Cells sense the overall chromosome congression state in metaphase to promote anaphase entry using RanGTP, which induces the reduction of two spindle assembly checkpoint proteins, BuGZ and Bub3, via the E3 ligase Ubr5.
Abstract
RanGTP is known to regulate the spindle assembly checkpoint (SAC), but the underlying molecular mechanism is unclear. BuGZ stabilizes SAC protein Bub3 through direct interaction and facilitates its mitotic function. Here we show that RanGTP promotes the turnover of BuGZ and Bub3 in metaphase, which in turn facilitates metaphase-to-anaphase transition. BuGZ and Bub3 interact with either importin-β or an E3 ubiquitin ligase, Ubr5. RanGTP promotes the dissociation of importin-β from BuGZ and Bub3 in metaphase. This results in increased binding of BuGZ and Bub3 to Ubr5, leading to ubiquitination and subsequent turnover of both proteins. We propose that elevated metaphase RanGTP levels use Ubr5 to couple overall chromosome congression to SAC silencing.
Introduction
The mitotic checkpoint complex (MCC), consisting mainly of Mad2, BubR1, Bub3, and Cdc20, functions to prevent premature chromosome segregation. Only after full chromosome alignment at the metaphase plate will the MCC become inactivated, resulting in spindle assembly checkpoint (SAC) silencing and subsequent anaphase onset (Musacchio and Salmon, 2007; Fang and Zhang, 2011; Kim and Yu, 2011; Musacchio, 2011). Cells use several means to ensure timely SAC silencing. Of these most are known to function in monitoring proper microtubule (MT)–kinetochore attachment. Accordingly, most SAC proteins, including Mad1, Mad2, BubR1, Bub3, and Mps1, are kinetochore associated (Howell et al., 2001; Habu et al., 2002; Xia et al., 2004; Mapelli et al., 2006; Griffis et al., 2007; Yang et al., 2007; Logarinho et al., 2008; Chan et al., 2009; Daum et al., 2009; Gaitanos et al., 2009; Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009; Barisic et al., 2010; Gassmann et al., 2010; Kim et al., 2010a,b; Liu et al., 2010).
Increasing RanGTP in Xenopus egg extracts or HeLa cells can lead to SAC silencing (Arnaoutov and Dasso, 2003; Li et al., 2007). Mathematical modeling, based on the mechanism of chromosome-mediated RanGTP production (Kaláb et al., 2002, 2006; Li et al., 2003; Li and Zheng, 2004), has shown that the highest RanGTP concentration is generated around fully aligned metaphase chromosomes (Li et al., 2007). A single misaligned chromosome not only experiences the lowest RanGTP but also reduces RanGTP concentrations generated around congressed chromosomes. Thus, the highest RanGTP concentration at metaphase could couple the completion of chromosome congression with SAC inactivation (Li et al., 2007). However, the mechanism by which Ran regulates SAC has been difficult to decipher.
By controlling interactions between importin-β and spindle assembly factors (SAFs), RanGTP coordinates the activity of SAFs to promote spindle assembly, spindle orientation, and spindle matrix assembly (Carazo-Salas et al., 1999, 2001; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Wilde et al., 2001; Goodman and Zheng, 2006; Tsai et al., 2006; O’Connell and Khodjakov, 2007; Ma et al., 2009; Goodman et al., 2010; Zheng, 2010; Kim et al., 2011; Wee et al., 2011; Kiyomitsu and Cheeseman, 2012). This suggests that the elevated RanGTP surrounding the metaphase chromosomes might maximally displace importin-β from regulators of SAC to promote anaphase entry. By studying BuGZ (Bub3 interacting and GLEBS motif containing ZNF207), which functions as a chaperone for Bub3 (Jiang et al., 2014; Toledo et al., 2014), we report that RanGTP promotes the E3 ubiquitin ligase Ubr5-mediated turnover of BuGZ and Bub3 during metaphase to facilitate anaphase entry.
Results and discussion
BuGZ and Bub3 turnover may facilitate anaphase entry
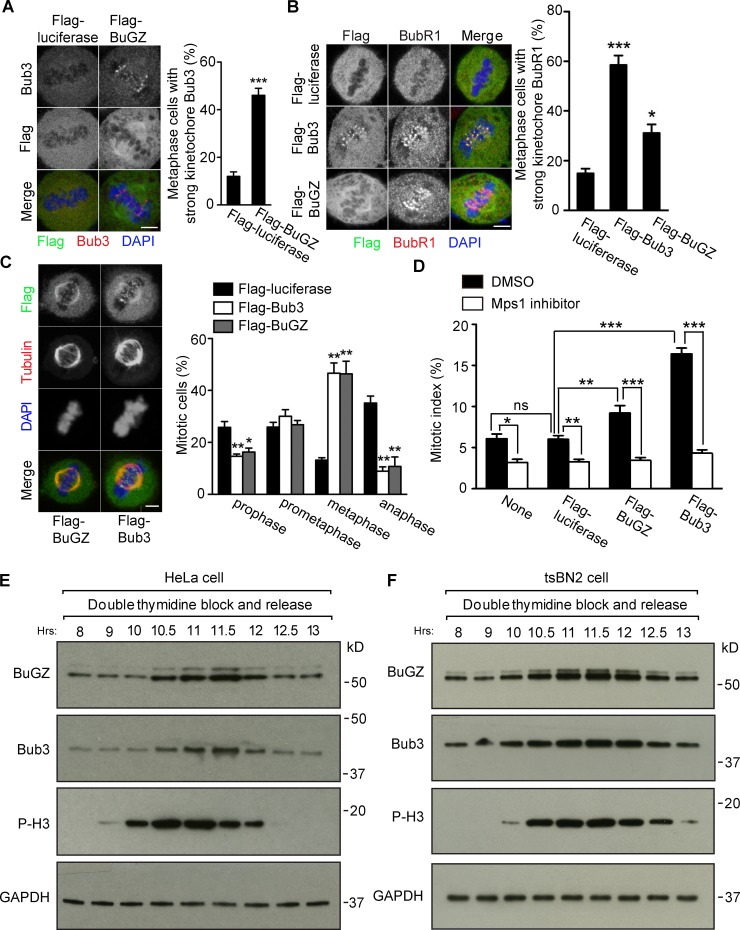
Expression of Flag-tagged BuGZ or Bub3 48 h after transfection caused increased metaphase cells with strong kinetochore Bub3 and BubR1 signals, compared with controls (Fig. 1, A and B). This resulted in a significant enrichment of metaphase cells at the expense of anaphase cells (Fig. 1 C), and an elevated mitotic index that could be ameliorated by 1 µM of Mps1 inhibitor (NMS-P715) 28 h after transfection (Fig. 1 D). Analyzing HeLa or tsBN2 cells synchronized by double thymidine block (DTB) showed that BuGZ and Bub3 rose upon mitotic entry and fell upon mitotic exit (Fig. 1, E and F).
Figure 1.
Bub3 and BuGZ levels affect mitosis. (A–C) Overexpression of Flag-BuGZ or Flag-Bub3 increased metaphase kinetochore Bub3 (A) or BubR1 (B), respectively, and the percentage of metaphase cells (C). Approximately 50 metaphase (A and B) or 100 mitotic cells (C) were counted per experiment. Bars, 5 µm. (D) Mps1 inhibition alleviated mitotic block caused by Flag-BuGZ or Flag-Bub3 overexpression. Approximately 500 cells were counted per experiment. (E and F) Bub3 and BuGZ rose and fell in HeLa (E) or tsBN2 (F) cells after release from DTB. Phospho-Histone H3 (P-H3) marks mitosis. GAPDH, loading control. Error bars indicate SEM. Student’s t test: ns, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001 from three experiments.
To determine precisely when BuGZ and Bub3 declined, HeLa cells released from DTB were blocked in prometaphase by nocodazole or metaphase by MG132 and then released. Immunostaining and Western blotting revealed that BuGZ and Bub3 declined before or upon anaphase onset (Fig. S1, A–D). Consistently (Howell et al., 2004), live imaging of HeLa cells expressing a moderate level of GFP-Bub3 (Jiang et al., 2014) showed that the GFP-Bub3 levels were reduced in metaphase, but persisted in prometaphase (Fig. S1, E and F). Thus, BuGZ and Bub3 undergo turnover during metaphase.
RanGTP-stimulated metaphase turnover of BuGZ and Bub3 facilitates anaphase entry
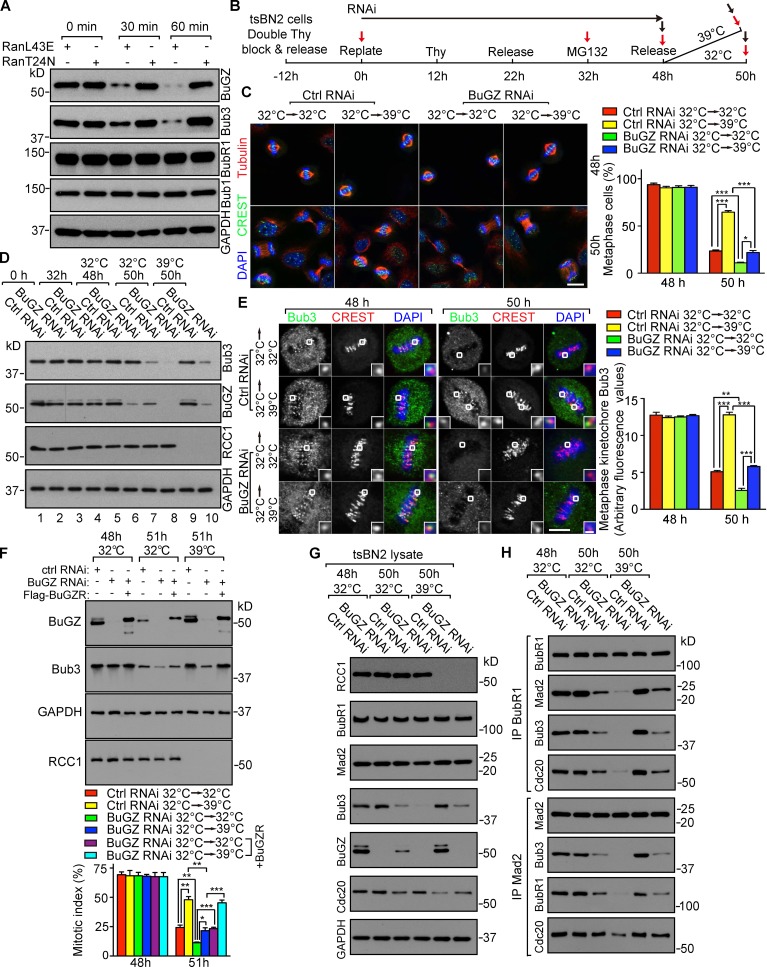
Because a high BuGZ level stabilizes the SAC component Bub3 and promotes chromosome alignment in prometaphase (Jiang et al., 2014), the rise of both proteins in prometaphase and their decline in metaphase would, respectively, facilitate chromosome congression and anaphase entry. Consistently, RanL43E (mimicking the GTP-bound activated Ran) silenced SAC in SAC-activated Xenopus egg extracts, whereas the inactive RanGDP-mimicking RanT24N failed to do so, and RanL43E-induced SAC override resulted in a reduction of BuGZ and Bub3 without affecting BubR1 and Bub1 (Figs. 2 A and S2, A and B).
Figure 2.
RanGTP-mediated BuGZ/Bub3 turnover promotes anaphase entry. (A) RanGTP specifically induced BuGZ and Bub3 degradation in SAC-activated Xenopus egg extract. (B) Synchronization and RNAi of tsBN2 cells. Black and red arrows indicate collection of cells for immunostaining or Western blotting (after mitotic shake-off), respectively. (C) Cells were labeled for MTs, chromosomes (DAPI), and centromeres (CREST). Approximately 100 mitotic cells were quantified per experiment. Bar, 10 µm. (D) RCC1 depletion elevated Bub3 levels in mitotic cells. (E) RCC1 depletion enhanced metaphase kinetochore Bub3. 50 kinetochores in 5–10 cells were counted per experiment. Outlined kinetochores are enlarged to show details (insets). Bars: (main panels) 5 µm; (enlargements) 0.5 µm. (F) Effects of BuGZ depletion were rescued by BuGZR, the RNAi-insensitive BuGZ. (G and H) Analyses of MCC components by immunoprecipitation (IP). Error bars indicate SEM. Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001 from three experiments.
To test whether RanGTP could promote BuGZ and Bub3 turnover in mitosis in vivo, we used tsBN2 cells, whose Ran can be inactivated after a 2-h temperature shift from 32°C to 39°C due to the degradation of its nucleotide exchange factor RCC1 (Nishitani et al., 1991; Li and Zheng, 2004). When mitotic cells were collected from tsBN2 cells that were released from DTB for 10 h and incubated for another 2 h at 32°C or 39°C, RCC1 degradation at 39°C was indeed accompanied by elevated BuGZ and Bub3 (Fig. S2 C).
To study whether RanGTP is required for BuGZ and Bub3 turnover in metaphase and whether this facilitates anaphase entry, we need to synchronize cells in metaphase while maintaining Bub3 levels before shifting cells to 39°C to degrade RCC1. Because the BuGZ depletion–induced Bub3 degradation could be blocked by MG132 (Jiang et al., 2014), we tested the time course of BuGZ and Bub3 reduction upon BuGZ RNAi to identify a time of MG132 addition that would stabilize Bub3. tsBN2 cells were synchronized and treated by control or BuGZ RNAi (see Fig. S2 D). Mitotic shake-off was used to collect mitotic cells as indicated (Fig. S2 D, red arrows). In the absence of MG132, a clear reduction of BuGZ and Bub3 occurred after 36 h of BuGZ RNAi, whereas MG132 stabilized Bub3 up to 48 h of BuGZ RNAi when the BuGZ reduction was apparent (Fig. S2 E).
BuGZ facilitates efficient chromosome alignment by stabilizing Bub3 and promoting Bub3′s kinetochore loading (Jiang et al., 2014). Thus, despite Bub3 stabilization by MG132, the BuGZ depletion by RNAi would still compromise the efficiency of Bub3 kinetochore loading and chromosome alignment. Indeed, analysis of the percentage of cells with misaligned chromosomes in cells treated by the scheme in Fig. 2 B showed an increased chromosome misalignment after 40 and 44 h of BuGZ RNAi regardless of MG132 addition, compared with control RNAi (Fig. S2 F). However, after 16 h of MG132 block (corresponding to 48 h of BuGZ RNAi), chromosome misalignment was reduced to a similar level as seen in control RNAi-treated tsBN2 cells without MG132 (Fig. S2 F). Thus, prolonged MG132 block provided time for chromosome alignment in BuGZ-depleted cells.
Using the conditions defined above, we synchronized tsBN2 cells and treated them with control or BuGZ siRNA as shown in Fig. 2 B. Mitotic cells (Fig. 2 B, red arrows) were used for Western blotting, whereas cells grown on coverslips were used for immunostaining (Fig. 2 B, black arrows). Incubation at 39°C reduced anaphase entry of control siRNA-treated cells (Fig. 2 C), coinciding with the lack of BuGZ and Bub3 turnover (Fig. 2 D, compare lanes 5, 7, and 9). BuGZ siRNA-treated cells, however, entered anaphase at 39°C (Fig. 2 C). In the BuGZ RNAi cells, RCC1 depletion no longer prevented Bub3 turnover (Fig. 2 D, compare lanes 6 and 9 with 10). Consistently, BuGZ RNAi-treated cells at 39°C entered anaphase less efficiently than those at 32°C (Fig. 2 C, compare the green and blue bars at 50 h), coinciding again with different Bub3 (and BuGZ) levels (Fig. 2 D, compare lanes 8 and 10). RCC1 depletion at 39°C also resulted in stronger kinetochore Bub3 in both control and BuGZ RNAi-treated cells than those incubated at 32°C (Fig. 2 E), correlating with their inefficient anaphase entry (Fig. 2 C). The inefficient anaphase entry of tsBN2 cells as judged by elevated mitotic index could be fully rescued by the RNAi-insensitive mouse Flag-BuGZ (Fig. 2 F) and was correlated with a reduction of Bub3 and BubR1 in the MCC as judged by immunoprecipitation using BubR1 or Cdc20 antibodies (Fig. 2, G and H). Thus, RanGTP promotes SAC silencing by inducing BuGZ and Bub3 turnover in metaphase.
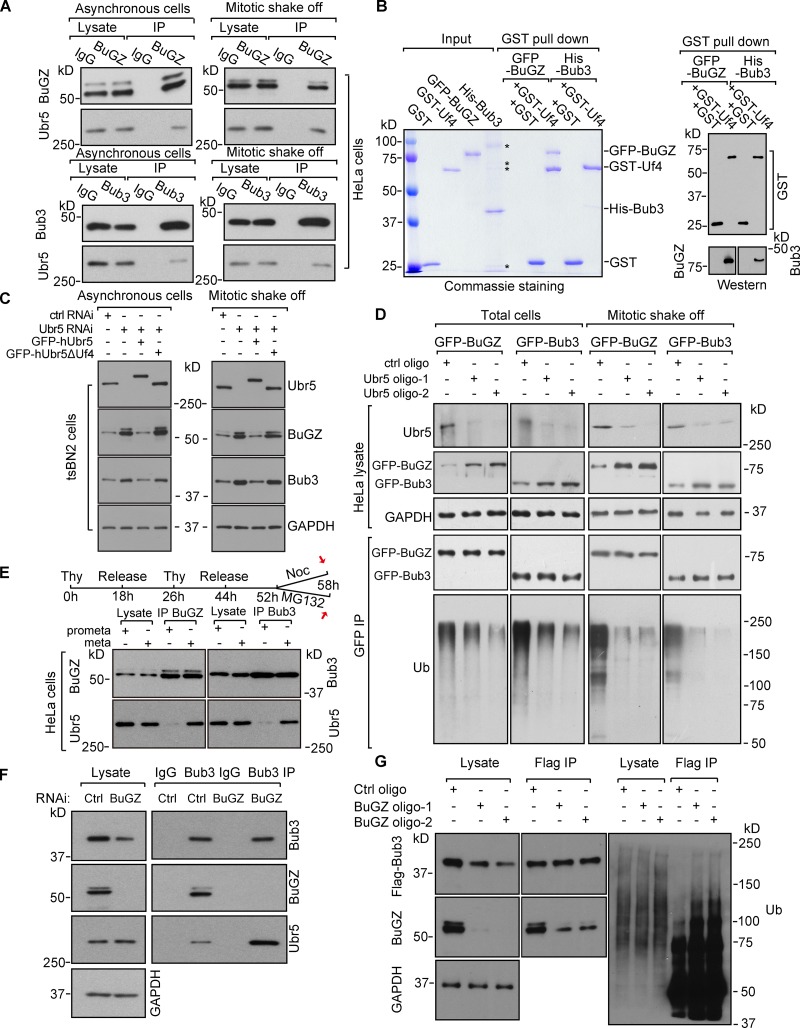
Ubr5 binds BuGZ and Bub3 to mediate their ubiquitination and turnover
Among the top putative BuGZ-interacting proteins (Jiang et al., 2014), we noticed a HECT (homologous to the E6-AP C terminus) domain–containing E3 ubiquitin ligase called Ubr5. Ubr5 can regulate DNA damage response (Honda et al., 2002; Henderson et al., 2006; Munoz et al., 2007; Ling and Lin, 2011; Watts and Saunders, 2011; Smits, 2012; Zhang et al., 2014), but its role in mitosis is unclear. Flag-BuGZ or Flag-Bub3 pulled down Ubr5 in HeLa cells (Fig. S2 G). Endogenous BuGZ and Bub3 also associated with Ubr5 in asynchronous or mitotic HeLa cells and in cytostatic factor (CSF)–arrested Xenopus laevis egg extracts (Figs. 3 A and S2 H). Truncation analyses (Fig. S2 I; Jiang et al., 2011; Benavides et al., 2013) revealed that the purified Ubr5 fragment GST-Uf4, containing the substrate binding HECT domain, pulled down purified BuGZ or Bub3 (Fig. 3 B). Thus, Ubr5 directly binds to BuGZ and Bub3.
Figure 3.
Ubr5 targets BuGZ and Bub3 for ubiquitination and turnover. (A) IP Ubr5 by BuGZ or Bub3 antibodies in HeLa cells. (B) The HECT domain of Ubr5 (Uf4) directly bound to BuGZ and Bub3. Asterisks indicate contaminating bands. (C) Ubr5 depletion stabilized Bub3 and BuGZ in tsBN2 cells. GFP-hUbr5, but not GFP-hUbr5ΔUf4, rescued the effect. GAPDH, loading control. (D) Ubr5 depletion reduced ubiquitinated GFP-Bub3 or GFP-BuGZ in HeLa cells. (E) Stronger interaction between Ubr5 and BuGZ/Bub3 in metaphase than in prometaphase. HeLa cells were collected as shown by red arrows. (F and G) BuGZ depletion led to elevated Bub3-Ubr5 association and Bub3 ubiquitination. Mitotic HeLa cells were used for IP. In G, Flag-Bub3–expressing cells were treated by MG132 before mitotic shake-off.
Ubr5 depletion by RNAi led to higher BuGZ and Bub3 levels in asynchronous or mitotic tsBN2 or HeLa cells than controls (Figs. 3 C and S2 J). Expressing the human GFP-Ubr5 (GFP-hUbr5), which is insensitive to the siRNA oligo targeting hamster Ubr5 in tsBN2 cells, reduced BuGZ and Bub3 levels in the Ubr5 RNAi-treated tsBN2 cells, whereas expressing the GFP-hUbr5 deleted of Uf4 (hUbr5ΔUf4) had no effect (Fig. 3 C). We then tested whether Ubr5 could mediate BuGZ and Bub3 ubiquitination. Because BuGZ and Bub3 antibody pulldown gave rise to high background in the high molecular regions (unpublished data), we used HeLa cells expressing GFP-BuGZ or GFP-Bub3 to allow the detection of polyubiquitinated proteins by GFP antibody pulldown. HeLa cells were treated by control or Ubr5 RNAi for 66 h followed by incubation with MG132 for 6 h. Total cells or mitotic cells (collected by mitotic shake-off) were used for GFP pulldown and Western blotting. Both GFP-BuGZ and GFP-Bub3 were polyubiquitinated, which was reduced upon Ubr5 depletion (Fig. 3 D).
Next we synchronized HeLa cells (see Fig. 3 E). Mitotic cells collected by mitotic shake-off were used for immunoprecipitation by BuGZ or Bub3 antibodies. Ubr5 exhibited greater interaction with BuGZ and Bub3 in metaphase (MG132 block) than in prometaphase (nocodazole block; Fig. 3 E). We then treated HeLa cells with control or BuGZ siRNA for 60 h, collected the mitotic cells by mitotic shake-off, and performed Bub3 antibody pulldown. BuGZ depletion promoted the Bub3–Ubr5 interaction in mitotic cells (Fig. 3 F). Next Flag-Bub3–expressing HeLa cells were treated with control or BuGZ siRNA for 54 h followed by incubation with MG132 for 6 h. Flag antibody pulldown assays using cells collected by mitotic shake-off showed that BuGZ depletion enhanced Bub3 polyubiquitination (Fig. 3 G). Thus Ubr5 mediates the turnover of BuGZ followed by Bub3 in metaphase to facilitate anaphase entry.
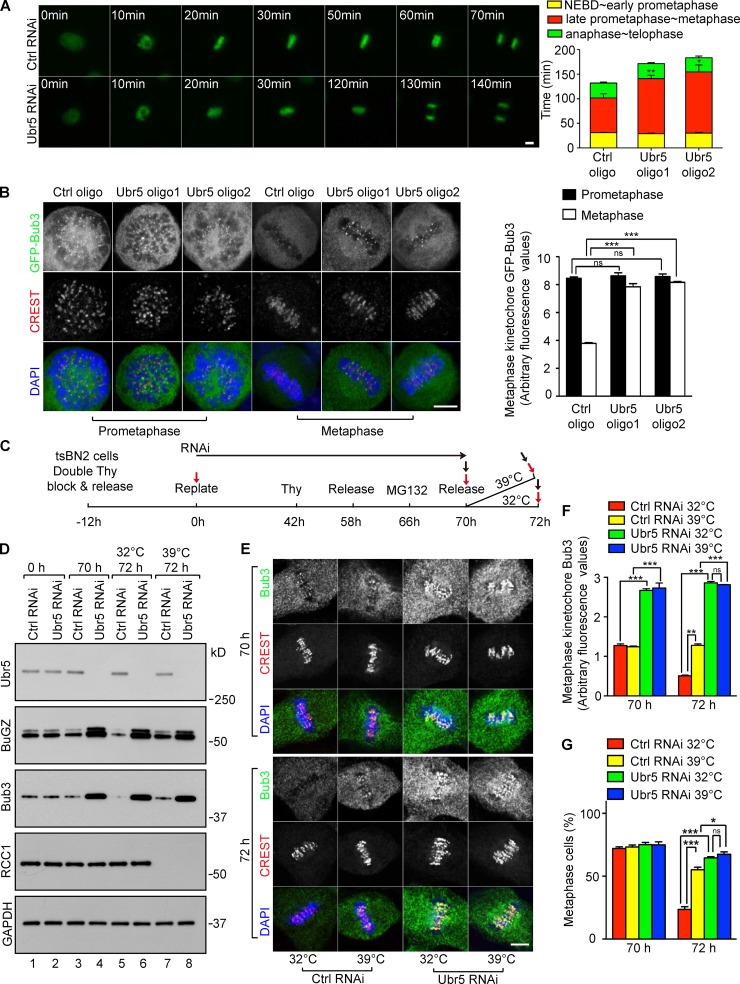
Ubr5 mediates the turnover of Bub3 and BuGZ induced by RanGTP in metaphase
We treated HeLa cells expressing Histone H2B-GFP with Ubr5 siRNA for 60 h followed by live imaging. Ubr5 RNAi led to longer metaphase than controls, without affecting other mitotic phases (Fig. 4 A). Ubr5 depletion by 72 h of RNAi led to increased metaphase cells compared with controls (Fig. S3 A). This metaphase block was similar to those seen in cells overexpressing BuGZ or Bub3 (see Fig. 1 C). Consistently, Ubr5-depleted HeLa and tsBN2 cells had an elevated mitotic index, and the elevated mitotic index in tsBN2 cells was ameliorated by expressing GFP-hUbr5, but not by expressing GFP-hUbr5ΔUf4 (Fig. S2 K). Metaphase kinetochores in Ubr5-deleted cells exhibited increased Bub3, BubR1, and Mad1, whereas Bub3 appeared the same on prometaphase kinetochores compared with controls (Figs. 4 B and S3, B and C). Codepletion of Ubr5 with Bub3 reduced the metaphase kinetochore BubR1 and Mad1 (Fig. S3, B and C). Thus, Ubr5 facilitates efficient SAC silencing in metaphase.
Figure 4.
Ubr5-mediated Bub3 turnover contributes to RanGTP-induced SAC inactivation. (A) Ubr5 depletion caused delayed anaphase onset. Approximately 20 mitotic HeLa cells expressing histone H2B-GFP were quantified per experiment. (B) Ubr5 depletion increased metaphase kinetochore Bub3. Quantifications were based on 50 kinetochores from ∼5–10 cells per experiment. (C) Scheme of manipulating tsBN2 cells. Cells were collected for Western blotting (red arrows, mitotic shake-off) or immunostaining (black arrows, no mitotic shake-off). (D) Ubr5 mediated the turnover of BuGZ and Bub3 induced by RanGTP in metaphase. GAPDH, loading control. (E–G) Ubr5 depletion increased metaphase kinetochore Bub3 (E and F) and promoted metaphase block (G) independent of RCC1 degradation. tsBN2 cells were stained by DAPI, CREST sera, and Bub3 antibody (E). 50 kinetochores in ∼5–10 cells (F) or ∼100 mitotic cells (G) were analyzed per experiment. Bar, 5 µm. Error bars indicate SEM. Student’s t test: ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 from three experiments.
We then treated synchronized tsBN2 cells by Ubr5 RNAi (see Fig. 4 C). Mitotic cells collected by mitotic shake-off (Fig. 4 C, red arrows) were used for Western blotting. Total cells (Fig. 4 C, black arrows) were processed for immunostaining. As expected (Fig. 2), RCC1 mediated BuGZ and Bub3 turnover in control mitotic cells (Fig. 4 D, lane 5 vs. 7). The levels of BuGZ and Bub3 markedly increased in metaphase cells upon Ubr5 depletion (Fig. 4 D, lane 3 vs. 4). Importantly, in the Ubr5-depleted mitotic cells, BuGZ and Bub3 were no longer sensitive to RCC1 (Fig. 4 D, lanes 4 and 6 vs. 8). Ubr5 depletion also increased metaphase kinetochore Bub3 and inhibited anaphase entry (Fig. 4, E–G). Thus, Ubr5 aids RanGTP-induced BuGZ and Bub3 turnover and anaphase entry.
RanGTP promotes Ubr5-mediated turnover of BuGZ and Bub3 by displacing importin-β
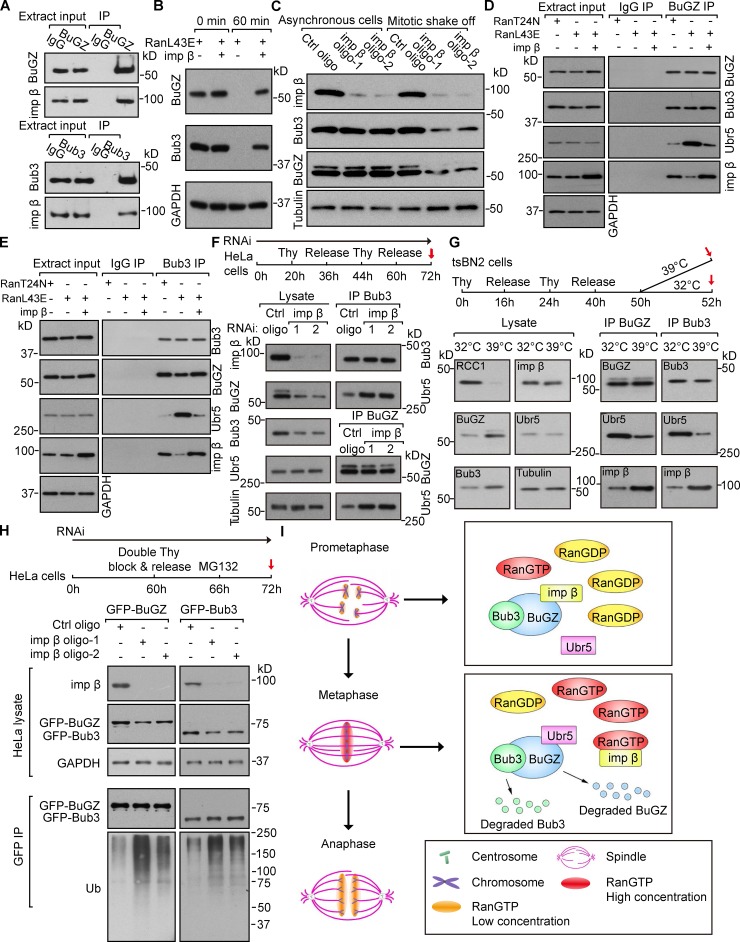
BuGZ and Bub3 pulled down importin-β in CSF Xenopus egg extracts and HeLa cells (Figs. 5 A and S3, D and E). Purified importin-β alleviated BuGZ and Bub3 turnover induced by RanL43E in the SAC-activated egg extracts (Fig. 5 B). Depleting importin-β by RNAi reduced BuGZ and Bub3 in mitotic cells but not in asynchronous cells (Fig. 5 C). BuGZ and Bub3 failed to rise and fall in mitosis in HeLa cells depleted of importin-β (Fig. S3 F). Because the binding of Ubr5 to BuGZ and Bub3 is stronger in metaphase than in prometaphase (see Fig. 3 E), the highest RanGTP levels surrounding metaphase chromosomes could displace importin-β from BuGZ and Bub3, thereby enhancing Ubr5 binding to BuGZ and Bub3. Indeed RanL43E, but not RanT24N, enhanced the binding of Ubr5 to BuGZ and Bub3, while importin-β inhibited such interaction (Fig. 5, D and E). Next, we synchronized the importin-β siRNA-treated HeLa cells and collected mitotic cells by mitotic shake-off (see Fig. 5 F). Pulldown assays showed that importin-β depletion led to increased binding of Ubr5 to BuGZ and Bub3 in mitosis (Fig. 5 F).
Figure 5.
RanGTP and importin-β antagonistically regulate the binding of Ubr5 to BuGZ and Bub3. (A) Importin-β (imp β) associated with BuGZ or Bub3 in SAC-activated Xenopus egg extract. (B) Purified imp β inhibited RanL43E-induced BuGZ and Bub3 turnover in egg extract. GAPDH, loading control. (C) Imp β depletion enhanced BuGZ and Bub3 turnover in mitotic HeLa cells. Tubulin, loading control. (D and E) Imp β inhibited RanL43E-stimulated association of Ubr5 with BuGZ and Bub3 in egg extract. (F) Imp β depletion enhanced the binding of Ubr5 to Bub3 or BuGZ in mitosis. Cells treated as shown were collected by mitotic shake-off (red arrow). Tubulin, loading control. (G) RCC1 depletion affected associations of Ubr5 and imp β with BuGZ or Bub3 in mitotic tsBN2 cells. Tubulin, loading control. (H) Imp β depletion increased ubiquitination of GFP-BuGZ and GFP-Bub3 in mitotic HeLa cells. GAPDH, loading control. (I) A model for RanGTP-regulated anaphase entry mediated by Ubr5-induced BuGZ and Bub3 turnover in metaphase.
We then synchronized tsBN2 cells and collected mitotic cells (Fig. 5 G, red arrows). Pulldown assays showed that degradation of RCC1 (at 39°C) in mitosis diminished Ubr5-BuGZ/Bub3 binding, but enhanced the interaction between importin-β and BuGZ/Bub3 (Fig. 5 G). Finally, we treated HeLa cells expressing GFP-Bub3 or GFP-BuGZ with control or importin-β siRNA followed by synchronization in metaphase. Cells collected by mitotic shake-off (see Fig. 5 H, red arrow) were used for GFP antibody pulldown. Importin-β depletion led to increased polyubiquitination of BuGZ and Bub3 (Fig. 5 H). Thus, RanGTP promotes the Ubr5-mediated polyubiquitination of BuGZ and Bub3 by displacing importin-β in metaphase.
Our findings suggest that before metaphase, importin-β binding stabilizes BuGZ and Bub3. In metaphase, the high RanGTP levels around metaphase chromosomes increase the displacement of importin-β from BuGZ and Bub3, allowing the binding of Ubr5 to BuGZ and Bub3 for polyubiquitination (Fig. 5 I). Because BuGZ stabilizes Bub3, Ubr5 may first target BuGZ for degradation followed by Bub3 during metaphase. The reduction of Bub3 then leads to reduced MCC activity, which contributes to SAC silencing and metaphase to anaphase transition (Fig. 5 I). The N terminus of BuGZ has a predicted NLS. Thus, importin-β may only bind to BuGZ to block the binding of Ubr5 to BuGZ. Upon the Ubr5-mediated BuGZ degradation, the Ubr5-binding site on Bub3 would become exposed, leading to Bub3 turnover. However, sequence analyses suggest that Bub3 may also contain an NLS. Further mapping the importin-β binding sites on BuGZ and Bub3 will reveal how importin-β hinders the binding of Ubr5 to BuGZ and Bub3.
Unlike the SAC silencing mechanisms that rely on sensing MT–kinetochore interactions (Musacchio and Salmon, 2007; Fang and Zhang, 2011; Kim and Yu, 2011; Musacchio, 2011), RanGTP-mediated SAC silencing could offer the cell a means to detect the overall state of chromosome alignment. This global sensing could coordinate SAC silencing on individual kinetochores to signal the final transition into anaphase. Interestingly, even after apparent satisfaction of bipolar attachment of all kinetochores, mammalian cells linger in metaphase for minutes before anaphase onset. RanGTP-stimulated polyubiquitination and degradation of BuGZ and Bub3 may contribute to the extra time the cell spent in metaphase, which would provide additional time to ensure complete chromosome congression. RanGTP-induced Bub3 reduction at metaphase may also inhibit MCC to usher in orderly chromosome segregation and cytokinesis.
Materials and methods
Construction of expression vectors
Plasmids expressing His-mBub3 (bacteria), Flag-mBuGZ (and the RNAi-insensitive version), Flag-mBub3, GFP-mBuGZ, and GFP-mBub3 (mammalian cells) have been described previously (Jiang et al., 2014). Refer to Table S2 for detailed information. GST-hUbr5 fragments (Uf), Uf1, Uf2, Uf3, Uf4, GFP-hUbr5, and GFP-hUbr5ΔUf4 (see Fig. S2 I) were amplified by PCR amplification using the MBP-hUbr5 plasmid (Maddika and Chen, 2009) as a template (see Table S2 for all primers used and detailed cloning strategies).
Cell culture and manipulations
The tsBN2 cell line (from T. Nishimoto, Japan) containing a temperature-sensitive RCC1 protein was derived from the BHK cell line. HeLa and tsBN2 cells were cultured in DMEM medium (Thermo Fisher Scientific) supplemented with 15% (vol/vol) FBS (Biochrom). HeLa cells were grown at 37°C in a 5% CO2 humidified chamber, whereas tsBN2 cells were maintained at 32°C in a 5% CO2 humidified chamber. Cells were transfected using FuGENE 6 Transfection Reagent (Roche) to express BuGZ, Bub3, or Ubr5 (from J. Chen, Department of Therapeutic Radiology, Yale University School of Medicine, New Haven, CT). For RNAi, cells were transfected using Lipofectamine RNAiMAX (Invitrogen) and 0.05 µM siRNA oligo for 60 h (BuGZ RNAi) or 72 h (Ubr5, importin-β or Ubr5 plus Bub3 RNAi).
The sequences of siRNA oligos used
Oligos used in HeLa cells: BuGZ oligo1, 5′-GCCUGCUACACUUACAACAACUAGU-3′; BuGZ oligo2, 5′-CGAUGGGAAUGAGACCUCCUGUAAU-3′; Bub3 oligo, 5′-CCGGUUCUAACGAGUUCAAGCUGAA-3′; Ubr5 oligo1, 5′-CAACUUAGAUCUCCUGAAA-3′; Ubr5 oligo2, 5′-GCAGUGUUCCUGCCUUCUU-3′; Importin-β oligo1, 5′-UCGGUUAUAUUUGCCAAGAUA-3′; Importin-β oligo2, 5′-CAAGAACUCUUUGACAUCUAA-3′; control oligo (for BuGZ, Bub3, or Ubr5 RNAi), Life Technologies, catalogue no. 12935-300; control oligo (for importin-β RNAi), QIAGEN, catalogue no. SI03650318. Oligos used in tsBN2 cells: BuGZ oligo2, 5′-CGAUGGGAAUGAGACCUCCUGUAAU-3′; Ubr5 oligo3, 5′-CCGGAACAUUCUGCAUGCUUGCGUA-3′; control oligo (for BuGZ or Ubr5 RNAi), Life Technologies, catalogue no. 12935-300.
Cell synchronization and manipulation of SAC
To obtain mitotic cells, cells were treated with double thymidine (Sigma-Aldrich) block and release, followed by nocodazole (Sigma-Aldrich) or MG132 (Sigma-Aldrich) treatment whenever indicated. For DTB, 30% confluent HeLa or tsBN2 cells were treated with 2 mM thymidine for 16 h and then released and incubated for 8 h. After another 16-h incubation with 2 mM thymidine, the cells were washed with PBS and released into fresh medium, followed by collection at the indicated time points or by additional treatments. In our hands, >70% cells entered into mitosis ∼11 h after the DTB and release. To further synchronize cells into prometaphase or metaphase, 8 h after the DTB and release, the cells were treated with nocodazole (final concentration 330 nM) or MG132 (final concentration 10 µM) for 6 h and collected at the indicated time after nocodazole/MG132 washout for Western blotting or immunostaining. To abrogate the SAC signal, 1 µM Mps1 inhibitor NMS-P715 was added to cells for 20 h followed by fixation and analyses.
Immunofluorescence microscopy and quantification
Cells were fixed with 4% paraformaldehyde in PBS at 37°C for 7–10 min followed by permeabilization with 0.5% Triton X-100 in PBS for 10 min. The cells were then blocked with 4% BSA in PBS for >1 h followed by incubation with primary and secondary antibodies. An Eclipse TE2000-U microscope (Nikon) driven by MetaMorph software (Molecular Devices) or a TCS SP5 microscope (Leica) driven by LAS AF software was used for imaging. For confocal microscopy, a 63× 1.4 NA oil objective lens (Leica) was used and the cells were imaged by scanning optical sections at ∼0.5-µm intervals at room temperature. For live imaging, cells were incubated in DMEM medium supplemented with 15% (vol/vol) FBS and imaged with a 20× 0.45 NA lens under a microscope (Eclipse TE2000-U) in a stage incubator (Pathology Devices, Inc.) at 37°C with 5% CO2 and 70% humidity. Mitotic index was determined by dividing the number of cells in mitosis as judged by DAPI staining with the total number of cells counted.
The percentage of cells with misaligned chromosomes was defined as previously described (Meraldi and Sorger, 2005). Cells that formed the metaphase plate were included in the analyses. Mitotic chromosomes found outside the rectangular area encompassing the central 30% of the spindle were considered as misaligned chromosomes. The intensity of kinetochore proteins was measured as described previously (Jiang et al., 2014). In brief, computer-generated 9 × 9 and 13 × 13 pixel regions made with Photoshop software (Adobe) were centered over each kinetochore and the total integrated fluorescence counts were obtained for the 9 × 9 and 13 × 13 pixel regions. Because the 9 × 9 pixel region was typically large enough to contain 90% of kinetochore fluorescence, the intensity difference between 9 × 9 and 13 × 13 pixel regions was used as the background intensity. For details of antibodies, dilutions, and fluorochromes used, see Table S1.
Assays in Xenopus egg extracts
Only CSF egg extracts (Murray, 1991) that were tested to form sperm-induced spindles were used for experiments.
To assay whether RanL43E or RanT24N functioned in SAC inactivation in CSF egg extracts, we followed the protocol described previously (Arnaoutov and Dasso, 2003) with some modifications. In brief, 25 µl of the egg extract was added with energy mix (final concentrations: 10 mM creatine phosphate, 0.5 mM Na-ATP, 0.5 mM MgCl2, and 50 µM EGTA, pH 7.7), nocodazole (10 µg/ml final), and sperm nuclei (10,000/µl final) plus purified RanL43E, RanT24N, or BSA (∼1 mg/ml final). After incubation at 4°C for 30 min, 0.6 mM Ca2+ were added into the egg extract and then incubated at room temperature at the indicated time. 5-µl egg extracts were collected for either staining with Hoechst to examine the nuclear morphology or Western blotting to probe for Cyclin-B2.
To assay for the effect of RanL43E and RanT24N on the stability of Bub3 and BuGZ, 25 µl of the egg extract was supplemented with energy mix, nocodazole, sperm nuclei, and purified RanL43E or RanT24N as above. After the incubation at 4°C for 30 min, the egg extracts were then gently rotated at room temperature for the indicated time, followed by Western blotting using antibodies to BuGZ, Bub3, BubR1, Bub1, and GAPDH.
To assay whether importin-β could rescue the RanL43E-induced degradation of BuGZ and Bub3, the egg extract with energy mix, nocodazole, sperm nuclei, and purified RanL43E as above were further supplemented with importin-β (0.5 mg/ml final) or XB buffer (10 mM Hepes, 50 mM sucrose, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 5 mM EGTA, pH 7.7). After the incubation at 4°C for 30 min, the egg extracts were then gently rotated at room temperature for 60 min followed by Western blots probing for the indicated proteins.
To determine whether Ran and importin-β regulate the interaction between Ubr5 and BuGZ or Bub3, 100 µl of the CSF egg extracts were supplemented with energy mix, nocodazole, sperm, and different combinations of RanL43E, RanT24N, and importin-β as indicated in the figure (the final concentration of each supplement was the same as described above). After incubation at 4°C for 30 min, BuGZ or Bub3 were immunoprecipitated at 4°C using 30 µg of BuGZ antibody, Bub3 antibody, or control IgG for 30 min, followed by Western blots probing for BuGZ, Bub3, importin-β, Ubr5, and GAPDH.
To study the interaction between importin-β and Bub3/BuGZ, 100 µl of the CSF egg extracts supplemented with nocodazole and sperm (the final concentration for each supplement was the same as described above) were incubated at 4°C for 30 min, followed by immunoprecipitations by the BuGZ or Bub3 antibody as above. For details of antibodies and dilutions used see Table S1.
Protein expression, purification, and interactions
GST and GST-Uf4 (or other GST-Uf proteins) were expressed and purified from BL21 (DE3) according to the manufacturer’s instructions. For each protein expression, we used a single Escherichia coli colony to inoculate 3 ml of Lysogeny broth (LB) culture. After shaking at 37°C for 12 h, the culture was added into 2 liters of prewarmed LB medium for further incubation at 37°C with shaking. Once the OD600 reached 0.7, IPTG (final concentration 1 mM) was added to the medium to induce protein expression by incubating with shaking at 16°C for 18 h followed by collecting the cells by centrifugation at 5,000 rpm for 20 min at 4°C. The bacterial pellet was resuspended in 50 ml of ice-cold PBST (PBS plus 1% Triton X-100) containing 1 mM PMSF and a 1:100 dilution of the protease cocktail (AEBSF, Bestatin, E-64, Pepstain A, and Phosphoramidon; Sigma-Aldrich), and pipetted on ice to resuspend the bacteria. The mixture was then sonicated on ice until the lysate became clear (∼15 min), followed by centrifugation at 12,000 rpm for 30 min at 4°C using a rotor (JA20; Beckman Coulter). 4 ml of the 50% slurry of Glutathione Sepharose 4B was then added to the supernatant and incubated with rotation at 4°C for 6 h. The glutathione resin was then packed in a column and washed with 200 ml of PBS. 5 ml of the glutathione elution buffer (50 mM Tris-HCl and 10 mM reduced glutathione, pH 8.0) was added into the column and incubated at 4°C for 1 h to elute the protein. The eluted protein was exchanged into 2 ml of XB buffer with a PD10 column (GE Healthcare), and further concentrated to 1.2 mg/ml (GST-Uf4) or 4.2 mg/ml (GST) using an Amicon Ultra 30K device (Millipore) at 4°C. The concentrated proteins were divided into 15-µl aliquots, snap frozen in liquid nitrogen, and stored at −80°C for interaction studies.
The same protocol as above was used to induce the expression of His-GFP-mBuGZ and His-mBub3 in bacteria. The bacterial pellet was resuspended in 50 ml of the ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 5 mM imidazole, pH 7.8) containing 1 mM PMSF and 1:100 dilution of the protease cocktail (see above) by pipetting on ice. After the sonication and centrifuging under the same conditions as above, the supernatant was supplemented with 4 ml of the 50% slurry of Ni-NTA beads (QIAGEN) and rotated at 4°C for 4 h, followed by packing the beads into a column. The column was washed with 100 ml of wash buffer (50 mM NaH2PO4 and 300 mM NaCl, pH 7.8) containing 10 mM imidazole followed by 100 ml of wash buffer containing 20 mM imidazole and then eluted with 5 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 300 mM imidazole, pH 7.8) into 0.5-ml fractions. The peak fractions were exchanged into 2 ml of XB buffer using a PD10 column, and further concentrated to 0.5 mg/ml (His-mBub3) or 5.2 mg/ml (His-GFP-mBuGZ) with the Amicon Ultra 30K device at 4°C, followed by dividing into 15-µl aliquots. They were then snap frozen in liquid nitrogen and stored at −80°C.
To study the direct interaction between the HECT domain of Ubr5 (GST-Uf4) with BuGZ or Bub3, 4 µg of purified GST or GST-Uf4 were incubated with 30 µl of the 50% slurry of Glutathione Sepharose 4B and rotated for 2 h at 4°C, followed by washing the beads with 1 ml of ice-cold lysis buffer (as above) and 1 ml of wash buffer (as above) three times each. The beads were incubated with 4 µg of purified His-GFP-mBuGZ or His-mBub3, and rotated at 4°C for another 4 h, followed by washing the beads with 1 ml of lysis buffer and 1 ml of wash buffer for another four times each. 10% of GST, GST-Uf4, His-GFP-mBuGZ, or His-mBub3 (∼0.4 µg for each protein) was loaded as an input sample for Coomassie blue staining, whereas 25% or 5% of each immunoprecipitates were loaded for Coomassie staining or Western blotting, respectively. Antibodies used for Western blotting can be found in Table S1.
Ubiquitination assay
To assay for the effect of Ubr5 on the ubiquitination of BuGZ and Bub3, 2 × 106 (for collecting total cells) or 1 × 107 (for collecting mitotic cells by mitotic shake-off) HeLa cells stably expressing GFP-Bub3 or GFP-BuGZ were plated 1 d before transfection. The cells were transfected with the control or Ubr5 siRNA using Lipofectamine RNAiMAX (Invitrogen). 66 h after transfection, the cells were treated with 10 µM MG132 for 6 h and collected with or without mitotic shake-off, followed by incubation with 650 µl of the low-salt lysis buffer (as above). After clarification at 12,000 rpm for 30 min at 4°C, 60 µl of anti-GFP (Allele Biotech) bead slurry was added into the supernatant and rotated for 2.5 h at 4°C, followed by washing the beads with 1 ml of low-salt lysis buffer and 1 ml of low-salt wash buffer four times each. The bead-bound proteins were eluted with 25 µl of glycine buffer (0.1 M glycine, pH 2.5). The immunoprecipitates were used for Western blotting analyses probing for ubiquitin and other proteins.
To study the effect of importin-β on the ubiquitination of BuGZ and Bub3, 2 × 106 HeLa cells stably expressing GFP-Bub3 or GFP-BuGZ were plated 1 d before transfection. The cells were transfected with the control or importin-β siRNA using Lipofectamine RNAiMAX (Invitrogen) each time when they were changed into a new medium. 20 h after the first round of transfection, the cells were treated with 2 mM thymidine for 16 h. 8 h after the release from the thymidine block, the cells were blocked with 2 mM thymidine for another 16 h. 6 h after releasing from the second thymidine release, the cells were incubated in the fresh medium containing 10 µM MG132 for another 6 h followed by mitotic shake-off. The cells were subjected to immunoprecipitation as described using anti-GFP beads, followed by Western blotting.
To study the effect of BuGZ on Bub3 ubiquitination, 1 × 107 HeLa cells were plated 1 d before transfection. The control or BuGZ siRNA and the plasmid expressing Flag-Bub3 were cotransfected using Lipofectamine RNAiMAX (Invitrogen). 54 h after transfection, 10 µM MG132 was added into the medium for 6 h, followed by collecting the mitotic cells by mitotic shake-off. The cells were subjected to immunoprecipitation using 30 µl of anti-Flag bead slurry, followed by Western blotting.
Time-lapse microscopy
Mitotic HeLa cells stably coexpressing RFP-Histone H2B and GFP-Bub3 or GFP were imaged live using a 63× objective lens on an UltraVIEW VoX Imaging Station (PerkinElmer) at 6-min intervals for 2 h to capture full mitosis. The laser intensity and the exposure time were the same for all cells. Images were processed using ImageJ and converted to QuickTime movies (Apple) for further analyses.
To quantify the total or mean GFP or GFP-Bub3, we selected four images representing four different mitotic phases. (1) Prometaphase: 18 min before the first sign of full chromosome congression at the metaphase plate. (2) Early metaphase: the first sign of full chromosome congression. (3) Late metaphase: the image right before chromosome separation. (4) Anaphase: the first image of chromosome separation. For cells treated with nocodazole, the mitotic cells imaged at 0, 24, 48, and 72 min were selected for quantification. For quantification, cell outline was drawn based on the ImageJ-processed images. The total and mean intensity (total intensity/the cell area) were obtained using the measurement tool in ImageJ. Background intensity measured from the area next to each cell was subtracted. At least 15 mitotic cells were quantified for each sample and condition.
To analyze the effect of Ubr5 depletion on mitotic progression by live imaging, HeLa cells expressing H2B-GFP (Vong et al., 2005) were transfected with Ubr5 siRNA for 60 h and imaged live using a 10× objective lens for another 20–40 h in a stage incubator (Pathology Devices, Inc.) at 37°C with 5% CO2 and 70% humidity. Images were captured at 5-min intervals with an ECLIPSE TE2000-U microscope controlled with MetaMorph software (Molecular Devices). NEBD to early prometaphase: from nuclear envelope breakdown to the frame before the first sign of full chromosome congregation. Late prometaphase to metaphase: from the first sign of full chromosome congregation to the frame right before chromosome separation. Anaphase to telophase: from the first sign of chromosome segregation to the first sign of nuclear reformation.
Online supplemental material
Fig. S1 shows BuGZ and Bub3 turnover during cell division. Fig. S2 shows SAC regulation by RanGTP and analyses of protein–protein interactions. Fig. S3 shows the analyses of the role of Ubr5 in SAC inactivation. Table S1 provides a list of antibodies, their sources, and dilutions used in either Western blotting or immunostaining. Table S2 provides a list of primers used to construct the indicated expression vectors. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503122/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Nishimoto for tsBN2 cells, Dr. Junjie Chen for MBP-hUbr5 plasmid, Ona Martin for technical support, the shared confocal microscope at Carnegie Institution’s Embryology Department, and members of the Zheng laboratory for critical reading.
This work was supported by the Chinese Academy of Sciences (XDA01010107 to X. Zhu), the Ministry of Science and Technology of China (2014CB964803 to X. Zhu), the National Science Foundation of China (31420103916 to X. Zhu and Y. Zheng), and the National Institutes of Health (GM056312, GM110151, and GM06023 to Y. Zheng).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BuGZ
- Bub3 interacting and GLEBS motif containing ZNF207
- CSF
- cytostatic factor
- DTB
- double thymidine block
- HECT
- homologous to the E6-AP C terminus
- MCC
- mitotic checkpoint complex
- MT
- microtubule
- SAC
- spindle assembly checkpoint
References
- Arnaoutov A., and Dasso M.. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell. 5:99–111. 10.1016/S1534-5807(03)00194-1 [DOI] [PubMed] [Google Scholar]
- Barisic M., Sohm B., Mikolcevic P., Wandke C., Rauch V., Ringer T., Hess M., Bonn G., and Geley S.. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 21:1968–1981. 10.1091/mbc.E09-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides M., Chow-Tsang L.-F., Zhang J., and Zhong H.. 2013. The novel interaction between microspherule protein Msp58 and ubiquitin E3 ligase EDD regulates cell cycle progression. Biochim. Biophys. Acta. 1833:21–32. 10.1016/j.bbamcr.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., and Mattaj I.W.. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181. 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Gruss O.J., Mattaj I.W., and Karsenti E.. 2001. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3:228–234. 10.1038/35060009 [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H.A., Gerlich D.W., Nigg E.A., and Santamaria A.. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874. 10.1083/jcb.200812167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum J.R., Wren J.D., Daniel J.J., Sivakumar S., McAvoy J.N., Potapova T.A., and Gorbsky G.J.. 2009. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr. Biol. 19:1467–1472. 10.1016/j.cub.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., and Zhang P.. 2011. Aneuploidy and tumorigenesis. Semin. Cell Dev. Biol. 22:595–601. 10.1016/j.semcdb.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., and Nigg E.A.. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., and Desai A.. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971. 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B., and Zheng Y.. 2006. Mitotic spindle morphogenesis: Ran on the microtubule cytoskeleton and beyond. Biochem. Soc. Trans. 34:716–721. 10.1042/BST0340716 [DOI] [PubMed] [Google Scholar]
- Goodman B., Channels W., Qiu M., Iglesias P., Yang G., and Zheng Y.. 2010. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. J. Biol. Chem. 285:35238–35244. 10.1074/jbc.M110.140749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., and Vale R.D.. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015. 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 104:83–93. 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Habu T., Kim S.H., Weinstein J., and Matsumoto T.. 2002. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21:6419–6428. 10.1093/emboj/cdf659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson M.J., Munoz M.A., Saunders D.N., Clancy J.L., Russell A.J., Williams B., Pappin D., Khanna K.K., Jackson S.P., Sutherland R.L., and Watts C.K.W.. 2006. EDD mediates DNA damage-induced activation of CHK2. J. Biol. Chem. 281:39990–40000. 10.1074/jbc.M602818200 [DOI] [PubMed] [Google Scholar]
- Honda Y., Tojo M., Matsuzaki K., Anan T., Matsumoto M., Ando M., Saya H., and Nakao M.. 2002. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem. 277:3599–3605. 10.1074/jbc.M104347200 [DOI] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., and Salmon E.D.. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964. 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., Xiong Y., Guan K.-L., and Zhao S.. 2011. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 43:33–44. 10.1016/j.molcel.2011.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., He X., Wang S., Jia J., Wan Y., Wang Y., Zeng R., Yates J. III, Zhu X., and Zheng Y.. 2014. A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev. Cell. 28:268–281. 10.1016/j.devcel.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R.T., and Dasso M.. 1999. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9:481–484. 10.1016/S0960-9822(99)80213-9 [DOI] [PubMed] [Google Scholar]
- Kaláb P., Weis K., and Heald R.. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 295:2452–2456. 10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- Kaláb P., Pralle A., Isacoff E.Y., Heald R., and Weis K.. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 440:697–701. 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Kim S., and Yu H.. 2011. Mutual regulation between the spindle checkpoint and APC/C. Semin. Cell Dev. Biol. 22:551–558. 10.1016/j.semcdb.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Sun H., Ball H.L., Wassmann K., Luo X., and Yu H.. 2010a Phosphorylation of the spindle checkpoint protein Mad2 regulates its conformational transition. Proc. Natl. Acad. Sci. USA. 107:19772–19777. 10.1073/pnas.1009000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Holland A.J., Lan W., and Cleveland D.W.. 2010b Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 142:444–455. 10.1016/j.cell.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Sharov A.A., McDole K., Cheng M., Hao H., Fan C.-M., Gaiano N., Ko M.S.H., and Zheng Y.. 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 334:1706–1710. 10.1126/science.1211222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.Y., and Zheng Y.. 2004. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18:512–527. 10.1101/gad.1177304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.Y., Wirtz D., and Zheng Y.. 2003. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160:635–644. 10.1083/jcb.200211004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.Y., Ng W.P., Wong C.H., Iglesias P.A., and Zheng Y.. 2007. Coordination of chromosome alignment and mitotic progression by the chromosome-based Ran signal. Cell Cycle. 6:1886–1895. 10.4161/cc.6.15.4487 [DOI] [PubMed] [Google Scholar]
- Ling S., and Lin W.-C.. 2011. EDD inhibits ATM-mediated phosphorylation of p53. J. Biol. Chem. 286:14972–14982. 10.1074/jbc.M110.182527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., and Lampson M.A.. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820. 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Resende T., Torres C., and Bousbaa H.. 2008. The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol. Biol. Cell. 19:1798–1813. 10.1091/mbc.E07-07-0633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tsai M.-Y., Wang S., Lu B., Chen R., Iii J.R.Y., Zhu X., and Zheng Y.. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256. 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddika S., and Chen J.. 2009. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11:409–419. 10.1038/ncb1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Filipp F.V., Rancati G., Massimiliano L., Nezi L., Stier G., Hagan R.S., Confalonieri S., Piatti S., Sattler M., and Musacchio A.. 2006. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 25:1273–1284. 10.1038/sj.emboj.7601033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., and Sorger P.K.. 2005. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 24:1621–1633. 10.1038/sj.emboj.7600641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz M.A., Saunders D.N., Henderson M.J., Clancy J.L., Russell A.J., Lehrbach G., Musgrove E.A., Watts C.K.W., and Sutherland R.L.. 2007. The E3 ubiquitin ligase EDD regulates S-phase and G(2)/M DNA damage checkpoints. Cell Cycle. 6:3070–3077. 10.4161/cc.6.24.5021 [DOI] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Musacchio A. 2011. Spindle assembly checkpoint: the third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3595–3604. 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., and Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Maresca T.J., Salmon W.C., Waterman-Storer C.M., Heald R., and Weis K.. 2001. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106. 10.1016/S0092-8674(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Nishitani H., Ohtsubo M., Yamashita K., Iida H., Pines J., Yasudo H., Shibata Y., Hunter T., and Nishimoto T.. 1991. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J. 10:1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.B., and Khodjakov A.L.. 2007. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 120:1717–1722. 10.1242/jcs.03442 [DOI] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., and Nishimoto T.. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 284:1356–1358. 10.1126/science.284.5418.1356 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Nelson C.R., and Biggins S.. 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19:1182–1187. 10.1016/j.cub.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits V.A.J. 2012. EDD induces cell cycle arrest by increasing p53 levels. Cell Cycle. 11:715–720. 10.4161/cc.11.4.19154 [DOI] [PubMed] [Google Scholar]
- Toledo C.M., Herman J.A., Olsen J.B., Ding Y., Corrin P., Girard E.J., Olson J.M., Emili A., DeLuca J.G., and Paddison P.J.. 2014. BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Dev. Cell. 28:282–294. 10.1016/j.devcel.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.-Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., and Zheng Y.. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 311:1887–1893. 10.1126/science.1122771 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., and Hardwick K.G.. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181. 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong Q.P., Cao K., Li H.Y., Iglesias P.A., and Zheng Y.. 2005. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 310:1499–1504. 10.1126/science.1120160 [DOI] [PubMed] [Google Scholar]
- Watts C.K., and Saunders D.N.. 2011. Effects of EDD on p53 function are context-specific. J. Biol. Chem. 286:le13 10.1074/jbc.L110.182527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee B., Johnston C.A., Prehoda K.E., and Doe C.Q.. 2011. Canoe binds RanGTP to promote Pins(TPR)/Mud-mediated spindle orientation. J. Cell Biol. 195:369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M.S., Adam S.A., Merdes A., and Zheng Y.. 2001. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science. 291:653–656. 10.1126/science.1057661 [DOI] [PubMed] [Google Scholar]
- Wilde A., and Zheng Y.. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 284:1359–1362. 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- Wilde A., Lizarraga S.B., Zhang L., Wiese C., Gliksman N.R., Walczak C.E., and Zheng Y.. 2001. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3:221–227. 10.1038/35060000 [DOI] [PubMed] [Google Scholar]
- Xia G., Luo X., Habu T., Rizo J., Matsumoto T., and Yu H.. 2004. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23:3133–3143. 10.1038/sj.emboj.7600322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li B., Tomchick D.R., Machius M., Rizo J., Yu H., and Luo X.. 2007. p31comet blocks Mad2 activation through structural mimicry. Cell. 131:744–755. 10.1016/j.cell.2007.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Cronshaw J., Kanu N., Snijders A.P., and Behrens A.. 2014. UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proc. Natl. Acad. Sci. USA. 111:12091–12096. 10.1073/pnas.1400230111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. 2010. A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 11:529–535. 10.1038/nrm2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.