Transmission EM methods reveal that cell–cell fusion of individual myoblasts with growing Drosophila flight muscles is a stepwise process in which the cell adhesion and branched actin machineries mediate tight apposition and formation of multiple contacts and pores between the surfaces of the fusing cells.
Abstract
Fusion of individual myoblasts to form multinucleated myofibers constitutes a widely conserved program for growth of the somatic musculature. We have used electron microscopy methods to study this key form of cell–cell fusion during development of the indirect flight muscles (IFMs) of Drosophila melanogaster. We find that IFM myoblast–myotube fusion proceeds in a stepwise fashion and is governed by apparent cross talk between transmembrane and cytoskeletal elements. Our analysis suggests that cell adhesion is necessary for bringing myoblasts to within a minimal distance from the myotubes. The branched actin polymerization machinery acts subsequently to promote tight apposition between the surfaces of the two cell types and formation of multiple sites of cell–cell contact, giving rise to nascent fusion pores whose expansion establishes full cytoplasmic continuity. Given the conserved features of IFM myogenesis, this sequence of cell interactions and membrane events and the mechanistic significance of cell adhesion elements and the actin-based cytoskeleton are likely to represent general principles of the myoblast fusion process.
Introduction
The fusion of individual myoblasts into a single syncytial unit constitutes a highly conserved program underlying the formation of body wall and limb muscle fibers and serves as a primary example of the use of cell–cell fusion as a means for conferring specialized tissue properties (Rochlin et al., 2010; Aguilar et al., 2013). Study of this fundamental aspect of myogenic differentiation has benefited greatly from investigation in model organisms amenable to genetic analysis (Richardson et al., 2008; Abmayr and Pavlath, 2012; Millay et al., 2013). In particular, the study of myoblast fusion during Drosophila melanogaster embryogenesis, which gives rise to the musculature of the fly larva, has led to the identification and characterization of a wide spectrum of molecular players involved in the fusion process (Chen and Olson, 2004; Önel and Renkawitz-Pohl, 2009; Haralalka and Abmayr, 2010; Önel et al., 2014).
The larval muscles of Drosophila, a holometabolous insect, are destroyed during metamorphosis along with most other tissues and organs, and new muscle groups are generated to serve the distinct requirements of the adult fly (Fernandes et al., 1991; Fernandes and Keshishian, 1999; Roy and VijayRaghavan, 1999; Dutta and VijayRaghavan, 2006). Prominent among these are the dorsolongitudinal muscles (DLMs), a set of 12 large indirect flight muscle (IFM) fibers, which, together with the smaller, orthogonally positioned dorsoventral muscles, fill the adult thoracic cavity and power flight through connections to the cuticular exoskeleton (Dickinson, 2006; Fig. 1 A). The DLMs serve as a particularly attractive model for myogenesis. Both their developmental program, which relies on fusion of stem cell–derived migratory myoblasts with a set of target templates, and their myofibrillar organization after maturation, features that are unique among Drosophila muscles, resemble key aspects of vertebrate skeletal myogenesis, establishing DLM development as a myogenic program of general relevance (Fernandes et al., 1991; Bernstein et al., 1993; Gunage et al., 2014).
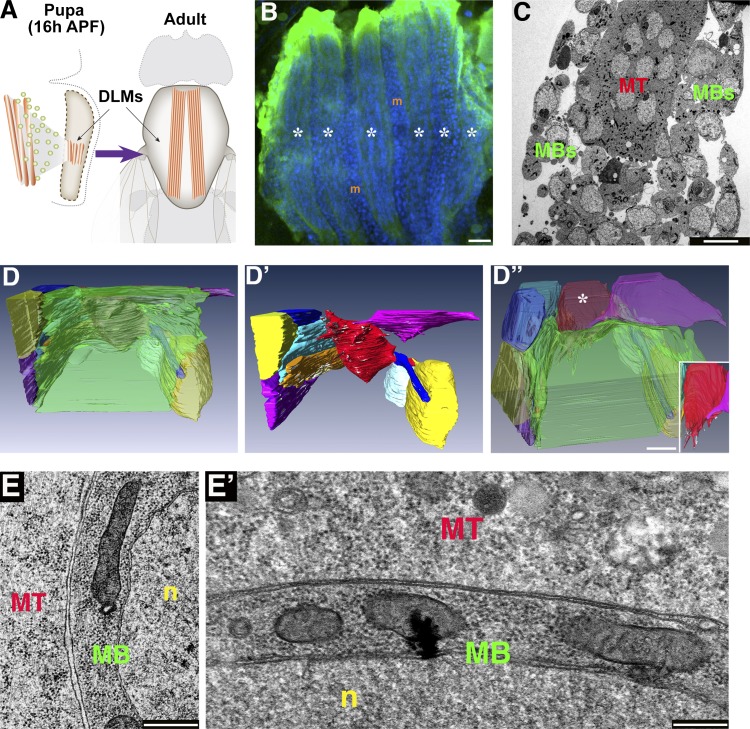
Figure 1.
FIB/SEM visualization reveals an extensive flat interface between DLM myotubes and associated myoblasts. (A) Schematics of an early pupa (left) and an adult fly (right) illustrating the position and relative size of the IFMs. A swarm of wing disc–derived myoblasts (green) fuses with a set of three persistent larval muscles in each pupal hemithorax, which go on to split and mature into a set of six DLMs. (B) A set of six nascent DLMs (asterisks) at 20 h APF visualized using mef2-GAL4>UAS-mCD8-GFP (green). DAPI-stained nuclei (blue) fill the syncytial muscles and also mark the positions of the mononucleated myoblasts (m) surrounding the muscle fibers. (C) Low magnification TEM of DLMs at 20 h APF. Mononucleated myoblasts (MBs) surround a syncytial myotube (MT). (D–D”) Reconstruction of a FIB/SEM dataset (see Video 1) of a DLM myotube and associated myoblasts. D displays the semitransparent myotube (green) and neighboring myoblasts (individually colored), whereas D’ shows the same view of the myoblasts alone, where the flattened surfaces of the myoblasts are readily apparent. D” displays the same cells as in D, at the same magnification, but from a different (tilted) angle, revealing a single myoblast (red, marked by an asterisk, and shown on its own in the inset) extending protrusions toward the myotube. (E and E’) Low (E) and high (E’) magnification views of the interface between a DLM myotube (MT) and an associated myoblast (MB), prepared for TEM using the hybrid CF and HPF/FS protocol (see Materials and methods section TEM), which allows for high quality preservation of smooth cell membranes and a rich cytoplasm. n, nucleus. Bars: (B) 20 µm; (C and D”) 10 µm; and (E and E’) 500 nm.
Despite the appeal of the Drosophila IFM model, study of various myogenic processes in this system, including myoblast fusion, has lagged behind the embryonic setting, primarily because of difficulties in applying genetic analysis to an advanced phase of development. Although generation of mosaic mutant clones has traditionally enabled the study of genetic requirements during late developmental events (Blair, 2003), the syncytial organization of muscles precludes the use of this powerful tool. The introduction of RNAi-based approaches, which can be applied in spatial- and temporal-specific fashions, now circumvents these problems to a large extent (Schnorrer et al., 2010), and these tools have been successfully used recently in the study of myoblast fusion in IFMs and other adult fly muscles (Mukherjee et al., 2011; Gildor et al., 2012).
Ultrastructural analysis using transmission EM (TEM) techniques has made important contributions to the elucidation of cellular mechanisms governing Drosophila embryonic myoblast fusion (Doberstein et al., 1997; Schröter et al., 2004; Estrada et al., 2007; Kim et al., 2007; Massarwa et al., 2007; Sens et al., 2010). DLM formation presents a particularly appropriate and unique setting for TEM-level analysis of Drosophila myoblast fusion, as it involves many hundreds of repeated fusion events between myoblasts and a set of identical myotubes over a period of only a few hours. Such reiterations hold the promise of observing and distinguishing between different phases of the process and producing a plausible interpretation for progress through individual fusion events from the snapshot nature of TEM datasets, which are generated from fixed material. Investigations of adult IFM formation using these approaches are rare, however, and limited to details of myofibril formation with minimal focus on the fusion process itself (Shafiq, 1963; Reedy and Beall, 1993). The perceived unique benefits of a TEM-based analysis of DLM myoblast fusion, coupled with the genetic manipulations now available for this system, prompted us to apply state-of-the-art TEM methods to this key myogenic setting.
Here, we provide an ultrastructural description and analysis of DLM myoblast fusion in which conventional TEM imaging is combined with 3D visualization methods, including focused ion beam (FIB)/scanning EM (SEM) and scanning transmission EM (STEM) tomography. Importantly, this analysis was performed on IFM samples prepared in a manner that successfully preserves both membrane integrity and cytoplasmic content and was applied to preparations from wild-type (WT) flies, as well as to preparations from flies in which the function of key contributors to the fusion process was disrupted by genetic means. In brief, our observations suggest that cell surface adhesion proteins mediate an initial ordered association between myoblasts and myotubes, while regulators of branched actin networks mediate subsequent flattening of myoblast surfaces, after which the two cell types become tightly apposed. This spatial configuration promotes formation of multiple sites of contact along the apposed surfaces, which give rise to nascent pores that will go on to expand so that full cytoplasmic continuity is achieved. Our results provide a high resolution description of IFM myoblast fusion and its mechanistic underpinnings, which is likely to be general to programs of somatic myogenesis.
Results
Myoblast membranes flatten onto the myotube surface
The early developmental stages of IFM formation are challenging for study as an intact tissue at the electron microscope level because the IFM set at this stage occupies a relatively small portion of the histolysing pupal thorax (Fig. 1 A) and is therefore difficult to identify and isolate. We addressed this problem by expressing GFP constructs specifically in the pupal musculature (Fig. 1 B), enabling straightforward in vivo identification of the developing IFMs, which were dissected out and processed for TEM visualization. Low magnification views of sectioned material from such preparations readily revealed the established arrangement of multinucleated IFM myotubes surrounded by a “swarm” of mononucleated myoblasts (Roy and VijayRaghavan, 1998) as they approach the myotubes and prepare to fuse with them (Fig. 1 C).
To obtain an appreciation for the spatial organization of the myogenic cells at the height of the fusion process (∼20 h after puparium formation [APF]), we subjected the IFM preparations to serial surface imaging (Bennett et al., 2009; Weiner et al., 2011; Villa et al., 2013). This method, which uses coordinated FIB milling and serial SEM imaging, enables 3D reconstruction of cell and tissue morphology in thick specimens and is therefore suitable for visualization of entire myotubes and their immediate environment at EM-level resolution. Our FIB/SEM analysis revealed that as much as 90% of the myotube surface is engaged in close interactions with its neighboring thoracic myoblasts (Fig. 1 D and Video 1). Although the myoblasts adopt a variety of morphologies, they share one prominent feature in that large portions of their cell bodies flatten onto the myotube surface (Fig. 1, D and D’). The outcome is an extensive arrangement of closely apposed cell surfaces between the myotube and the oval myotube-embracing myoblasts. The apposed surfaces at these cell–cell interfaces are almost invariably flat, but we also note the infrequent appearance of myoblasts bearing multiple protrusions that extend into the adjoining myotube (Fig. 1 D”).
Tight apposition of myotube–myoblast surfaces coincides with formation of distinct contact sites
To characterize the myoblast–myotube interaction at high resolution, we next turned to detailed, high magnification TEM examination of multiple single sections of growing and fusing IFM preparations. Toward this end, we used a hybrid chemical/cryofixation protocol (Sosinsky et al., 2008), which generated preparations showing a high degree of cytoplasm preservation coupled with a strikingly clear demarcation of continuous and smooth cell membranes (Fig. 1, E and E’; and see Materials and methods section TEM and Fig. S1 and accompanying text for a discussion of technical considerations).
Building upon the FIB/SEM-based observations, we initially focused our single-section TEM analysis on features and aspects of the close and extensive association between myotubes and myoblasts leading up to their fusion. Examination of multiple paired cell–cell interfaces revealed two distinct classes of cell appositions between myotubes and neighboring myoblasts. Roughly half (26/50) of these pairings were characterized by long (∼4 µm) stretches of apposed membranes, which, however, did not show any signs of direct contact (Fig. 2 A). Measurements of the width of the extracellular space separating such cell pairs revealed a spread of intermembrane separation distances, most of which fall into the 22–50-nm range (Fig. 2, B and H). In contrast, a second class of myotube–myoblast pairs exhibited similar lengthy surface membrane appositions, but these were studded with multiple sites of contact, where no resolvable gap between the surfaces was apparent (Fig. 2 C). Importantly, appearance of the contacts was associated with a significant overall reduction of intercellular distance, so that ∼60% of measured separations between the neighboring cells covered <22 nm of extracellular space (Fig. 2, D and H). We refer to this configuration as “tightly apposed” cell surfaces throughout the text. The observations identifying two distinct configurations of apposed myotube–myoblast pairs suggest an active transition by which neighboring cells are brought closely together to initiate contact that will eventually lead to fusion.
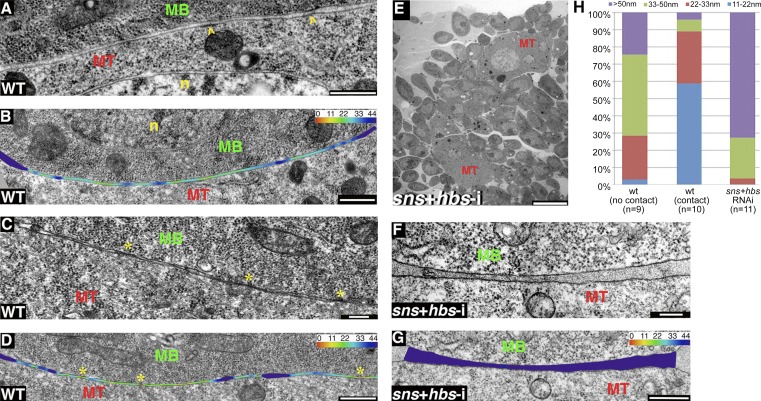
Figure 2.
DLM myotube–myoblast association and contact involves several distinct configurations. (A–D) WT DLMs. Views of myotube (MT)–myoblast (MB) interfaces before (A and B) and after (C and D) formation of contact sites (yellow asterisks). n, nucleus. Contact site formation appears to be associated with tighter apposition of the cells. B and D include a color-coded heat map of the distances between neighboring membranes (see Materials and methods section Cell surface distance analysis). Membrane-associated electron-dense plaques, reminiscent of a structure observed in Drosophila embryonic preparations (Doberstein et al., 1997), are occasionally observed (arrowheads in A, but their association with contact sites is not clear. (E–G) DLMs after simultaneous knockdown of the adhesion elements Sns and Hbs (mef2-GAL4>UAS-sns-i,UAS-hbs-i). (E) A low magnification view demonstrates the resulting fusion arrest phenotype with myoblasts congregating around thin myotubes (MT). (F and G) High magnification views reveal the relatively wide gap between myotubes and neighboring myoblasts, which do not flatten their apposed surface. (H) Bar graph showing the distribution of myotube–myoblast intermembrane distances (see Materials and methods section Cell surface distance analysis). n, number of cells analyzed in single TEM sections. A distance distribution profile in which most (50–70%) membrane separations are of intermediate value (22–50 nm; mean distance = 27.2 ± 15.5 nm) is observed in WT DLM preparations where the cells do not make contact (left bar). The profile is clearly biased toward smaller distances (0–22 nm; mean distance = 15.0 ± 4.7 nm) for WT cell pairs that are in contact with each other (middle bar) and toward larger distances (>50 nm; mean distance = 65.4 ± 33.0 nm) in adhesion-defective (sns + hbs RNAi) DLM preparations (right bar). Bars: (A, B, D, and G) 500 nm; (C and F) 200 nm; and (E) 10 µm.
Cell adhesion elements promote the initial phase of myoblast–myotube association
Molecular genetic analysis of myoblast fusion has repeatedly identified elements of two key cellular machineries, intercellular adhesion proteins and mediators of branched actin array formation, as prominent contributors to the fusion process (Simionescu and Pavlath, 2011). We thus reasoned that insight into the cellular mechanisms underlying close association and eventual contact and fusion between myoblast and nascent IFM myotubes could be obtained through study of intercellular adhesion and formation of branched actin arrays. Toward this end, we sought to examine IFM preparations from mutant pupae in which the function of elements mediating these key molecular machineries was compromised.
During Drosophila embryogenesis, fusion-related cell–cell adhesion involves cell type–specific Ig domain transmembrane proteins. These include the myotube-specific elements Kin-of-Irre/Dumbfounded (Kirre/Duf) and Roughest (Rst) and the myoblast-specific elements Sticks-and-stones (Sns) and Hibris (Hbs), which act to mediate myotube–myoblast association via heterotypic adhesive interactions (Ruiz-Gómez et al., 2000; Dworak and Sink, 2002; Galletta et al., 2004; Shelton et al., 2009). We had previously established that simultaneously attenuating the function of the myoblast-specific elements Sns and Hbs, via muscle-specific expression of relevant RNAi constructs, led to a strong fusion arrest (Gildor et al., 2012). We therefore examined the role of cell adhesion through EM analysis of IFM preparations (∼20 h APF) from such knockdown pupae.
Single sections of this material at low magnification revealed a large population of individual myoblasts congregated around thin myotube templates that displayed a relatively dilute cytoplasm, as expected from situations of fusion arrest (Fig. 2 E; compare with Fig. 1 C). Furthermore, and in marked difference to the WT arrangement, where many of the swarming myoblasts are found immediately adjacent to the myotubes, sns/hbs knockdown myoblasts were not able to complete their approach, with most myoblasts (∼72%) positioned >50 nm away from the myotube surface (Fig. 2, F–H). A complementary feature was that mutant myoblasts retained the ball-like morphology characteristic of WT myoblasts still distant from the myotube, with minimal flattening of the membrane facing the myotubes (∼2.5 µm of apposed surface membrane, on average; Fig. 2, F and G; and Fig. 3 M). These observations imply that adhesion elements, including Sns and Hbs, mediate a close-range phase of association between IFM myoblasts and myotubes that precedes tight apposition and cell–cell contact and is associated with a flattening of the myoblast surface.
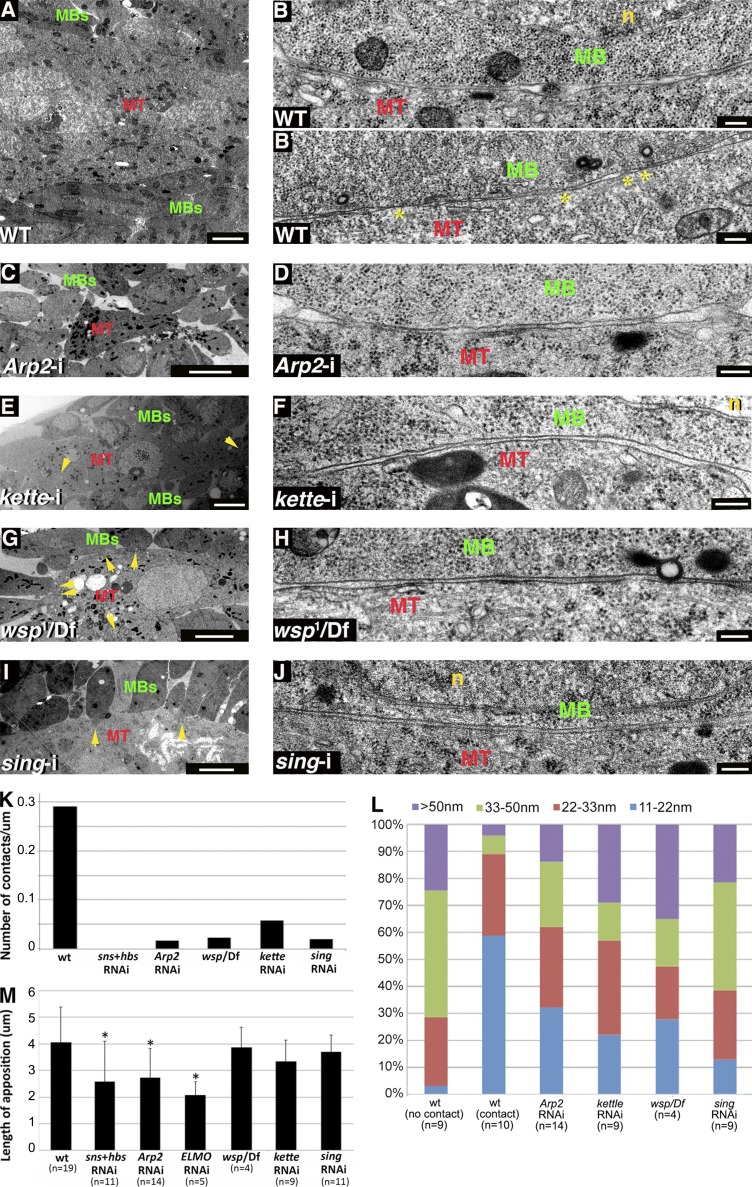
Figure 3.
Branched actin polymerization and Sing are necessary for tight apposition and contact between DLM myotubes and associated myoblasts. (A–B’) DLM preparations from WT pupae. A low magnification view (A) shows myoblasts (MBs) surrounding a multinucleated myotube (MT), and high magnification panels show myotube–myoblast interfaces before (B) and after (B’) establishment of contacts (asterisks). n, nucleus. (C–J) DLM preparations from knockdown and mutant pupae, including mef2-GAL4>UASArp2-i (C and D), mef2-GAL4>UASkette-i (E and F), wsp1/Df(3R)3450 (G and H), and mef2-GAL4>UASsing-i (I and J) pupae. Low magnification views (C, E, G, and I) reveal fusion arrest in all cases, with myoblasts congregating around a thin myotube containing few nuclei. Elongated myoblasts sending projections toward the myotube (arrows) are observed in kette knockdown (E), WASp mutant (G), and sing knockdown (I) pupae. High magnification views (D, F, H, and J) demonstrate incomplete apposition and lack of contacts between myoblast and myotube plasma membranes, similar to the WT panel (B). While kette knockdown (E), WASp mutant (H), and sing knockdown myoblasts (J) flatten their apposed surfaces, Arp2 (D) myoblasts fail to do so. (K) Bar graph showing the frequency of contact sites along apposed myotube–myoblast surfaces in different genetic backgrounds. The number of cell pairs examined and the total length of membrane surveyed for contact sites in the different genotypes were as follows. WT: 50 cell pairs, ∼200 µm; sns + hbs knockdown: 13 cell pairs, ∼20 µm; Arp2 knockdown: 18 cell pairs, ∼50 µm; WASp mutant: 21 cell pairs, ∼100 µm; kette knockdown: 14 cell pairs, ∼50 µm; and sing knockdown: 22 cell pairs, ∼100 µm. (L) Bar graph showing the distribution of myotube–myoblast intermembrane distances in different genetic backgrounds generated as in Fig. 2 H. n, number of cells analyzed in single TEM sections. WT bars are the same as in Fig. 2 H. Preparations in which the function of branched actin elements or Sing is compromised (four right bars) all display a distance distribution profile in which most (50–70%) membrane separations are of intermediate value (22–50 nm). A similar profile is characteristic of WT DLM preparations where the cells do not make contact (left bar), whereas establishment of contacts between WT cell pairs (second bar from left) is associated with a shift toward smaller distances (0–22 nm) and tight apposition. (M) Bar graph comparing the mean length of myoblast surface membrane apposed to a neighboring myotube in different genetic backgrounds. n, number of myoblast–myotube pairs analyzed. Standard deviation of the measurement range is shown. Asterisks mark bars that are distinct from the WT value in a statistically significant fashion (analysis of variance, F(6,56) = 3.79, P = 0.003). Longer appositions, matching a flattened appearance, are characteristic of both classes of WT myoblasts as well as WASp mutant (Dunnett’s test, P = 0.99), kette knockdown (P = 0.56), and sing knockdown myoblasts (P = 0.96) but are not achieved after knockdown of sns-hbs (P = 0.020), Arp2 (P = 0.011), or ELMO (P = 0.005). Bars: (A, E, G, and I) 2 µm; (B, B’, D, F, H, and J) 200 nm; (C) 5 µm.
The branched actin nucleation machinery mediates myoblast surface flattening and tight myoblast–myotube association
The Arp2/3 complex constitutes the primary microfilament nucleating system for generation of branched actin arrays in eukaryotic cells (Goley and Welch, 2006; Rotty et al., 2013) and is triggered by nucleation-promoting factors (NPFs), with the major ones belonging to the WASp and SCAR/WAVE protein families (Pollitt and Insall, 2009). These elements have been repeatedly recognized as essential mediators of myoblast fusion in flies and mice (Kim et al., 2007; Massarwa et al., 2007; Richardson et al., 2007; Berger et al., 2008; Gildor et al., 2009; Nowak et al., 2009; Mukherjee et al., 2011; Gruenbaum-Cohen et al., 2012). We therefore sought to determine which aspects of IFM myotube–myoblast association were dependent on generation of branched actin structures.
Full disruption of branched actin polymerization was achieved after muscle-specific expression of RNAi directed against Arp2, which encodes a core subunit of the Arp2/3 complex, resulting in a strong fusion-arrest phenotype (Fig. 3 C; Mukherjee et al., 2011). Close examination of the myotube–myoblast interfaces in Arp2 knockdown DLMs revealed a nearly complete failure in establishing points of membrane contact (Fig. 3, D and K). Furthermore, the myoblast and myotube surfaces are commonly positioned at least 20 nm away from each other, with a spread of intercellular distances resembling that observed in WT DLM preparations before tight apposition and formation of cell contacts (Fig. 3, B, D, and L). The Arp2 knockdown myoblasts are morphologically distinct, however, from similarly positioned WT myoblasts in that they maintain only relatively short lengths (∼2.5 µm) of surface membrane in apposition to the neighboring myotube (Fig. 3, D and M). Therefore, the involvement of branched actin polymerization during IFM myoblast fusion commences after the initial adhesion-mediated positioning of myoblasts near myotubes and is required both for flattening of the apposed myoblast surface and for establishing the tight myotube–myoblast surface apposition associated with contact formation.
Arp2/3 nucleation of branched actin polymerization can be stimulated by different NPFs in response to a variety of signal transduction cues (Goley and Welch, 2006). To address this issue in the context of DLM myoblast fusion, we separately disrupted the function of the two major Arp2/3 NPF systems, WASp and the SCAR/WAVE complex, and examined the consequences on DLM formation. To interfere with Drosophila WASp function, we made use of the strong loss-of-function allele wsp1. The maternal contribution of WASp is sufficient for embryonic development, allowing homozygous and hemizygous zygotic wsp mutant flies to survive to pupal stages of development where functional roles, including requirements during IFM myoblast fusion, become apparent (Ben-Yaacov et al., 2001; Mukherjee et al., 2011). This approach cannot be used to study the requirements for the SCAR/WAVE NPF system because mutations in SCAR itself, or in elements of its associated complex, result in embryonic/larval lethality. We therefore used for this purpose muscle-specific RNAi-based knockdown of kette (also known as Hem), which encodes the Drosophila Nap1 homologue, a SCAR/WAVE complex subunit that has been used extensively in the study of embryonic myoblast fusion (Schröter et al., 2004; Richardson et al., 2007; Berger et al., 2008).
As observed after knockdown of Arp2, disruption of either of the two NPFs results in a strong arrest of IFM myotube–myoblast fusion (Fig. 3, E and G), where the tight apposition of the myotube–myoblast interfaces is not achieved (Fig. 3, F, H, and L), and, correspondingly, contact sites are rare (Fig. 3 K), and no fusion pores are found. These observations confirm the essential involvement of branched actin polymerization in bringing the surface membranes into close proximity and suggest, furthermore, that the Arp2/3 complex requires activation by both NPF systems in this setting. A second observation is that, unlike Arp2 knockdown myoblasts, kette knockdown or wsp1 mutant myoblasts appear capable of flattening their surfaces (Fig. 3, F, H, and M). This may imply that the influence of branched actin on surface flattening can be achieved via either of the two NPF machineries. Surface flattening is likely to involve functional input from yet another actin regulator, ELMO(CED12)-DOCK1, a bipartite guanine exchange factor (GEF) for the Rac GTPase (Brugnera et al., 2002) and an established, conserved contributor to myoblast fusion (Geisbrecht et al., 2008; Laurin et al., 2008). We find that a failure to flatten myoblast surfaces is characteristic of the strong DLM fusion arrest observed after muscle-specific knockdown of ELMO (Fig. 3 M and Fig. S2).
Collectively, these observations suggest that branched actin polymerization drives the transition from a state of apposed but separated surfaces (at a distance of ∼20–50 nm), mediated by cell adhesion, to a tight myotube–myoblast surface membrane apposition of <10 nm, where intercellular distance is minimal and membrane contacts can be established. This transition appears to involve two distinct actin-based events, myoblast surface flattening and the tight apposition of neighboring membranes, leading to formation of cell–cell contact sites. Although actin microfilament networks are difficult to preserve for EM visualization, light microscopy studies have identified a large, transient actin focus that presages and marks fusion sites within both embryonic and pupal myoblasts (Kesper et al., 2007; Kim et al., 2007; Richardson et al., 2007; Gildor et al., 2009; Mukherjee et al., 2011). Establishment of this structure requires both Sns-Hbs–mediated adhesion and a functional Arp2/3 complex, but it can form in the absence of either of the two NPF systems (Fig. S3). It would appear that formation of the actin focus coincides with the functional requirements for branched actin established here, suggesting a framework for its involvement in myoblast fusion.
Shared requirements for Arp2/3 NPFs and the MARVEL domain element Singles-bar during IFM myoblast fusion
Mutations in singles-bar (sing) result in a strong myoblast fusion arrest during Drosophila embryogenesis (Estrada et al., 2007). Sing is a member of the MARVEL domain family of transmembrane proteins, which have been implicated as mediators of vesicle fusion and membrane apposition (Sánchez-Pulido et al., 2002). Sing may thus constitute a membrane structural element interacting with the various cellular machineries required to promote myoblast fusion. Muscle-specific RNAi-based knockdown of sing results in a strong fusion arrest during IFM formation, demonstrating that sing function is required for fusion in this setting as well (Fig. 3 I and Fig. S3; Brunetti et al., 2015). Interestingly, several features of tissue organization and myoblast morphology observed after knockdown of sing closely resemble those seen in WASp mutant and kette knockdown DLMs. As is common to all backgrounds in which branched actin regulatory factors are disrupted, myoblast and myotube surfaces in sing knockdown DLMs are positioned in close (∼20 nm) proximity, but tighter apposition is not obtained, and contacts are rarely observed (Fig. 3, J–L). Similar to WASp mutant and kette knockdown myoblasts, sing knockdown myoblasts flatten their myotube-apposed surface (Fig. 3, J and M). The broad similarities between the fusion arrest phenotypes suggest that Sing and the Arp2/3 NPFs operate in tandem as essential contributors to the progress of IFM myoblast fusion.
Myoblast surface protrusions follow functional disruption of Arp2/3 NPFs
To further characterize the consequences of fusion arrest resulting from disruption of Arp2/3 NPF activity, we turned again to FIB/SEM serial scanning, which was applied to DLMs isolated from WASp mutant pupae. This analysis revealed a mixed population of myoblasts: those with extensive flat surfaces apposing the myotubes, along with others whose myotube-facing interfaces display multiple protrusions (Fig. 4 A and Video 2), an uncommon feature among WT myoblasts (Fig. 1 D”). The full 3D structure of the WASp mutant projections can be appreciated from the reconstructions derived from serial scanning (Fig. 4 A). Measurements based on these data reveal a general similarity to the dimensions of the infrequent protrusions (Fig. 1 D”) emanating from WT myoblasts (∼2.3–2.5 µm in length, on average, and ∼0.4 µm in width), suggesting that they represent similar structures.
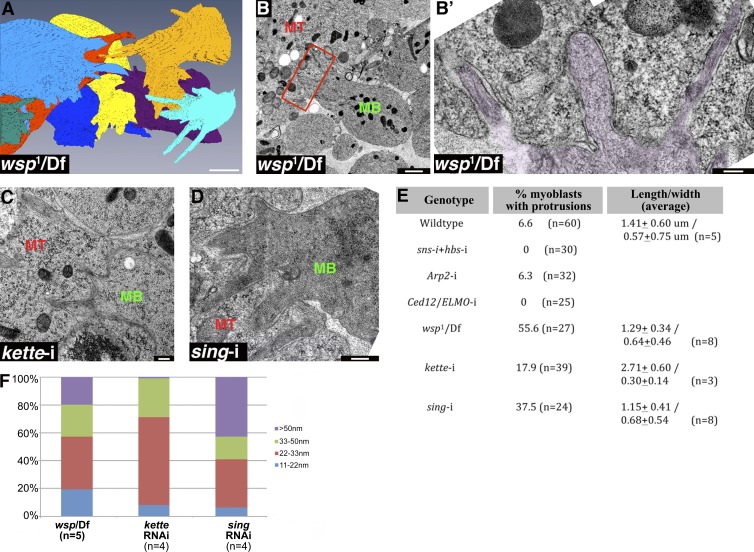
Figure 4.
Protrusive extensions are a shared feature of WASp mutant and sing knockdown myoblasts. (A) Reconstruction of a FIB/SEM dataset of DLM myotube-associated myoblasts from a WASp mutant pupa (see Video 2). A sizable portion of the mutant myoblasts display extensions, mostly oriented toward the myotube, which occupies the center of the panel and was left out of the image for clarity. (B and B') TEM section of WASp mutant DLMs, demonstrating typical myoblast (MB) fusion arrest phenotypes that include an abnormally elongated morphology and finger-shaped extensions (rectangle in B, magnified and false-colored purple in B’), which protrude into the neighboring myotube (MT). The myotube and myoblast surfaces maintain separation along the extensions, and no signs of contact between the cells are apparent. (C and D) TEM sections of kette (C) and sing knockdown DLMs, demonstrating the WASp mutant-like finger-shaped protrusions that myoblasts (MB) often extend toward the myotube (MT) in these mutant backgrounds. No signs of contact between the cells or of tight apposition are apparent along the extensions. (E) Quantification of the frequency and dimensions of surface protrusions in different genetic backgrounds. Although protrusions are a common feature of the myotube-associated myoblasts in WASp mutant, kette knockdown, and sing knockdown DLMs, they are rarely observed in WT, sns-hbs, Arp2, or Ced-12/ELMO knockdown DLMs. Protrusion dimensions are similar in the different backgrounds. Lengths are shorter than those computed from the FIB reconstructions, as sections do not capture the entire structure. (F) Bar graph showing the distribution of myotube–myoblast intermembrane distances along myoblast protrusions in different genetic backgrounds, generated as in Fig. 2 H. n, number of individual protrusions analyzed in single TEM sections. Surface-membrane separation profiles along the protrusions on WASp mutant, kette knockdown, and sing knockdown DLMs match those observed for the flat portions of the myoblast surface in these backgrounds (Fig. 3 L). Bars: (A) 2 µm; (B’, C, and D) 200 nm; (B) 1 µm.
The surface protrusions are readily observed upon examination of TEM sections of WASp mutant DLM preparations (Fig. 4, B and B’) and were also found to extend from the myotube-facing surfaces of kette knockdown and sing knockdown myoblasts using this method (Fig. 4, C and D). Measurements of protrusion dimensions using the TEM sections again demonstrated a general similarity between protrusions emanating from WT myoblasts and those from WASp mutant, kette knockdown, and sing knockdown myoblasts (Fig. 4 E). Furthermore, the characteristic intercellular distance of >20 nm from the myotube, observed for the flat portions of myotube surfaces, is maintained along myoblast surface extensions in both instances of Arp2/3 NPF functional disruption and after sing knockdown (Fig. 4 F), and no points of membrane contact with the myotube were observed.Surface protrusions are not a general feature of fusion arrest, as the surfaces of myoblasts in sns-hbs, Arp2, and Ced-12/ELMO knockdown DLM preparations, all of which fail to flatten, remain smooth, exhibiting few protrusions if at all (Fig. 4 E).
Myotube–myoblast fusion involves formation and expansion of multiple fusion pores
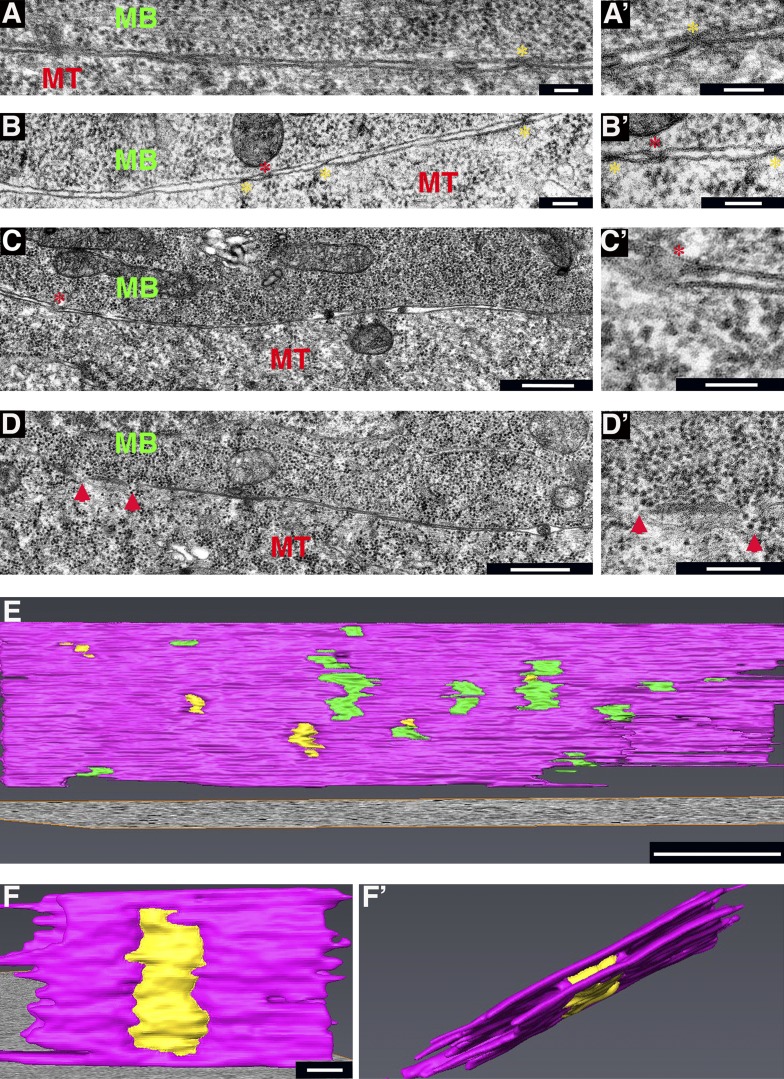
Having established an apparently stepwise process through which DLM myoblasts and myotubes achieve tight apposition and contact between their surface membranes before fusion, we sought to determine the manner by which subsequent cytoplasmic continuity between the two cell types is accomplished. Closer examination of the multiple cell–cell contacts described above (Fig. 2, C and D; and Fig. 3 B’), which form almost exclusively between tightly apposed myoblast and myotube membranes, suggested that the cell surfaces at these sites display one of two possible conformations of similar size (Fig. 5, A–C’). One is an X-shaped appearance of the contact site, in which the paired surfaces remain distinct but are drawn close together, to within a few nanometers of each other (Fig. 5, A’ and B’). The second conformation, observed less frequently, is characterized by a blurred contact interface bordered by membrane loops, where the two apposed surfaces appear to be joined together (Fig. 5, B’ and C’). The similar dimensions of these contacts, and the simultaneous presence of both configurations in individual sections, suggests that they represent successive steps in the formation of nascent fusion pores.
Figure 5.
Multiple contact sites and nascent fusion pores form between tightly apposed myotube and myoblast surfaces. (A–C’). Three examples of paired and tightly apposed myotube (MT)–myoblast (MB) surface membranes displaying points of contact. Primed panels represent high magnification views of regions in left panels. Yellow asterisks mark the X-shaped configuration, where both membranes remain intact, whereas red asterisks mark blurred interfaces bordered by membrane loops, presumably representing nascent fusion pores. (D and D’) Myotube–myoblast interface displaying small fusion pores (red arrowheads). (D’) Cytoplasmic continuity can be recognized by apparent flow of dark, round ribosomes through the pores. (E) Tomogram 3D reconstruction of a myotube–myoblast interface displaying multiple cell–cell contacts (green) and small pores (yellow). Membranes are colored pink. STEM was performed on 350–400-nm-thick sections. Also see Video 3. (F and F’). Tomogram 3D reconstruction of a single pore, viewed from two different angles. Also see Video 4. Bars: (A, A’, B, B’, and C’) 100 nm; (C and D) 500 nm; (D’ and E) 200 nm; (F and F’) 25 nm.
Myotube–myoblast interfaces exhibiting clear features of cell fusion and cytoplasmic continuity contain multiple membrane discontinuities (which we refer to as pores) along the paired surface membranes (Fig. 5, D and D’). Such pore-containing interfaces comprise more than half (14/24) of the surveyed population of myotube–myoblast pairs in close contact with each other. These pores, which were associated only with the tightly apposed, contact-studded class of membrane pairs, range in size (50–150 nm) and display looped membrane borders, suggesting that they form by further expansion of the initial cell–cell contact sites and nascent fusion pores. This interpretation is supported by the observation of apparent material flow through the enlarged pores (Fig. 5 D’), suggesting that the cell cytoplasms are beginning to mix.
To obtain a 3D appreciation for the features associated with the appearance of fusion pores, we subjected IFM blocks to dual-axis STEM tomography, a useful and informative method for ultrastructural visualization of thick specimens (Aoyama et al., 2008; Sousa and Leapman, 2012). STEM analysis revealed that the large and flat interfaces between template myotubes and closely associated myoblasts are pock-marked with a variety of surface discontinuities (contact sites and pores) of different sizes and dimensions, consistent with an ongoing process in which contacts arise at multiple locations and mature into fusion pores (Fig. 5 E and Video 3). 3D rendering of the tomograms accentuates the continuous nature of the pores, which span the area between the neighboring cell surfaces and gradually expand (Fig. 5, F and F’; and Video 4).
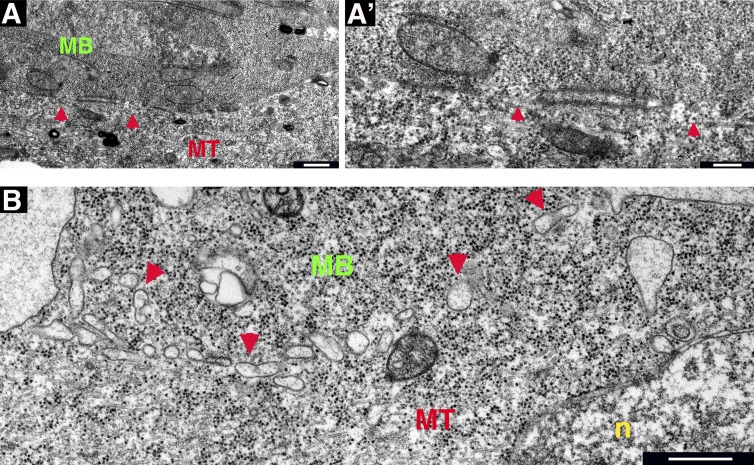
Further progress of the fusion process could be recognized in single TEM sections of interfaces containing larger pores in the 100–500-nm range (Fig. 6 A). In these sections, the overall lengths of the apposed stretches of surface membranes are considerably shorter, whereas the surfaces bounded by the loops often separate, generating oblong, sac-like structures (Fig. 6 A’). These enclosures appear to break up into smaller entities (Fig. 6 B), which disperse within the cytoplasm of the fusing cell pair, suggesting that the merged cell membranes are removed by vesiculation.
Figure 6.
Fusion pore expansion leads to cytoplasmic continuity between fusing myoblasts and myotubes. (A and A’). Myotube (MT)–myoblast (MB) interface displaying expanded fusion pores (red arrowheads). A’ is a higher magnification view of a portion of A. Cytoplasmic continuity is clearly discernible, and paired membranes between pores form an expanded enclosure. (B) Myotube–myoblast pair at an advanced phase of fusion. The cytoplasm is uniform in appearance, and the cell–cell interface appears as a series of vesicle-like structures (red arrowheads). n, nucleus. Bars: (A and B) 500 nm; (A’) 200 nm.
In summary, our ultrastructural analysis suggests that the incorporation of myoblasts into growing DLM myotubes involves progress through several forms of association between the two myogenic cells, followed by the establishment of a large number of small pores traversing the paired membranes, whose expansion leads to complete fusion.
Discussion
We have used advanced TEM methods for high resolution visualization of myoblast fusion during formation of the Drosophila IFMs. IFM development provides an informative setting for the study of this fundamental aspect of myogenesis, as it allows for application of powerful genetic approaches available in Drosophila to a muscle set that shares a variety of morphological and developmental features with the vertebrate somatic musculature, thereby holding the promise of uncovering conserved principles of the fusion process. Furthermore, this setting is characterized by a large number of repeated and identical fusion events that take place over a short time period, features that are critical for reliable interpretation of the underlying sequence of cellular events.
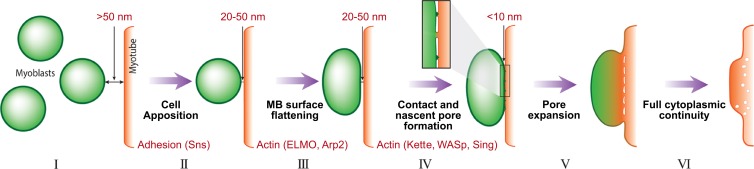
Our observations demonstrate that fusion between growing IFM myotubes and the myoblasts that surround them involves tight association between large surface domains of the two cell types and formation of numerous cell–cell contacts within the apposed surfaces. These points of contact appear to serve as precursors for small fusion pores, which go on to expand, leading to merging of the surface membranes at multiple sites, establishment of cytoplasmic continuity, and, finally, complete cell fusion. Use of informative mutant and knockdown backgrounds has further enabled us to view these observations as a progressive series of discrete cellular events, and propose a framework for the underlying developmental and molecular mechanisms (Fig. 7):
Figure 7.
Model for progress of IFM myoblast fusion. (I) Wing disc–derived myoblasts (green) migrate toward the nascent DLM myotubes (red) and congregate in their vicinity at a distance of 50 nm or more. (II) An initial phase of myoblast–myotube apposition is mediated by cell adhesion elements such as the myoblast cell surface protein Sns. Cell surfaces are positioned 20–50 nm from each other. (III) A branched actin–dependent cell shape change, also involving ELMO, flattens the myoblast surface. (IV) A second branched actin–based process, dependent on the activity of the NPFs WASp and SCAR/WAVE, as well as on the membrane protein Sing, brings the cells in close apposition (<10 nm). This tight association allows for the formation of multiple cell–cell contacts, which serve as sites for initiation of nascent fusion pores. (V and VI) The surface membranes merge and vesiculate as the pores expand, so that eventually full cytoplasmic continuity is obtained, and the myoblast is incorporated into the DLM myotube.
1. IFM growth initiates at an early phase of pupal development, when a swarm of wing disc–derived myoblasts, originating from muscle stem cells (Gunage et al., 2014), migrates toward the nascent DLM myotubes, a persistent set of larval muscles, and congregates in their vicinity (Fernandes et al., 1991; Roy and VijayRaghavan, 1999; Fig. 7 I). This initial, loose form of association between the two cell types triggers a program of myoblast differentiation involving down-regulation of Delta–Notch signaling between the myoblasts (Bernard et al., 2006) and leading to acquisition of fusion competence through the expression of key contributors to the myoblast fusion process, such as elements of the adhesion and branched actin machineries (Gildor et al., 2012).
2. Knockdown of the cell surface element Sns, together with its functional partner Hbs, allowed us to first assess the involvement of cell adhesion in initiating IFM myoblast fusion. The results of this analysis suggest that adhesive interaction between cell surface proteins promotes an ordered form of apposition between the myotube and the surrounding myoblasts, where the neighboring cell surfaces are brought to within ∼20–50 nm of each other (Fig. 7 II). This degree of separation between the two surfaces is in good agreement with the expected size of the Duf-Sns adhesion complex (45 nm) derived from the crystal structure of the Caenorhabditis elegans homologues SYG-1 and SYG-2 (Özkan et al., 2014).
3. Positioning of the cells via cell adhesion appears to be a key intermediate step that serves as a prerequisite for the events that follow. One of these is a flattening of the myoblast cell membrane facing the myotube, increasing the area of the apposed surfaces (Fig. 7 III). This process requires a functional Arp2/3 complex, implying involvement of the branched actin nucleation machinery. A second actin-related element implicated in myoblast surface flattening is the Rac-GEF subunit ELMO/CED-12. A possible molecular scenario (Haralalka and Abmayr, 2010) is that ELMO-dependent activation of Rac, possibly through cross talk with transmembrane cell surface adhesion proteins, leads to stimulation of the Arp2/3 NPF SCAR/WAVE in this context. However, the capacity of kette knockdown myoblasts to flatten their surface suggests that additional targets downstream of the ELMO GEF participate in this branched actin–dependent cell shape change.
4. A second actin-based process, which we view as a particularly critical step for initiation of fusion, brings the myoblast and myotube membranes in tight (<20 nm) apposition to each other (Fig. 7 IV). Initial intermembrane separation on the order of 10–20 nm is characteristic of pairs of biological membranes destined to fuse, and dedicated mechanisms are required to reduce the distance between them to the single nanometer range (Chernomordik and Kozlov, 2008; Martens and McMahon, 2008). Such short distances are compatible with formation of hemifusion intermediates between the outer leaflets of fusion membrane bilayers, a critical, unifying feature underlying vesicular, viral-cell and developmental cell–cell forms of membrane fusion (Chernomordik et al., 2006; Sapir et al., 2008; Leikina et al., 2013). We suggest that the multiple point-like contacts between myoblast and myotube surfaces, which form upon tight apposition, represent sites at which productive fusion between plasma membranes is initiated, in a process requiring the activity of the Arp2/3 NPFs WASp and SCAR/WAVE as well as the transmembrane MARVEL domain protein Sing.
IFM myoblast fusion appears, therefore, to repeatedly make use of molecular cross talk between transmembrane and branched actin nucleation elements to achieve the cell surface arrangements that characterize and promote the different phases of the fusion process. It is interesting to consider the manner by which such cross talk could promote closing of the gap between myoblasts and myotubes, so as to achieve a fusion-ready state. Recent studies have suggested diverse mechanisms by which actin contributes to reorganization of adhesion proteins and enhancement of adhesive interactions in epithelia (Herszterg et al., 2013; Wu et al., 2015). An attractive scenario in the context of myoblast fusion is that initial and relatively weak adhesion interactions, involving the Duf–Rst-Sns–Hbs system, stimulate stronger interactions (via enhanced adhesion between the same elements or incorporation of a distinct molecular adhesion system altogether) in a process mediated by the branched actin machinery.
5. We propose that the multiple and small cell–cell contacts serve as sites for initiation of nascent fusion pores. The surface membranes merge and vesiculate as the pores expand, so that full cytoplasmic continuity is eventually obtained (Fig. 7, V and VI), and the myoblast is incorporated into the DLM myotube. We previously identified a role for WASp in pore expansion during myoblast fusion in Drosophila embryos (Massarwa et al., 2007). The requirement for WASp at an earlier phase of the process in IFMs, in promoting tight surface membrane apposition, prevents direct assessment of its contribution to pore expansion in this context.
This proposed sequence of events for Drosophila IFM myoblast fusion shares several fundamental features with models of cell fusion in diverse settings (Doberstein et al., 1997; Shemer and Podbilewicz, 2000; Chen et al., 2008). One key aspect is that membrane interactions between the cells are initiated at multiple and presumably random sites of contact that are established along the apposed surfaces, giving rise to nascent pores. Additional common features include expansion of the small pores as a means of achieving cytoplasmic continuity and vesiculation of the merged cell membranes before their removal from the former cell–cell interface.
Importantly, initiation of IFM fusion via multiple membrane discontinuities agrees with the classic portrayal of somatic muscle myoblast fusion in Drosophila embryos (Doberstein et al., 1997) but differs substantially from more recent descriptions of myoblast fusion in this setting (Sens et al., 2010; Chen, 2011; Haralalka et al., 2011). These latter models propose a primary role for invasive, myoblast-based membrane extensions in promoting a fusion process based on expansion of a single pore. Although protrusive extensions of similar dimensions emanate from WT IFM myoblasts, they are only infrequently observed. Although no overt aspects of cell surface contact and fusion pore formation were found to be associated with these structures, their relative scarcity precludes a definitive assessment of their contribution to myoblast fusion in this setting. An excess of protrusions is found in WASp mutant, kette knockdown, and sing knockdown pupae. These protrusions are similar in appearance and dimension to those observed on WT myoblasts and maintain the same myoblast–myotube distance distribution profile as the flat portions of the cell surface. Notably, protrusions are absent from sns/hbs, Arp2, and ELMO fusion-arrested IFMs. Collectively, these observations may imply that the low level protrusive activity of IFM myoblasts, whose functional significance is presently unknown, commences upon myotube association and flattening of IFM myoblast membranes and becomes pronounced in the absence of a productive fusion process.
Building upon the involvement of the cell adhesion and branched actin machineries in IFM myoblast fusion established here, future studies can now focus on the roles that specific elements play during this process. A variety of fusion-related functions have been ascribed to the Arp2/3 system in mediating myoblast fusion during development of the somatic muscles of Drosophila embryos, including, among others, myoblast migration toward myotube targets, formation and dissolution of the actin focus, and establishment of invasive myoblast surface extensions. In this context, we also wish to note recent studies that have suggested roles for actin-based myosin motors in embryonic myoblast fusion (Bonn et al., 2013; Kim et al., 2015). All of these constitute intriguing mechanistic modes for using the actin-based cytoskeleton and are highly worthwhile of exploring in the context of the IFM system as well.
In summary, we propose a mechanistic framework at high resolution for myoblast fusion during Drosophila IFM formation. This program involves a stepwise process in which the plasma membranes of myoblasts destined to fuse are placed in tight apposition with the myotube, giving rise to multiple contact sites that resolve into expanding fusion pores. Specific roles are suggested for transmembrane and cytoskeletal elements in mediating this series of events. We expect this model to be of widespread significance as it pertains to a setting bearing general features and characteristics of myogenic differentiation.
Materials and methods
Drosophila IFM preparation and genetics
White (0–1 h APF) pupae were aged appropriately and dissected at room temperature on Sylgard plates (Dow Corning) in a 200-µl drop of Schneider’s Drosophila medium. A recombinant chromosome bearing both the mef2-GAL4 driver (Ranganayakulu et al., 1996) and a UAS-mCD8-GFP transgenic insertion (Lee and Luo, 1999) was present in all backgrounds to aid in IFM dissection by viewing the pupae under blue light with a fluorescence stereomicroscope (MZ16F; Leica). The isolated muscles were immediately placed in the appropriate fixative. The recombinant mef2-GAL4,UAS-mCD8-GFP chromosome also contained a UAS-Dicer2 element for enhancement of RNAi activity (Dietzl et al., 2007). RNAi-based knockdown of specific genes was achieved by crossing flies bearing the recombinant chromosome to flies bearing UAS-RNAi transgenes from the Vienna Drosophila Resource Center (VDRC) and Transgenic RNAi Project (TRiP) collections. TRiP lines were obtained from the Bloomington Drosophila Stock Center at Indiana University. UAS-RNAi lines used in the study include sns (VDRC GD877), hbs (VDRC GD27065), Arp2 (TRiP JF02785), kette/HEM (TRiP HMS02252), and sing (VDRC GD12202). IFMs isolated from the mef2-GAL4,UAS-mCD8-GFP stock served as the WT control. The strong wsp1 allele results from a small intragenic deletion within the WASp coding sequence (Ben-Yaacov et al., 2001).
Immunofluorescence
Dissected IFMs were fixed at room temperature in 4% paraformaldehyde/PBS for 20 min, washed, and stained overnight at 4°C with chicken anti-GFP antibodies (1:1,000; Abcam) and, after washes, for 2 h at room temperature with Alexa Fluor 488–conjugated goat-anti–chicken antibodies (1:1,000; Molecular Probes) to visualize myogenic cells. Microfilaments were visualized by incubation (together with the secondary antibodies) with either 5 µg/ml TRITC-phalloidin (Sigma-Aldrich) or Atto647N-phalloidin (Fluka), and nuclei were similarly visualized with DAPI. Images of immunofluorescent samples were obtained at room temperature using a confocal scanning system (LSM 710; Carl Zeiss) equipped with a microscope (Axio Vert; Carl Zeiss) and using a 40× water-immersion 1.1 NA lens. Initial image acquisition was performed using the imaging system Zen software, and images were processed using Photoshop (CS3; Adobe).
TEM
Isolated pupal IFMs were generally processed by a hybrid method using principles of both chemical fixation (CF) and high pressure freezing (HPF) followed by freeze substitution (FS). Muscles were chemically fixed using a mixture of 4% paraformaldehyde and 2.5% glutaraldehyde (Electron Microscopy Sciences) in sodium cacodylate buffer, pH 7.4, for 1 h at room temperature and washed three times for 10 min with buffer alone. Samples were then quickly placed in aluminum planchettes with 50-µm deep depressions (Wohlwend GmbH), filled with 10% BSA in Schneider’s Drosophila medium (Sigma-Aldrich), and covered with a flat disc. The sandwiched samples were cryoimmobilized in a HPF machine (HPM010; Bal-Tec). FS was performed in an automated FS apparatus (Leica). Frozen planchettes were transferred to precooled cryogenic vials containing FS medium (1% OSO4 and 0.1% uranyl acetate in anhydrous acetone). The samples were incubated at −90°C for 36 h, and the temperature was then slowly raised to −20°C over 24 h. Samples were then removed, kept at 0°C for 1 h, rinsed three times for 10 min with anhydrous acetone, incubated in 2% uranyl acetate for 30 min at room temperature, and rinsed twice with anhydrous acetone. The samples were infiltrated with a mixture of acetone and epon (Agar Scientific) at 30% for 4 h, 60% overnight, 90% for 4 h (4°C), and 100% two times for 12 h at room temperature and embedded and polymerized at 60°C for 48 h. Ultrathin sections were cut using an Ultracut apparatus (Ultracut UCT; Leica). Sections were poststained with 1% lead citrate and 2% uranyl acetate. Images were recorded using a transmission electron microscope (T12 spirit BioTWIN; FEI) operating at 120 kV and equipped with a charge-coupled device camera (Eagle 2K; FEI). Images were processed using Photoshop.
STEM tomography
Processed IFM samples were cut into 350–400-nm-thick sections using a microtome (Ultracut E; Leica). Both sides of the sections were decorated with 10-nm colloidal gold beads, which served as fiducial markers. Sections were subsequently stained and carbon coated. Dual-tilt series were acquired with a microscope (Tecnai F20; FEI) equipped with a field emission gun operating at 200 kV in scanning transmission mode and using a brightfield STEM detector (Gatan). Images were acquired using the SerialEM program for automated tilt series collection as previously described (Mastronarde, 2005) at angular intervals of 1.5° over a tilt range of 70° to −70° (2k × 2k pixel). Alignment and 3D reconstruction were performed using the IMOD image-processing package as previously described (Kremer et al., 1996). Tomograms were segmented and rendered using AVIZO 3D visualization software.
FIB/SEM imaging
Serial view imaging was performed on processed IFM samples as previously described (Weiner et al., 2011). In brief, resin blocks were mounted on stubs and placed into a dual beam system (Helios 600; FEI). Serial views of freshly exposed surfaces were acquired using the Auto Slice and View G2 software (FEI). An ion beam current of 0.92 NA at 30 kV was used for milling slices at a step size of 10 nm. ImageJ software (National Institutes of Health) was used to align images using the stackreg plugin as previously described (Schindelin et al., 2012). Amira (Visage Imaging) was used for segmentation (using the paint brush tool), rendering, and visualization of the data.
Cell surface distance analysis
FIJI and MATLAB software (MathWorks) were used to measure the distance between the myoblasts and myotubes during fusion. Membranes from both the cells at the contact zone were marked using the polygon section-copy-edit-selection-create mask (binary). Using MATLAB, all the pixels in the mask were added up, and the values were binned into the different categories.
Online supplemental material
Fig. S1 is a discussion of the features of the hybrid TEM method and comparison with other TEM techniques. Fig. S2 shows a presentation and analysis of ELMO mutant phenotypes. Fig. S3 is a visualization of fusing myoblasts and associated actin foci in WT and mutant backgrounds using fluorescent probes. Video 1 depicts a visualization of a WT DLM myotube and adjacent myoblasts using FIB/SEM serial view imaging. Video 2 shows a visualization of a WASp mutant DLM myotube and adjacent myoblasts using FIB/SEM serial view imaging. Video 3 portrays a visualization of a myotube–myoblast interface using STEM tomography. Video 4 shows a visualization of a single fusion pore from a myotube–myoblast interface using STEM tomography. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503005/DC1.
Supplementary Material
Acknowlegments
We wish to thank Beni Podbilewicz and Talila Volk for critical reading of the manuscript and Orit Bechar for the schemes used in Figs. 1 and 7. We are grateful to Allon Weiner and Katya Rechav for instruction and advice regarding serial surface imaging and for generating the 3D reconstructions accompanying Videos 1 and 2, and to Mugdha Sathe for advice and MATLAB-based analysis of cell–cell distances. We thank our colleagues in the Shilo and VijayRaghavan laboratories for their continuous input and encouragement.
This work was supported by a grant from the Israel Science Foundation to E.D. Schejter and B-Z. Shilo. B-Z. Shilo is an incumbent of the Hilda and Cecil Lewis Professorial Chair of Molecular Genetics.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APF
- after puparium formation
- CF
- chemical fixation
- DLM
- dorsolongitudinal muscle
- FIB
- focused ion beam
- FS
- freeze substitution
- GEF
- guanine exchange factor
- HPF
- high pressure freezing
- IFM
- indirect flight muscle
- NPF
- nucleation-promoting factor
- SEM
- scanning EM
- STEM
- scanning transmission EM
- TEM
- transmission EM
- WT
- wild type
References
- Abmayr S.M., and Pavlath G.K.. 2012. Myoblast fusion: lessons from flies and mice. Development. 139:641–656. 10.1242/dev.068353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar P.S., Baylies M.K., Fleissner A., Helming L., Inoue N., Podbilewicz B., Wang H., and Wong M.. 2013. Genetic basis of cell-cell fusion mechanisms. Trends Genet. 29:427–437. 10.1016/j.tig.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama K., Takagi T., Hirase A., and Miyazawa A.. 2008. STEM tomography for thick biological specimens. Ultramicroscopy. 109:70–80. 10.1016/j.ultramic.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Bennett A.E., Narayan K., Shi D., Hartnell L.M., Gousset K., He H., Lowekamp B.C., Yoo T.S., Bliss D., Freed E.O., and Subramaniam S.. 2009. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 5:e1000591 10.1371/journal.ppat.1000591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaacov S., Le Borgne R., Abramson I., Schweisguth F., and Schejter E.D.. 2001. Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J. Cell Biol. 152:1–13. 10.1083/jcb.152.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Schäfer G., Kesper D.A., Holz A., Eriksson T., Palmer R.H., Beck L., Klämbt C., Renkawitz-Pohl R., and Onel S.F.. 2008. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J. Cell Sci. 121:1303–1313. 10.1242/jcs.022269 [DOI] [PubMed] [Google Scholar]
- Bernard F., Dutriaux A., Silber J., and Lalouette A.. 2006. Notch pathway repression by vestigial is required to promote indirect flight muscle differentiation in Drosophila melanogaster. Dev. Biol. 295:164–177. 10.1016/j.ydbio.2006.03.022 [DOI] [PubMed] [Google Scholar]
- Bernstein S.I., O’Donnell P.T., and Cripps R.M.. 1993. Molecular genetic analysis of muscle development, structure, and function in Drosophila. Int. Rev. Cytol. 143:63–152. 10.1016/S0074-7696(08)61874-4 [DOI] [PubMed] [Google Scholar]
- Blair S.S. 2003. Genetic mosaic techniques for studying Drosophila development. Development. 130:5065–5072. 10.1242/dev.00774 [DOI] [PubMed] [Google Scholar]
- Bonn B.R., Rudolf A., Hornbruch-Freitag C., Daum G., Kuckwa J., Kastl L., Buttgereit D., and Renkawitz-Pohl R.. 2013. Myosin heavy chain-like localizes at cell contact sites during Drosophila myoblast fusion and interacts in vitro with Rolling pebbles 7. Exp. Cell Res. 319:402–416. 10.1016/j.yexcr.2012.12.005 [DOI] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S.F., Tosello-Trampont A.C., Macara I.G., Madhani H., Fink G.R., and Ravichandran K.S.. 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4:574–582. [DOI] [PubMed] [Google Scholar]
- Brunetti T.M., Fremin B.J., and Cripps R.M.. 2015. Identification of singles bar as a direct transcriptional target of Drosophila Myocyte enhancer factor-2 and a regulator of adult myoblast fusion. Dev. Biol. 401:299–309. 10.1016/j.ydbio.2015.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.H. 2011. Invasive podosomes and myoblast fusion. Curr Top Membr. 68:235–258. 10.1016/B978-0-12-385891-7.00010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.H., and Olson E.N.. 2004. Towards a molecular pathway for myoblast fusion in Drosophila. Trends Cell Biol. 14:452–460. 10.1016/j.tcb.2004.07.008 [DOI] [PubMed] [Google Scholar]
- Chen A., Leikina E., Melikov K., Podbilewicz B., Kozlov M.M., and Chernomordik L.V.. 2008. Fusion-pore expansion during syncytium formation is restricted by an actin network. J. Cell Sci. 121:3619–3628. 10.1242/jcs.032169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., and Kozlov M.M.. 2008. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15:675–683. 10.1038/nsmb.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Zimmerberg J., and Kozlov M.M.. 2006. Membranes of the world unite! J. Cell Biol. 175:201–207. 10.1083/jcb.200607083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M. 2006. Insect flight. Curr. Biol. 16:R309–R314. 10.1016/j.cub.2006.03.087 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Doberstein S.K., Fetter R.D., Mehta A.Y., and Goodman C.S.. 1997. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 136:1249–1261. 10.1083/jcb.136.6.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., and VijayRaghavan K. 2006. Metamorphosis and the formation of the adult musculature. In Muscle Development in Drosophila. Sink H., editor. Springer Science+Business Media, New York, NY: 125–142. [Google Scholar]
- Dworak H.A., and Sink H.. 2002. Myoblast fusion in Drosophila. BioEssays. 24:591–601. 10.1002/bies.10115 [DOI] [PubMed] [Google Scholar]
- Estrada B., Maeland A.D., Gisselbrecht S.S., Bloor J.W., Brown N.H., and Michelson A.M.. 2007. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev. Biol. 307:328–339. 10.1016/j.ydbio.2007.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.J., and Keshishian H.. 1999. Development of the adult neuromuscular system. Int. Rev. Neurobiol. 43:221–239. 10.1016/S0074-7742(08)60547-4 [DOI] [PubMed] [Google Scholar]
- Fernandes J., Bate M., and Vijayraghavan K.. 1991. Development of the indirect flight muscles of Drosophila. Development. 113:67–77. [DOI] [PubMed] [Google Scholar]
- Galletta B.J., Chakravarti M., Banerjee R., and Abmayr S.M.. 2004. SNS: Adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech. Dev. 121:1455–1468. 10.1016/j.mod.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Geisbrecht E.R., Haralalka S., Swanson S.K., Florens L., Washburn M.P., and Abmayr S.M.. 2008. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 314:137–149. 10.1016/j.ydbio.2007.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor B., Massarwa R., Shilo B.Z., and Schejter E.D.. 2009. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 10:1043–1050. 10.1038/embor.2009.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor B., Schejter E.D., and Shilo B.Z.. 2012. Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development. 139:4040–4050. 10.1242/dev.077495 [DOI] [PubMed] [Google Scholar]
- Goley E.D., and Welch M.D.. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7:713–726. 10.1038/nrm2026 [DOI] [PubMed] [Google Scholar]
- Gruenbaum-Cohen Y., Harel I., Umansky K.B., Tzahor E., Snapper S.B., Shilo B.Z., and Schejter E.D.. 2012. The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc. Natl. Acad. Sci. USA. 109:11211–11216. 10.1073/pnas.1116065109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunage R.D., Reichert H., and VijayRaghavan K.. 2014. Identification of a new stem cell population that generates Drosophila flight muscles. eLife. 3:e03126 10.7554/eLife.03126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S., and Abmayr S.M.. 2010. Myoblast fusion in Drosophila. Exp. Cell Res. 316:3007–3013. 10.1016/j.yexcr.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S., Shelton C., Cartwright H.N., Katzfey E., Janzen E., and Abmayr S.M.. 2011. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 138:1551–1562. 10.1242/dev.057653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszterg S., Leibfried A., Bosveld F., Martin C., and Bellaiche Y.. 2013. Interplay between the dividing cell and its neighbors regulates adherens junction formation during cytokinesis in epithelial tissue. Dev. Cell. 24:256–270. 10.1016/j.devcel.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Kesper D.A., Stute C., Buttgereit D., Kreisköther N., Vishnu S., Fischbach K.F., and Renkawitz-Pohl R.. 2007. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev. Dyn. 236:404–415. 10.1002/dvdy.21035 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Ren Y., Ng W.P., Li S., Son S., Kee Y.S., Zhang S., Zhang G., Fletcher D.A., Robinson D.N., and Chen E.H.. 2015. Mechanical tension drives cell membrane fusion. Dev. Cell. 32:561–573. 10.1016/j.devcel.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Shilagardi K., Zhang S., Hong S.N., Sens K.L., Bo J., Gonzalez G.A., and Chen E.H.. 2007. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev. Cell. 12:571–586. 10.1016/j.devcel.2007.02.019 [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Laurin M., Fradet N., Blangy A., Hall A., Vuori K., and Côté J.F.. 2008. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc. Natl. Acad. Sci. USA. 105:15446–15451. 10.1073/pnas.0805546105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., and Luo L.. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Leikina E., Melikov K., Sanyal S., Verma S.K., Eun B., Gebert C., Pfeifer K., Lizunov V.A., Kozlov M.M., and Chernomordik L.V.. 2013. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J. Cell Biol. 200:109–123. 10.1083/jcb.201207012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S., and McMahon H.T.. 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9:543–556. 10.1038/nrm2417 [DOI] [PubMed] [Google Scholar]
- Massarwa R., Carmon S., Shilo B.Z., and Schejter E.D.. 2007. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev. Cell. 12:557–569. 10.1016/j.devcel.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Millay D.P., O’Rourke J.R., Sutherland L.B., Bezprozvannaya S., Shelton J.M., Bassel-Duby R., and Olson E.N.. 2013. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 499:301–305. 10.1038/nature12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P., Gildor B., Shilo B.Z., VijayRaghavan K., and Schejter E.D.. 2011. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Development. 138:2347–2357. 10.1242/dev.055012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak S.J., Nahirney P.C., Hadjantonakis A.K., and Baylies M.K.. 2009. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J. Cell Sci. 122:3282–3293. 10.1242/jcs.047597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önel S.F., and Renkawitz-Pohl R.. 2009. FuRMAS: triggering myoblast fusion in Drosophila. Dev. Dyn. 238:1513–1525. 10.1002/dvdy.21961 [DOI] [PubMed] [Google Scholar]
- Önel S.F., Rust M.B., Jacob R., and Renkawitz-Pohl R.. 2014. Tethering membrane fusion: common and different players in myoblasts and at the synapse. J. Neurogenet. 28:302–315. 10.3109/01677063.2014.936014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E., Chia P.H., Wang R.R., Goriatcheva N., Borek D., Otwinowski Z., Walz T., Shen K., and Garcia K.C.. 2014. Extracellular architecture of the SYG-1/SYG-2 adhesion complex instructs synaptogenesis. Cell. 156:482–494. 10.1016/j.cell.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt A.Y., and Insall R.H.. 2009. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J. Cell Sci. 122:2575–2578. 10.1242/jcs.023879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G., Schulz R.A., and Olson E.N.. 1996. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 176:143–148. 10.1006/dbio.1996.9987 [DOI] [PubMed] [Google Scholar]
- Reedy M.C., and Beall C.. 1993. Ultrastructure of developing flight muscle in Drosophila. I. Assembly of myofibrils. Dev. Biol. 160:443–465. 10.1006/dbio.1993.1320 [DOI] [PubMed] [Google Scholar]
- Richardson B.E., Beckett K., Nowak S.J., and Baylies M.K.. 2007. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 134:4357–4367. 10.1242/dev.010678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B.E., Nowak S.J., and Baylies M.K.. 2008. Myoblast fusion in fly and vertebrates: new genes, new processes and new perspectives. Traffic. 9:1050–1059. 10.1111/j.1600-0854.2008.00756.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K., Yu S., Roy S., and Baylies M.K.. 2010. Myoblast fusion: when it takes more to make one. Dev. Biol. 341:66–83. 10.1016/j.ydbio.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., Wu C., and Bear J.E.. 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14:7–12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Roy S., and VijayRaghavan K.. 1998. Patterning muscles using organizers: larval muscle templates and adult myoblasts actively interact to pattern the dorsal longitudinal flight muscles of Drosophila. J. Cell Biol. 141:1135–1145. 10.1083/jcb.141.5.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., and VijayRaghavan K.. 1999. Muscle pattern diversification in Drosophila: the story of imaginal myogenesis. BioEssays. 21:486–498. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gómez M., Coutts N., Price A., Taylor M.V., and Bate M.. 2000. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 102:189–198. 10.1016/S0092-8674(00)00024-6 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pulido L., Martín-Belmonte F., Valencia A., and Alonso M.A.. 2002. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem. Sci. 27:599–601. 10.1016/S0968-0004(02)02229-6 [DOI] [PubMed] [Google Scholar]
- Sapir A., Avinoam O., Podbilewicz B., and Chernomordik L.V.. 2008. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev. Cell. 14:11–21. 10.1016/j.devcel.2007.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Schönbauer C., Langer C.C., Dietzl G., Novatchkova M., Schernhuber K., Fellner M., Azaryan A., Radolf M., Stark A., et al. 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 464:287–291. 10.1038/nature08799 [DOI] [PubMed] [Google Scholar]
- Schröter R.H., Lier S., Holz A., Bogdan S., Klämbt C., Beck L., and Renkawitz-Pohl R.. 2004. kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 131:4501–4509. 10.1242/dev.01309 [DOI] [PubMed] [Google Scholar]
- Sens K.L., Zhang S., Jin P., Duan R., Zhang G., Luo F., Parachini L., and Chen E.H.. 2010. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J. Cell Biol. 191:1013–1027. 10.1083/jcb.201006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiq S.A. 1963. Electron microscopic studies on the indirect flight muscles of Drosophila melanogaster. II. Differentiation of myofibrils. J. Cell Biol. 17:363–373. 10.1083/jcb.17.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C., Kocherlakota K.S., Zhuang S., and Abmayr S.M.. 2009. The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development. 136:1159–1168. 10.1242/dev.026302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G., and Podbilewicz B.. 2000. Fusomorphogenesis: cell fusion in organ formation. Dev. Dyn. 218:30–51. [DOI] [PubMed] [Google Scholar]
- Simionescu A., and Pavlath G.K.. 2011. Molecular mechanisms of myoblast fusion across species. Adv. Exp. Med. Biol. 713:113–135. 10.1007/978-94-007-0763-4_8 [DOI] [PubMed] [Google Scholar]
- Sosinsky G.E., Crum J., Jones Y.Z., Lanman J., Smarr B., Terada M., Martone M.E., Deerinck T.J., Johnson J.E., and Ellisman M.H.. 2008. The combination of chemical fixation procedures with high pressure freezing and freeze substitution preserves highly labile tissue ultrastructure for electron tomography applications. J. Struct. Biol. 161:359–371. 10.1016/j.jsb.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A.A., and Leapman R.D.. 2012. Development and application of STEM for the biological sciences. Ultramicroscopy. 123:38–49. 10.1016/j.ultramic.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E., Schaffer M., Plitzko J.M., and Baumeister W.. 2013. Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr. Opin. Struct. Biol. 23:771–777. 10.1016/j.sbi.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Weiner A., Dahan-Pasternak N., Shimoni E., Shinder V., von Huth P., Elbaum M., and Dzikowski R.. 2011. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell. Microbiol. 13:967–977. 10.1111/j.1462-5822.2011.01592.x [DOI] [PubMed] [Google Scholar]
- Wu Y., Kanchanawong P., and Zaidel-Bar R.. 2015. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev. Cell. 32:139–154. 10.1016/j.devcel.2014.12.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.