tomography reveals that prion aggregate structure depends on the levels of Hsp70 chaperones, the protein remodeling ATPase Hsp104, and the Hsp70 nucleotide exchange factor/disaggregase Sse1 (yeast Hsp110).
Abstract
Prions consist of misfolded proteins that have adopted an infectious amyloid conformation. In vivo, prion biogenesis is intimately associated with the protein quality control machinery. Using electron tomography, we probed the effects of the heat shock protein Hsp70 chaperone system on the structure of a model yeast [PSI+] prion in situ. Individual Hsp70 deletions shift the balance between fibril assembly and disassembly, resulting in a variable shell of nonfibrillar, but still immobile, aggregates at the surface of the [PSI+] prion deposits. Both Hsp104 (an Hsp100 disaggregase) and Sse1 (the major yeast form of Hsp110) were localized to this surface shell of [PSI+] deposits in the deletion mutants. Elevation of Hsp104 expression promoted the appearance of this novel, nonfibrillar form of the prion aggregate. Moreover, Sse1 was found to regulate prion fibril length. Our studies reveal a key role for Sse1 (Hsp110), in cooperation with Hsp104, in regulating the length and assembly state of [PSI+] prion fibrils in vivo.
Introduction
Protein quality control is essential for cellular function and viability (Dobson, 2003; Sontag et al., 2014; Wolff et al., 2014). The capacity of the quality control systems can be exceeded under certain conditions, resulting in the accumulation of misfolded proteins (Balch et al., 2008; Hartl et al., 2011). Protein misfolding and/or mislocalization are implicated in human neurodegenerative disease (Soto and Estrada, 2008; Aguzzi and Rajendran, 2009). In the amyloid diseases, the misfolded proteins associate into amyloid assemblies, formed by a self-templating β sheet–rich conformation (Carrell and Lomas, 1997; Fändrich, 2007). Prions, which occur in fungi and mammals, constitute a transmissible form of amyloid.
Cells have evolved diverse mechanisms to prevent and reverse protein misfolding and aggregation (Sontag et al., 2014; Wolff et al., 2014). In yeast, nonamyloid amorphous aggregates accumulate in various quality control compartments (Kaganovich et al., 2008; Wang et al., 2008; Gong et al., 2012; Escusa-Toret et al., 2013; Miller et al., 2015b). Deposits found in the nucleus (intranuclear quality control, previously described as juxtanuclear; Kaganovich et al., 2008) contain soluble, highly mobile, and ubiquitinated proteins that are likely destined for proteasomal degradation (Miller et al., 2015a). In contrast, other aggregates, such as prions, which may be more resistant to disaggregation and proteolysis, are sequestered into relatively immobile compartments for terminally aggregated proteins such as the insoluble protein deposit (Kaganovich et al., 2008). Many of the cellular factors that target the different aggregates to these functionally distinct compartments are known (Sontag et al., 2014).
Yeast prions provide a favorable model for assessing amyloid formation and its interactions with the cellular quality control network (Liebman and Chernoff, 2012). Yeast possess many prion proteins, although perhaps the best characterized is that of the prion form of Sup35, [PSI+] (Liebman and Derkatch, 1999; Serio et al., 1999; Tuite and Cox, 2006; Crow and Li, 2011). Sup35 consists of a C-terminal translation termination factor GTPase domain after the prion-determining N-terminal (N) and middle (M) domains. The N domain, essential for [PSI+] induction and propagation, is rich in glutamine and asparagine (Q/N) residues and constitutes the minimal prion/fibril-forming region (Glover et al., 1997; King et al., 1997; Paushkin et al., 1997; Baxa et al., 2011). The function of the charged M domain is unknown, although it may mediate an interaction with the heat shock protein Hsp104 disaggregase, a protein remodeling factor essential for yeast prion propagation (Helsen and Glover, 2012). Fibril assembly by the prion-forming domain sequesters soluble Sup35 into insoluble amyloid aggregates, impairing efficient cellular translation termination (Liebman and Chernoff, 2012).
The key molecular chaperones involved in processing and propagation of yeast prions are Hsp70 and its cofactors Hsp40, Hsp104, and Hsp110/Sse1 (Liebman and Chernoff, 2012; Tyedmers, 2012; Winkler et al., 2012a; Chernova et al., 2014; Sontag et al., 2014). Although the small heat shock proteins Hsp42 and Hsp26 can influence de novo [PSI+] prionogenesis (Duennwald et al., 2012), it is the Hsp70/Hsp40 system that is believed to target the prion fragmentation machinery in yeast. Growing evidence suggests that Hsp70s such as Ssa1 or Ssa2, together with Sis1 (Hsp40), initially bind prion aggregates and recruit Hsp104 for remodeling of the prion aggregates (Misselwitz et al., 1998; Higurashi et al., 2008; Tipton et al., 2008; Kampinga and Craig, 2010). Sse1 (Hsp110), a nucleotide exchange factor for Hsp70, regulates de novo [PSI+] induction and functions in prion propagation by modulating the nucleotide occupancy of Hsp70 (Fan et al., 2007; Kryndushkin and Wickner, 2007; Sadlish et al., 2008). In addition, Sse1 may promote prion nucleation as well as interact directly with prion aggregates (Bagriantsev et al., 2008; Sadlish et al., 2008).
Mammals lack an Hsp104 homologue, but they have a disaggregase system composed of Hsp70, Hsp40, and Hsp110 (Shorter, 2011; Rampelt et al., 2012). Furthermore, mammalian Hsp110 has been shown to reverse the toxicity associated with mutant superoxide dismutase 1 aggregation in an axonal transport model (Song et al., 2013).
For yeast prions, the balance between fibril self-assembly, growth, and chaperone-mediated fragmentation/disassembly determines the outcome of prion propagation in vivo. We therefore sought to determine the spatial relationship between Hsp70-related molecular chaperones and prion aggregates in a [PSI+] model. We used a well-characterized overexpression system consisting of a fusion of the Sup35 NM domain to YFP (NM-YFP) to amplify the prion aggregate structure in our strains (Tyedmers, 2012). This allowed us to use correlative light and cryoelectron microscopy to probe the conformation of NM-YFP prion aggregates in vivo. We used these organized [PSI+] deposits (Kawai-Noma et al., 2010; Tyedmers et al., 2010; Saibil et al., 2012) to investigate the cellular determinants of prion assembly and processing in yeast. Using individual chaperone deletions, we modulated the balance between assembly and disassembly of NM-YFP dot aggregates and revealed a novel, nonfibrillar form of prion aggregate. This work demonstrates a key role for Sse1 (Hsp110), in cooperation with Hsp104, in regulating the length and assembly state of [PSI+] prion fibrils in vivo.
Results
Correlative microscopy and electron tomography of a [PSI+] prion model
We exploited the well-characterized yeast [PSI+] prion expression model, NM-YFP, to visualize aggregates of [PSI+] in vivo. This was constitutively overexpressed in cells lacking the endogenous NM region of SUP35 (Tyedmers et al., 2010; Saibil et al., 2012), so that NM-YFP aggregates were propagated while leaving the endogenous Sup35p C-terminal translation termination domain free from toxic recruitment into the prion (Chernoff et al., 1992, 1993; Vishveshwara et al., 2009). The NM-YFP aggregates in our model system assembled into one large focus or “dot” structure (Fig. S1 A, top). To study the 3D structural organization of these dot aggregate species in cells, we used a combination of high-pressure freezing, freeze substitution, resin embedding, and ultramicrotomy to generate well-preserved stained sections for electron tomography. In parallel, we used cryoelectron tomography of unstained vitrified cell sections to confirm that the observed structures reflect the true assemblies in their native hydrated state (Al-Amoudi et al., 2004a,b). In both instances, the YFP fluorescence was maintained by the preparation procedures, enabling direct correlation with EM of the aggregates (Materials and methods). The NM-YFP dot aggregates formed ordered fibrillar arrays (Fig. 1 A and Fig. S1, B–E), in agreement with previous studies (Kawai-Noma et al., 2010; Tyedmers et al., 2010; Baxa et al., 2011; Saibil et al., 2012).
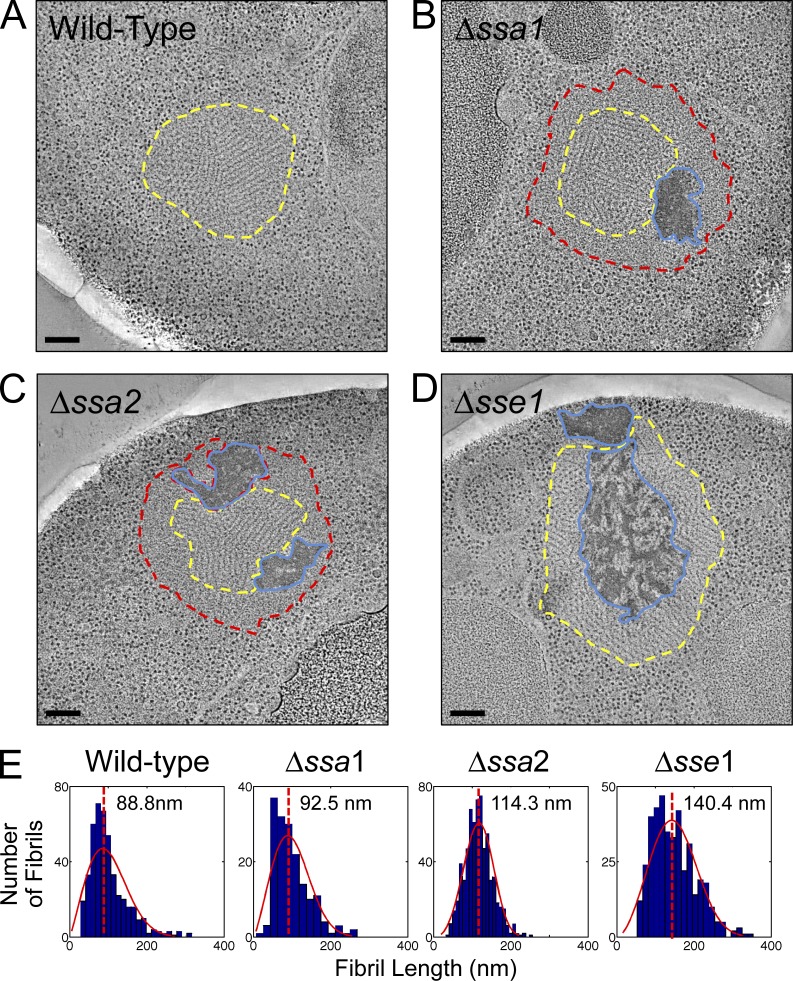
Figure 1.
Aggregate remodeling in chaperone knockout strains. Representative tomographic slices through reconstructions of NM-YFP dots in HM20-embedded cell sections from cells with a wild-type chaperone complement and those with Δssa1, Δssa2, and Δsse1 deletions, as indicated. Fibril-containing regions are outlined in yellow, unstructured perimeter zones are outlined in red, and dark amorphous aggregates are outlined in blue. These structures were visible in 100% of the aggregates of each respective strain and were from at least 20 independent NM-YFP dot observations (tomograms and projections). Bars, 200 nm. (A) Wild-type chaperone complement. The yellow outline around the fibril aggregate was manually traced by following the interface between the fibrils and the cytosolic ribosomes. (B) Δssa1. The yellow outline around the fibrils was manually traced at the interface between the fibrils and the unstructured peripheral layer. The red outline was traced at the interface between the unstructured zone and the cytosolic ribosomes. The blue outline was drawn around the perimeter of the dark amorphous material. (C) Δssa2. The red, yellow, and blue outlines were defined as in B. (D) Δsse1. The yellow outline was defined as in A and the blue outline was defined as in B. (E) Distribution plots of individual fibril lengths in tomograms of NM-YFP aggregates from wild-type, Δssa1, Δssa2, and Δsse1 cells. Fibrils were measured in at least three independent dot assemblies for each strain. The total number of fibrils was 450 for the wild type, 171 for Δssa1, 668 for Δssa2, and 407 for the Δsse1 aggregates. For each histogram, the number of bins was equal to the square root of the number of elements in the data set. A Weibull distribution was calculated for the data and plotted with each histogram.
Deletion of individual molecular chaperones causes aggregate remodeling
Transient inactivation of Hsp70 chaperones can influence the organization of [PSI+] assemblies (Saibil et al., 2012) and their association with chaperones (Winkler et al., 2012b). To systematically probe the chaperone actions, we constructed single Hsp70 (Δssa1 and Δssa2) or Hsp110/Sse1 (Δsse1) chaperone knockouts of our [PSI+] prion model strain (Materials and methods), and we assessed the resulting alterations to the aggregate structures in these cells by correlative fluorescence and electron tomography. There was no obvious alteration in the fluorescence pattern of NM-YFP aggregates in any of the single chaperone deletion strains compared with cells with a wild-type chaperone complement (Fig. S1 A). However, electron tomography of the deletion strains showed striking deviations from the normal dot arrangement (Fig. 1, A–D; and Fig. S2, A–E). The deletions introduced in both Δssa1 or Δssa2 strains resulted in the appearance of featureless perimeter zones surrounding the fibrillar core of the aggregate (Fig. 1, B and C; and Fig. S2, A and C). Occasionally, the dot aggregates entirely consisted of this nonfibrillar zone (Fig. S2 B), although this could be a result of the section plane not passing through the center of the dot. Where present, the fibrils in the core region were similar in length to NM-YFP fibrils formed with a wild-type chaperone background (Fig. 1 E, wild type, Δssa1, and Δssa2) and had a normal packing arrangement (Fig. 1, A–C). In addition to the nonfibrillar perimeter shells, clumps of dark-staining amorphous material were often seen interspersed with the NM-YFP fibril assemblies in these two Hsp70 deletion strains. All of these features could be readily identified in cryotomograms of vitreous sections of these cells, precluding any significant artifact from our sample preparation method (Fig. S2, E and F).
The deletion of SSE1 slowed the growth of our model strain, as reported elsewhere (Mukai et al., 1993; Trott et al., 2005; Abrams et al., 2014). Here, we noted extensive areas of dark amorphous material intercalated with large but otherwise normal-looking dot fibril arrays (Fig. 1 D and Fig. S2 D). This amorphous material formed a coarse meshwork when present toward the center of the NM-YFP fibril assemblies (Fig. 1 D and Fig. S2 D). Although packing within these fibril arrays was similar to those in cells with a wild-type chaperone complement, the individual fibrils themselves were found to be approximately 60% longer (Fig. 1 E, wild type and Δsse1).
These large structural modifications to the NM-YFP dot aggregates in each of the three chaperone deletions strains (Δssa1, Δssa2, and Δsse1) did not affect the stability of prion propagation. We monitored the presence of the fluorescent dot aggregates in these strains in successive progeny (50 generations for Δssa1 and Δssa2, and 32 generations for Δsse1) and found them to persist just as stably as dots in cells with a wild-type chaperone complement (Fan et al., 2007; Kryndushkin and Wickner, 2007; Sharma and Masison, 2008).
Similar to other insoluble protein deposit substrates (Kaganovich et al., 2008; Kawai-Noma et al., 2009; Weisberg et al., 2012; Yang et al., 2013), our overexpressed NM-YFP foci were immobile when analyzed by FRAP (Fig. S3). NM-YFP foci in each of the three individual chaperone deletion strains (Δssa1, Δssa2, and Δsse1) displayed similar recovery curves (Fig. S3), regardless of whether the bleached region of interest was located in the center or more toward the periphery of the dot (not depicted). This indicated that despite the structural differences in these NM-YFP assemblies, the cells were still sequestering both the fibrillar and nonfibrillar aggregates into discrete foci with very little cytosolic exchange.
Chaperone deletion strains sustain a pool of small NM-YFP oligomers
The NM-YFP expression level in each of the individual chaperone deletion strains (Δssa1, Δssa2, and Δsse1) was similar to that observed in cells with a wild-type complement of chaperones, whether NM-YFP was in the [PSI+] prion state or [psi−] soluble form (Fig. 2 A). Several lower molecular mass bands could also be seen in the blots. These bands were found to be specific to NM-YFP because an otherwise isogenic strain devoid of the NM-YFP cassette had no detectable signal on the Western blots (Fig. 2 A, control lane). These additional bands were likely attributable to proteolytic degradation; accordingly, soluble NM-YFP present in the [psi−] background appeared more susceptible to this phenomenon than the NM-YFP present in the amyloid form.
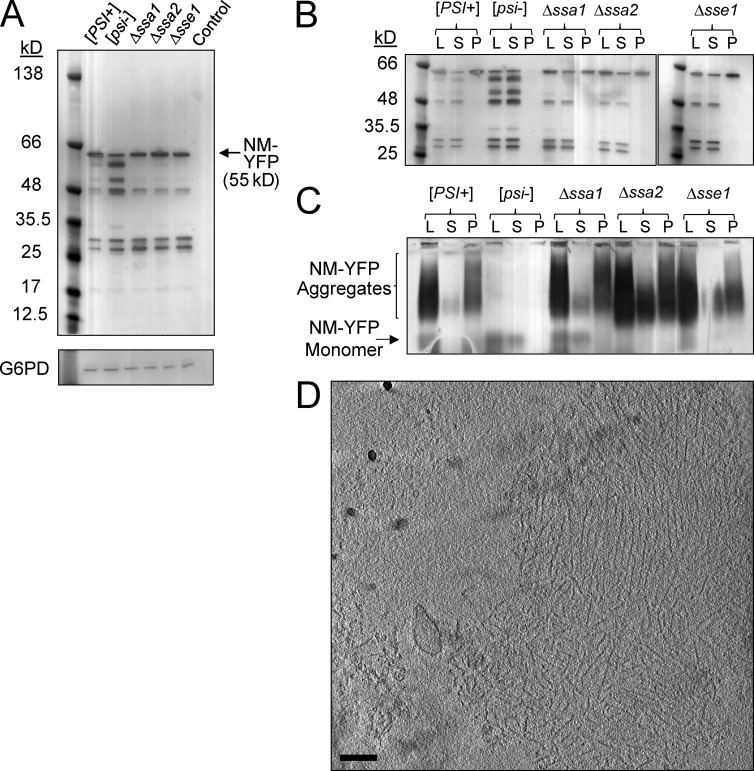
Figure 2.
Chaperone deletion alters the size distribution of NM-YFP aggregates. (A) Western blot of cell lysates using an α-YFP antibody showing NM-YFP expression levels in [PSI+], [psi−], Δssa1, Δssa2, and Δsse1 cells. The parental wild-type strain, devoid of the NM-YFP expression cassette, but otherwise isogenic, is included as a control. An α-Glucose-6-phosphate dehydrogenase (G6PD) antibody was used to verify equal loading of the lysates. (B) Ultracentrifugation sedimentation of NM-YFP aggregates in lysates of [PSI+], [psi−], Δssa1, Δssa2, and Δsse1 cells, analyzed by Western blot using an α-YFP antibody. Lysate (L), supernatant (S), and pellet (P) fractions are indicated. (C) SDD-AGE analysis and immunoblotting of the ultracentrifugation fractions described in B. (D) Slice through a representative cryotomogram of an NM-YFP dot aggregate isolated from the pellet fraction of the ultracentrifuged [PSI+] cell lysate. Of seven individual pelleted dots observed, 43% had preserved the fibril array architecture, as shown, whereas the remainder consisted of disordered clusters of NM-YFP fibrils. The disorder in fibril packing for these dots had been introduced by sample handling, because all dot assemblies from five independent dot aggregates observed directly in the lysates, without ultracentrifugation, had a well-preserved fibril array arrangement consistent with the example shown. Bar, 200 nm.
The oligomeric properties of the NM-YFP aggregates in lysates from each of these strains were assessed by ultracentrifugation (Fig. 2 B). As expected for aggregates formed with a wild-type chaperone complement, a substantial part of the full-length NM-YFP was in the pellet fraction (Fig. 2 B, [PSI+]). We analyzed this fraction by correlative cryofluorescence/cryoelectron tomography and found that the normal fibrillar array organization was partly preserved for many of the dot aggregates (Fig. 2 D). For lysates from each of the Δssa1, Δssa2, and Δsse1 strains, the full-length NM-YFP population was clearly observed in both the supernatant and pellet fractions after ultracentrifugation (Fig. 2 B). These supernatant pools of NM-YFP were composed of a smaller oligomeric species, but not monomeric NM-YFP, as assessed by semidenaturating detergent agarose gel electrophoresis (SDD-AGE; Fig. 2 C). The presence of these small oligomeric aggregates coincides with the appearance of the amorphous material in dot assemblies in these mutant backgrounds as described earlier (Fig. 1, B–D).
Hsp104 is localized to the periphery of NM-YFP aggregates
To relate the observed structures to changes in chaperone levels, we monitored global Hsp104, Hsp70 (SSA), and Hsp110 (Sse1) expression levels in each of our single chaperone deletion strains. Global Hsp70 and Sse1 expression was not significantly altered in the deletion strains relative to their levels in cells with a wild-type complement of chaperones (Fig. 3 A). A weak Hsp110 signal was detectable in our ΔSSE1 strain, which was likely attributable to Sse2 (which has 76% identity to Sse1), although Sse2 does not functionally substitute for Sse1 in [PSI+] prion biology (Moran et al., 2013). In contrast, Hsp104 levels were significantly altered in the single chaperone deletions (Fig. 3, A and B). In the Hsp70 deletion backgrounds (Δssa1 and Δssa2), Hsp104 levels increased three- to fivefold, whereas no statistically significant increase was observed in the Δsse1 strain. Therefore, it appears that Hsp104 is increased to compensate for the loss of a particular Hsp70 (Ssa1 or Ssa2), as described elsewhere (Jung et al., 2000).
Figure 3.
NM-YFP assemblies have a nonuniform distribution of molecular chaperones. (A) Western blot of cell lysates showing Hsp104, Hsp70, and Sse1 expression levels in cells with a wild-type chaperone complement, which were either [PSI+] (NM-YFP aggregate dots) or [psi−] (soluble NM-YFP expressed but not converted to the prion state) and Δssa1, Δssa2, and Δsse1 cells. Glucose-6-phosphate dehydrogenase (G6PDH) was used as a loading control. (B) Plot of the quantitation of three independent immunoblots with the αHsp104 antibody, as indicated. The abundance of Hsp104 for each of the lysates is expressed relative to its level in the [PSI+] lysate and error bars correspond to the SD between the replicates. (C) Representative confocal Z-sections showing the distribution of mCherry-tagged Hsp104, Ssa1, Ssa2, and Sis1 on NM-YFP aggregates in cells with a wild-type complement of chaperones. The data are representative of at least 20 different NM-YFP dots for each labeled chaperone from two independent experiments. Imaging was optimized to visualize the fluorescence from the dot aggregates. A line plot illustrating the colocalization of Hsp104-mCherry with the NM-YFP dot is shown in Fig. S4 A. Bars, 2 µm. (D) Representative confocal Z-sections showing the distribution of mCherry-tagged Hsp104, Ssa1, and Sis1 on NM-YFP aggregates in Δssa1 cells. Δssa2 cells displayed a similar chaperone distribution and thus have been omitted here. The data are representative of at least 20 different NM-YFP dots for each labeled chaperone from two independent experiments. Imaging was optimized to visualize the fluorescence from the dot aggregates. A line plot illustrating the colocalization of Hsp104-mCherry with the NM-YFP dot in this strain is shown in Fig. S4 B. (E) Representative confocal Z-sections showing the distribution of mCherry-tagged Hsp104 and Hsp70 (Ssa1 and Ssa2) on NM-YFP aggregates in Δsse1 cells. We were unable to obtain a viable Sis1-tagged clone in this deletion strain. The data are representative of at least 20 different NM-YFP dots for each labeled chaperone from two independent experiments. Imaging was optimized to visualize the fluorescence from the dot aggregates. A line plot illustrating the colocalization of Hsp104-mCherry with the NM-YFP dots in this strain is shown in Fig. S4 C.
Molecular chaperones are abundant in yeast prion aggregates (Chernoff et al., 1995; Allen et al., 2005; Krzewska and Melki, 2006; Bagriantsev et al., 2008; Tipton et al., 2008; Tyedmers et al., 2010; Saibil et al., 2012; Yang et al., 2013). To assess the spatial distribution of molecular chaperones within the NM-YFP dot aggregates in our deletion strains, we created a series of mCherry fusions. C-terminally tagged Hsp104, Hsp70 (Ssa1 and Ssa2), and Hsp40 (Sis1) derivatives were constructed by integrating a copy of mCherry in frame with the endogenous coding regions. Confocal microscopy was then used to assess their respective cellular distributions. Hsp70 (Ssa1 and Ssa2) and Hsp40 (Sis1) were distributed evenly throughout NM-YFP dot aggregates in cells with a wild-type chaperone complement and in those with single Hsp70 deletions (Δssa1 or Δssa2; Fig. 3, C and D). Although Hsp104 was also seen to colocalize with the NM-YFP dot assemblies in these cells, it was concentrated around the periphery of the dots (Fig. 3, C and D). Although the Hsp104-mCherry intensity was very weak in cells with a wild-type chaperone complement (Fig. 3 C), the peripheral signal was still slightly higher than the central area (Fig. S4 A). However, the strong Hsp104-mCherry fluorescence in the Hsp70 deletion strains (Δssa1 and Δssa2) clearly showed its peripheral distribution (Fig. 3 D and Fig. S4 B). In the Δsse1 strain, Hsp104 was almost exclusively confined to the perimeter of the dots (Fig. 3 E and Fig. S4 C), with the mCherry signal toward the center at background level. Hsp70 (Ssa1 and Ssa2) also showed a preferred localization toward the periphery of NM-YFP dots in the Δsse1 strain (Fig. 3 E).
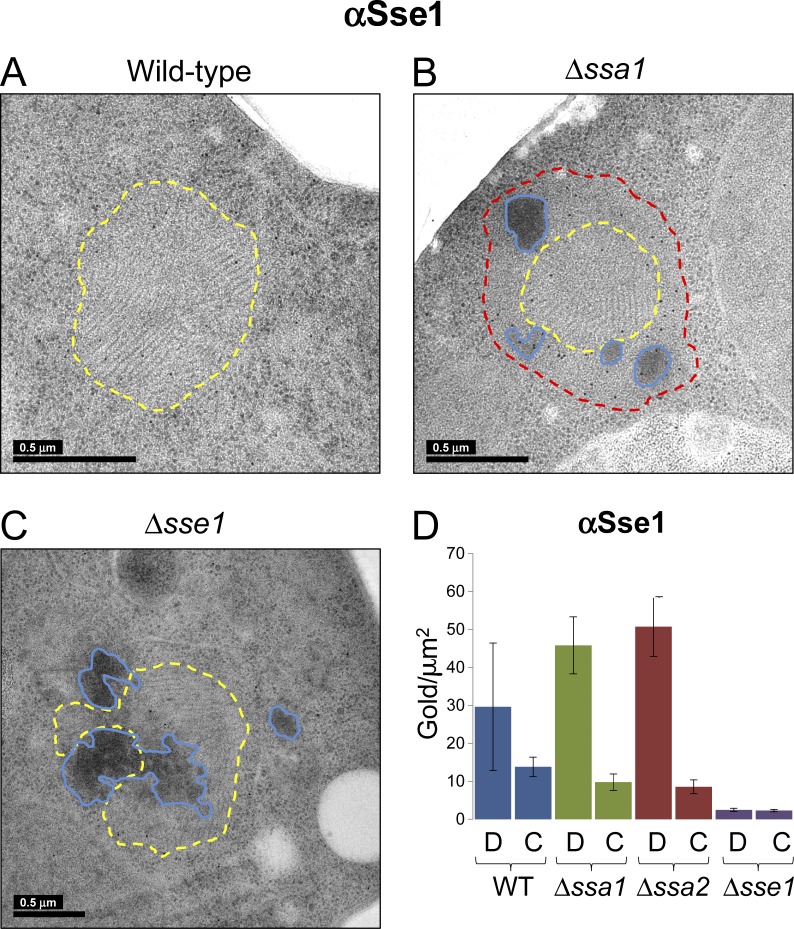
Amorphous shells surrounding NM-YFP fibrils are enriched in Hsp110
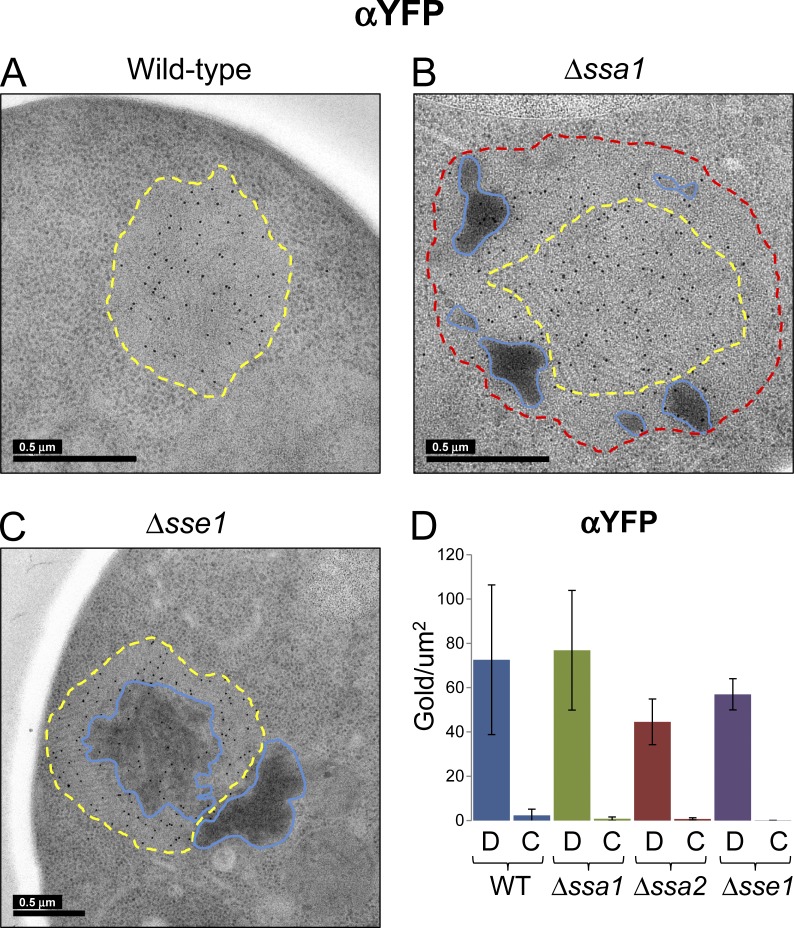
We used immunogold EM on freeze-substituted sections to define the chaperone distributions in NM-YFP dots on the nanoscale. An anti–YFP antibody clearly localized NM-YFP in all of the cell sections (Fig. 4). As in the confocal analysis, NM-YFP was confined to the dot assemblies with little or no cytosolic labeling (Fig. 4). In the Hsp70 deletions (Δssa1 or Δssa2), NM-YFP was also detected in the nonfibrillar perimeter shells and the dark amorphous material of the dot aggregates (Fig. 4 B). However, the dark amorphous material observed in dot aggregates in the Δsse1 strain was sparsely labeled (if at all), suggesting either poor accessibility of the antibody or that this material is composed of other aggregates (Fig. 4 C).
Figure 4.
Immunogold labeling of NM-YFP in dot assemblies in Hsp70 and Hsp110 knockout cell sections. Representative EM projection images of immunogold-labeled cell sections of cells with a wild-type chaperone complement and those from Δssa1 and Δsse1 cells, as indicated. Δssa2 cell sections exhibited similar labeling as Δssa1 and thus projections of these were omitted from the figure. An α-YFP antibody was used to detect and label the NM-YFP prion protein fusion and the images are representative of at least 10 independent NM-YFP dots observed in each strain. Bars, 0.5 µm. (A) Wild-type chaperone complement. Fibril-containing regions are outlined in yellow as described in Fig. 1 A. (B) Δssa1. Fibril-containing regions are outlined in yellow, unstructured perimeter zones are outlined in red, and dark amorphous aggregates are outlined in blue, as described in Fig. 1 B. (C) Δsse1. Fibril-containing regions are outlined in yellow and dark amorphous aggregates are outlined in blue, as in A and B. (D) Graphs illustrate the labeling efficiency with this antibody on the NM-YFP dot aggregates or in the cytosol. The data are representative of at least five independent observations and the error bars correspond to the SD in the data. C, cytosol; D, dot aggregate; WT, wild type.
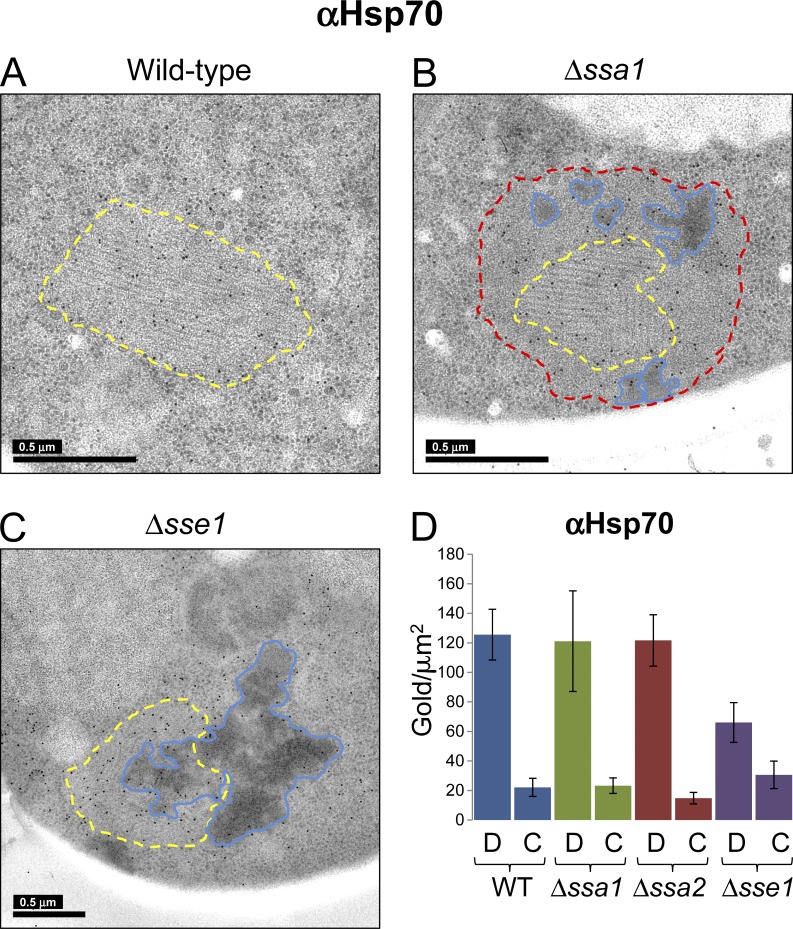
Regarding the distribution of Hsp70 in cell sections, we noted a sixfold higher concentration in the dots compared with cytosolic levels of Hsp70 in strains with a wild-type chaperone complement and the Hsp70 deletions, Δssa1 and Δssa2 (Fig. 5). In cells with a single Hsp70 deletion, the remaining Hsp70s were present in the fibrillar core, the nonfibrillar perimeter shells, and the dark amorphous clumps (Fig. 5 B, yellow, red, and blue outlines, respectively). For the Δsse1 strain, only a modest (approximately twofold) increase in Hsp70 labeling was observed on the dot aggregates compared with the cytosol (Fig. 5 D). Again, the mesh-like amorphous material in these strains was poorly labeled with the α-Hsp70 antibody (Fig. 5 C, blue outline).
Figure 5.
Immunogold detection of Hsp70 on NM-YFP assemblies in Hsp70 and Hsp110 knockout cell sections. Same as for Fig. 4, but detection was with an α-Ssa antibody to label Hsp70. C, cytosol; D, dot aggregate; WT, wild type.
Sse1 was not significantly concentrated in the dots in cells with a wild-type complement of chaperones (Fig. 6 A). In contrast, in Δssa1 or Δssa2 strains, there was a more than fourfold increase in the levels of Sse1 on the dot assemblies (Fig. 6, B and D). This redistribution of Sse1 was remarkable, given that Sse1 was not significantly up-regulated in these strains (Fig. 3 A). Furthermore, Sse1 was predominantly located in the nonfibrillar peripheral zones and the amorphous clumps with little fibril labeling (Fig. 6 B). As expected, barely detectable background labeling was observed with α-Sse1 labeling of sections from the Δsse1 strain, which was presumably attributable to Sse2, as described earlier (Fig. 6, C and D).
Figure 6.
Immunogold detection of Sse1 on NM-YFP assemblies in Hsp70 and Hsp110 knockout cell sections. Same as for Fig. 4, except that detection was with an α-Sse1 antibody. C, cytosol; D, dot aggregate; WT, wild type.
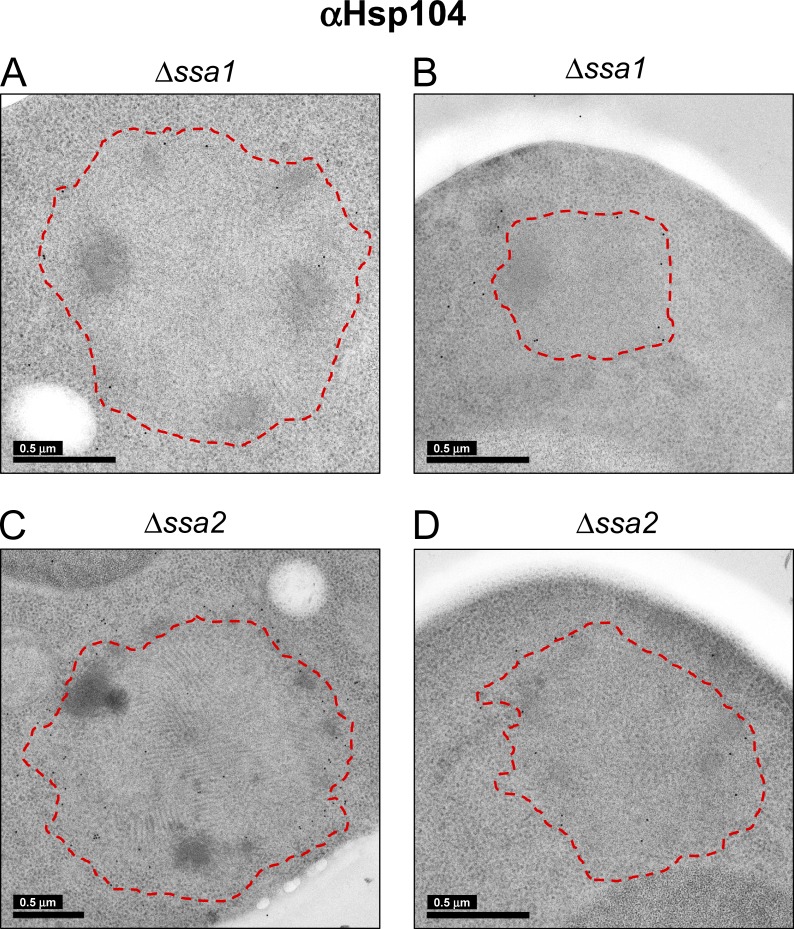
Consistent with the fluorescence observations (Fig. 3 B), our Western blots revealed a low abundance of Hsp104 in NM-YFP–expressing cells with a wild-type chaperone complement (Fig. 3 A), making it undetectable by immunogold EM. For cell sections of the Hsp70 deletions (Δssa1 and Δssa2), however, the elevated Hsp104 expression levels facilitated specific immunogold detection of Hsp104 (albeit with low labeling efficiency; Fig. 7, A–D), confirming the peripheral localization seen by confocal microscopy (Fig. 3 C).
Figure 7.
Immunogold detection of Hsp104 on NM-YFP assemblies in Hsp70 knockout cell sections. (A–D) Representative EM projection images of immunogold-labeled cell sections of Δssa1 (A and B) and Δssa2 (C and D) cells, as indicated. Detection was with an α-Hsp104 antibody. The red outlines, encompassing entire NM-YFP dot aggregates, were manually traced at the interface between the peripheral unstructured zone of the aggregates and the ribosomes in the cytosol.
The perimeter shells thus contained Hsp70 (Ssa1/Ssa2; Fig. 3 C and Fig. 5 B), Hsp40/Sis1 (Fig. 3 C), and Hsp104 (Fig. 3 C and Fig. 7, A–D) and were significantly enriched for Hsp110/Sse1 (i.e., all of the major components of the yeast disaggregase machinery; Fig. 6, B and D). Therefore, these nonfibrillar zones define a region of NM-YFP aggregates with an altered balance between aggregation and disaggregation.
Hsp104 and Sse1 can independently influence NM-YFP fibril assembly
Our immunogold labeling experiments suggested that NM-YFP fibrils are remodeled in regions enriched in Hsp104 and/or Sse1. To further test this hypothesis, we transiently overexpressed either Hsp104 or Sse1 individually (on plasmids pHsp104 or pSse1, respectively) in an otherwise wild-type chaperone background and examined the resulting NM-YFP dot assemblies by electron tomography.
In cells transiently overexpressing Hsp104, 80% of dot structures (40 dots examined) contained disorganized or amorphous regions (Fig. 8, A and B). The remaining dots contained large but normal NM-YFP fibrillar arrays (not shown). In the altered dot aggregates, large nonfibrillar zones were usually present, reminiscent of those seen in the Δssa1 and Δssa2 strains (Fig. 1, B and C; and Fig. 8 A). Both Hsp104 and Sse1 could be detected within the nonfibrillar zones in these dot aggregates (Fig. 8, C and D), consistent with their combined presence in these regions in the Δssa1 and Δssa2 strains. Other abnormal dot aggregates in this population comprised regions of highly fragmented and randomly arranged small fibrils that were often accompanied by dark-staining amorphous aggregates (Fig. 8 B).
Figure 8.
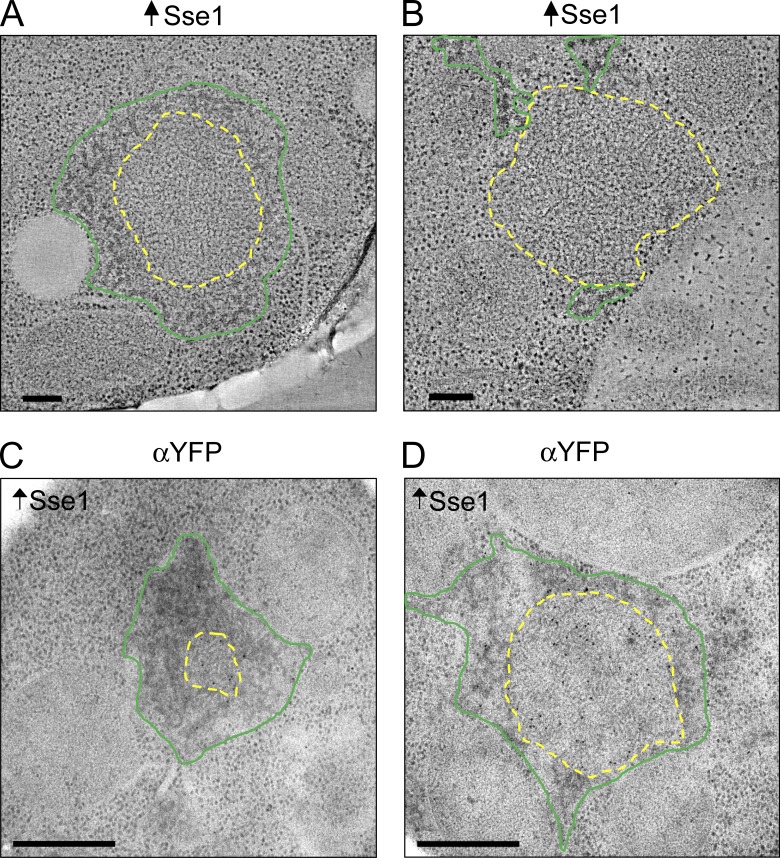
Hsp104 overexpression disrupts NM-YFP aggregate organization. (A and B) Representative tomographic slices from reconstructions of NM-YFP dots in cell sections of HM20-embedded cells with a wild-type chaperone complement and overexpressing Hsp104 from plasmid pHsp104. The interface between the cytosolic ribosomes and the aggregate surface was traced manually and outlined in red. Dark amorphous aggregates were manually outlined in blue. Bars, 200 nm. (C and D) Representative EM projection images of immunogold-labeled HM20-embedded cell sections of cells with a wild-type chaperone complement and overexpressing Hsp104. Sections were labeled with either α-Hsp104 (C) or α-Sse1 (D) antibodies, as indicated. The labeling shows the combined presence of Hsp104 and Sse1 in the unstructured perimeter zones. Fibril-containing regions were manually traced around the interface between the fibrils and the surrounding unstructured zone and are outlined in yellow. The unstructured material is outlined in red and was manually traced at the interface between the unstructured zone and the cytosolic ribosomes. Bars, 0.5 µm.
Transient overexpression of Sse1 also resulted in a majority of abnormal NM-YFP aggregate assemblies. Here, ∼75% were disorganized, whereas the remaining assemblies were large but normal-looking dots. The disrupted dots had an extremely disordered fibrillar core surrounded by a web of darkly stained amorphous material (Fig. 9, A and B). This material did not contain NM-YFP (Fig. 9, C and D) and was reminiscent of the dark amorphous aggregates seen occasionally in fibril assemblies in the Δsse1 strain, as described earlier.
Figure 9.
Sse1 overexpression disrupts NM-YFP aggregate organization. (A and B) Representative tomographic slices from reconstructions of NM-YFP dots in cell sections of HM20-embedded cells with a wild-type chaperone complement and overexpressing Sse1 from plasmid pSse1. Normal fibrillar regions are outlined in yellow, manually traced at the interface between the fibrils and the surrounding cytosolic ribosomes and/or mesh-like amorphous aggregates. The mesh-like amorphous material was manually outlined in green. Bars, 200 nm. (C and D) Representative EM projection images of immunogold-labeled HM20-embedded cell sections of cells with a wild-type chaperone complement and overexpressing Sse1. Sections were labeled with an α-YFP antibody to detect NM-YFP. The mesh-like amorphous material was sparsely labeled, suggesting that it contained mostly other aggregates. Fibrils are outlined in yellow and the amorphous material is outline in green, as described for A and B. Bars, 0.5 µm.
Discussion
We have explored the role of the Hsp70 chaperone system in the in situ organization of a [PSI+] prion model. We describe a novel nonfibrillar surface layer of amyloid-containing dot aggregates that appears to result from an altered balance between fibril assembly and disassembly. This material was indistinguishable from the “normal” fibrillar NM-YFP aggregates in our cells by fluorescence and could only be discerned by electron tomography.
We used a model system with a highly overexpressed but nontoxic form of the [PSI+] prion. This enabled us to visualize the 3D structure of the aggregates by correlative fluorescence and electron tomography and to probe the effects of modulating the major Hsp70-related chaperones. Although the fluorescent foci appear small and dispersed under native expression conditions (Satpute-Krishnan and Serio, 2005; Song et al., 2005; Park et al., 2014), it seems likely that similar molecular interactions would occur in the smaller aggregates.
Although yeast prion proteins can assemble into amyloid fibrils in vitro in the absence of additional cofactors, interactions with the protein quality control machinery are required for their assembly, deposition, and propagation in yeast cells (Tuite et al., 2011; Liebman and Chernoff, 2012; Tyedmers, 2012). Perturbing proteostasis through selective Hsp70 deletion revealed the nonfibrillar aggregates of NM-YFP, giving a nanoscale structural view of processes underlying previous observations of the cellular roles of Hsp70s on prion propagation (Sharma and Masison, 2008; Sharma et al., 2009). Our single Hsp70 deletions stably propagated dot aggregates of NM-YFP, but changes in expression level of the Hsp104 disaggregase appear to account for the observed dramatic structural changes. First, although the Δssa1 and Δssa2 strains maintained global Hsp70 and Sse1 expression levels, there was an up-regulation of Hsp104 (Fig. 3 A). Moreover, Hsp104 was primarily located around the periphery of NM-YFP dots (Fig. 3, C–E; and Fig. 7). In addition, there was a marked recruitment of Sse1 to this boundary region in Δssa1 and Δssa2 cells (Fig. 6 B). Our findings indicate that recruitment of Hsp104 and Hsp110/Sse1, rather than loss of a specific Hsp70 function, plays a key role in the structural changes, possibly by aggregate remodeling.
It is well established that Hsp104 is essential for [PSI+] propagation (Chernoff et al., 1995), most likely by fibril trimming/fragmentation in cooperation with Hsp70/Hsp40, to generate new seeds (Misselwitz et al., 1998; Ferreira et al., 2001; Wegrzyn et al., 2001; Higurashi et al., 2008; Tessarz et al., 2008; Tipton et al., 2008; Kampinga and Craig, 2010; Moosavi et al., 2010; Winkler et al., 2012b; Park et al., 2014). Although there is evidence that Hsp110/Sse1 is also involved in these processes, its precise role has been unclear. As a major nucleotide exchange factor for Hsp70, Sse1 can stimulate amyloid fragmentation by Hsp104 both in vivo and in vitro (Fan et al., 2007; Kryndushkin and Wickner, 2007; Sadlish et al., 2008; Shorter, 2011). Recent studies show that Sse1 (and Hsp110 in mammals), together with Hsp70/Hsp40, acts as a disaggregase (Shorter, 2011; Rampelt et al., 2012; Song et al., 2013; Torrente and Shorter, 2013), although the in vitro reaction with Sup35 NM fibrils was extremely slow (Duennwald et al., 2012; Torrente and Shorter, 2013).
What is the function of Sse1 in the nonfibrillar, peripheral regions of NM-YFP assemblies in our strains? This question was addressed by examining the morphology of dot aggregates generated in the absence of Sse1. Although deletion of SSE1 causes slow growth in yeast (Mukai et al., 1993; Trott et al., 2005; Abrams et al., 2014), our Δsse1 strain stably propagated NM-YFP dot aggregates. Interestingly, the fibrils within NM-YFP dots from Δsse1 cells were approximately 60% longer than those from Δssa1, Δssa2, and cells with a wild-type chaperone complement (Fig. 1 E). Unlike an Hsp104 knockout, which cannot propagate [PSI+] (Chernoff et al., 1995), it would appear that removal of Sse1 alters the balance toward [PSI+] fibril growth over cleavage, resulting in aggregates with longer fibrils. By contrast, the Hsp70 deletions (Δssa1 and Δssa2) did not affect the lengths of the remaining fibrils (Fig. 1 E), despite the elevated Hsp104 levels in these mutants. We hypothesize that in these strains, remodeling of the outer layer of the aggregates most likely results from elevated Hsp104 progressively disassembling fibrils in initially normal dots, starting from the outer surface.
The alterations in aggregate morphology suggest direct functional roles for both Hsp104 and Sse1 in fragmentation and disassembly of NM-YFP fibrils in vivo. But what are the individual contributions of these two molecular chaperones to this process? In our [PSI+] model, we found that transient overexpression of either Hsp104 or Sse1 separately had a significant effect on the resulting NM-YFP dot morphology. Hsp104 overexpression reproduced the nonfibrillar zones seen in aggregates in the Hsp70 deletions (Fig. 8 A) with Sse1 recruited to these amorphous regions (Fig. 8 D). Overexpression of Sse1 produced more disorganized NM-YFP dot assemblies, often surrounded by mesh-like, amorphous, unidentified aggregates (Fig. 9, A and B). The Hsp104 overexpression result supports the notion that it simply disassembles NM-YFP fibrils. This conclusion was originally based on the reduction in aggregate size observed by SDD-AGE, a method that does not distinguish between individual polymers and clumps (Kryndushkin et al., 2003) Sse1, however, appears to regulate fibril length and organization in a more measured type of fragmentation.
In summary, our findings reveal the in situ roles of Hsp104 and Sse1 as principal determinants of the in situ amyloid structure. Our manipulations revealed an active zone in which these chaperones were enriched, likely remodeling the aggregate species found there. Deletion of SSE1 did not result in a nonfibrillar zone despite the clear localization of Hsp104 to the dot periphery (Fig. 3 E and Fig. S4 C). It therefore seems that both chaperone components are required to efficiently process fibrils of NM-YFP. Notably, these structural changes did not affect prion stability, still generating prion seeds that maintain epigenetic inheritance. This work has allowed us, for the first time, to directly visualize products generated by the yeast disaggregation machinery in vivo. How reduced Ssa and/or elevated Hsp104 levels trigger the observed redistribution of Sse1 and whether or not there is a direct interaction between Hsp104 and Sse1 is unclear. In addition, the precise role played by Sse1 in disaggregation warrants further investigation. A thorough understanding of the interplay between Hsp110s, such as Sse1, and their partner chaperones in dealing with amyloid aggregates in cells could potentiate strategies for controlling protein conformational disorders.
Materials and methods
Yeast strains media and plasmids
Yeast strains for the analysis of [PSI+] aggregates were derived from the 74D-694 background (MATα, ade1-14 ura3-52 leu2-3,112 trp1-289 his3-Δ200; Chernoff et al., 1995), which was manipulated to constitutively propagate an NM-YFP fusion in the prion state (Tyedmers et al., 2010). The NM domain–encoding region of the endogenous SUP35 gene was deleted in this parental strain and an NM-YFP fusion cassette, expressed from the constitutive GPD promoter, was integrated at the TRP locus. Strains were grown using standard culturing techniques in YPD media (1% yeast extract, 2% peptone, and 2% glucose) or synthetic dropout media (0.7% yeast nitrogen base without amino acids, 2% glucose) lacking the relevant nutrient for auxotrophic plasmid selection. Excess adenine, at 100 mg/l, was included in media when growing 74D-694 derivatives. Where appropriate, antibiotic selection was achieved using G418 (200 µg/ml) or hygromycin B (50 µg/ml).
Construction of the Δssa1, Δssa2, and Δsse1 deletion derivatives of our NM-YFP parental strain was achieved using a standard PCR-based deletion strategy (Baudin et al., 1993; Wach et al., 1994). In brief, the kanMX4 cassette from plasmid pDH6 (Yeast Resource Centre; Wach et al., 1997) was amplified by PCR, incorporating the appropriate flanking homology sequences for targeted integration and gene deletion by homologous recombination, using the oligonucleotide combinations listed in Table S1. Cells were transformed with these PCR products using a standard lithium acetate transformation protocol and were selected for resistance to G418. The integrity of the deletions was confirmed by colony PCR.
Fluorescent protein labeling of Hsp104, Ssa1, Ssa2, and Sis1 for colocalization studies with NM-YFP was conducted by a C-terminal fusion of the respective chromosomal coding sequence with mCherry (Shaner et al., 2004) using standard PCR targeting (Wach et al., 1997). The integration cassettes were amplified from plasmid pBS35 (Yeast Resource Centre) and contained the mCherry chromosomal coding sequence, the hygromycin B resistance cassette, and the appropriate flanking homology sequences for targeted integration. The oligonucleotide primer pairs used are listed in Table S1. Cells were transformed with these PCR products using a standard lithium acetate transformation protocol and were selected for resistance to hygromycin B. Correct integration was confirmed by PCR and the resulting fusions were verified by sequencing.
Plasmids pHsp104 and pSse1 were constructed by PCR amplification of HSP104 and SSE1, respectively, and digestion with BamHI-SalI followed by ligation to p425Gal1 (Mumberg et al., 1995), which had been digested with the same restriction endonuclease pair. Oligonucleotide pairs used are listed in Table S1. Both plasmids were verified by sequencing. Galactose induction of the Gal1 promoter on p425Gal1 was conducted by growing cultures initially in SD media (lacking leucine) with 2% glucose to mid-late log phase. Cells were then pelleted by centrifugation (3400 rpm, 10 min), resuspended in media with 2% galactose, and grown for a further 6 h. Overexpression of Hsp104 and Sse1 was verified by Western blotting.
Fluorescence microscopy of yeast cells
For routine live-cell imaging, yeast cells were mounted on polysine slides (VWR International LLC) with coverslips (0.13- to 0.16-mm thick) in growth media. Conventional epifluorescence images were acquired using an Axioscope A1 microscope (Carl Zeiss), equipped with an X-Cite Series 120Q lamp and an Orca R2 charge-coupled device (CCD) camera (Hamamatsu Photonics), using a 63× Plan-Apochromat oil immersion objective lens (NA1.4). Imaging was performed with 500/20-nm and 535/30-nm excitation and emission filters, respectively, for YFP and 546/12-nm and LP 590-nm excitation and emission filters, respectively, for mCherry. Image analysis and processing were performed using Axiovision (Carl Zeiss) and Fiji (Schindelin et al., 2012).
For colocalization studies, cells were grown to mid-late log phase, fixed with 4% PFA (Electron Microscopy Sciences) in growth media, and immobilized on 1% agarose pads with Mowiol on glass slides with coverslips. Confocal Z-stacks were acquired using a Leica TCS SP5 microscope equipped with an HCX Plan-Apochromat lambda blue 63× oil UV objective lens (NA1.4). For detection of the YFP signal, the 514-nm laser line was used and emission was collected within a band pass range of 527–581 nm. Excitation of mCherry was with the 594-nm laser line and emission was collected at 613–716 nm. Clear spectral separation of YFP and mCherry signals was ensured by the confocal settings, and sequential scanning was used during acquisition to minimize fluorophore cross talk. The photomultiplier tube voltage for both YFP and mCherry signals was optimized for detection of the fluorescence in the dot aggregate assemblies. Image analysis and processing were performed using Fiji (Schindelin et al., 2012).
FRAP analysis was performed on live cells grown to mid-late log phase using the Leica TCS SP5 microscope and objective lens described in the preceding paragraph. Cells were immobilized on 1% agarose pads in growth media on glass slides with coverslips and imaged immediately. A 0.35-µm2 region of interest, typically corresponding to between 5% and 10% of the total area of an NM-YFP dot, was bleached with five 0.147-s pulses using the 513-nm laser line at full power. Bleaching conditions typically resulted in 90% loss in fluorescence in the bleached area. Recovery from photobleaching was then monitored every 0.2 s for 10 s and then every 1 s for 30 s. Background noise and total fluorescence in the NM-YFP dot were also monitored during the time course and used for normalization. The data were normalized using the easyFRAP program to account for differences in the starting intensities and for acquisition photobleaching during the time course (Rapsomaniki et al., 2012). The data presented for each strain were the mean of at least seven independent dot aggregates. Error bars in the graph correspond to the SD in the data.
EM sample preparation
Yeast cells grown to mid-late log phase were pelleted by centrifugation (3,400 rpm, 10 min) and resuspended in growth medium to achieve 500- to 1000-fold concentration. Aliquots were immediately transferred a Leica type A aluminum specimen carrier (0.2-mm indentation), covered with the flat side of a type B specimen carrier, and high-pressure frozen using the EM HPM100 system (Leica). Cryofixed samples were then freeze substituted and embedded with Lowicryl HM20 (Agar Scientific) using an EM AFS2 device (Leica) according to a protocol adapted from Hawes et al. (2007). In brief, cells were freeze substituted in dry acetone containing 0.1% uranyl acetate and 1% methanol at −90°C for 16 h. Samples were then warmed to −50°C at 20°C/h and washed twice in 100% ethanol for 30 min, before five exchanges with 1-h incubations into increasing concentrations of HM20 (25%, 50%, 75%, 100%, and 100%). A further exchange into 100% HM20 was then performed and incubation was continued at −50°C for 18 h. After a fresh HM20 exchange and a second 18-h incubation, the HM20 was polymerized under UV light at −50°C for 48 h. The samples were then warmed to 20°C at 10°C/h and polymerization was continued under UV illumination for an additional 48 h.
HM20-embedded sample blocks were then sectioned at RT on an EM UC7 Ultramicrotome (Leica) using an Ultra 45° diamond knife (Diatome). For tilt-series acquisition, cell sections (200- to 250-nm thick) were cut and mounted on carbon-coated 200-mesh copper finder grids (Agar Scientific). After fluorescence imaging (see below), sections on grids were poststained with 2% uranyl acetate in 70% methanol for 5 min, followed by 3 min in lead citrate (Venable and Coggeshall, 1965), and were subsequently coated on both sides with protein A gold (Electron Microscopy Sciences). Cell sections (100-nm thick) were mounted on carbon-coated 300-mesh copper grids (Agar Scientific) for immunogold labeling experiments.
For cryotomography, yeast cells were cryofixed in growth media containing 20% high molecular mass dextran using the EM HPM100 with the copper tube system (Leica). 65- to 100-nm-thick cryo-sections were cut at −140°C using an EM UC7 Ultramicrotome equipped with an EM FC7 cryochamber using a cryo 25°C diamond knife (Diatome). Cryosection ribbons were mounted on custom-made C-flat carbon-coated 200-mesh Gilder finder grids (Protochips Inc.) that had been covered with a 5-nm continuous carbon layer and coated with protein A gold. Cryosections were stored under liquid nitrogen for future use.
Correlative cryo-fluorescence imaging
HM20-embedded cell sections mounted on EM grids were visualized by wet mount as described elsewhere (Kukulski et al., 2011). 0.2-µm Blue FluoSpheres (365/415; Life Technologies) were applied to grids as fluorescent fiducials for correlation with EM projection images. Fluorescence microscopy was performed using an Axioscope A1 microscope, equipped with an X-Cite Series 120Q lamp and an Orca R2 CCD camera, and using a 63× Plan-Apochromat oil immersion objective lens (NA1.4). YFP signal was monitored using 546/512-nm and LP 590-nm excitation and emission filters, respectively. Blue FluoSpheres were imaged using G 365-nm and 445/450-nm excitation and emission filters, respectively. Correlation between fluorescence and low magnification EM images of the same sample was performed using the “cpselect,” “cp2tform,” and “tformfwd” functions from the image processing toolbox in MATLAB (The MathWorks, Inc.). Affine transformations accounting for translation, rotation, and scaling differences between the two images were calculated using the coordinate positions of the blue FluoSpheres as control point pairs. The coordinate positions of NM-YFP dots on fluorescence images were then transformed onto the corresponding low-magnification EM projection images. This process was repeated for EM images of a magnification series up to that used for tilt-series acquisition.
Cryofluorescence imaging of vitreous cryosections was performed with the same microscope described above equipped with a cryostage (Linkam Scientific Instruments) operated at −195°C, using a 100× LD EC Epiplan-Neofluor objective lens (NA0.75). Correlation to NM-YFP dots in fluorescently mapped cryosections was performed in real time on the electron microscope using a software routine written in MATLAB and described elsewhere (van Driel et al., 2009). The four grid bar corners of a single EM grid square were used as landmarks for the registration of fluorescence images with a view of the same area on the electron microscope. The EM stage coordinates of these four known points were recorded by the software, which then calculated, by affine transformation, the stage coordinates corresponding to an NM-YFP dot location on the fluorescence image. The software was then used to set the EM stage to the location of the NM-YFP dot in the sample. This method had an expected accuracy of 0.5 µm (van Driel et al., 2009). NM-YFP dots typically occupied an area of 0.7–1.7 µm2 in cell sections, so the EM field of view of 5.3 µm2 used for tilt-series acquisition (see below) was chosen so as to always include the feature of interest. Precise correlation validation was subsequently performed using the MATLAB image processing toolbox, as outlined above for resin cell sections. The pattern of holes in the C-flat support film and autofluorescence from the cryosections (recorded using G 365-nm and 445/450-nm excitation and emission filters) facilitated accurate correlation.
Immunogold labeling of HM20-embedded cell sections
100-nm cell sections mounted on carbon-coated 300-mesh copper grids were used for on-section gold labeling. Grids were initially floated section-side down on 30-µl droplets of blocking buffer (1% BSA in PBS with 0.01% Tween 20) for 5 min. They were then transferred onto 30-µl droplets of the appropriate primary antibody diluted in blocking buffer and incubated at RT for 60 min. The grids were washed by six 2-min exchanges onto 60-µl droplets of blocking buffer before being transferred to 30-µl droplets of Protein-A gold, diluted 1:10 with blocking buffer. Incubation was continued at RT for 30 min. The grids were then washed with three changes of blocking buffer and three changes of PBS, for 2 min each. Labeling on the sections was then fixed by floating on 30-µl droplets of 1% buffered glutaraldehyde for 5 min, followed by four 1-min washes in milliQ water. NM-YFP was labeled with a mouse monoclonal α-YFP antibody (Takara Bio Inc.) at a working concentration of 20 µg/ml. A rabbit polyclonal α-Hsp104 antibody (Abcam) was used for Hsp104 labeling at a concentration of 50 µg/ml. Hsp70 labeling was with a rabbit polyclonal α-Ssa1 antibody diluted 1:50 (Winkler et al., 2012b). Sse1 was labeled with a rabbit polyclonal α-Sse1 antibody diluted 1:50.
EM data collection and processing
Grids mounted with HM20-embedded cell sections were placed into a Model 2040 dual-axis tomography holder (Fischione Instruments) and imaged using a Tecnai F20 microscope (FEI) operated at 200 kV with a 4k CCD camera (Ultrascan US4000; Gatan). Dual-axis tilt series were collected using SerialEM (Mastronarde, 2005) typically over a tilt range of ±60° with 2° increments at an unbinned pixel size of 0.585 nm. The nominal defocus was 0.5 µm. For reconstruction, tilt-series images were binned to a pixel size of 1.17 nm and dual-axis tomograms were generated using the IMOD package, version 4 (Kremer et al., 1996; Mastronarde, 1997). For plotting of individual fibril lengths in NM-YFP dots, fibrils were measured using the model tools of the 3dmod software package implemented in IMOD (version 4; Kremer et al., 1996). The data were analyzed using MATLAB.
Imaging of immunogold-labeled cell sections was conducted on Tecnai T10 and T12 microscopes (FEI), operated at full voltage, and equipped with 1K Multiscan CCD cameras (Gatan). Images were collected at various magnifications within a calibrated pixel size range of between 1.5 and 4.3 nm. Quantification of labeling was performed using Fiji (Schindelin et al., 2012).
Vitrified cryosections were imaged using a Tecnai Polara microscope (FEI) operated at 300 kV with a 4K CCD camera (Ultrascan US4000; Gatan). Tilt series were collected using SerialEM (Mastronarde, 2005) over a tilt range of ±60° with 1.5° increments at nominal defoci of between 8.0 and 10.0 µm and an unbinned pixel size of 0.563 nm. Low-dose conditions were implemented to apply a total dose of 60 e/Å2 for each tilt series. Tomogram reconstructions were performed using IMOD (version 4; Kremer et al., 1996).
Yeast lysate preparation, Western blotting, and SDD-AGE
Yeast cells were grown in the appropriate growth media to mid-late log phase, pelleted (3400 rpm, 10 min), and resuspended in lysis buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.5 mM EDTA) with protease inhibitors (P8215; Sigma-Aldrich). Resuspended cells were lysed by explosive decompression with two passages through a French Pressure Cell (Aminco; Thermo Fisher Scientific) operated at 10,000 pounds per square inch. The resulting crude lysates were centrifuged to remove unlysed cells and cellular debris (2,700 rpm, 10 min) and the clarified lysates were then stored at −80°C for future use.
SDS-PAGE and Western blots were performed using standard techniques. For Western analysis, 3 µg of each lysate was run on gels and equal loading was monitored with an α-glucose-6-phosphate dehydrogenase–HRP conjugate antibody (ab34601; Abcam). α-YFP (Takara Bio Inc.), α-Ssa1 (Winkler et al., 2012b), and α-Sse1 antibodies were used at a dilution of 1:10,000. α-Hsp104 (Abcam) was used at a dilution of 1:5,000. Band intensities were quantified using Fiji (Schindelin et al., 2012) and analyzed in MATLAB.
SDD-AGE analysis was performed as previously described (Kryndushkin et al., 2003; Halfmann and Lindquist, 2008). In brief, 40 µg of the lysates was incubated for 5 min at 37°C in 0.5 M Tris-acetate-EDTA, 2% SDS, 5% glycerol, and 0.05% bromophenol blue and subjected to electrophoresis in 1.5% agarose with 0.1% SDS, without heating the samples or the gel during running. Prion protein was detected by immunoblotting with α-YFP.
Sedimentation profiling of yeast lysates was performed by centrifuging 1 ml of each lysate, adjusted to 0.5 mg/ml in lysis buffer, at 65,000 rpm in a TLA110 fixed angle rotor for 50 min using a Beckman Optima TLX Preparative Ultracentrifuge. The supernatant fractions were collected and the pellets were then resuspended in 1 ml of lysis buffer. Fractions were separated by SDS-PAGE or SDD-AGE and analyzed by Western blotting.
Database depositions
Representative tomograms have been deposited in the Electron Microscopy Databank with accession codes EMD-3118 (Fig. S2 E), EMD-3119 (Fig. S1 C), and EMD-3124 (Fig. 2 D).
Online supplemental material
Fig. S1 shows live-cell fluorescence/differential interference contrast microscopy and cryoelectron tomography of NM-YFP dot aggregates. The fluorescent dot aggregates are resolved as bundles of hexagonally packed fibril arrays by electron tomography of both freeze-substituted and vitreous cryosections. In Fig. S2, tomographic sections of dot aggregates in yeast strains Δssa1, Δssa2, and Δsse1 show marked structural changes. The fibril arrays are disrupted or absent in Δssa1 and Δssa2 strains, and dark, amorphous material is associated with the aggregates. These features are confirmed in tomograms of cryosections. In the Δsse1 strain, large aggregates with long fibrils are overlaid by a meshwork of amorphous material. Fig. S3 shows FRAP curves reporting on molecular mobility in NM-YFP dot aggregates. In all of the above strains, the prion protein is immobile, including the ones with nonfibrillar regions. Fig. S4 contains line traces of fluorescence images of dots in wild-type, Δssa2, and Δsse1 strains, showing that Hsp104 is located around the periphery of the dot aggregates. The oligonucleotides used in this study are listed in Table S1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505104/DC1.
Supplementary Material
Acknowledgments
We thank David Houldershaw, Yanni Goudetsidis, and Richard Westlake for providing computing support, and Luchun Wang for EM technical assistance. We are very grateful to Bram Koster, Linda van Driel, and Frank Faas for installation of their correlative software package on our transmission EM and Richard Hayward for critical reading of the manuscript. We especially thank Jens Tyedmers for invaluable advice on yeast genetics, provision of background strains and cloning vectors, and critical reading of the manuscript. Finally, we thank members of the Birkbeck EM group, past and present, for helpful discussions.
This work was supported by Wellcome Trust Program and equipment grants 079605, 086018, 089050, and 101488.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- SDD-AGE
- semidenaturating detergent agarose gel electrophoresis
References
- Abrams J.L., Verghese J., Gibney P.A., and Morano K.A.. 2014. Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289:13155–13167. 10.1074/jbc.M113.530014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A., and Rajendran L.. 2009. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 64:783–790. 10.1016/j.neuron.2009.12.016 [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A., Chang J.J., Leforestier A., McDowall A., Salamin L.M., Norlén L.P., Richter K., Blanc N.S., Studer D., and Dubochet J.. 2004a Cryo-electron microscopy of vitreous sections. EMBO J. 23:3583–3588. 10.1038/sj.emboj.7600366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A., Norlen L.P., and Dubochet J.. 2004b Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J. Struct. Biol. 148:131–135. 10.1016/j.jsb.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Allen K.D., Wegrzyn R.D., Chernova T.A., Müller S., Newnam G.P., Winslett P.A., Wittich K.B., Wilkinson K.D., and Chernoff Y.O.. 2005. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics. 169:1227–1242. 10.1534/genetics.104.037168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev S.N., Gracheva E.O., Richmond J.E., and Liebman S.W.. 2008. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol. Biol. Cell. 19:2433–2443. 10.1091/mbc.E08-01-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., and Kelly J.W.. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919. 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., and Cullin C.. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. 10.1093/nar/21.14.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U., Keller P.W., Cheng N., Wall J.S., and Steven A.C.. 2011. In Sup35p filaments (the [PSI+] prion), the globular C-terminal domains are widely offset from the amyloid fibril backbone. Mol. Microbiol. 79:523–532. 10.1111/j.1365-2958.2010.07466.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell R.W., and Lomas D.A.. 1997. Conformational disease. Lancet. 350:134–138. 10.1016/S0140-6736(97)02073-4 [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Inge-Vechtomov S.G., Derkach I.L., Ptyushkina M.V., Tarunina O.V., Dagkesamanskaya A.R., and Ter-Avanesyan M.D.. 1992. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast. 8:489–499. 10.1002/yea.320080702 [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Derkach I.L., and Inge-Vechtomov S.G.. 1993. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 24:268–270. 10.1007/BF00351802 [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist S.L., Ono B., Inge-Vechtomov S.G., and Liebman S.W.. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 268:880–884. 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- Chernova T.A., Wilkinson K.D., and Chernoff Y.O.. 2014. Physiological and environmental control of yeast prions. FEMS Microbiol. Rev. 38:326–344. 10.1111/1574-6976.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow E.T., and Li L.. 2011. Newly identified prions in budding yeast, and their possible functions. Semin. Cell Dev. Biol. 22:452–459. 10.1016/j.semcdb.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.M. 2003. Protein folding and misfolding. Nature. 426:884–890. 10.1038/nature02261 [DOI] [PubMed] [Google Scholar]
- Duennwald M.L., Echeverria A., and Shorter J.. 2012. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 10:e1001346 10.1371/journal.pbio.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S., Vonk W.I., and Frydman J.. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15:1231–1243. 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Park K.W., Du Z., Morano K.A., and Li L.. 2007. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 177:1583–1593. 10.1534/genetics.107.077982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fändrich M. 2007. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 64:2066–2078. 10.1007/s00018-007-7110-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P.C., Ness F., Edwards S.R., Cox B.S., and Tuite M.F.. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40:1357–1369. 10.1046/j.1365-2958.2001.02478.x [DOI] [PubMed] [Google Scholar]
- Glover J.R., Kowal A.S., Schirmer E.C., Patino M.M., Liu J.J., and Lindquist S.. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 89:811–819. 10.1016/S0092-8674(00)80264-0 [DOI] [PubMed] [Google Scholar]
- Gong H., Romanova N.V., Allen K.D., Chandramowlishwaran P., Gokhale K., Newnam G.P., Mieczkowski P., Sherman M.Y., and Chernoff Y.O.. 2012. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet. 8:e1002634 10.1371/journal.pgen.1002634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R., and Lindquist S.. 2008. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp. 17:838 10.3791/838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., and Hayer-Hartl M.. 2011. Molecular chaperones in protein folding and proteostasis. Nature. 475:324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hawes P., Netherton C.L., Mueller M., Wileman T., and Monaghan P.. 2007. Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells. J. Microsc. 226:182–189. 10.1111/j.1365-2818.2007.01767.x [DOI] [PubMed] [Google Scholar]
- Helsen C.W., and Glover J.R.. 2012. A new perspective on Hsp104-mediated propagation and curing of the yeast prion [PSI+]. Prion. 6:234–239. 10.4161/pri.19913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T., Hines J.K., Sahi C., Aron R., and Craig E.A.. 2008. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA. 105:16596–16601. 10.1073/pnas.0808934105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Jones G., Wegrzyn R.D., and Masison D.C.. 2000. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 156:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., and Frydman J.. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095. 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., and Craig E.A.. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Noma S., Pack C.G., Tsuji T., Kinjo M., and Taguchi H.. 2009. Single mother-daughter pair analysis to clarify the diffusion properties of yeast prion Sup35 in guanidine-HCl-treated [PSI] cells. Genes Cells. 14:1045–1054. 10.1111/j.1365-2443.2009.01333.x [DOI] [PubMed] [Google Scholar]
- Kawai-Noma S., Pack C.G., Kojidani T., Asakawa H., Hiraoka Y., Kinjo M., Haraguchi T., Taguchi H., and Hirata A.. 2010. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J. Cell Biol. 190:223–231. 10.1083/jcb.201002149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C.Y., Tittmann P., Gross H., Gebert R., Aebi M., and Wüthrich K.. 1997. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA. 94:6618–6622. 10.1073/pnas.94.13.6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kryndushkin D., and Wickner R.B.. 2007. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2149–2154. 10.1091/mbc.E07-02-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D.S., Alexandrov I.M., Ter-Avanesyan M.D., and Kushnirov V.V.. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278:49636–49643. 10.1074/jbc.M307996200 [DOI] [PubMed] [Google Scholar]
- Krzewska J., and Melki R.. 2006. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 25:822–833. 10.1038/sj.emboj.7600985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W., Schorb M., Welsch S., Picco A., Kaksonen M., and Briggs J.A.. 2011. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 192:111–119. 10.1083/jcb.201009037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S.W., and Chernoff Y.O.. 2012. Prions in yeast. Genetics. 191:1041–1072. 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S.W., and Derkatch I.L.. 1999. The yeast [PSI+] prion: making sense of nonsense. J. Biol. Chem. 274:1181–1184. 10.1074/jbc.274.3.1181 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 1997. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120:343–352. 10.1006/jsbi.1997.3919 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Miller S.B., Ho C.T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., et al. . 2015a Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34:778–797. 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.B., Mogk A., and Bukau B.. 2015b Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J. Mol. Biol. 427:1564–1574. 10.1016/j.jmb.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Misselwitz B., Staeck O., and Rapoport T.A.. 1998. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell. 2:593–603. 10.1016/S1097-2765(00)80158-6 [DOI] [PubMed] [Google Scholar]
- Moosavi B., Wongwigkarn J., and Tuite M.F.. 2010. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI(+)] prion but not for prion propagation. Yeast. 27:167–179. 10.1002/yea.1742 [DOI] [PubMed] [Google Scholar]
- Moran C., Kinsella G.K., Zhang Z.R., Perrett S., and Jones G.W.. 2013. Mutational analysis of Sse1 (Hsp110) suggests an integral role for this chaperone in yeast prion propagation in vivo. G3 (Bethesda). 3:1409–1418. 10.1534/g3.113.007112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai H., Kuno T., Tanaka H., Hirata D., Miyakawa T., and Tanaka C.. 1993. Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene. 132:57–66. 10.1016/0378-1119(93)90514-4 [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller R., and Funk M.. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 156:119–122. 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- Park Y.N., Zhao X., Yim Y.I., Todor H., Ellerbrock R., Reidy M., Eisenberg E., Masison D.C., and Greene L.E.. 2014. Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot. Cell. 13:635–647. 10.1128/EC.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov V.V., Smirnov V.N., and Ter-Avanesyan M.D.. 1997. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 277:381–383. 10.1126/science.277.5324.381 [DOI] [PubMed] [Google Scholar]
- Rampelt H., Kirstein-Miles J., Nillegoda N.B., Chi K., Scholz S.R., Morimoto R.I., and Bukau B.. 2012. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31:4221–4235. 10.1038/emboj.2012.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsomaniki M.A., Kotsantis P., Symeonidou I.E., Giakoumakis N.N., Taraviras S., and Lygerou Z.. 2012. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 28:1800–1801. 10.1093/bioinformatics/bts241 [DOI] [PubMed] [Google Scholar]
- Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andréasson C, Lindquist S, and Bukau B. 2008. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 3:e1763. doi:http://dx.doi.org/10.1371/journal.pone.0001763 [DOI] [PMC free article] [PubMed]
- Saibil H.R., Seybert A., Habermann A., Winkler J., Eltsov M., Perkovic M., Castaño-Diez D., Scheffer M.P., Haselmann U., Chlanda P., et al. . 2012. Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc. Natl. Acad. Sci. USA. 109:14906–14911. 10.1073/pnas.1211976109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., and Serio T.R.. 2005. Prion protein remodelling confers an immediate phenotypic switch. Nature. 437:262–265. 10.1038/nature03981 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio T.R., Cashikar A.G., Moslehi J.J., Kowal A.S., and Lindquist S.L.. 1999. Yeast prion Ψ+ and its determinant, Sup35p. Methods Enzymol. 309:649–673. 10.1016/S0076-6879(99)09043-6 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., and Tsien R.Y.. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Sharma D., and Masison D.C.. 2008. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics. 179:1301–1311. 10.1534/genetics.108.089458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Martineau C.N., Le Dall M.T., Reidy M., Masison D.C., and Kabani M.. 2009. Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS ONE. 4:e6644 10.1371/journal.pone.0006644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. 2011. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 6:e26319 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Wu Y.X., Jung G., Tutar Y., Eisenberg E., Greene L.E., and Masison D.C.. 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell. 4:289–297. 10.1128/EC.4.2.289-297.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Nagy M., Ni W., Tyagi N.K., Fenton W.A., López-Giráldez F., Overton J.D., Horwich A.L., and Brady S.T.. 2013. Molecular chaperone Hsp110 rescues a vesicle transport defect produced by an ALS-associated mutant SOD1 protein in squid axoplasm. Proc. Natl. Acad. Sci. USA. 110:5428–5433. 10.1073/pnas.1303279110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E.M., Vonk W.I., and Frydman J.. 2014. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr. Opin. Cell Biol. 26:139–146. 10.1016/j.ceb.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., and Estrada L.D.. 2008. Protein misfolding and neurodegeneration. Arch. Neurol. 65:184–189. 10.1001/archneurol.2007.56 [DOI] [PubMed] [Google Scholar]
- Tessarz P., Mogk A., and Bukau B.. 2008. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 68:87–97. 10.1111/j.1365-2958.2008.06135.x [DOI] [PubMed] [Google Scholar]
- Tipton K.A., Verges K.J., and Weissman J.S.. 2008. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell. 32:584–591. 10.1016/j.molcel.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente M.P., and Shorter J.. 2013. The metazoan protein disaggregase and amyloid depolymerase system: Hsp110, Hsp70, Hsp40, and small heat shock proteins. Prion. 7:457–463. 10.4161/pri.27531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott A., Shaner L., and Morano K.A.. 2005. The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics. 170:1009–1021. 10.1534/genetics.105.043109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M.F., and Cox B.S.. 2006. The [PSI+] prion of yeast: a problem of inheritance. Methods. 39:9–22. 10.1016/j.ymeth.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Tuite M.F., Marchante R., and Kushnirov V.. 2011. Fungal prions: structure, function and propagation. Top. Curr. Chem. 305:257–298. 10.1007/128_2011_172 [DOI] [PubMed] [Google Scholar]
- Tyedmers J. 2012. Patterns of [PSI+] aggregation allow insights into cellular organization of yeast prion aggregates. Prion. 6:191–200. 10.4161/pri.18986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J., Treusch S., Dong J., McCaffery J.M., Bevis B., and Lindquist S.. 2010. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc. Natl. Acad. Sci. USA. 107:8633–8638. 10.1073/pnas.1003895107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel L.F., Valentijn J.A., Valentijn K.M., Koning R.I., and Koster A.J.. 2009. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur. J. Cell Biol. 88:669–684. 10.1016/j.ejcb.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Venable J.H., and Coggeshall R.. 1965. A simplified lead citrate stain for use in electron microscopy. J. Cell Biol. 25:407–408. 10.1083/jcb.25.2.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N., Bradley M.E., and Liebman S.W.. 2009. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol. Microbiol. 73:1101–1114. 10.1111/j.1365-2958.2009.06836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., and Philippsen P.. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808. 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., and Philippsen P.. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- Wang Y., Meriin A.B., Zaarur N., Romanova N.V., Chernoff Y.O., Costello C.E., and Sherman M.Y.. 2008. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 23:451–463. 10.1096/fj.08-117614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn R.D., Bapat K., Newnam G.P., Zink A.D., and Chernoff Y.O.. 2001. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21:4656–4669. 10.1128/MCB.21.14.4656-4669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.J., Lyakhovetsky R., Werdiger A.C., Gitler A.D., Soen Y., and Kaganovich D.. 2012. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc. Natl. Acad. Sci. USA. 109:15811–15816. 10.1073/pnas.1205829109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J., Tyedmers J., Bukau B., and Mogk A.. 2012a Chaperone networks in protein disaggregation and prion propagation. J. Struct. Biol. 179:152–160. 10.1016/j.jsb.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Winkler J., Tyedmers J., Bukau B., and Mogk A.. 2012b Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 198:387–404. 10.1083/jcb.201201074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S., Weissman J.S., and Dillin A.. 2014. Differential scales of protein quality control. Cell. 157:52–64. 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Yang Z., Hong J.Y., Derkatch I.L., and Liebman S.W.. 2013. Heterologous Gln/Asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet. 9:e1003236 10.1371/journal.pgen.1003236 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.