The cotranslational transfer of nascent membrane proteins to the SecYEG translocon is facilitated by a reorientation of the SecY-bound signal recognition particle (SRP) receptor, FtsY, which accompanies the formation of a quaternary targeting complex consisting of SecYEG, FtsY, SRP, and the ribosome.
Abstract
Cotranslational protein targeting delivers proteins to the bacterial cytoplasmic membrane or to the eukaryotic endoplasmic reticulum membrane. The signal recognition particle (SRP) binds to signal sequences emerging from the ribosomal tunnel and targets the ribosome-nascent-chain complex (RNC) to the SRP receptor, termed FtsY in bacteria. FtsY interacts with the fifth cytosolic loop of SecY in the SecYEG translocon, but the functional role of the interaction is unclear. By using photo-cross-linking and fluorescence resonance energy transfer measurements, we show that FtsY–SecY complex formation is guanosine triphosphate independent but requires a phospholipid environment. Binding of an SRP–RNC complex exposing a hydrophobic transmembrane segment induces a rearrangement of the SecY–FtsY complex, which allows the subsequent contact between SecY and ribosomal protein uL23. These results suggest that direct RNC transfer to the translocon is guided by the interaction between SRP and translocon-bound FtsY in a quaternary targeting complex.
Introduction
The signal recognition particle (SRP) is responsible for cotranslational protein targeting in both prokaryotic and eukaryotic cells (Kudva et al., 2013; Denks et al., 2014). SRP recognizes and binds to hydrophobic signal sequences with high affinity (Bernstein et al., 1989; Bornemann et al., 2008; Holtkamp et al., 2012; Akopian et al., 2013b) and delivers the ribosome-nascent-chain complex (RNC) to the membrane-bound SRP receptor, which in bacteria consists of the single GTPase subunit FtsY (Luirink et al., 1994; Valent et al., 1998; Angelini et al., 2005). FtsY is crucial for SRP-dependent targeting because it guides the SRP–RNC complex to the SecYEG translocon or, alternatively, to the YidC insertase for subsequent insertion of the nascent chain into the membrane (Koch et al., 1999; Jagath et al., 2000; Angelini et al., 2005; Braig et al., 2011; Welte et al., 2012). The FtsY–SRP complex dissociates on GTP hydrolysis by both partners after RNCs have been delivered to SecYEG (Kusters et al., 1995; Egea et al., 2004) and this GTP hydrolysis is influenced by the lipid and protein environment (de Leeuw et al., 2000; Angelini et al., 2006). Details on the mechanism by which the RNCs are transferred from the SRP–RNC–FtsY complex to the SecYEG translocon are as yet undefined.
FtsY comprises three domains: The universally conserved N and G domains are required for GTP hydrolysis and SRP interaction, and the N-terminal A domain is involved in membrane binding (de Leeuw et al., 1997; Montoya et al., 1997; Fig. 1 A). The A domain of Escherichia coli FtsY binds to negatively charged phospholipids (Parlitz et al., 2007; Braig et al., 2009; Stjepanovic et al., 2011) and to the conserved cytoplasmic loops C4 and C5 of SecY, which is the channel-forming subunit of the SecYEG translocon (Kuhn et al., 2011). By site-specific in vivo cross-linking, residue 357 within the C5 loop was shown to contact FtsY, SecA, and the ribosomal tunnel exit protein uL23 (formerly named L23; Kuhn et al., 2011; Ban et al., 2014; Fig. 1 B). This could indicate that FtsY occupies the ribosome-binding site of the SecYEG translocon until it is relocated by the binding of an SRP–RNC complex. In such a scenario, FtsY could guide SRP–RNC complexes directly to the SecYEG translocon, bypassing the need for the transfer of the RNC from a lipid-bound FtsY–SRP–RNC complex to SecYEG.
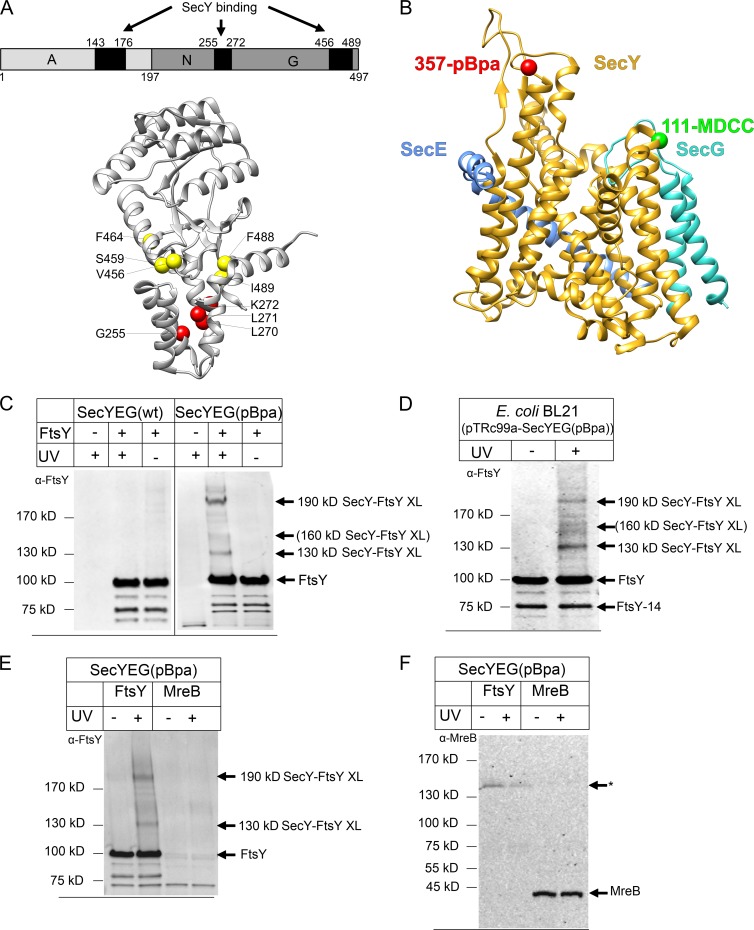
Figure 1.
A reconstituted system for analyzing the SecY–FtsY interaction. (A) Domain structure of E. coli FtsY (top). The FtsY regions that were cross-linked to SecY are indicated by black boxes. Crystal structure of the FtsY NG domain with the membrane targeting sequence (Stjepanovic et al., 2011; PDB accession no. 2YHS; bottom). The FtsY residues within the N domain that were cross-linked to SecY are indicated by red spheres and those within the G domain by yellow spheres. (B) Crystal structure of E. coli SecYEG (Park et al., 2014; PDB accession no. 3J45). Position 357 of SecY, where pBpa was incorporated, is indicated in red and position 111, at which the fluorophore MDCC was attached, is indicated in green. (C) SecYEG(pBpa)-PLs (SecYEG(pBpa); 10 nM SecYEG) were incubated with FtsY (1.2 µM) or buffer (−). After UV treatment for activating pBpa, the sample was extracted with Na2CO3 for removing excess FtsY and separated by SDS-PAGE. After Western transfer, the blot was decorated with polyclonal α-FtsY antibodies. Indicated are FtsY and the SecY–FtsY cross-link products at 130 and 190 kD. Weak cross-linking products at ∼160 kD are indicated in brackets. As a control, PLs containing SecYEG without pBpa [SecYEG(wt)] were analyzed. (D) For comparison, an in vivo cross-linking assay of E. coli cells expressing SecYEG(pBpa) was performed. After UV exposure of whole cells, SecYEG(pBpa) and its cross-link products were purified and separated by SDS-PAGE. The UV-dependent cross-link products are indicated. Independently of UV exposure, FtsY co-purifies with SecY and is visible as full-length protein and N-terminally truncated derivative (FtsY-14). (E) SecYEG(pBpa)-PLs were incubated with 1.2 µM FtsY or 1.2 µM MreB and treated as described in A. After Western transfer, the blot was decorated with polyclonal α-FtsY antibodies. (F) The material described in E was decorated with polyclonal α-MreB antibodies. The asterisk indicates an unidentified protein that is nonspecifically recognized by α-MreB in the FtsY-containing sample.
In this study, we have developed a purified system for monitoring the interaction between SecY and FtsY in the presence of RNCs, nontranslating ribosomes, or SecA. Our data are consistent with the formation of a quaternary SecY–FtsY–SRP–RNC complex, in which, upon binding of an RNC–SRP complex, a cross-linker at position 357 of SecY moves away from FtsY and instead contacts ribosomal protein uL23. These data support a model for RNC transfer to the SecYEG translocon in which the interaction between SRP and FtsY induces the mutual exposure of the ribosome-binding site on SecY and the SecY-binding site on the ribosome. This facilitates the delivery of RNCs to the ribosome-binding site of the SecYEG translocon.
Results
Interaction between the SecYEG translocon and the SRP receptor monitored by UV light–induced cross-linking
Site-specific in vivo cross-linking using the photoreactive phenylalanine derivative p-benzoyl-l-phenylalanine (pBpa; Ryu and Schultz, 2006) is a powerful tool for monitoring SecY interactions in the unbiased setting of living cells. However, cross-linking in vivo is unsynchronized, which makes it difficult to analyze the individual effects of ribosomes, RNCs, or SecA on the SecY–FtsY contact.
To overcome this problem, we developed an in vitro system in which purified SecYEG carrying pBpa at position 357 of SecY (SecYEG[pBpa]) was reconstituted into liposomes. Residue 357 was selected for incorporation of pBpa because this residue was shown to be in contact with FtsY, SecA, and ribosomal protein uL23 in vivo (Kuhn et al., 2011). The incorporation of pBpa at this position had no effect on SecY function in vivo because the corresponding plasmid was able to complement the cold-sensitive phenotype of secY mutant strains (Kuhn et al., 2011).
When proteoliposomes (PLs) containing SecYEG(pBpa) were incubated with purified FtsY and exposed to UV light, two UV-dependent cross-linking products of ∼130 and 190 kD were detected by α-FtsY antibodies, which were not visible in the absence of FtsY or when unmodified SecYEG lacking pBpa was reconstituted into liposomes (Fig. 1 C). Consistent with previous studies, the 130- and 190-kD SecY–FtsY cross-link products were also observed in vivo, that is, when whole cells expressing SecYEG(pBpa) were exposed to UV irradiation and the resulting cross-link products were purified via metal-affinity chromatography (Kuhn et al., 2011; Fig. 1 D). Previously, the 190-kD product was identified by mass spectrometry (MS) as a cross-link between SecY and residues 143–176 of the flexible N-terminal A domain of FtsY (Kuhn et al., 2011; Fig. 1 A). Using high-resolution MS, we now identified the 130-kD product as a cross-link between SecY and the C-terminal GTPase domain of FtsY (Fig. 1 A and Table 1), which confirms previous proposals that FtsY contains at least two binding sites for SecY (de Leeuw et al., 2000; Angelini et al., 2006; Kuhn et al., 2011). A position-dependent different mobility of pBpa cross-linking products on SDS-PAGE have been observed before (Das and Oliver, 2011; Kuhn et al., 2011; Sachelaru et al., 2013) and are probably the result of differences in the 3D structure. In addition to the 130- and 190-kD SecY–FtsY cross-link products, we observed weak cross-link products at ∼160 kD both in vivo and in vitro (Fig. 1, C and D), which were also further identified by MS. The 160-kD product corresponds to a cross-link between SecY and the N domain of FtsY (Fig. 1 A and Table 1). However, the 160-kD cross-link product was formed in small amounts both in vivo and in vitro and was not always detectable. An additional weak cross-link product was occasionally observed on top of the 190-kD band (Fig. 1 C). Because these two cross-linking products were not always observed, they were not included in our analyses. In summary, our data demonstrate that the same contacts between SecY and FtsY can be observed in living cells and in the purified in vitro system. Our data also show that FtsY can contact SecY via its N-terminal A domain or via the C-terminal GTPase domain.
Table 1. Interlinked peptides of FtsY with SecY(pBpa) identified by MS .
| Mass (MH+)/Da | Peptide of FtsY | Peptide of SecY | P valuea | Intensity | Bandb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sequencec | Site | P valued | Sequencec | Site | P valued | ||||
| 3011.5843 | K.IITNLTEGASRK.Q | G255 | 4.4 × 10−6 | K.SGAFVPGIUPGEQTAK.Y | R357 | 6.2 × 10−7 | 2.7 × 10−12 | 1.3 × 10+6 | ×2 |
| 3801.921 | R.DAEALYGLLKEEMGEILAK.V | K272 | 2.8 × 10−2 | K.SGAFVPGIUPGEQTAK.Y | R357 | 4.0 × 10−8 | 1.2 × 10−9 | 1.1 × 10+7 | ×2 |
| 3817.9328 | R.DAEALYGLLKEEM*GEILAK.V | K272 | 3.1 × 10−6 | K.SGAFVPGIUPGEQTAK.Y | R357 | 7.4 × 10−3 | 2.3 × 10−8 | 7.4 × 10+6 | ×2 |
| 3801.9419 | R.DAEALYGLLKEEMGEILAK.V | L270 | 1.2 × 10−4 | K.SGAFVPGIUPGEQTAK.Y | R357 | 5.9 × 10−4 | 7.0 × 10−8 | 2.0 × 10+6 | ×2 |
| 3801.9302 | R.DAEALYGLLKEEMGEILAK.V | L271 | 6.2 × 10−6 | K.SGAFVPGIUPGEQTAK.Y | R357 | 7.4 × 10−3 | 4.6 × 10−7 | 1.4 × 10+6 | ×2 |
| 3384.7577 | K.GGVIFSVADQFGIPIR.Y | V456 | 4.8 × 10−7 | K.SGAFVPGIUPGEQTAK.Y | R357 | 3.1 × 10−6 | 1.5 × 10−12 | 3.6 × 10+6 | ×1 |
| 3384.7606 | K.GGVIFSVADQFGIPIR.Y | S459 | 4.0 × 10−6 | K.SGAFVPGIUPGEQTAK.Y | R357 | 6.5 × 10−10 | 2.6 × 10−15 | 3.6 × 10+6 | ×1 |
| 3384.7611 | K.GGVIFSVADQFGIPIR.Y | F464 | 2.4 × 10−4 | K.SGAFVPGIUPGEQTAK.Y | R357 | 1.4 × 10−4 | 3.3 × 10−8 | 3.3 × 10+6 | ×1 |
| 2976.4774 | K.ADDFIEALFAR.E | F488/I489 | 2.1 × 10−4 | K.SGAFVPGIUPGEQTAK.Y | R357 | 6.6 × 10−3 | 1.4 × 10−6 | 8.4 × 10+7 | ×1 |
Sites of cross-linking are underlined. SecYEG(pBpa) with pBpa incorporated at position 357 of SecY was expressed in vivo and purified after UV exposure of whole cells. Cross-linked FtsY peptides were analyzed as described in the Materials and methods. Residues 198–280 of FtsY correspond to the N domain, and residues 281–497 of FtsY correspond to the GTPase (G) domain.
P value for cross-linked peptide.
Bands from SDS gel at 130 kD (×1) and 160 kD (×2).
Underlined residues signify site of cross-link; M*, oxidized methionine.
Subscores per peptide.
The cross-linking experiments were routinely performed with PLs containing ∼10 nM SecYEG(pBpa) in the presence of 1.2 µM FtsY. The large excess of FtsY was used because a substantial portion of FtsY is bound to the phospholipid surface of PLs (Braig et al., 2009). Although the presence of identical SecY–FtsY cross-link products in living E. coli cells and in vitro supports a specific interaction between the two proteins, the large excess of FtsY in the in vitro system could in principle favor nonspecific cross-linking. To exclude nonspecific interactions, we performed in vitro pBpa cross-linking experiments in the presence of MreB, the bacterial actin homolog (Graumann, 2007). Like FtsY, MreB associates with the bacterial membrane via an N-terminal helix and is largely found in close contact with the membrane in vivo (Nurse and Marians, 2013; Strahl et al., 2014). When SecYEG(pBpa)-PL were incubated with 1.2 µM of FtsY or MreB, we observed the 130- and 190-kD SecY–FtsY cross-link products but no SecY–MreB cross-link products (Fig. 1, E and F). This demonstrates that the cross-links between SecY and FtsY are specific and not the result of random interactions in the in vitro system.
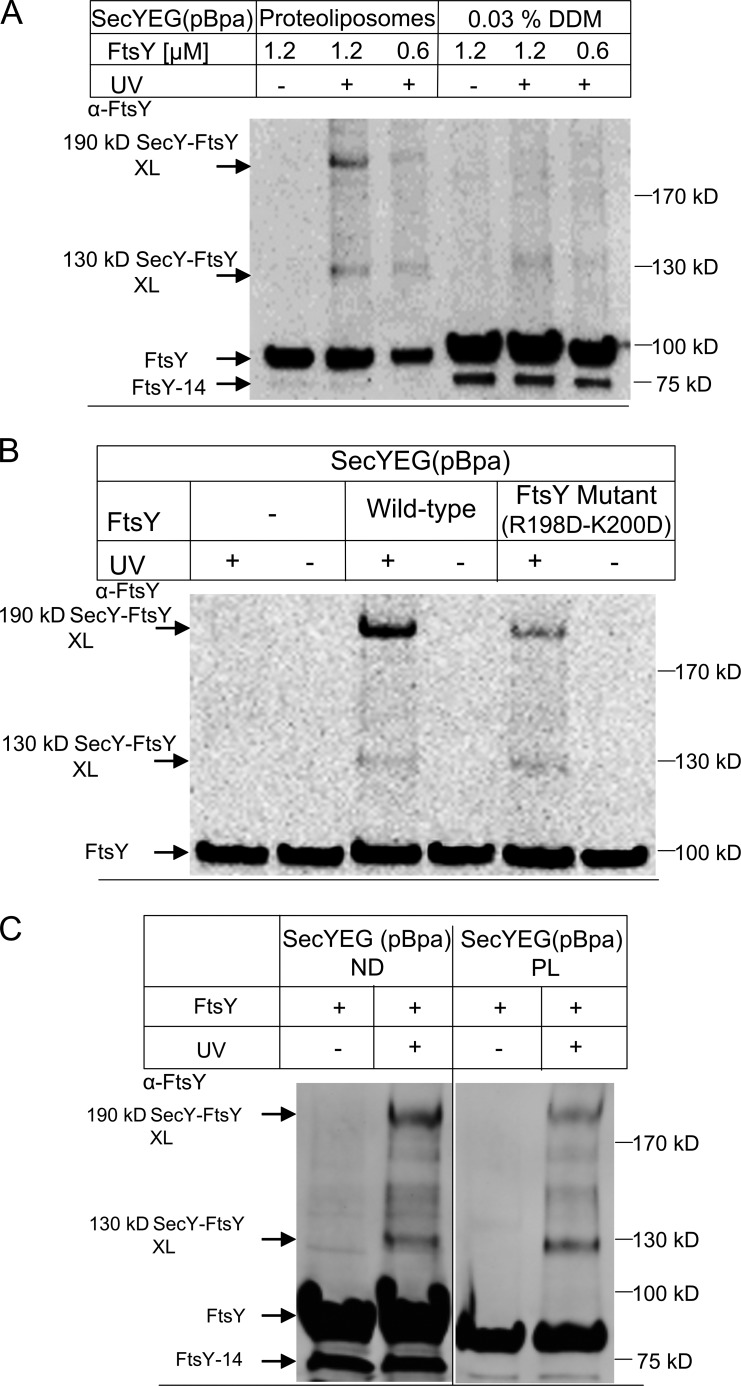
FtsY contains two lipid-binding helices, and lipid contact of FtsY stimulates the FtsY–SRP interaction (Parlitz et al., 2007; Braig et al., 2009; Lam et al., 2010; Stjepanovic et al., 2011). To examine whether lipid contact is also required for the interaction between FtsY and SecY, we compared the cross-linking pattern of SecYEG(pBpa) in detergent (0.03% dodecyl-maltoside [DDM]) with the cross-linking pattern of SecYEG(pBpa)-PL. In PLs, both the 130- and 190-kD FtsY cross-link products were visible in an FtsY concentration–dependent manner (Fig. 2 A), whereas SecYEG(pBpa) in DDM showed no 190-kD cross-linking product and only a weak and diffuse 130-kD product (Fig. 2 A). The SecYEG(pBpa)-PL sample was routinely extracted with sodium carbonate after UV exposure to remove the non–membrane-bound portion of FtsY. Therefore, the Western blot signal for non–cross-linked FtsY is weaker in the SecYEG(pBpa)-PL sample, although the same amount of FtsY was present in the PLs and detergent sample during UV exposure. To exclude the possibility that carbonate extraction influenced the outcome of this experiment, we also analyzed the samples without carbonate extraction and found cross-links only in the PL sample (Fig. S1). The possibility that an efficient FtsY–SecY contact requires the lipid environment was further verified by analyzing the cross-linking pattern between SecYEG(pBpa)-PL and the FtsY(R198D-K200D) mutant, which is impaired in lipid binding (Weiche et al., 2008; Braig et al., 2009). The FtsY(R198D-K200D) mutant showed a significantly reduced 190-kD cross-link, but no major change in the 130-kD cross-link, compared with wild-type FtsY (Fig. 2 B).
Figure 2.
The SecY–FtsY interaction requires the presence of lipids. (A) SecYEG(pBpa)-PL or SecYEG(pBpa) in detergent solution (0.03% DDM) were incubated with different FtsY concentrations and UV activated. SecYEG(pBpa)-PLs were Na2CO3 treated as described in the legend to Fig. 1, and SecYEG(pBpa) in detergent was directly precipitated with TCA. Pellets after centrifugation were loaded on SDS-PAGE and further treated as in Fig. 1. FtsY-14 does not efficiently interact with membranes (Weiche et al., 2008) and is therefore not visible in the carbonate-treated sample. (B) SecYEG(pBpa)-PL were incubated with wild-type FtsY or the FtsY(R198D-K200D) mutant. The conditions for cross-linking and the detection of cross-links were as in A. (C) The SecY–FtsY interaction was monitored in both PL and nanodiscs. Treatment and conditions were identical as in Fig. 1, except that cross-linking products of SecYEG(pBpa)-nanodiscs were not carbonate extracted.
Finally, we also analyzed cross-linking of SecYEG(pBpa) that was inserted into nanodiscs as described previously (Ge et al., 2014). Nanodiscs are planar phospholipid bilayer discs held together by membrane scaffold protein derived from apolipoprotein B of high-density lipoproteins (Alami et al., 2007). This allows highly purified, biochemically well-defined translocons to be studied in a native-like membrane environment. In the presence of SecYEG(pBpa)-nanodiscs, we observed the same cross-linking pattern as with PLs (Fig. 2 C). As with SecYEG(pBpa) in DDM, SecYEG(pBpa)-nanodiscs are not carbonate resistant and therefore the nanodisc samples after cross-linking were TCA precipitated directly. Thus, although the cross-linking experiments with nanodiscs and PLs were performed in the presence of the same amounts of FtsY, the Western blot signal of non–cross-linked FtsY in the PL sample was weaker because a substantial portion of the lipid-bound FtsY was removed by the carbonate extraction.
SecY–FtsY interaction is nucleotide independent and persists in the presence of SRP
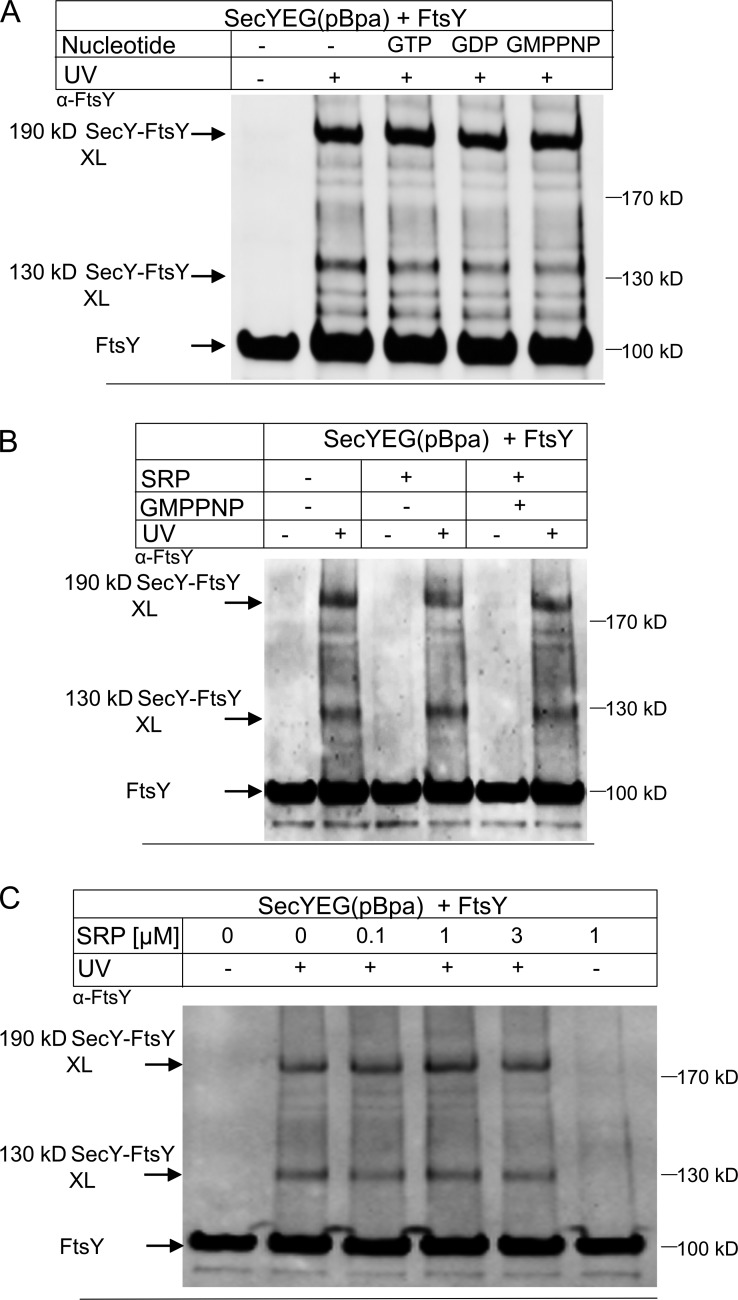
FtsY exhibits GTPase activity (Kusters et al., 1995), and we therefore analyzed whether the cross-link between FtsY and SecY was influenced by nucleotides. The addition of GTP, GDP, or the nonhydrolyzable GTP analogue guanosine 5′-(β,γ-imido) triphosphate (GMP-PNP) at a concentration of 50 µM did not significantly change the appearance of the 190- and 130-kD SecY–FtsY cross-link products (Fig. 3 A). This indicates that the SecY–FtsY interaction is largely independent of the presence of guanine nucleotides. This conclusion is supported by the equilibrium titrations described in the following section.
Figure 3.
The SecY–FtsY cross-links are nucleotide independent and not influenced by SRP. (A) SecYEG(pBpa)-PL (10 nM SecYEG) were incubated with FtsY (1.2 µM), either in the absence or presence of nucleotides (50 µM final concentration). Samples were processed as in Fig. 1. Indicated are FtsY and the SecY–FtsY cross-link products at 130 and 190 kD. (B) SecYEG(pBpa)-PL were incubated with FtsY as in A. When indicated SRP (1 µM) and GMP-PNP (50 µM) were added before UV exposure. (C) SecYEG(pBpa)-PL were incubated with FtsY as described in A and increasing SRP concentrations together with GMP-PNP (50 µM). Samples were further processed as in A.
We also tested whether the addition of SRP influenced the FtsY–SecY interaction. SecYEG(pBpa)-PL was incubated with FtsY and 1 µM SRP in the presence of GMP-PNP, which stabilizes the FtsY–SRP complex (Jagath et al., 2000; Shan et al., 2004). We did not observe a significant effect of SRP on the FtsY–SecY cross-linking pattern (Fig. 3 B). The SecY–FtsY cross-link was also not influenced when the SRP concentration was varied between 0.1 and 3 µM in the presence of GMP-PNP (Fig. 3 C). To demonstrate that SRP forms a complex with FtsY under these conditions, we performed blue native PAGE analyses (Fig. S2). These data confirm the presence of an FtsY–SRP complex and a SecYEG–FtsY–SRP complex in the presence of GMP-PNP. Thus, the interaction between SecY and FtsY is preserved even in the presence of SRP.
SecYEG-FtsY complex formation monitored by fluorescence resonance energy transfer (FRET)
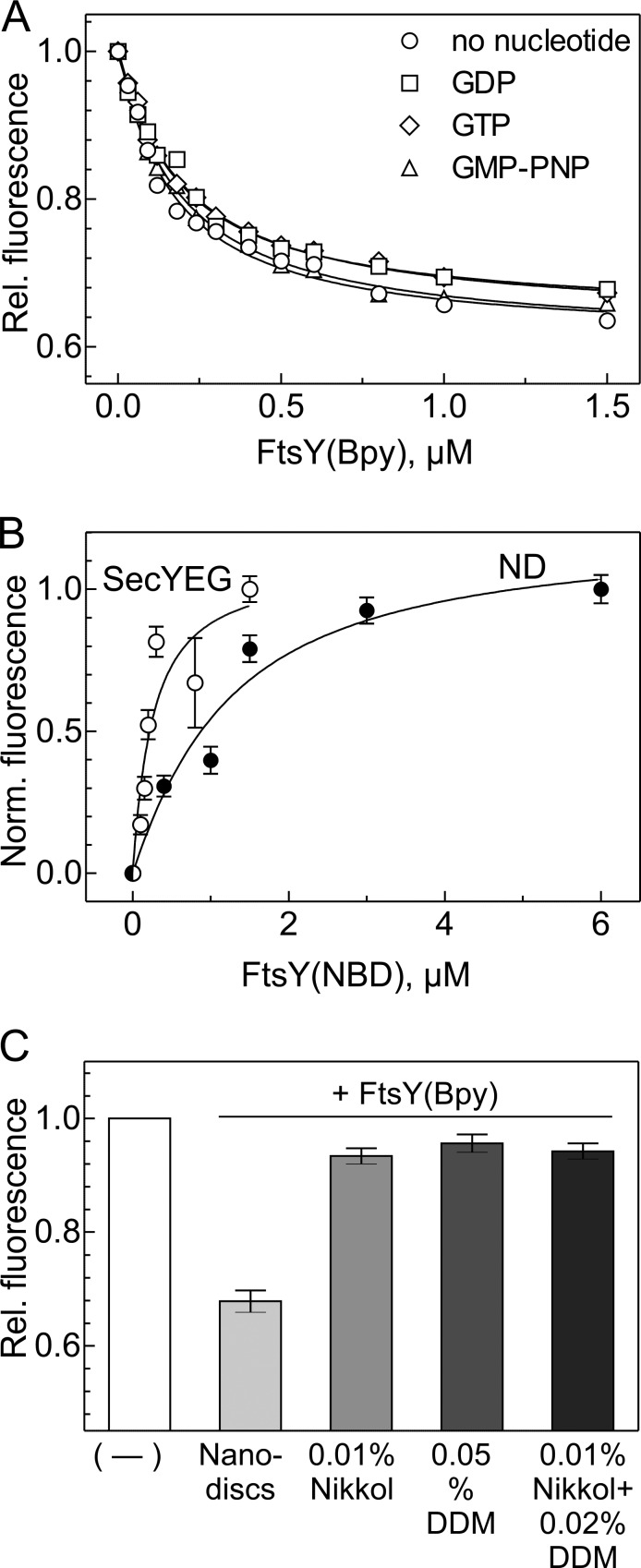
To quantitate FtsY binding to the translocon, we have performed equilibrium titrations, monitoring FRET between SecYEG labeled with the donor fluorophore 7-diethylamino-3-((((2-maleimidyl)ethyl) amino)-carbonyl)coumarin (MDCC) at position 111 of SecY; this position is outside the FtsY binding site (Kuhn et al., 2011) and, among several positions tested in that region, provided the largest FRET change upon complex formation with labeled FtsY. FtsY was labeled with BODIPY FL (Bpy; acceptor; Invitrogen) at position 196 near the C-terminus of the A domain; the latter position was chosen as the most suitable from several label positions in the A domain. The FRET approach monitors complex formation by the decrease of donor fluorescence owing to energy transfer from the excited donor to the acceptor. The FRET couple MDCC and Bpy are characterized by a Förster radius of about 3 nm (i.e., the FRET efficiency is 50% when the two labels are 3 nm apart). For these experiments, the translocon was embedded in nanodiscs (Ge et al., 2014). Complex formation led to an ∼40% decrease in MDCC fluorescence (Fig. 4 A), indicating that donor and acceptor approached one another in the complex. The titration yielded a Kd for the SecY–FtsY complex of 0.18 ± 0.02 µM. About the same Kd value was measured when complex formation was monitored by the fluorescence increase of the fluorophore 7-nitrobenzofurazan (NBD), which is particularly sensitive to its environment, attached to cysteine engineered into position 26 of FtsY (Fig. 4 B). The NBD label also allowed for monitoring complex formation with nanodiscs not containing SecYEG, resulting in a Kd value of 1.2 ± 0.4 µM (Fig. 4 B). Thus, the affinity of the complex of FtsY with SecYEG embedded in a phospholipid bilayer is dominated by the interaction between FtsY and SecY. Complex formation was independent of the presence of a guanine nucleotide, as about the same Kd values were obtained in the absence of nucleotides and in the presence of GTP, GMP-PNP, or GDP (Fig. 4 A). This is in line with the cross-link data obtained with PLs (Fig. 3 A).
Figure 4.
FtsY binding to SecYEG monitored by fluorescence. (A) Equilibrium titrations of SecYEG-FtsY complex formation monitored by FRET. SecYEG embedded into nanodiscs was titrated with FtsY in the absence of guanine nucleotide (○) or in the presence of 0.5 mM each of GDP (□), GTP (⋄), or GMP-PNP (△). SecY was labeled with the donor fluorophore MDCC at position 111 and FtsY with the acceptor fluorophore BODIPY FL at position 196 [FtsY(Bpy)]. Donor fluorescence is plotted relative to the initial fluorescence measured before FtsY(Bpy) addition and set to 1.0. Plotted are mean values from two titrations; SEMs were ≤5%. Kd values were obtained by nonlinear fitting using equation 1 (see Materials and methods); errors are SEMs of the fits. (B) Equilibrium titrations of FtsY binding to SecYEG in nanodiscs (○) and to empty nanodiscs (●) monitored by the fluorescence increase of an NBD label attached to position 26 of FtsY, corrected for the linear signal increase measured upon titrating FtsY(NBD) into buffer (Materials and methods). Plotted are mean values from two titrations each; error bars represent SEM. Upon complex formation, the fluorescence signal of FtsY(NBD) increased 6–8-fold; to facilitate comparison, the fluorescence increase is plotted in normalized numbers. Kd values of 0.16 ± 0.02 µM (SecYEG in nanodiscs) and 1.2 ± 0.4 µM (empty nanodiscs) were determined by nonlinear fitting using equation 1 (Materials and methods); errors are SEMs of the fits. (C) Inhibition of FtsY binding to SecYEG in the presence of nonionic detergents. The fluorescence of SecYEG(MDCC) (0.05 µM) embedded in nanodiscs or solubilized by detergent, as indicated, was monitored on addition of FtsY(Bpy) (1 µM). The fluorescence signal measured after the addition of FtsY(Bpy) is plotted relative to the respective initial signal set to 1.0. The error bars indicate the SD (n = 3).
We observed no FRET change when labeled FtsY was added to labeled SecYEG in the presence of the nonionic detergents DDM or Nikkol (Fig. 4 C), indicating that no complex was formed, in keeping with the lack of cross-links under these conditions (Fig. 2 A). These results indicate that efficient binding of FtsY to SecYEG requires the presence of phospholipids and that it is impaired in the presence of nonionic detergents.
The SecY–FtsY cross-link is abolished by RNCs exposing a nascent membrane protein
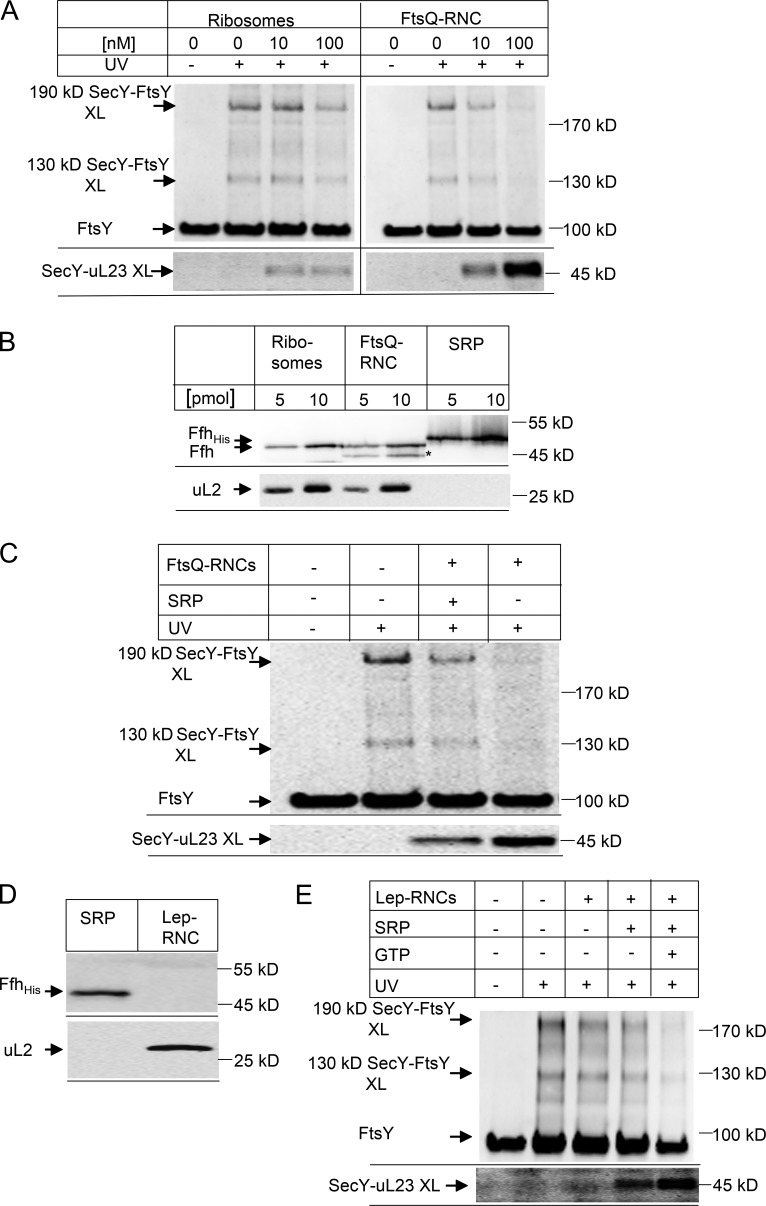
Position 357 in SecY is part of the ribosome-binding site in SecY, and the presence of FtsY close to this position potentially allows for the alignment of the incoming SRP–RNC complex with the SecY channel. For analyzing the SecY–FtsY interaction in response to the addition of the SRP–RNC complex, we incubated SecYEG(pBpa)-PLs with FtsY in the presence of vacant ribosomes or with RNCs of the single spanning membrane protein FtsQ (FtsQ-RNCs), which is a typical substrate for SRP-dependent targeting to SecYEG (Scotti et al., 1999; van der Laan et al., 2001). The addition of 10-nM vacant ribosomes did not substantially influence the appearance of the 130- and 190-kD SecY–FtsY cross-links (Fig. 5 A). Only when vacant ribosomes were added at a concentration of 100 nM was the intensity of both cross-link bands reduced (Fig. 5 A). When the same experiment was performed with FtsQ-RNCs, we observed significantly weaker FtsY–SecY cross-link bands already with 10 nM RNCs and an almost complete disappearance of the cross-link products when the RNC concentration was increased to 100 nM (Fig. 5 A). The disappearance of the FtsY–SecY cross-links indicates that, in the presence of the FtsQ–RNC, FtsY loses contact with position 357 of SecY. The effect apparently is stronger with the RNC compared with vacant ribosomes, although the binding affinities with Kd values of 10 nM for RNC (see following discussion) and 20 nM (for vacant ribosomes) are not much different (Wu et al., 2012; Ge et al., 2014). This indicates that the respective complexes are structurally different.
Figure 5.
Nascent membrane proteins abolish SecY–FtsY cross-linking. (A) SecYEG(pBpa)-PL (10 nM final concentration SecYEG) were incubated with 1.2 µM FtsY and 50 µM GTP in the absence or presence of different concentrations of FtsQ ribosome nascent chains of 102 amino acid length (FtsQ-RNCs) or nontranslating ribosomes. After SDS-PAGE and Western transfer, the membrane was horizontally cut and the upper part was decorated with polyclonal α-FtsY antibodies (top) and the lower part with polyclonal α-uL23 antibodies (bottom). (B) Different amounts of the ribosomes and FtsQ-RNCs used in (A) were separated by SDS-PAGE and after Western transfer decorated with antibodies against Ffh, the protein component of the bacterial SRP. Purified His-tagged SRP was used as reference, and antibodies against the ribosomal protein uL2 were used to determine the ribosome concentration. A band nonspecifically recognized by α-Ffh antibodies in the FtsQ-RNC sample is marked with an asterisk. (C) SecYEG(pBpa)-PL were incubated with FtsY as in A in the presence of 50 µM GTP. When indicated, 100 nM FtsQ-RNC was added without additional SRP or with 100 nM SRP. (D) RNCs of leader peptidase (Lep-RNCs, 94 amino acid length, 10 pmol) were separated by SDS-PAGE and after Western transfer were decorated with antibodies against uL2 and Ffh. His-tagged Ffh (10 pmol) served as a control. (E) SRP-free Lep-RNCs (100 nM) were incubated with SecYEG(pBpa)-PL and FtsY as in A. When indicated, SRP (1 µM) and GTP (50 µM) were added. The top panel was decorated with antibodies against FtsY and the bottom panel with antibodies against uL23.
If the change of the SecY–FtsY interaction is part of a functional targeting cycle, one could expect that the disappearance of the SecY–FtsY cross-link is accompanied by the appearance of a SecY-ribosome cross-link. This possibility was analyzed by probing the cross-linked material with antibodies against ribosomal protein uL23, which is located at the ribosomal tunnel exit and a major contact site for both SecY (Kuhn et al., 2011) and SRP (Gu et al., 2003; Halic et al., 2006). In the presence of FtsQ-RNCs, we observed a concentration-dependent increase of a SecY–uL23 cross-link product (Fig. 5 A, bottom). Because vacant ribosomes also bind to SecY (Prinz et al., 2000; Welte et al., 2012; Ge et al., 2014) and show a weak ability to displace FtsY from SecY (Fig. 5 A), we also tested for potential SecY–uL23 cross-links in the presence of vacant ribosomes. We observed very low amounts of SecY–uL23 cross-link products (Fig. 5 A, bottom), suggesting that although vacant ribosomes can partially dislocate FtsY from SecY, they do not establish a prominent contact with SecY via the uL23 protein. It is likely that in the absence of a nascent chain the ribosome-SecY contact shows a certain degree of flexibility, which reduces the extent of cross-linking from position 357 of SecY.
The relocation of FtsY by RNCs did not require the addition of SRP, and we therefore analyzed whether SRP was present in sufficient amounts in the RNC and ribosome preparation. The amount of ribosomes/RNCs in the cross-linking assay corresponded to 4 pmol (at 10 nM final concentration) and 40 pmol (at 100 nM final concentration), respectively. By Western blotting with antibodies against the SRP protein Ffh, we determined the SRP content in ribosomes and RNCs and compared it to 5 and 10 pmol purified SRP. The data show that both ribosomes and RNCs contained sufficient amounts of SRP to mediate cotranslational targeting (Fig. 5 B). In a control experiment, we also probed for ribosomal protein uL2 (Fig. 5 B). When comparing the SRP content of ribosomes/RNCs with purified SRP, we estimate that ∼25% of the ribosomes/RNCs had SRP bound.
Adding purified SRP to a final concentration of 100 nM to the cross-linking assay did not enhance FtsY dislocation and cross-linking to uL23 but instead slightly reduced FtsY dislocation and the SecY–uL23 cross-link product (Fig. 5 C). This indicates that in our experimental system SRP is not limiting for cotranslational targeting of FtsQ-RNCs to the FtsY–SecYEG complex. The potentially inhibitory effect of increasing SRP concentrations was not further analyzed but supports in vivo data showing that high levels of Ffh or its M domain interfere with ribosome activity, probably by occupying proteins bL20 and uL24 (Yosef et al., 2010), which are located close to the uL23 protein at the tunnel exit (Halic et al., 2004; Schaffitzel et al., 2006).
The effect of SRP on the RNC-induced relocation of FtsY was further analyzed by using in vitro synthesized and SRP-free Lep75-RNCs, exposing the N-terminal portion of leader peptidase encompassing the signal-anchor sequence (Fig. 5 D). In the absence of SRP, Lep-RNCs induced only a weak relocation of FtsY (Fig. 5 E). However, in the presence of SRP and GTP we observed complete relocation of FtsY and strong contact between SecY and uL23 (Fig. 5 E). These data demonstrate that SRP is required for efficient relocation of FtsY by RNCs.
Binding of RNC or SRP induces a rearrangement of the SecYEG-FtsY complex
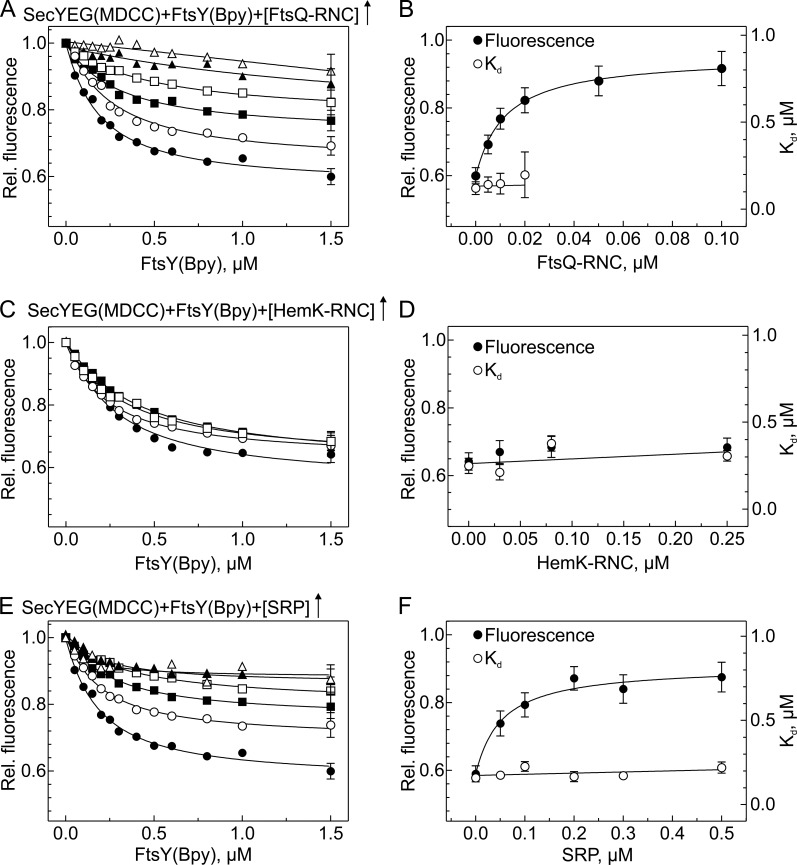
The disappearance of the SecY–FtsY cross-link products in the presence of RNCs can be explained by dissociation of FtsY from the SecYEG translocon or by a conformational change that abolishes SecY–FtsY cross-linking. We used the FRET approach to examine whether the loss of cross-links between FtsY and the translocon observed upon the addition of FtsQ-RNC was attributable to a loss of the complex (i.e., competitive binding) or to a different arrangement of FtsY in the respective ternary complex compared with the binary SecYEG-FtsY complex. We titrated SecYEG with FtsY in the presence of increasing amounts of FtsQ-RNC. We observed a strong reduction in the FRET amplitude (Fig. 6 A) with no change in Kd (Fig. 6 B), indicating concurrent, noncompetitive binding. The control experiment with HemK-RNC, which lacks a hydrophobic signal sequence in the nascent chain, showed no change of the FRET amplitude or in the Kd (Fig. 6, C and D), indicating that the rearrangement induced by the FtsQ-RNC was a result of the signal sequence. A similar rearrangement was observed when increasing amounts of SRP were present in the titration of SecYEG with FtsY (Fig. 6 E). The substantially smaller FRET changes observed in the presence of FtsQ-RNC (Fig. 6 A) or SRP (Fig. 6 E) indicate that concurrent binding of those two ligands is accompanied by a structural change of the SecYEG-FtsY complex, which increases the distance between donor and acceptor, thereby diminishing FRET. From the dependence of the donor fluorescence at saturation on the concentration of FtsQ-RNC (Fig. 6 B), a Kd of 8 nM was estimated assuming noncompetitive ternary complex formation, as suggested by the fact that the Kd of FtsY binding remained unchanged (Fig. 6 B). This value is very close to the 10-nM Kd observed for the binding of Lep75-RNC to SecYEG (Ge et al., 2014). Similarly, the 40-nM Kd for the binding of SRP to SecYEG-bound FtsY, as estimated from the data depicted in Fig. 6 F, is similar to previously reported values (Jagath et al., 2000; Peluso et al., 2000; Bornemann et al., 2008).
Figure 6.
SecYEG-FtsY complex formation in the presence of FtsQ-RNC, HemK-RNC, or SRP. (A) SecYEG-FtsY complex formation in the presence of FtsQ-RNC. MDCC-labeled SecYEG in nanodiscs was titrated with FtsY(Bpy) in the presence of increasing concentrations of FtsQ-RNC (µM): 0, ●; 0.005, ○; 0.01, ■; 0.02, □; 0.05, ▲; 0.1, △. For clarity, representative error bars (SEM; n = 2) are indicated only on the last titration point. Kd values of ∼0.18 µM were determined by nonlinear fitting using equation 1 (Materials and methods); at concentrations of FtsQ-RNC >0.02 µM, the fluorescence change was too small to allow for the estimation of reliable Kd values. (B) Effect of FtsQ-RNC binding on the SecYEG-FtsY complex. Kd values from A are plotted against the concentration of FtsQ-RNC (○; right Y-axis). Donor (MDCC) fluorescence measured at saturation with FtsY(Bpy) (●, left Y-axis) is plotted relative to the initial fluorescence of SecYEG(MDCC) measured before the addition of FtsY(Bpy); error bars represent SEM from (A). Nonlinear fitting to equation 2 (Materials and methods) yielded an apparent Kd = 8 ± 1 nM for the binding of FtsQ-RNC to the SecYEG-FtsY complex. (C) SecYEG-FtsY complex formation in the presence of HemK-RNC. Titrations were performed as in A in the presence of increasing concentrations of HemK-RNC (µM): 0, ●; 0.03, ○; 0.08, ■; 0.25, □. (D) No effect of HemK-RNC on SecYEG-FtsY complex formation. (E) SecYEG-FtsY complex formation in the presence of SRP. MDCC-labeled SecYEG was titrated with FtsY(Bpy) in the presence of increasing concentrations of SRP (µM): 0, ●; 0.05, ○; 0.1, ■; 0.2, □; 0.3, ▲; 0.5, △. Apparent Kd values of ∼0.2 µM (○, right Y-axis) were determined by nonlinear fitting as in A. (F) Effect of SRP on the SecYEG-FtsY complex. Kd values from C are plotted against the SRP concentration (○). Donor (MDCC) fluorescence measured at saturation (●, left Y-axis) is plotted relative to the initial fluorescence measured before the addition of FtsY(Bpy). Nonlinear fitting as in B yielded an apparent Kd of 40 ± 10 nM for the binding of SRP to the SecYEG-FtsY complex.
In summary, these results show that an RNC exposing a signal peptide can reorient FtsY at the C5-loop of SecY. This supports a model in which the SRP receptor occupies the ribosome-binding site of the SecY translocon until it is dislocated by RNC binding to the translocon. Dislocation of FtsY by nontranslating ribosomes is much less efficient and leads to a different arrangement of ribosomes on SecYEG because the cross-link of SecY to protein uL23 is significantly weaker (Fig. 5 A).
FtsY and SecA compete for SecY binding
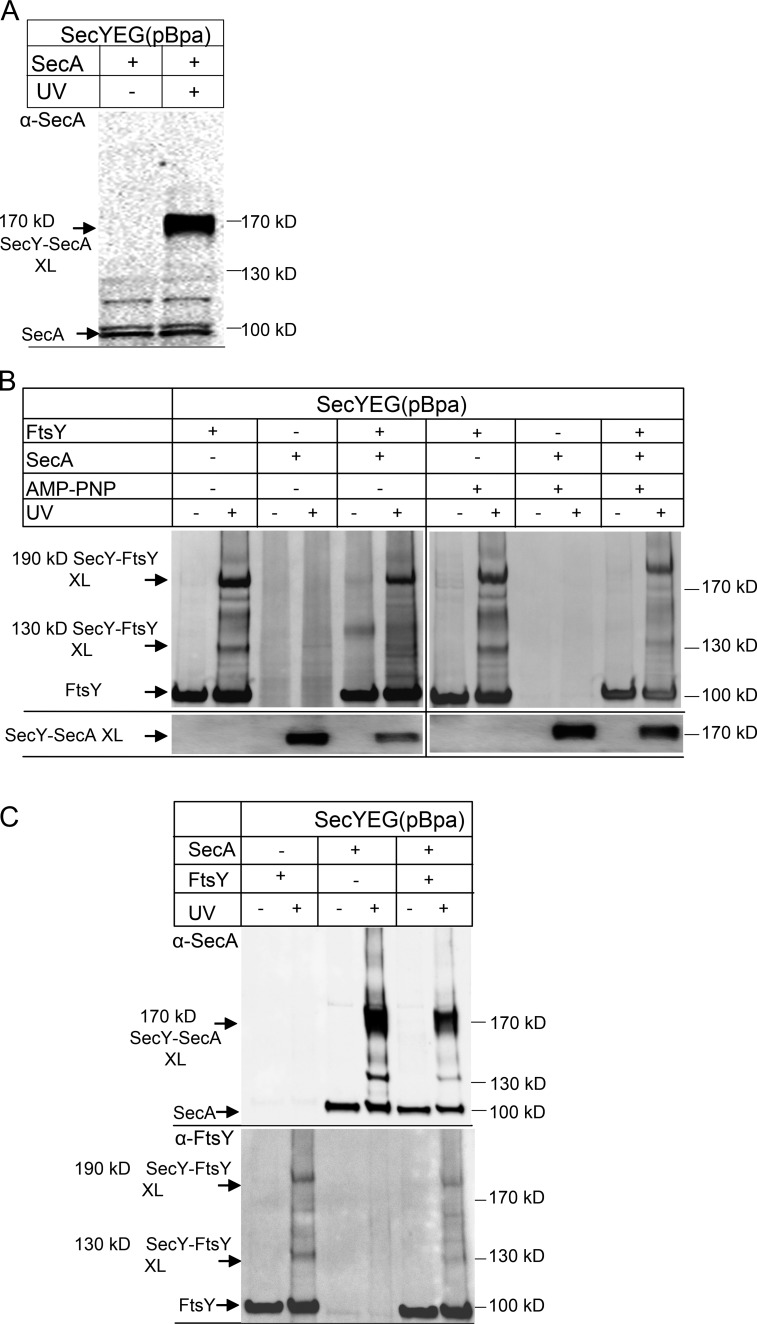
The C5-loop of SecY also binds SecA, which serves as receptor and motor protein for post-translational protein targeting (Mori and Ito, 2006; Das and Oliver, 2011; Kudva et al., 2013). When SecYEG(pBpa)-PLs were incubated with purified SecA, we observed a strong UV-dependent cross-link product at ∼170 kD (Fig. 7 A). This cross-link product was also observed in vivo and was previously identified as SecY–SecA cross-linking product by high-resolution MS (Kuhn et al., 2011). We then tested whether the appearance of the SecY–FtsY cross-linking products would be influenced by the addition of SecA. In the absence of SecA, the two SecY–FtsY cross-linking products were readily visible (Fig. 7 B). In the presence of just SecA, the FtsY antibodies did not recognize any specific bands after UV exposure, indicating no cross-reactivity of the α-FtsY antibodies. The α-SecA antibody, on the other hand, recognized the strong SecY–SecA cross-linking product at 170 kD (Fig. 7 B, bottom). When SecYEG(pBpa)-PLs were incubated with FtsY in the presence of about equimolar concentrations of SecA, both SecY–FtsY and the SecY–SecA cross-links became weaker, suggesting that the two proteins compete for binding to the SecYEG channels present in limiting amounts. We repeated the experiment in the presence of the nonhydrolyzable ATP analogue adenosine 5′-(β,γ-imido)triphosphate, which stabilizes the SecY–SecA interaction (Taufik et al., 2013). Under these conditions, SecA appeared to have an advantage over FtsY in binding to SecY because we found slightly more of the SecY–SecA cross-linking product and less of the SecY–FtsY cross-linking products compared with the nucleotide-free sample (Fig. 7 B).
Figure 7.
SecA and FtsY compete for access to the SecYEG translocon. (A) SecYEG(pBpa)-PL (10 nM SecYEG final concentration) were incubated with 1.0 µM SecA and UV exposed, when indicated. After Na2CO3 extraction the sample was separated by SDS-PAGE and, after Western transfer, decorated with polyclonal α-SecA antibodies. Indicated are SecA and the 170 kD SecY–SecA cross-link product. (B) SecYEG(pBpa)-PL were incubated with FtsY or SecA or with both proteins and UV exposed. When both FtsY and SecA were present, FtsY was added before SecA. After SDS-PAGE and Western transfer, the membrane was decorated with polyclonal α-FtsY antibodies (top). After stripping of the membrane by treatment with SDS/DTT, the membrane was decorated with polyclonal α-SecA antibodies (bottom). (C) As in B, but when both FtsY and SecA were present, SecA was added before FtsY. The blot was decorated with polyclonal α-SecA antibodies (top) or polyclonal α-FtsY antibodies (bottom).
The competition between FtsY and SecA was also visible when SecA was added first to SecYEG(pBpa)-PLs and only then FtsY was added (Fig. 7 C). As before, we noticed a reduction of both the SecY–SecA cross-linking product and the SecY–FtsY cross-linking products.
Discussion
The mechanism and the timing of RNC transfer from the FtsY–SRP complex to the SecYEG translocon have been a largely unresolved issue in cotranslational protein transport. In vivo cross-linking and biochemical studies have revealed that FtsY interacts directly with SecY, which suggests that SRP delivers RNCs directly to the translocon-bound FtsY (Angelini et al., 2005, 2006; Kuhn et al., 2011). However, the determinants for the FtsY–SecY interaction and the consequences for the targeting reaction were unknown. By combining qualitative in vitro site-directed cross-linking with quantitative FRET analyses, we now demonstrate that FtsY binds to SecY rather strongly, with a Kd of 0.18 µM. The FtsY–SecY interaction is GTP independent but requires the presence of phospholipids. Anionic phospholipids are required for membrane binding of FtsY (de Leeuw et al., 2000; Braig et al., 2009; Erez et al., 2010) and influence FtsY–SRP complex formation and stability (Lam et al., 2010; Braig et al., 2011). In E. coli, anionic phospholipids are enriched at the SecYEG translocon (Gold et al., 2010) and these lipids induce a random coil-to-helix transition of the essential and conserved lipid-binding helix of FtsY (Stjepanovic et al., 2011). This lipid-dependent conformational switch is nucleotide independent (Stjepanovic et al., 2011) and was suggested to enhance the interaction of FtsY with specific partner proteins, such as SecY. This is supported by our current observation that mutating the lipid-binding helix interferes with FtsY–SecY cross-linking. The GTP-independent lipid–SecY interaction of FtsY also agrees with the almost exclusive membrane localization of FtsY that has been observed in E. coli cells (Mircheva et al., 2009). In proteobacteria, FtsY contains a second lipid-binding helix at the N-terminus of the nonessential A domain, which additionally contributes to the stability of membrane binding (Weiche et al., 2008; Braig et al., 2009). In E. coli FtsY, one SecY-binding site is sandwiched between the two lipid-binding helices, and this interaction gives raise to the 190-kD cross-linking product (Kuhn et al., 2011). On the other hand, our MS data identify the 130-kD cross-linking product as a cross-link between the G domain of FtsY and SecY. The latter cross-linking product is weakly detectable even in the absence of lipids or when one of the two lipid-binding helices of FtsY is mutated. Thus, the G domain can interact with SecY even when the interaction of SecY with the A domain of FtsY is impaired. This probably explains why the A domain is not essential for FtsY function in E. coli and absent in many bacterial FtsY homologues (Bibi et al., 2001; Eitan and Bibi, 2004; Weiche et al., 2008).
Previous work has demonstrated that the SecA-SecYEG interaction also depends on anionic lipids (Lill et al., 1990; Benach et al., 2003). Like FtsY, SecA has a high-affinity binding site for SecY and a low-affinity binding site for anionic phospholipids (Dapic and Oliver, 2000). Thus, FtsY, the receptor of SRP in the cotranslational targeting pathway, and SecA, the receptor in post-translational targeting, probably bind to SecY in similar ways. This is also reflected by the similar affinities of FtsY (Kd = 0.18 µM) and SecA (Kd = 0.05 µM; Douville et al., 1995) for SecYEG and the observation that FtsY and SecA compete for SecY binding, as monitored by cross-linking. However, a significant portion of both SecA and FtsY is bound to the phospholipid surface of PLs (Ulbrandt et al., 1992; Braig et al., 2009) and therefore the exact amounts of FtsY or SecA that are available for SecY binding might differ. Furthermore, in our current analyses we monitored interactions only at SecY residue 357, although both FtsY and SecA have been cross-linked to other SecY residues (Mori and Ito, 2006; Kuhn et al., 2011). Nevertheless, despite these limitations, the molar SecA/FtsY ratio in vivo is estimated to be ∼1:5 (Kudva et al., 2013), which could indicate that FtsY has more frequent access to the SecY translocon. This is in line with the concept of SRP–FtsY–dependent cotranslational targeting in bacteria, which primarily delivers aggregation-prone membrane proteins to the SecYEG translocon, whereas SecA handles less aggregation-prone periplasmic and outer membrane proteins.
FtsY binds to the C4 and C5 loops of SecY (Kuhn et al., 2011), which also constitute major binding sites for ribosomes. The C4 loop reaches into the ribosomal peptide tunnel, contacts helices H6/24/50 of 23S rRNA, and is close to a loop of ribosomal protein uL23 (Frauenfeld et al., 2011); the latter is involved in sensing the presence of a nascent peptide chain within the tunnel (Bornemann et al., 2008). These close ribosome-SecY contacts are possible only if RNCs or ribosomes are able to reposition FtsY from the ribosome-binding site of the SecYEG translocon. Our data now demonstrate that FtsQ-RNCs relocate FtsY at the C5 loop in such a way that FtsY loses contact with the conserved SecY residue 357. This residue, in turn, is then occupied by the ribosomal uL23 protein. In keeping with this observation, FRET data indicate that RNC binding induces a conformational change of the FtsY–SecY complex and that FtsY is not dissociated from the SecYEG translocon. This finding suggests the presence of a quaternary complex consisting of SRP–RNC, SecYEG, and FtsY. The possible existence of such a quaternary complex has been proposed before (Shen et al., 2012; Saraogi et al., 2014), although our data suggest a different sequence of interactions. Saraogi et al. (2014) reported the existence of a soluble ternary RNC–SRP–FtsY complex that subsequently interacts with SecYEG. This interaction was suggested to displace the SRP NG domain from the ribosomal tunnel exit and to facilitate the subsequent SecY–uL23 interaction (Saraogi et al., 2014). According to that model, the signal sequence is then transferred from the SRP M domain to SecYEG and the FtsY–SRP complex is relocated to the distal site of the SRP RNA, where GTP hydrolysis leads to FtsY–SRP dissociation (Akopian et al., 2013a; Voigts-Hoffmann et al., 2013). Although we cannot exclude the existence of a ternary RNC–SRP–FtsY complex, our current and previous data (Mircheva et al., 2009; Braig et al., 2011) indicate that most of SRP–RNCs are targeted to SecYEG- or lipid-bound FtsY. The difference between our study and that of Saraogi et al. could be explained by the fact that the latter study used SecYEG in detergent whereas we used SecYEG embedded in phospholipids. As our data show, the presence of phospholipids is required for efficient SecY-FtsY interaction. Furthermore, considering that >90% of FtsY in E. coli is membrane bound and that a ternary RNC–SRP–FtsY complex is only inefficiently targeted to SecY (Mircheva et al., 2009), RNC–SRP–FtsY complexes that are not translocon-bound probably represent a minor species during targeting.
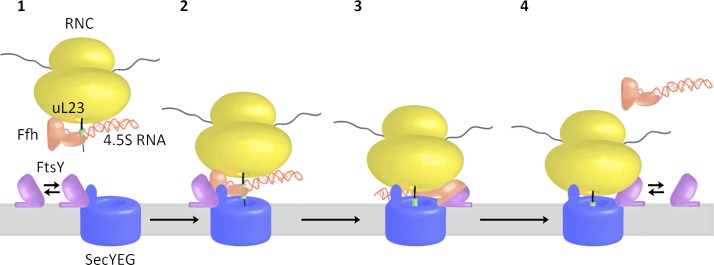
In summary, our data suggest a sequential handover at the C5 loop of SecY during cotranslational targeting (Fig. 8). FtsY is bound to the C5 loop of SecY and can recruit SRP–RNCs directly to the SecYEG translocon (Fig. 8, steps 1 and 2). SRP–RNC binding to FtsY induces a conformational change at the SecY-FtsY interface that allows SecY to contact uL23 at the ribosomal tunnel exit (Fig. 8, step 3). The conformational change at the SecY–FtsY interface is matched by a conformational change at the SRP–ribosome interface upon FtsY binding. This is indicated by cryo-EM and cross-linking studies, which suggest movement of the SRP NG domain (Halic et al., 2006) and demonstrate the loss of the SRP-uL23 cross-link (Pool et al., 2002) upon binding of the SRP receptor (Fig. 8, step 3). Collectively, these data support a model in which the interaction between SRP and FtsY results in a reciprocal conformational change that simultaneously exposes the SecY-binding site of the ribosome and the ribosome-binding site on SecY. This allows for the subsequent RNC–SecY interaction and for the insertion of the growing polypeptide into the membrane. These reciprocal conformational changes probably also facilitate subsequent GTP hydrolysis and dissociation of the FtsY–SRP complex (Fig. 8, step 4), which could include the relocation of the FtsY–SRP complex to the distal end of the 4.5S RNA, as recently suggested (Voigts-Hoffmann et al., 2013).
Figure 8.
Model for cotranslational targeting of SRP–RNCs to the SecYEG translocon. For details, see the text.
Materials and methods
Protein purification
The SecYEG complex was purified from BL21 pSup-BpaRS-6TRN cells expressing pTRc99aSecY(His)EG carrying an amber stop codon at codon 357 of SecY (Kuhn et al., 2011). In that strain, SecY(His)EG is under the control of the lac promoter in plasmid pTRc99a as backbone. Plasmid pSUP-pBpaRS-6TRN encodes six copies of the TAG suppressor tRNA under the control of the proK promoter and a mutant Methanocaldococcus jannaschii tyrosyl-tRNA synthetase under the control of the glnS promoter (Ryu and Schultz, 2006). Cells were grown in minimal medium supplemented with 1 mM pBpa at 30°C and 220 rpm to an OD at 600 nm (OD600) of 1.0 and induced with 1 mM IPTG for 4 h. Purification by metal-affinity chromatography followed basically the protocol published previously (Braig et al., 2011). In brief, after cell breakage using a French press cell, a crude membrane fraction was isolated and solubilized with S1 buffer for 1 h at 4°C (S1 buffer: 20 mM Tris/HCl, pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 1% DDM, 5 mM imidazole, and complete protease inhibitor [Roche]). After centrifugation for 25 min at 30.000 rpm in a Ti50.2 rotor, the supernatant was added to preequilibrated TALON beads (Clontech Laboratories) and incubated for 1 h at 4°C. After washing with S1 buffer containing 0.03% DDM, SecYEG was eluted with S1 buffer containing 0.03% DDM and 200 mM imidazole.
SecA containing an N-terminal (His)10-tag (pET19b-SecA) was constructed by cloning the chromosomal secA gene after NcoI and XhoI digestion into the equally digested pET19b vector backbone. In that construct secA expression is under T7 promoter control. FtsY containing a C-terminal (His)6-tag (pTrc99a-FtsY) was constructed by amplifying FtsY from plasmid pTP37 (Powers and Walter, 1997) and digesting the PCR product with NcoI and EcoRI and cloned into the equally digested pTrc99a as vector backbone. ftsY expression is under lac promoter control. Both proteins were purified from BL21(DE3)/BL21 cells as published previously for FtsY (Braig et al., 2011), with slight modifications. Cells were grown in LB medium at 37°C and 180 rpm and induced at an OD600 of 0.6 with 0.5 mM IPTG for 4 h (SecA) and 2 h (FtsY) at 37°C. Protein purification was performed using Ni-NTA FF crude (GE Healthcare) and an ÄKTA chromatography system (GE Healthcare). The equilibration/wash buffer (buffer A) contained 50 mM Hepes-KOH, pH 7.5, 1 M ammonium acetate, 10 mM magnesium acetate, 20 mM imidazole, and 10% glycerol, and His-tagged proteins were eluted with a linear gradient from 20 to 500 mM imidazole (in buffer A). FtsY was further purified by gel filtration (buffer A without imidazole) using a Superdex S200 column (GE Healthcare). Purified proteins were rebuffered in TSM5 buffer (100 mM triethanolamine acetate, pH 8.0, 250 mM sucrose, 5 mM magnesium acetate, and 1 mM DTT). Ffh, the protein component of the bacterial SRP, was purified from DH5α harboring pTrc99a-Ffh(His), expressing ffh under control of the lac promoter in vector pTRc99a (Braig et al., 2011). Cells were grown at 37°C and 180 rpm to an OD600 of 0.5, and cells were induced with 1 mM IPTG for 4 h. Ffh was purified via TALON beads after cell breakage (buffer A with 15% glycerol). After elution Ffh was rebuffered into HT buffer + 50% glycerol (50 mM Hepes, pH 7.6, 100 mM potassium acetate, pH 7.5, 10 mM magnesium acetate, 1 mM DTT) using a PD-10 column (GE Healthcare) and stored at −20°C. MreB was purified from BL21(DE3) expressing pET3c-mreB, which was constructed by amplifying the MreB ORF from E. coli DNA using the primers 5′-CGACATATGTTGAAAAAATTTCGTGGCATG-3′ and 5′-GACAGCTTATCATCGATAAGCTTTAATGC-3′. The PCR product was digested with NdeI and HindIII restriction enzymes and ligated into similarly digested pET3c (Novagen) DNA. Purification of MreB followed a published protocol (Nurse and Marians, 2013). For cross-linking, the protein buffer was exchanged with TSM5 buffer.
Preparation of PLs
Purified E. coli phospholipids were purchased from Avanti Polar Lipids. Liposomes were formed by sonication using final concentrations of 7 mg/ml phosphatidyl-ethanolamine, 2.5 mg/ml phosphatidyl-glycerol, and 0.5 mg/ml cardiolipin in 50 mM triethanolamine acetate, pH 7.5, 1 mM DTT, yielding a total lipid concentration of 10 mg/ml. For the preparation of PLs, final concentrations ∼0.13 mg/ml lipids, 0.85% octylglycoside, and 0.15 µM purified, DDM-solubilized SecYEG(pBpa) were mixed, incubated for 20 min at 4°C, and dialyzed (Spectra/Por membrane tubing, 6–8 kD; SpectrumLabs) against 50 mM triethanolamine acetate, pH 7.5, 1 mM DTT. The PLs were pelleted (1 h at 210,000 g) and resuspended in 50 mM triethanolamine acetate, pH 7.5, 1 mM DTT, to a final protein concentration of 0.25 to 0.5 µM, verified by semi-quantitative Western blotting. Proteoliposomes were briefly sonicated before use.
In vitro and in vivo cross-linking
In vivo cross-linking followed published protocols (Kuhn et al., 2011; Sachelaru et al., 2013). For cross-linking with PLs in vitro, SecYEG (10 nM), FtsY (1.2 µM), MreB (1.2 µM), RNCs/ribosomes (10–100 nM), SecA (1.2 µM) and SRP (0.1–3 µM) were incubated for 20 min at 25°C in TSM5 buffer (see Protein purification). When indicated nucleotides were present during cross-linking at a final concentration of 50 µM. Cross-linking was performed by UV exposure on ice for 30 min in a total volume of 400 µl (365 nm; Vilber Lourmat VL-6.L). PLs were treated with 0.5 M Na2CO3, pH 11.5 (final concentration 0.2 M) for 30 min on ice and pelleted (15 min, 264,000 g). The pellet was loaded on a gradient gel (5–12%). The cross-links were visualized by Western blotting using polyclonal antibodies against FtsY, SecA, or MreB (provided by P. de Boer, Case Western University, Chicago, IL) as primary antibodies and HRP-coupled anti–rabbit secondary antibodies. Cross-links to ribosomal proteins were detected by peroxidase-coupled anti–goat secondary antibodies.
Ribosome and RNC preparation
Ribosomes were purified by sucrose gradient centrifugation as described before (Koch et al., 1999) and N-terminally His-tagged RNCs carrying the first 102 amino acids of FtsQ followed by an HA tag and a TnaC stalling sequence (pftsQ-tna and KC6 were provided by R. Beckmann, Ludwig Maximilian University of Munich, Munich, Germany) were expressed in vivo in KC6 and purified essentially as described elsewhere (Bischoff et al., 2014). In brief, KC6 pftsQ-tna cells were grown to an OD of 0.5 and induced with 1 mM IPTG for 1 h. Cells were harvested and resuspended in buffer A containing 1 mM tryptophan and 250 µg/ml chloramphenicol. Cell lysis was achieved by a French press, and cell debris was removed by centrifugation for 20 min at 16,000 rpm in an SS34 rotor. The lysate was further purified via a sucrose cushion (750 mM sucrose in buffer A) and then purified via TALON beads as described earlier. Ribosomes and FtsQ-RNC were pelleted for 2 h at 86,000 g and resuspended in TSM5 buffer. Lep-RNC (carrying the N-terminal 75 amino acids of leader peptidase and comprising a strong SRP-specific signal sequence) and HemK-RNC (carrying the nascent chain of 75 N-terminal amino acids of HemK, a cytosolic enzyme without a signal sequence) were prepared by translation of the respective truncated mRNA in vitro (Bornemann et al., 2008).
Preparation of fluorescence-labeled SecYEG
SecYEG for fluorescence labeling was expressed from a pTrc99a construct containing N-terminally His6-tagged SecE. All native cysteine residues were mutated to serine by site-directed mutagenesis using the Phusion polymerase protocol (Thermo Fisher Scientific). SecY(S111C)EG was generated by using the same protocol. The protein was expressed in Lemo21(DE3) E.coli strain (New England Biolabs), expression was induced at OD600 of 0.6 by 0.4 mM IPTG for 4 h. SecY(S111C)EG was purified as described previously (Ge et al., 2014) by metal-affinity chromatography, followed by cation-exchange chromatography. Labeling with MDCC maleimide was performed by following the protocol provided by the manufacturer (Invitrogen), using labeling buffer (20 mM Hepes, pH 7.0, 150 mM KCl, 10% [wt/vol] glycerol, and 0.03% DDM). The efficiency of labeling was >90%, as determined photometrically.
Incorporation of SecYEG into nanodiscs
The procedure used to assemble nanodiscs containing a monomer of MDCC-labeled SecYEG was described previously (Ge et al., 2014). In brief, the mixture of one equivalent of SecYEG and two equivalents of membrane scaffold protein in DDM was incubated at 4°C for 15 h with a mixture of E. coli total membrane lipids (Avanti Polar Lipids) in the presence of Biobeads (Bio-Rad). After removal of the beads, SecYEG-containing nanodiscs were isolated by gel filtration (Superdex 200; GE Healthcare). Empty nanodiscs were assembled by the same procedure, except that SecYEG was omitted.
Fluorescence labeling of FtsY
Mutant FtsY(F196C) and FtsY(A26C) were generated by site-directed mutagenesis using the Phusion polymerase protocol (Thermo Fisher Scientific) and expressed and purified as described for the wild-type protein. Fluorescence labeling with Bpy or N,N′-dimethyl-N-(iodacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (also known as NBD; Invitrogen) was performed as described earlier using labeling buffer (20 mM Hepes, pH 7.0, 150 mM KCl, 10% [wt/vol] glycerol). The labeling efficiency was >90%, as determined photometrically.
Fluorescence titrations
The binding of FtsY(Bpy) to SecYEG(MDCC) was measured upon adding increasing amounts of FtsY(Bpy) in binding buffer (20 mM Hepes, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 10% [wt/vol] glycerol) at 25°C. Complex formation was monitored by FRET between the MDCC donor and the Bpy acceptor. The decrease of donor fluorescence at 460 nm (excitation at 420 nm) was measured and is plotted relative to the initial fluorescence measured before the addition of FtsY(Bpy); buffer blanks were <1% of the fluorescence signal and are subtracted throughout. The contribution of acceptor fluorescence at the wavelength of donor emission was negligible.
Binding of FtsY(NBD) to nanodiscs or SecYEG in nanodiscs was also measured by adding increasing amounts of FtsY(NBD) to 0.5 µM nanodiscs or 0.05 µM SecYEG in nanodiscs. The titrations were corrected by subtracting the linear fluorescence increase because of the increase of the FtsY(NBD) concentration in buffer measured in parallel; the resulting difference values were plotted in normalized form for easier comparison.
Titration curves were evaluated in terms of Kd using a quadratic equation: F = F0 + (Fmax – F0) × 0.5 × {(Pt + Lt + Kd) − sqrt[(P + Lt + Kd)2 – 4PtLt)]} (1), where F is the fluorescence change, F0 the initial fluorescence, Fmax the final fluorescence level, Pt the total protein concentration, Lt the added titrant concentration, and Kd the dissociation constant of the complex of P and L.
Titrations of SecYEG(MDCC) with FtsY(Bpy) in the presence of increasing amounts of FtsQ-RNC or SRP were evaluated additionally by plotting the fluorescence endpoints at saturation with FtsY(Bpy) relative to the initial fluorescence of SecYEG(MDCC) before the addition of FtsY(Bpy) and fitting the hyperbolic curves in terms of the apparent Kd of binding either ligand according to the following equation (Segel, 1993): F(B) = F0/[1 + ([B]/K(B)] (2). F(B) is the fluorescence end point in the presence of a second ligand, B; F0 is the fluorescence end point in the absence of B; [B] is the concentration of B; K(B) is the apparent Kd for the binding of ligand B to the binary complex.
Western blot analyses
After SDS-PAGE, proteins were electro-transferred by tank blotting to a nitrocellulose membrane (GE Healthcare). The α-uL23 and α-uL2 antibodies were raised in goat against the purified full-length proteins (gift from R. Brimacombe, Max Planck Institute for Molecular Genetics, Berlin, Germany). All other antibodies (α-FtsY, α-Ffh, α-SecA, α-MreB) were raised in rabbits against the purified full-length proteins. HRP-coupled goat anti–rabbit antibodies or sheep anti–goat from Caltech Laboratories were used as secondary antibodies, and ECL reagent (GE Healthcare) was used as detection substrate. Membranes were stripped by submerging them in stripping buffer (50 mM DTT, 2% SDS, 62.5 mM Tris/HCl, pH 6.5) for 20 min at 60°C. After three washing steps with TBS plus 0.1% Tween 20, the membrane was blocked again with 5% dried milk in TBS plus 0.1% Tween 20.
Mass spectrometric analyses
Protein bands were cut out and subjected to in-gel digestion using trypsin as described elsewhere (Cristodero et al., 2013). In brief, after reduction of cysteine residues with DTT and alkylation with iodoacetamide, proteins were digested with trypsin and the resulting peptide mixtures analyzed by liquid chromatography/MS using an UltiMate 3000 RSLCnano HPLC system (Thermo Fisher Scientific) and an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptide mixtures were washed and preconcentrated on a 5–mm × 0.3 mm PepMap C18 µ-precolumn (Thermo Fisher Scientific) followed by separation on a 50–cm × 75 µm C18 reversed-phase nano liquid chromatography column (Acclaim PepMap RSLC column; 2-µm particle size; 100-Å pore size; Thermo Fisher Scientific) using a binary solvent system consisting of 0.1% (vol/vol) formic acid (solvent A) and 50% (vol/vol) methanol/30% (vol/vol) acetonitrile in 0.1% formic acid (solvent B). After washing the analytical column with 5% solvent B for 5 min, peptides were eluted applying a 45-min gradient ranging from 5% to 62% solvent B, followed by an increase to 95% B within 5 min at a flow rate of 250 nl/min. Full mass spectrographic scans (m/z 370–1,700) were acquired in the orbitrap at a resolution of 120,000 (at m/z 400) with an automatic gain control (AGC) of 1 × 106 ions and a maximum fill time of 200 ms. Simultaneously, up to 15 of the most intense multiply-charged precursor ions (top 15 method) were fragmented by collision-induced dissociation in the linear ion trap at a normalized collision energy of 35%, an activation q of 0.25, an activation time of 10 ms, an AGC of 5 × 103 ions, and a maximum fill time of 150 ms. The dynamic exclusion time for previously fragmented precursor ions was 45 s. To obtain high-resolution MS/MS data, an aliquot of each sample was additionally analyzed by higher-energy collision-induced dissociation (HCD) using a top 10 method. HCD spectra were acquired in the orbitrap at a resolution of 15,000 applying a normalized collision energy of 35%, an activation time of 0.1 ms, an AGC of 5 × 104 ions, and a maximum fill time of 200 ms. Proteins were identified by database searches using the program OMSSA (version 2.1.9) as described previously (Kuhn et al., 2011). Searches were performed against the organism-specific UniProt database (UniProt Consortium, 2014) for E. coli and a set of common contaminants. Enzyme specificity was set to trypsin with up to two missed cleavages. Oxidation of methionine and carbamidomethylation of cysteine residues were considered as variable and fixed modification, respectively. The mass tolerance was set to 6 ppm for precursor ions and to 0.5 D or 0.1 D for fragment ion spectra measured in the linear ion trap or in the orbitrap. For peptide quantification based on signal intensities in MS1 scans, the program MaxQuant/Andromeda (version 1.3.0.5; Cox and Mann, 2008; Cox et al., 2011) was used with parameters as described earlier, except for a mass tolerance of 20 ppm for HCD spectra. The “match between runs” feature was applied with a tolerance in retention time of 2 min. For the identification of cross-linked peptides, precursor masses of MS/MS spectra were searched against accurate masses computed for pairs of tryptic peptides potentially generated by a cross-link from pBpa at the known position to any amino acid in sequences of SecY and FtsY using a program written in PHP. For those peptide pairs matching an accurate precursor mass, theoretical fragment ions were computed for all possible sites of cross-linking, filtered to the m/z-range of the MS/MS spectrum and compared with the experimental list of fragment ions. A P value giving the probability of finding at least the number of matched out of the number of theoretical masses by chance was calculated according to the Andromeda score (Cox et al., 2011). Separate P values were calculated for each peptide of the cross-linked pair, taking into account only theoretical fragments from this peptide. Candidate cross-linked peptide spectrum matches were validated by manual interpretation. Quantitative analysis of peptide spectrum matches was performed on the basis of the intensities of peptide features in the allPeptides.txt result file from the MaxQuant program. For each peptide spectrum match, the sum of intensities was retrieved for features within 6 ppm tolerance for m/z-values of precursor ions and a tolerance window of ±1 min for the retention time at which the MS/MS spectrum was recorded.
Blue native–PAGE analysis
SecYEG(pBpa) PLs (final SecYEG concentration 10 nM) were incubated with FtsY (1.2 µM) in the absence or in the presence of SRP (1 µM) and GMP-PNP (0.5 mM). Samples were solubilized with 0.2% DDM for 15 min at 4°C and centrifuged for 15 min at 17.000 g. Blue native–PAGE loading dye was added to a final concentration of 10% (vol/vol), and the samples were loaded on 4–16% native PAGE Novex Bis-Tris Protein Gels (Life Technology). After blotting on a nitrocellulose membrane, the protein complexes re detected by α-Ffh and α-FtsY antibodies and peroxidase-coupled anti–rabbit secondary antibodies.
Online supplemental material
Fig. S1 shows that the detection of cross-links between SecYEG(pBpa) and FtsY in PLs and in detergent is not influenced by carbonate extraction. Fig. S2 shows the formation of FtsY–SRP and SecYEG–FtsY–SRP complexes by blue native PAGE. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502103/DC1.
Supplementary Material
Acknowledgments
We gratefully acknowledge Roland Beckmann for providing the FtsQ plasmid and E. coli strain KC6, Richard Brimacombe for antibodies against ribosomal proteins, Piet de Boer for antibodies against E. coli MreB, Anna Pfeifer and Bettina Knapp for expert technical assistance, and Renuka Kudva for discussion.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (FOR 929, FOR 967, and GRK1478 to H.-G. Koch), the German Academic Exchange Service (to N.-A. Petriman), and the Excellence Initiative of the German Federal and State Governments (GSC-4 Spemann Graduate School for Biology and Medicine to H-G. Koch and EXC 294 BIOSS Centre for Biological Signaling Studies to B. Warscheid).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AGC
- automatic gain control
- Bpy
- BODIPY FL
- DDM
- dodecyl-maltoside
- FRET
- fluorescence resonance energy transfer
- GMP-PNP
- guanosine 5′-(β,γ-imido) triphosphate
- HCD
- higher-energy collision-induced dissociation
- MDCC
- 7-diethylamino-3-((((2-maleimidyl)ethyl) amino)-carbonyl)coumarin
- MS
- mass spectrometry
- NBD
- 7-nitrobenzofurazan
- pBpa
- p-benzoyl-l-phenylalanine
- PL
- proteoliposome
- RNC
- ribosome-nascent-chain complex
- SRP
- signal recognition particle
References
- Akopian D., Dalal K., Shen K., Duong F., and Shan S.O.. 2013a SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 200:397–405. 10.1083/jcb.201208045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D., Shen K., Zhang X., and Shan S.O.. 2013b Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82:693–721. 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami M., Dalal K., Lelj-Garolla B., Sligar S.G., and Duong F.. 2007. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26:1995–2004. 10.1038/sj.emboj.7601661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S., Deitermann S., and Koch H.G.. 2005. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 6:476–481. 10.1038/sj.embor.7400385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S., Boy D., Schiltz E., and Koch H.G.. 2006. Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J. Cell Biol. 174:715–724. 10.1083/jcb.200606093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M., et al. . 2014. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach J., Chou Y.T., Fak J.J., Itkin A., Nicolae D.D., Smith P.C., Wittrock G., Floyd D.L., Golsaz C.M., Gierasch L.M., and Hunt J.F.. 2003. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J. Biol. Chem. 278:3628–3638. 10.1074/jbc.M205992200 [DOI] [PubMed] [Google Scholar]
- Bernstein H.D., Poritz M.A., Strub K., Hoben P.J., Brenner S., and Walter P.. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 340:482–486. 10.1038/340482a0 [DOI] [PubMed] [Google Scholar]
- Bibi E., Herskovits A.A., Bochkareva E.S., and Zelazny A.. 2001. Putative integral membrane SRP receptors. Trends Biochem. Sci. 26:15–16. 10.1016/S0968-0004(00)01711-4 [DOI] [PubMed] [Google Scholar]
- Bischoff L., Wickles S., Berninghausen O., van der Sluis E.O., and Beckmann R.. 2014. Visualization of a polytopic membrane protein during SecY-mediated membrane insertion. Nat. Commun. 5:4103 10.1038/ncomms5103 [DOI] [PubMed] [Google Scholar]
- Bornemann T., Jöckel J., Rodnina M.V., and Wintermeyer W.. 2008. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15:494–499. 10.1038/nsmb.1402 [DOI] [PubMed] [Google Scholar]
- Braig D., Bär C., Thumfart J.O., and Koch H.G.. 2009. Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J. Mol. Biol. 390:401–413. 10.1016/j.jmb.2009.04.061 [DOI] [PubMed] [Google Scholar]
- Braig D., Mircheva M., Sachelaru I., van der Sluis E.O., Sturm L., Beckmann R., and Koch H.G.. 2011. Signal sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol. Biol. Cell. 22:2309–2323. 10.1091/mbc.E11-02-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Cristodero M., Mani J., Oeljeklaus S., Aeberhard L., Hashimi H., Ramrath D.J.F., Lukeš J., Warscheid B., and Schneider A.. 2013. Mitochondrial translation factors of Trypanosoma brucei: elongation factor-Tu has a unique subdomain that is essential for its function. Mol. Microbiol. 90:744–755. 10.1111/mmi.12397 [DOI] [PubMed] [Google Scholar]
- Dapic V., and Oliver D.. 2000. Distinct membrane binding properties of N- and C-terminal domains of Escherichia coli SecA ATPase. J. Biol. Chem. 275:25000–25007. 10.1074/jbc.M001100200 [DOI] [PubMed] [Google Scholar]
- Das S., and Oliver D.B.. 2011. Mapping of the SecA·SecY and SecA·SecG interfaces by site-directed in vivo photocross-linking. J. Biol. Chem. 286:12371–12380. 10.1074/jbc.M110.182931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E., Poland D., Mol O., Sinning I., ten Hagen-Jongman C.M., Oudega B., and Luirink J.. 1997. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 416:225–229. 10.1016/S0014-5793(97)01238-6 [DOI] [PubMed] [Google Scholar]
- de Leeuw E., te Kaat K., Moser C., Menestrina G., Demel R., de Kruijff B., Oudega B., Luirink J., and Sinning I.. 2000. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 19:531–541. 10.1093/emboj/19.4.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denks K., Vogt A., Sachelaru I., Petriman N.A., Kudva R., and Koch H.G.. 2014. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31:58–84. 10.3109/09687688.2014.907455 [DOI] [PubMed] [Google Scholar]
- Douville K., Price A., Eichler J., Economou A., and Wickner W.. 1995. SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem. 270:20106–20111. 10.1074/jbc.270.34.20106 [DOI] [PubMed] [Google Scholar]
- Egea P.F., Shan S.O., Napetschnig J., Savage D.F., Walter P., and Stroud R.M.. 2004. Substrate twinning activates the signal recognition particle and its receptor. Nature. 427:215–221. 10.1038/nature02250 [DOI] [PubMed] [Google Scholar]
- Eitan A., and Bibi E.. 2004. The core Escherichia coli signal recognition particle receptor contains only the N and G domains of FtsY. J. Bacteriol. 186:2492–2494. 10.1128/JB.186.8.2492-2494.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez E., Stjepanovic G., Zelazny A.M., Brugger B., Sinning I., and Bibi E.. 2010. Genetic evidence for functional interaction of the Escherichia coli signal recognition particle receptor with acidic lipids in vivo. J. Biol. Chem. 285:40508–40514. 10.1074/jbc.M110.140921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J., Gumbart J., Sluis E.O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., and Beckmann R.. 2011. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18:614–621. 10.1038/nsmb.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Draycheva A., Bornemann T., Rodnina M.V., and Wintermeyer W.. 2014. Lateral opening of the bacterial translocon on ribosome binding and signal peptide insertion. Nat. Commun. 5:5263 10.1038/ncomms6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold V.A., Robson A., Bao H., Romantsov T., Duong F., and Collinson I.. 2010. The action of cardiolipin on the bacterial translocon. Proc. Natl. Acad. Sci. USA. 107:10044–10049. 10.1073/pnas.0914680107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L. 2007. Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 61:589–618. 10.1146/annurev.micro.61.080706.093236 [DOI] [PubMed] [Google Scholar]
- Gu S.Q., Peske F., Wieden H.J., Rodnina M.V., and Wintermeyer W.. 2003. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 9:566–573. 10.1261/rna.2196403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M., Becker T., Pool M.R., Spahn C.M., Grassucci R.A., Frank J., and Beckmann R.. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 427:808–814. 10.1038/nature02342 [DOI] [PubMed] [Google Scholar]
- Halic M., Gartmann M., Schlenker O., Mielke T., Pool M.R., Sinning I., and Beckmann R.. 2006. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 312:745–747. 10.1126/science.1124864 [DOI] [PubMed] [Google Scholar]
- Holtkamp W., Lee S., Bornemann T., Senyushkina T., Rodnina M.V., and Wintermeyer W.. 2012. Dynamic switch of the signal recognition particle from scanning to targeting. Nat. Struct. Mol. Biol. 19:1332–1337. 10.1038/nsmb.2421 [DOI] [PubMed] [Google Scholar]
- Jagath J.R., Rodnina M.V., and Wintermeyer W.. 2000. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J. Mol. Biol. 295:745–753. 10.1006/jmbi.1999.3427 [DOI] [PubMed] [Google Scholar]
- Koch H.G., Hengelage T., Neumann-Haefelin C., MacFarlane J., Hoffschulte H.K., Schimz K.L., Mechler B., and Müller M.. 1999. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell. 10:2163–2173. 10.1091/mbc.10.7.2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudva R., Denks K., Kuhn P., Vogt A., Müller M., and Koch H.G.. 2013. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164:505–534. 10.1016/j.resmic.2013.03.016 [DOI] [PubMed] [Google Scholar]
- Kuhn P., Weiche B., Sturm L., Sommer E., Drepper F., Warscheid B., Sourjik V., and Koch H.G.. 2011. The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic. 12:563–578. 10.1111/j.1600-0854.2011.01167.x [DOI] [PubMed] [Google Scholar]
- Kusters R., Lentzen G., Eppens E., van Geel A., van der Weijden C.C., Wintermeyer W., and Luirink J.. 1995. The functioning of the SRP receptor FtsY in protein-targeting in E. coli is correlated with its ability to bind and hydrolyse GTP. FEBS Lett. 372:253–258. 10.1016/0014-5793(95)00997-N [DOI] [PubMed] [Google Scholar]
- Lam V.Q., Akopian D., Rome M., Henningsen D., and Shan S.O.. 2010. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J. Cell Biol. 190:623–635. 10.1083/jcb.201004129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Dowhan W., and Wickner W.. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 60:271–280. 10.1016/0092-8674(90)90742-W [DOI] [PubMed] [Google Scholar]
- Luirink J., ten Hagen-Jongman C.M., van der Weijden C.C., Oudega B., High S., Dobberstein B., and Kusters R.. 1994. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13:2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mircheva M., Boy D., Weiche B., Hucke F., Graumann P., and Koch H.G.. 2009. Predominant membrane localization is an essential feature of the bacterial signal recognition particle receptor. BMC Biol. 7:76 10.1186/1741-7007-7-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya G., Svensson C., Luirink J., and Sinning I.. 1997. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 385:365–368. 10.1038/385365a0 [DOI] [PubMed] [Google Scholar]
- Mori H., and Ito K.. 2006. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl. Acad. Sci. USA. 103:16159–16164. 10.1073/pnas.0606390103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., and Marians K.J.. 2013. Purification and characterization of Escherichia coli MreB protein. J. Biol. Chem. 288:3469–3475. 10.1074/jbc.M112.413708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Ménétret J.F., Gumbart J.C., Ludtke S.J., Li W., Whynot A., Rapoport T.A., and Akey C.W.. 2014. Structure of the SecY channel during initiation of protein translocation. Nature. 506:102–106. 10.1038/nature12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlitz R., Eitan A., Stjepanovic G., Bahari L., Bange G., Bibi E., and Sinning I.. 2007. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J. Biol. Chem. 282:32176–32184. 10.1074/jbc.M705430200 [DOI] [PubMed] [Google Scholar]
- Peluso P., Herschlag D., Nock S., Freymann D.M., Johnson A.E., and Walter P.. 2000. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 288:1640–1643. 10.1126/science.288.5471.1640 [DOI] [PubMed] [Google Scholar]
- Pool M.R., Stumm J., Fulga T.A., Sinning I., and Dobberstein B.. 2002. Distinct modes of signal recognition particle interaction with the ribosome. Science. 297:1345–1348. 10.1126/science.1072366 [DOI] [PubMed] [Google Scholar]
- Powers T., and Walter P.. 1997. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16:4880–4886. 10.1093/emboj/16.16.4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz A., Behrens C., Rapoport T.A., Hartmann E., and Kalies K.U.. 2000. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 19:1900–1906. 10.1093/emboj/19.8.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y., and Schultz P.G.. 2006. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods. 3:263–265. 10.1038/nmeth864 [DOI] [PubMed] [Google Scholar]
- Sachelaru I., Petriman N.A., Kudva R., Kuhn P., Welte T., Knapp B., Drepper F., Warscheid B., and Koch H.G.. 2013. YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J. Biol. Chem. 288:16295–16307. 10.1074/jbc.M112.446583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraogi I., Akopian D., and Shan S.O.. 2014. Regulation of cargo recognition, commitment, and unloading drives cotranslational protein targeting. J. Cell Biol. 205:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C., Oswald M., Berger I., Ishikawa T., Abrahams J.P., Koerten H.K., Koning R.I., and Ban N.. 2006. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 444:503–506. 10.1038/nature05182 [DOI] [PubMed] [Google Scholar]
- Scotti P.A., Valent Q.A., Manting E.H., Urbanus M.L., Driessen A.J., Oudega B., and Luirink J.. 1999. SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274:29883–29888. 10.1074/jbc.274.42.29883 [DOI] [PubMed] [Google Scholar]
- Segel I.H. 1993. Enyzme kinetics. John Wiley and Sons, Hoboken, NJ: 993 pp. [Google Scholar]
- Shan S.O., Stroud R.M., and Walter P.. 2004. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2:e320 10.1371/journal.pbio.0020320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Arslan S., Akopian D., Ha T., and Shan S.O.. 2012. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature. 492:271–275. 10.1038/nature11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stjepanovic G., Kapp K., Bange G., Graf C., Parlitz R., Wild K., Mayer M.P., and Sinning I.. 2011. Lipids trigger a conformational switch that regulates signal recognition particle (SRP)-mediated protein targeting. J. Biol. Chem. 286:23489–23497. 10.1074/jbc.M110.212340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H., Bürmann F., and Hamoen L.W.. 2014. The actin homologue MreB organizes the bacterial cell membrane. Nat. Commun. 5:3442 10.1038/ncomms4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taufik I., Kedrov A., Exterkate M., and Driessen A.J.. 2013. Monitoring the activity of single translocons. J. Mol. Biol. 425:4145–4153. 10.1016/j.jmb.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Ulbrandt N.D., London E., and Oliver D.B.. 1992. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J. Biol. Chem. 267:15184–15192. [PubMed] [Google Scholar]
- UniProt Consortium 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42:D191–D198. 10.1093/nar/gkt1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent Q.A., Scotti P.A., High S., de Gier J.W., von Heijne G., Lentzen G., Wintermeyer W., Oudega B., and Luirink J.. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17:2504–2512. 10.1093/emboj/17.9.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Houben E.N., Nouwen N., Luirink J., and Driessen A.J.. 2001. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2:519–523. 10.1093/embo-reports/kve106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigts-Hoffmann F., Schmitz N., Shen K., Shan S.O., Ataide S.F., and Ban N.. 2013. The structural basis of FtsY recruitment and GTPase activation by SRP RNA. Mol. Cell. 52:643–654. 10.1016/j.molcel.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiche B., Bürk J., Angelini S., Schiltz E., Thumfart J.O., and Koch H.G.. 2008. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J. Mol. Biol. 377:761–773. 10.1016/j.jmb.2008.01.040 [DOI] [PubMed] [Google Scholar]
- Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Müller M., Warscheid B., Drepper F., and Koch H.G.. 2012. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell. 23:464–479. 10.1091/mbc.E11-07-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.C., de Keyzer J., Kedrov A., and Driessen A.J.. 2012. Competitive binding of the SecA ATPase and ribosomes to the SecYEG translocon. J. Biol. Chem. 287:7885–7895. 10.1074/jbc.M111.297911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I., Bochkareva E.S., and Bibi E.. 2010. Escherichia coli SRP, its protein subunit Ffh, and the Ffh M domain are able to selectively limit membrane protein expression when overexpressed. MBio. 1:e00020-10. 10.1128/mBio.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.