In Caenorhabditis elegans, the RAL-1 GTPase regulates multivesicular body formation and fusion with the plasma membrane, thereby promoting exosome secretion; in mammals, RalA and RalB are required for the secretion of exosome-like vesicles.
Abstract
Exosomes are secreted vesicles arising from the fusion of multivesicular bodies (MVBs) with the plasma membrane. Despite their importance in various processes, the molecular mechanisms controlling their formation and release remain unclear. Using nematodes and mammary tumor cells, we show that Ral GTPases are involved in exosome biogenesis. In Caenorhabditis elegans, RAL-1 localizes at the surface of secretory MVBs. A quantitative electron microscopy analysis of RAL-1–deficient animals revealed that RAL-1 is involved in both MVB formation and their fusion with the plasma membrane. These functions do not involve the exocyst complex, a common Ral guanosine triphosphatase (GTPase) effector. Furthermore, we show that the target membrane SNARE protein SYX-5 colocalizes with a constitutively active form of RAL-1 at the plasma membrane, and MVBs accumulate under the plasma membrane when SYX-5 is absent. In mammals, RalA and RalB are both required for the secretion of exosome-like vesicles in cultured cells. Therefore, Ral GTPases represent new regulators of MVB formation and exosome release.
Introduction
Most cells secrete extracellular vesicles (EVs), which are released outside of the organism or within internal fluids. When EVs are taken up by a distant cell, their content (proteins, mRNAs, and microRNAs) can induce a cellular response (Raposo and Stoorvogel, 2013). Over the last decade, EVs have been implicated in a growing number of processes, such as signal transduction during development, extracellular matrix generation, host–pathogen communication, and tumor–stroma communication (Raposo and Stoorvogel, 2013).
Different types of EVs have been described, including exosomes (50–100 nm diameter), which originate from multivesicular bodies (MVBs), and microvesicles (50–1,000 nm diameter), which directly bud from the plasma membrane. MVBs are well-characterized endosomal precursors of the lysosomal degradation pathway, which can also fuse with the plasma membrane (Klumperman and Raposo, 2014). In this case, their intraluminal vesicle (ILV) content is released in the extracellular space and they are then called exosomes. Although the protein and RNA content of exosomes has been extensively characterized (Kalra et al., 2012; Kim et al., 2013), the molecular mechanisms leading to exosome biogenesis and secretion are only emerging (Kowal et al., 2014).
Two broad types of mechanisms have been involved in exosome biogenesis. The first comprises mechanisms related to the endosomal sorting complex required for transport (ESCRT) machinery (Tamai et al., 2010; Baietti et al., 2012; Gross et al., 2012; Abrami et al., 2013; Colombo et al., 2013; Hoshino et al., 2013), including the PLD2–syntenin–ALIX axis (Ghossoub et al., 2014), whereby four complexes control the inward budding of the MVB limiting membrane, leading to ILV formation. The second type consists of mechanisms unrelated to the ESCRT machinery, such as the ceramide pathway (Trajkovic et al., 2008).
Thus far, eight different RAB GTPases have been linked to exosome secretion (RAB-2B, RAB-5A, RAB-7, RAB-9A, RAB-11, RAB-27A, RAB-27B, and RAB-35). Among them, RAB-27 is the best characterized (Savina et al., 2005; Hsu et al., 2010; Ostrowski et al., 2010; Baietti et al., 2012), although the precise mechanism of action of these GTPases remains elusive (Kowal et al., 2014). In addition, the SNARE proteins VAMP-7 and YKT-6 are required for exosome release (Fader et al., 2009; Gross et al., 2012; Hong and Lev, 2014) and act at a step that remains to be defined. Finally, we found that the V0 complex of the H+-vacuolar ATPase promotes exosome secretion by epithelial cells in the nematode Caenorhabditis elegans independently of its role as a proton pump (Liégeois et al., 2006). Indeed, mutations in VHA-5, the largest subunit of the V0 complex, induce abnormally enlarged MVBs to accumulate, suggesting that this complex controls the final steps of MVB docking and fusion with the plasma membrane.
Here, we first identify C. elegans RAL-1 as a new regulator of MVB formation and exosome secretion. We show that RAL-1 acts independently of its common effector, the exocyst, but through the t-SNARE SYX-5 at the plasma membrane. Furthermore, we demonstrate the conservation of RAL GTPase function in mammals.
Results and discussion
RAL-1 and the exocyst are required for alae formation
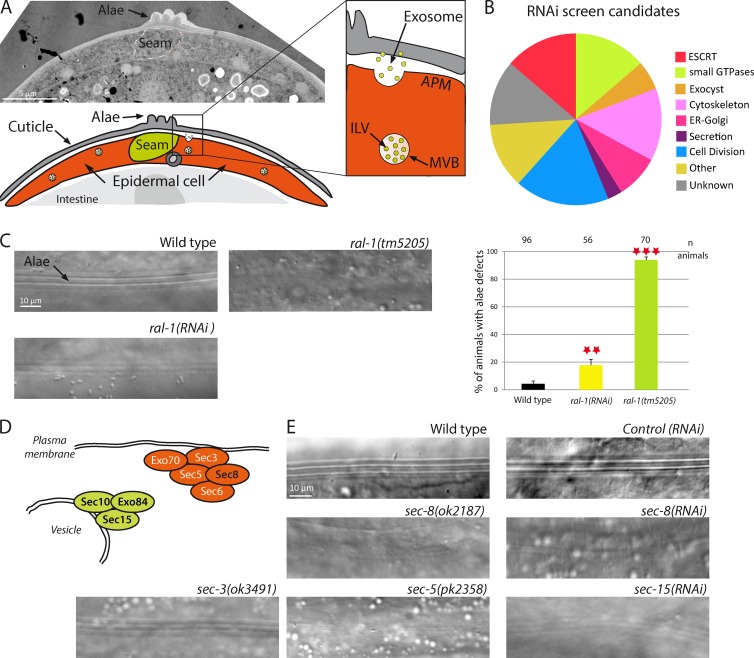
The C. elegans cuticle is a highly organized extra-cellular matrix mainly composed of cross-linked collagens, insoluble glycoproteins, and lipids, which is renewed at the end of each larval stage (Page and Johnstone, 2007). Two types of epithelial cells located below the cuticle, the Hyp and seam cells, secrete cuticular components. We previously showed that exosomes are secreted by epidermal Hyp cells and contribute to the formation of a specific cuticular structure, the alae (Fig. 1 A; Liégeois et al., 2006). The epithelial seam cells located under the alae also contribute to alae formation. To identify new genes required for exosome secretion, we conducted an RNAi-based screen for alae defects. We screened over a thousand genes predicted or previously linked to vesicular trafficking in C. elegans (Frand et al., 2005; Balklava et al., 2007; Kinchen et al., 2008), as well as all kinases and phosphatases for which a RNAi-inducing clone was available. We identified 73 genes affecting alae formation (Fig. 1 B and Table S1). Although some of them might indirectly affect alae formation (e.g., by impairing seam cell division), several others encode homologues of proteins found to affect exosome biogenesis in mammalian cultured cells (ESCRT components; RAB GTPases RAB-2, RAB-11, RAB-27, and RAB-35; Savina et al., 2005; Hsu et al., 2010; Ostrowski et al., 2010; Colombo et al., 2013).
Figure 1.
RAL-1 GTPase or exocyst deficiency induces alae defects. (A) C. elegans epidermal cells contain MVBs, which can fuse with the apical plasma membrane and liberate exosomes. These exosomes are integrated in the cuticle and contribute to the formation of the alae. (B) An RNAi-based screen identified 73 genes required for alae formation. (C) Disruption of ral-1 by RNAi, or by the null allele ral-1(tm5205), leads to alae defects. The number of animals is shown at the top of the graph. **, P < 0.05; ***, P < 0.01. (D) Schematic representation of the exocyst complex involved in plasma membrane attachment of secretory vesicles. Subunits found in the screen are in black. (E) Alae defects observed after disruption of several members of the exocyst complex in mutants or by RNAi.
Interestingly, we found that loss of the GTPase RAL-1, which plays key roles in secretion in various models (Kawato et al., 2008; Lopez et al., 2008), induces alae defects (Fig. 1 C). We confirmed the RNAi results using a previously characterized null ral-1 mutant, ral-1(tm5205) (Armenti et al., 2014), which displayed severe alae defects in 94% of the animals (Fig. 1 C, Fig. S1, and Table S2). To determine whether the GTPase activity of RAL-1 is involved in alae formation, we expressed constitutively active (CA; G23V) and dominant negative (DN, S28N) forms of RAL-1, specifically in epidermal cells. We found that both mutants led to alae defects when overexpressed in wild-type (WT) animals (Fig. S1 and Table S2). When expressed at a lower level (1 ng instead of 10 ng), RAL-1(CA), but not RAL-1(DN), partially rescued ral-1(tm5205) mutants at levels comparable to RAL-1(WT) (Fig. S1 and Table S2). The partial rescue is consistent with the observation that YFP::RAL-1 could not fully rescue the sterility of ral-1(tm5205) mutants (Armenti et al., 2014), which might reflect a requirement for a tight regulation of RAL-1 expression. These data suggest that RAL-1 functions in alae formation and that its GTPase activity is required.
In addition to RAL-1, we found four members of the exocyst complex in our screen (Fig. 1, B, D, and E; and Table S1). The exocyst has been shown to control the secretion of different types of vesicles downstream of RAL in many species, including C. elegans (Brymora et al., 2001; Moskalenko et al., 2002; Sugihara et al., 2002; Armenti et al., 2014). We confirmed the screen results, using previously characterized mutants for five of the eight exocyst subunits: sec-3, sec-5, sec-8, exoc-7, and exoc-8 (Jiu et al., 2012; Armenti et al., 2014). Indeed, all of them displayed alae defects, suggesting that they could affect exosome secretion (Fig. 1 E, Fig. S1, and Table S2).
RAL-1 directly controls MVB formation and exosome secretion
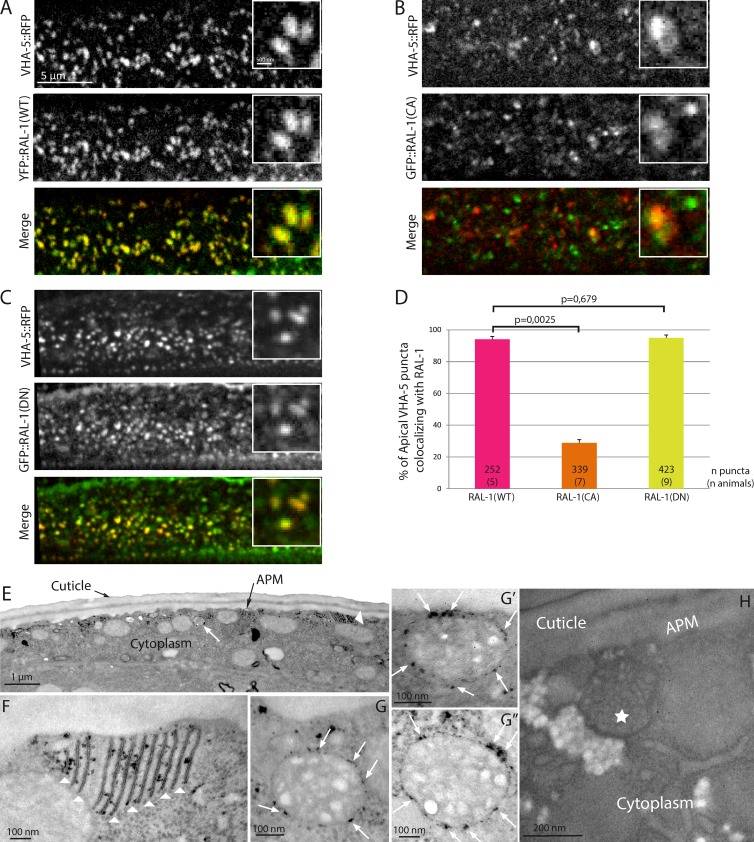
We first analyzed RAL-1 localization in C. elegans, showing that it has a punctate distribution in the epidermis (Fig. 2 A). Because no purely specific marker of the exosomal pathway has been identified thus far in any system, we used VHA-5, a V-ATPase component found on MVBs and at apical plasma membrane foldings called membrane stacks (Liégeois et al., 2006), as a nonexclusive marker of the exosomal pathway. We found that almost 100% of RAL-1 apical puncta colocalize with VHA-5 in the epidermis (Fig. 2, A and D), suggesting that it could localize at the surface of MVBs.
Figure 2.
RAL-1 localizes at the surface of MVBs. (A–D) WT (RAL-1(WT); A) and dominant negative (RAL-1(DN); C) versions of RAL-1, but not the constitutively active version (RAL-1(CA); B), colocalize fully with VHA-5 in the epidermis at the time of alae formation, as shown in the quantification (D). In D, the numbers inside the bars indicate the number of puncta (number of animals). (E–H) APEX::RAL-1(DN) shows DAB staining both at apical membrane stacks (E and F, arrowheads) and at the external surface of MVBs (E and G–G’’, arrows). Animals expressing no APEX tag treated with DAB show no staining (H). The star indicates a MVB. APM, apical plasma membrane.
To assess whether the GTPase activity of RAL-1 influences its localization, we analyzed the localization of RAL-1 mutants within the epidermis. We observed that the GTP-bound form RAL-1(CA) did not colocalize with VHA-5 as well as the WT form RAL-1(WT) or the GDP-bound form RAL-1(DN) (Fig. 2, A–D). This suggests that RAL-1 needs to cycle between GTP- and GDP-bound forms to properly localize in the epidermis. To establish that RAL-1 localizes to MVBs in the epidermis, we used the recently developed APEX tag, which catalyzes the local oxidation of DAB into an electron dense deposit visible by electron microscopy (Martell et al., 2012). After chemical fixation and DAB treatment, electron microscopy revealed a specific staining of animals expressing APEX-tagged RAL-1 (Fig. 2 E) at the external surface of MVBs (Fig. 2, G–G″) as well as on apical membrane stacks (Fig. 2 F). Collectively, our results clearly demonstrate that RAL-1 can localize at the surface of MVBs.
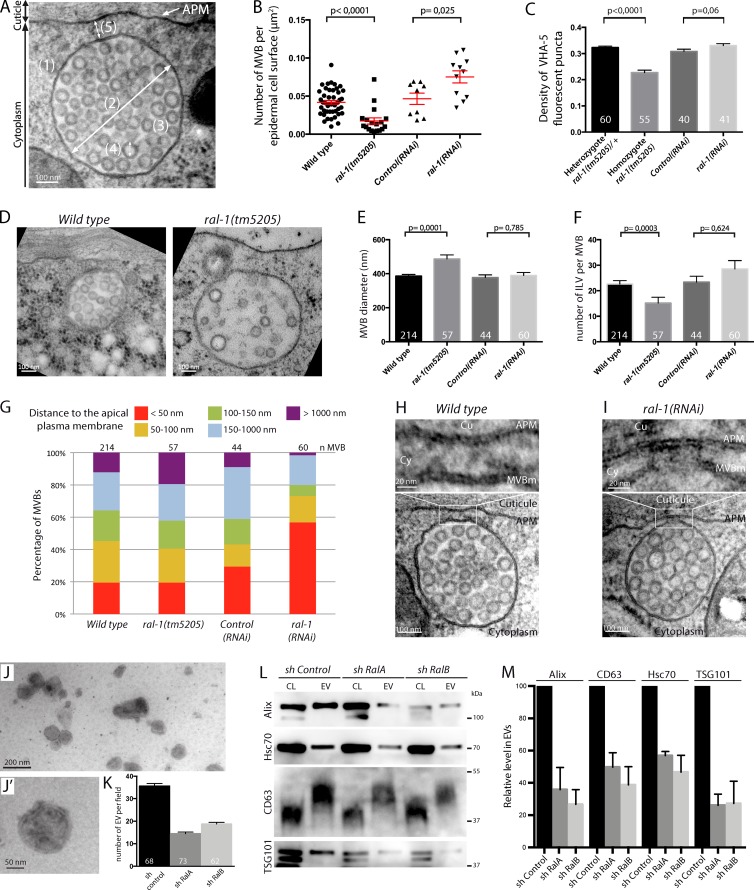
To determine how RAL-1 could affect MVBs and exosome secretion, we used a systematic quantitative analysis of MVBs by electron microscopy after high-pressure freezing (HPF; Fig. 3 A). We considered three types of compartments: light MVBs from which exosomes are likely to originate (Liégeois et al., 2006), dark MVBs (putative intermediate toward the lysosomes), and endolysosomes (Fig. S2). We quantified the density of each of these organelles (number per epidermal cell surface), their distance to the apical plasma membrane, their diameter, and the number and diameter of ILVs (Fig. 3 A). Strikingly, in ral-1(tm5205) null mutants, light MVBs were severely affected: (a) their density was drastically reduced (Fig. 3 B), (b) their mean size was increased (Fig. 3, D and E), and (c) the number of ILVs per MVB was decreased (Fig. 3, D and F). Consistent with a decrease in MVB density, that of VHA-5 fluorescent puncta also decreased in transgenic animals homozygous for ral-1(tm5205) compared with heterozygotes (Fig. 3 C). These results suggest that RAL-1 could either promote MVB formation or inhibit MVB degradation through the lysosomal pathway. We favor the first possibility, because the density of dark MVBs and endolysosomes also decreased in ral-1(tm5205) mutants compared with controls (Fig. S2). Thus, we suggest that the entire pathway from MVBs to lysosomes was affected, and that RAL-1 functions early in the process of MVB formation and generally in ILV biogenesis.
Figure 3.
RAL-1 affects different steps of exosome secretion. (A) Quantitative electron microscopy analysis of MVBs in epidermal cells: (1 and 2) density (1) and diameter (2) of MVBs, (3) number of ILVs per MVB, (4) ILV diameter, and (5) distance between MVB and the APM. (B) MVB density is decreased in ral-1(tm5205) compared with the WT and is increased in ral-1(RNAi) compared with control(RNAi). (C) The density of VHA-5::RFP puncta is decreased in ral-1(tm5205) compared with the WT but is unaffected in ral-1(RNAi). (D–F) In ral-1(tm5205) mutants, MVBs have an abnormal size (E) and ILV content (F). (G) In ral-1(RNAi) animals, 57% of MVBs are within 50 nm of the apical plasma membrane, compared with 20% in control animals. (H and I) Two MVBs in proximity of the apical plasma membrane from control (H) and ral-1(RNAi) (I) animals. MVBs from ral-1(RNAi) animals can form a hemifusion diaphragm (I) with the apical plasma membrane. (J and J’) EVs purified from 4T1 mammalian cells and observed by electron microscopy. (K) Depletion of either RalA or RalB by shRNA leads to a decrease in the number of EVs observed by electron microscopy compared with control shRNA (P < 0.0001 between sh control and either sh RalA or sh RalB; pool of four independent purifications, Mann-Whitney test). (L and M) Western blot of cell lysates and secreted EVs. One representative experiment (L) and pooled quantification (M) of four independent purifications (P < 0.03 between sh control and either sh RalA or sh RalB for each marker, Mann-Whitney test). Numbers in or above the bars indicate the number of animals (C), MVBs (E, F, and M) or fields (K) analyzed. APM, apical plasma membrane; CL, cell lysate; Cu, cuticle; Cy, cytoplasm; MVBm, MVB outer membrane. Errors bars, SEM.
To examine whether RAL-1 can also control later steps of MVB fate, we used RNAi to induce a mild depletion of ral-1, which induced lighter alae defects (Fig. 1 C) and reduced ral-1 transcripts by almost 60% based on quantitative RT-PCR assays (Fig. S1C). We found that, in this case, the density of MVBs was increased compared with either WT or control(RNAi) animals (Fig. 3 B and Fig. S2). The discrepancy between the null mutant and the RNAi phenotype was confirmed using a VHA-5::RFP integrated line. In the total absence of ral-1, the density of VHA-5 puncta decreased by 30%, whereas after depletion by RNAi, it remained comparable to the control (Fig. 3 C). Furthermore, electron microscopy analysis showed that MVBs were closer to the apical plasma membrane in ral-1(RNAi) animals compared with both controls (Fig. 3 G and Fig. S2N). A careful analysis of the MVBs revealed that 45% of them have a direct connection with the apical plasma membrane (ranging from a single link to a hemifusion diaphragm; Fig. 3 I), compared with 9% in WT and 11% in control(RNAi) animals (Fig. 3 H and Fig. S2 O). Therefore, we suggest that when RAL-1 becomes limiting, MVB fusion with the apical plasma membrane is hampered, likely because additional fusion factors are not recruited, but membrane attachment remains possible. Altogether, our data show that RAL-1 affects MVBs at different steps: in their formation and in the last steps before exosome secretion.
To directly assess the effect of RAL on exosome secretion (which is not possible in our system, because the cuticle prevents their collection), and to determine whether RAL function is conserved throughout evolution, we analyzed the role of the two ral-1 homologues in mammals, RalA and RalB. We first purified the EVs secreted by 4T1 mammary tumor cells by differential centrifugation. Combining electron microscopy and Western blot analysis, we observed that they display an exosomal size (mean diameter 84 ± 0.8 nm, n = 2,386 vesicles; Fig. 3, J and J″) and are enriched in four exosomal markers (Fig. 3 L). 4T1 cells knocked down with shRNA for either RalA or RalB (Fig. S1 D) secreted significantly fewer exosome-like vesicles than cells expressing a control shRNA (Fig. 3, K–M). This directly demonstrates that, like in C. elegans, the Ral GTPase is required for secretion of exosome-like vesicles in mammals. The precise mechanism of action of RalA and RalB remains to be determined in mammals.
RAL-1 controls exosome secretion independently of the exocyst
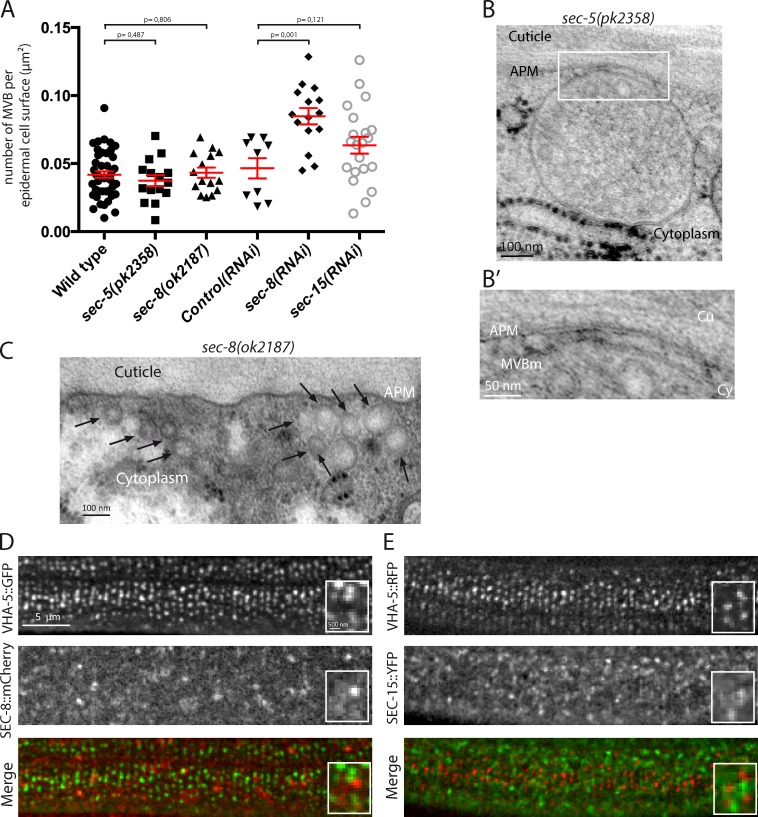
To assay whether RAL-1 and the exocyst act together, we analyzed the properties of MVBs present in null mutants of two exocyst subunits, sec-5(pk2358) and sec-8(ok2187). In contrast with the situation observed in ral-1(tm5205) null mutants, we found that the density of MVBs in these mutants was similar to that in control animals (Fig. 4 A). Occasionally, we observed some MVBs attached to the plasma membrane, indicating that this attachment occurred independently of the exocyst tethering complex (Fig. 4 B and Fig. S2 O). In addition, we found an accumulation of small vesicles (<100 nm diameter) under the plasma membrane of sec-5 and sec-8 mutants (Fig. 4 C), which are likely to be exocytic vesicles unable to attach to the plasma membrane, according to the known functions of the exocyst.
Figure 4.
The exocyst regulates alae formation independently of RAL-1. (A) Quantitative electron microscopy analysis reveals that the MVB density is similar in sec-5(pk2358) and sec-8(ok2187) null mutants and WT controls. MVB density is increased after sec-8(RNAi) but not sec-15(RNAi) compared with control(RNAi) (P values, Mann-Whitney test). (B) Example of an MVB attached to the apical plasma membrane in sec-5(pk2358) (enlarged in B’). (C) Small vesicles (<100 nm diameter; black arrows) accumulating under the plasma membrane in sec-8(ok2187) mutants. (D and E) The exocyst subunits SEC-8 (D) and SEC-15 (E) do not colocalize with VHA-5 in the epidermis. The scale bars in D apply to E. APM, apical plasma membrane; Cu, cuticle; Cy, cytoplasm; MVBm, MVB outer membrane.
To determine whether a complete lack of the exocyst would mask its requirement at other steps in MVB biogenesis, we used a mild depletion of two exocyst subunits. We found that depletion of sec-8, but not sec-15, by RNAi leads to a significant increase in MVB density compared with either WT or control(RNAi) animals, in an extent similar to that observed in ral-1(RNAi) animals (Fig. 3 A and Fig. S2). However, after RNAi against sec-8 or sec-15, the MVBs were not enriched in the vicinity of the apical plasma membrane, in contrast with what we observed after ral-1(RNAi) (Fig. S2 N).
Finally, we investigated the localization of two exocyst members, one on the target membrane side (SEC-8) and the other on the vesicle side (SEC-15), using integrated transgenic lines, which were previously shown to rescue mutant phenotypes and thus be functional (Armenti et al., 2014). Interestingly, although both proteins are expressed in the epidermis with a punctate localization (Fig. 4, D and E), neither SEC-8 nor SEC-15 significantly colocalized with VHA-5, whether in larval or adult animals (2%–5% colocalization in >50 animals, two independent strains in each case).
Altogether, the fact that exocyst depletion leads to MVB phenotypes different from RAL-1 depletion, added to the absence of colocalization between the exocyst and VHA-5, argues that RAL-1 and the exocyst function independently in alae formation. We propose that the exocyst is not directly involved in tethering MVBs at the plasma membrane. Thus, the alae defects observed in exocyst mutants reflect either an indirect effect on exosome secretion or a different secretory function, independent of exosome secretion.
Syntaxin-5 is involved in the fusion between the MVB membrane and the plasma membrane, downstream of RAL-1
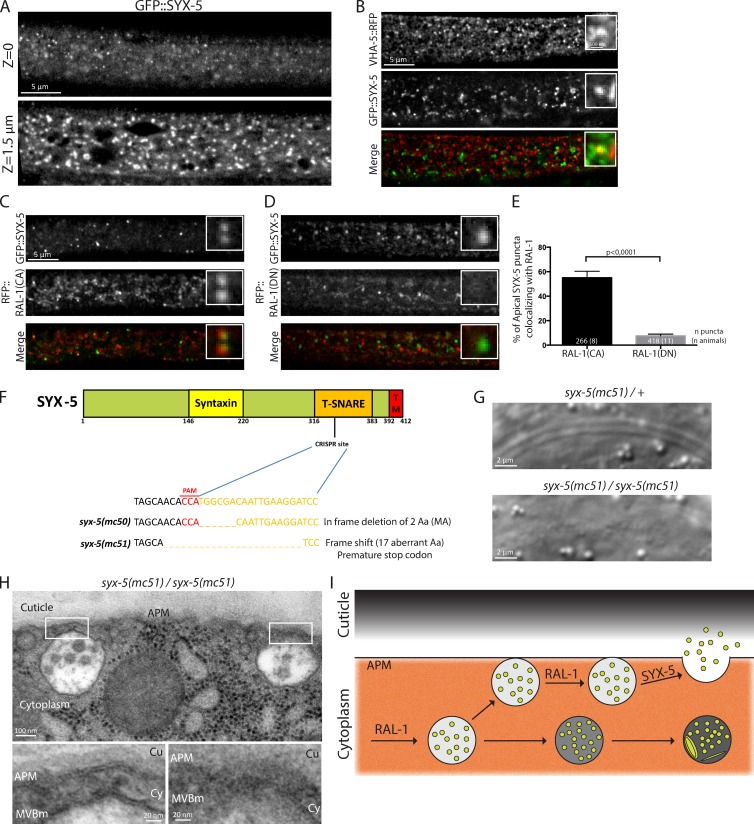
We next wanted to elucidate the molecular mechanisms acting downstream of RAL-1 to allow MVB fusion with the apical plasma membrane and exosome secretion. In our screen, we identified syx-5, a gene belonging to the syntaxin family (Table S1), whose closest mammalian homologue, syntaxin-5 (41% identity with SYX-5), is a t-SNARE mediating vesicle fusion in anterograde ER-Golgi trafficking (Hardwick and Pelham, 1992; Malsam and Söllner, 2011). Accordingly, we found that SYX-5 localizes in large round structures potentially corresponding to the epidermal Golgi apparatus. In addition, we observed more discrete puncta localizing at the most apical side of the epidermis (Fig. 5 A). These puncta occasionally colocalized with VHA-5 (Fig. 5 B). To determine whether SYX-5 could function downstream of RAL-1, we assessed its localization with RAL-1(CA) and RAL-1(DN) mutant forms. We found that SYX-5 small apical puncta partially colocalize with RAL-1(CA), but not with RAL-1(DN) (Fig. 5, C–E). Thus, an active form of RAL-1 could activate or recruit SYX-5 at the level of the apical plasma membrane to promote MVB fusion.
Figure 5.
SYX-5 controls MVB fusion with the apical plasma membrane. (A) In the epidermis, SYX-5 localizes in both large cytoplasmic puncta and smaller apical puncta. (B) SYX-5 displays some colocalization with VHA-5 in the epidermis at the time of alae formation. (C–E) SYX-5 colocalizes with RAL-1(CA) (C), but not with RAL-1(DN) (D), as revealed by quantification (E). The scale bars in B apply to C and D. The numbers inside the bars indicate the number of puncta (number of animals). (F) Generation of two mutant alleles for syx-5 using CRISPR/Cas9 technology. (G) Homozygote mutants for syx-5(mc51) show alae defects at the L1 stage. (H) Electron micrographs of two MVBs showing attachment to the apical plasma membrane in syx-5(mc51) mutant larva. (I) Model for the role of RAL-1 and SYX-5 in exosome biogenesis (see text). Errors bars, SEM.
To further address the function of SYX-5, we generated two syx-5 mutants using CRISPR-Cas9 technology (Dickinson et al., 2013). We targeted the SNARE domain of SYX-5, the characteristic coiled-coil motif responsible for bringing opposing membranes together and catalyzing their fusion (Hong and Lev, 2014). We recovered two alleles, syx-5(mc50) (in-frame deletion of two amino acids, including a methionine conserved throughout evolution) and syx-5(mc51) (frameshift adding 17 ectopic amino acids and a premature stop codon; Fig. 5 F). Both mutant alleles induced a premature larval arrest, likely because of an absence of molting, precluding a study of the role of SYX-5 in alae formation in adults. However, because L1 stage larvae also have alae, we tested whether syx-5 alleles could affect their formation. We found that 100% of syx-5 L1 mutants have absent or defective alae (Fig. 5 G and Fig. S1). This defect was partially rescued by the expression of a WT version of SYX-5 fused to GFP (Fig. S1 and Table S2). In addition, overexpression in WT animals of WT or mutant SYX-5 forms induced alae defects at the adult stage (Fig. S1 and Table S2). Finally, electron microscopy analysis on L1 syx-5(mc51) larvae showed that 62% of the MVBs present in the epidermis are within 50 nm of the apical plasma membrane (77 MVBs in three animals), compared with 24% in WT larvae (67 MVBs in four animals; Fig. 5 H). Accordingly, the mean distance between MVBs outer membrane and the apical plasma membrane is reduced in syx-5(mc51) mutants (68 ± 14 nm) compared with WT animals (152 ± 30 nm; P < 0.01, Student’s t test).This result suggests that in the absence of syx-5, the MVB outer membrane can no longer fuse with the plasma membrane.
Our work demonstrates a new role for the Ral GTPase in exosome secretion in mammals and nematodes. It further establishes this GTPase as an important player in cell–cell communication, because RalA was previously shown to promote the formation of tunneling nanotubes (Hase et al., 2009). Our data collectively suggest a model (Fig. 5 I) whereby C. elegans RAL-1 acts both at the initial step of MVB formation and at the late stage of fusion between the MVB membrane and the plasma membrane. One possibility could be that RAL-1 recruits different effectors involved in ILV budding or membrane fusion (Gentry et al., 2014). In particular, Arf6 and PLD are known RalA targets (Luo et al., 1998; Bhattacharya et al., 2004; Vitale et al., 2005; Corrotte et al., 2010) that were recently shown to modulate ILV budding through Alix–syntenin and, as a consequence, exosome secretion (Laulagnier et al., 2004; Trajkovic et al., 2008; Strauss et al., 2010; Ghossoub et al., 2014). Whether RAL-1 functions with the Alix–syntenin pathway to generate MVBs remains to be determined; an RAL-1–independent mechanism is likely to exist because MVBs still contain ILVs in its absence. One important finding of this study is that RAL-1 functions independently of the exocyst complex in MVB tethering and fusion at the plasma membrane. Instead, we suggest that it recruits a yet-to-be-identified tethering factor or complex, as well as the syntaxin SYX-5 at the site of fusion. How the V0-ATPase (including VHA-5) contributes to membrane fusion together with syntaxins remains to be investigated. Another important finding of this study is that the partial loss of RAL-1 tends to block MVBs attached to the plasma membrane and is sometimes locked in a state resembling hemifusion. We speculate that RAL-1 might respond to the local concentration of SNARE complexes required to promote fusion (Xu et al., 2005) or orchestrate their action (Hernandez et al., 2014). Alternatively, RAL-1 could be more directly involved in pore fusion resolution by recruiting important fusion partners acting downstream of SNARE complexes, such as VHA-5. Because MVBs are large flat structures compared with small vesicles, which are likely to oppose a higher energy barrier to fusion (Hernandez et al., 2014; Risselada et al., 2014), and because their ILV content presumably requires a larger fusion pore to be released compared with simple peptides, the final step of MVB fusion could require a tight regulation involving several proteins.
Materials and methods
Strains and CRISPR/Cas9
Strains were propagated and handled as described previously (Brenner, 1974). A complete list of mutants and fluorescent reagents is presented in Table S3. All mutants were characterized previously (Jiu et al., 2012; Armenti et al., 2014), except syx-5(mc50) and syx-5(mc51) mutants, which were obtained using CRISPR/Cas9 technology, based on the protocol described by Dickinson et al. (2013). The SNARE coding domain of syx-5 was targeted using the single guide RNA (sgRNA; 5′-GGATCCTTCAATTGTCGCCATGG-3′) site present on the reverse DNA strand and covering a restriction site for the NcoI enzyme. The syx-5 sgRNA sequence was introduced into the pDD162 plasmid by PCR (Dickinson et al., 2013). A DNA mix containing the following plasmids was injected in adult animals: pDD162 (Cas9-sgRNA, 50 ng/µl), PGH8 (prab-8::mCherry, 10 ng/µl), PcFJ104 (pmyo-3::mCherry, 5ng/µl), PCFJ90 (pmyo-2::mCherry, 2.5 ng/µl), and pBLuescript (80 ng/µl; Dickinson et al., 2013). 384 F1 animals carrying the injection markers were screened by PCR followed by NcoI digestion, leading to the identification of two syx-5 mutant alleles. Both alleles displayed similar phenotypes of larval arrest and were balanced to be maintained as heterozygous. We eliminated the coinjection markers by selecting nonfluorescent animals.
Cloning and transgenesis
All plasmids were generated by PCR, with digestion by restriction enzymes or Gibson cloning (NEB), using a Phusion High-Fidelity DNA polymerase (Thermoscientific). RAL-1(CA) (G23V) and RAL-1(DN) (S28N) forms were generated based on previously reported single amino-acid mutations in the GTPase pocket of RAL in diverse species (Jiang et al., 1995; Ohta et al., 1999; Goi et al., 2000; Moskalenko et al., 2002). Specific expression in the epidermal cells of transgenic animals was obtained using the dpy-7 promoter (Gilleard et al., 1997). The APEX2 tag (Martell et al., 2012) was adapted for C. elegans codon usage (in vitro synthesis by Sigma-Aldrich). For transgenesis, DNA microinjection was performed with 5 ng/μl of coinjection markers (pmyo-2::gfp or pmyo-2::mcherry) and 10 ng/µl for constructs of interest (except for the rescue of ral-1(tm5205) by pdpy-7::gfp::ral-1(CA) and pdpy-7::gfp::ral-1(DN), which was performed at 1 ng/µl), and completed to 150 ng/µl with pBLuescript.
RNAi and alae defects screen
RNAi by feeding was performed using standard procedures, with 100 µg/ml ampicillin/1 mM IPTG (Sigma-Aldrich) using clones from the Ahringer-MRC library (Kamath et al., 2003); plasmids were sequenced and retransformed into HT115 (DE3). Bacteria containing an empty L4440 RNAi vector were used as a control. The selection of genes for the screen was based on positive hits from previous screens for endocytic genes performed in C. elegans (Frand et al., 2005; Balklava et al., 2007; Kinchen et al., 2008). In addition, all putative kinases and phosphatases, as well as all available rab and escrt genes, were screened. Mothers were bleached on feeding plates; animals were scored and analyzed the following days. Adult animals were scored for alae defects (see following paragraph). The screening was repeated twice. Genes positive in both rounds of screening were considered as positive hits (Table S1). The efficiency of ral-1(RNAi) was measured by performing quantitative RT-PCR (in triplicate) on RNA extracted from young adult animals treated with ral-1(RNAi) or control(RNAi) since hatching. Three reference genes (eif-3f, act-1, and Y45F10D.4) were used to normalize ral-1 mRNA levels.
Light microscopy and quantification
Live young adult animals were immobilized with 0.1% tricaine/0.01% tetramisole and mounted on 2% agarose pads at 20°C. Alae defects phenotypes were observed by differential interference contrast using a Leica DMRXA2 6000 equipped with a Coolsnap HQ (Roper Scientific) camera with a 100× objective (oil; NA 1.4). To follow the localization of fluorescent proteins, Z-stacks (0.3-µm spacing) were acquired using a DMI6000 (Leica) spinning disk (Yokogawa CSU22 with an Andor iXonEM+ 897 camera) with a 63× objective (oil; NA 1.4). Leica Type F immersion medium was used. Young adults were observed (right after alae formation and before formation of the first embryos). Identical settings were used for control and mutant animals. Image analysis was performed using ImageJ software (National Institutes of Health). Colocalization (Fig. 1, Fig. 2, and Fig. 4) was quantified using a semiautomated method (comparison of local intensity maxima obtained in each channel) and a manual method based on scan line analysis (comparison of fluorescence intensity profile along a line crossing a fluorescent punctum in a given channel with the profile obtained from the same line in the other channel), which was previously described (Hyenne et al., 2012).
Transmission electron microscopy and APEX tag
Animals were processed for HPF, freeze substitution, and flat embedding as previously described (Kolotuev et al., 2010). In brief, animals were transferred to 200-µm-deep flat carriers and processed in the EMPACT-2 HPF apparatus (Leica). Freeze substitution was performed in the Leica electron microscopy AFS in 2% osmium tetroxide, 0.25% uranyl acetate, 0.5% glutaraldehyde, and 1% water in pure acetone; the temperature was then increased and the samples were embedded in epon resin (Sigma-Aldrich). Samples were flat embedded in epon resin between two aclar sheets, and laser carving (10× objective; LMD 6000; Leica) was used to precisely localize the region of interest. Young adults were sectioned transversally, at the same position (∼80 µm anterior to the vulva). Thin sections were stained with uranyl acetate (4% for 10 min) and lead citrate (for 4 min). Images were acquired with an Orius 1000 charge-coupled device camera (Gatan) mounted on a Philips CM12 microscope operated at 80 kV. For systematic quantitative analysis of MVBs, three to eight animals were analyzed for each genotype (except for control(RNAi), in which only one animal was recovered and analyzed; however, control(RNAi) and WT animals were statistically identical for each and every parameter measured [P > 0.3, Mann-Whitney test]), using three to five thin sections (nonconsecutive sections, spaced by 600 nm) for each animal. For each thin section, the total surface of the epidermal cell was measured and a picture of each MVB (dark and light) and endolysosome was taken at high magnification. Measurements of surfaces and lengths were performed using ImageJ software.
For photooxidation of the APEX tag, animals were fixed in 2.5% glutaraldehyde, cut in two pieces, and fixed on ice for at least 2 h. After they were rinsed five times in cold cacodylate buffer (0.1 M, pH 7.4), they were incubated for 30 min in blocking solution (10 mM KCN, 5 mM aminotriazole, and 50 mM glycine in cacodylate buffer) on ice. Photooxidation was performed with freshly prepared DAB (1 mg/ml) combined with 10 mM H2O2 in blocking buffer on ice. Animals were rinsed five times in cacodylate buffer, embedded in epon resin, and processed for thin sectioning and observation. Control animals were processed similarly, but without the DAB treatment.
Mammalian cells and EV purification
4T1 mammary tumor cells were chosen for their ability to secrete exosomes in a rab-27 dependent manner (Bobrie et al., 2012). For stable inhibition of RalA or RalB expression, cells infected with shRNA expressing lentiviruses were selected and maintained in medium containing 1 µg/ml puromycin. Sequences for shRNA (mRalA: 5′-CCGGGTGCAGATCGACATCTTAGATCTCGAGATCTAAGATGTCGATCTGCACTTTTTG-3′; mRalB: 5′-CCGGCCTGGTACTTCACAAGGTCATCTCGAGATGACCTTGTGAAGTACCAGGTTTTTG-3′; and control scramble: 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3′) were inserted into the AgeI to EcoRI sites of pLKO.1 vector (a gift from D. Root, Broad Institute of MIT and Harvard, Cambridge, MA; plasmid 10878; Addgene). Cells were cultured in exosome-depleted medium (by overnight ultracentrifugation at 100,000 g) for 24 h before supernatant collection. Extracellular medium was concentrated using a Centricon Plus-70 centrifugal filter (10k; Millipore) and EVs were purified by successive centrifugation at 4°C as follows: 5 min at 300 g, 10 min at 2,000 g, 30 min at 10,000 g, and 70 min at 100,000 g (using a XL-70 and a SW28 rotor; Beckman Coulter). Pellets were washed in PBS, centrifuged again at 100,000 g for 70 min, taken up in PBS and stored at −80°C.
For electron microscopy analysis, 3 µl of EV extracts secreted from similar cell number were allowed to dry on grids for 20 min and were fixed in 3% PFA for 10 min, rinsed in water, and contrasted in a uranyl acetate (0.4%)/methylcellulose (2%) mix for 10 min on ice. For quantification, images were acquired as described above at 53,000× magnification and EVs were counted and measured in 15–20 fields per condition.
For Western blotting analysis, extracts corresponding to similar cell numbers were loaded on 4%–20% polyacrylamide gels (Bio-Rad Laboratories, Inc.). All extracts were run under denaturing conditions, except for CD63. The following antibodies were used: Alix (mouse, 611621; BD), Hsc70 (rat, ADI-SPA-815D; Enzo Life Sciences), CD63 (mouse D623-3; MBL), TSG101 (mouse GTX70255; GeneTex), RalA (mouse, 610221; BD), RalB (mouse, 04037; Millipore), α-tubulin (mouse, CP06; Millipore), gapdh (goat, AB006720; Sicgen). Acquisitions were done using a PXi system (Syngene). Intensities were normalized over cellular tubulin or gapdh levels.
Statistical analysis
Statistical significance of the results was analyzed using GraphPad Prism software. For data following a Gaussian distribution, the statistical difference of the mean was analyzed using the unpaired two-tailed Student’s t test. For data not following a Gaussian distribution, the nonparametric Mann-Whitney test was used. In the graphs, error bars represent SEMs, except for Fig. S1, where they represent SDs. P values are indicated directly in the graphs or in the figure legends.
Online supplemental material
Fig. S1 shows the classification of alae defects in various genetic backgrounds. It also displays the efficiency of ral-1 partial depletion by RNAi in C. elegans and the efficiency of RalA and RalB knock-downs by shRNA in mammalian cells. Fig. S2 shows the quantitative electron microscopy analysis of MVBs and endolysosomes in different nematode strains. Table S1 is a list of the genes identified in the RNAi screen for alae defects. Table S2 is a quantification of alae defects in various backgrounds. Table S3 is a list of the nematode strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504136/DC1.
Supplementary Material
Acknowledgments
We thank J. Nance for strains and constructs; J. Jäntti, the Japanese consortium, and the Caenorhabditis Genetics Center for strains; P. Schultz for access to the EMPACT2 high-pressure system; and M. Koch, P. Kessler, and J. Pontabry for imaging advice. We also thank S. Quintin, C. Gally, and Roland Le Borgne for critical reading of the manuscript and Clothilde Théry for useful discussions.
This work was supported by a fellowship from the Fondation ARC pour la Recherche sur le Cancer (V. Hyenne); grants from the Institut National du Cancer (to M. Labouesse), the Agence Nationale de la Recherche (ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir labeled ANR-10-IDEX-0002-02 to the IGBMC), and the Institut National du Cancer and Roche (to J.G. Goetz); and institutional funds from the Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, and University of Strasbourg.
The authors declare no competing financial interests.
Author contributions: V. Hyenne and M. Labouesse planned the project. V. Hyenne designed and conducted most of the experiments. A. Apaydin carried out the initial RNAi screen. D. Rodriguez and V. Hyenne contributed to the cloning. S. Hoff-Yoessle and V. Hyenne generated the syx-5 mutants by CRISPR/Cas9. V. Hyenne carried out most electron microscopy experiments with training and help from Y. Schwab and C. Spiegelhalter. S. Tak carried out ral-1 mRNA quantification. M. Diem helped in cloning and analyzing RAL-1 mutants. O. Lefebvre generated the knocked down mammalian cells. V. Hyenne and M. Labouesse wrote the manuscript. J.G. Goetz contributed to mammalian experiment design and to the manuscript.
Footnotes
Abbreviations used in this paper:
- CA
- constitutively active
- DN
- dominant negative
- ESCRT
- endosomal sorting complex required for transport
- EV
- extracellular vesicle
- HPF
- high-pressure freezing
- ILV
- intraluminal vesicle
- MVB
- multivesicular body
- sgRNA
- single guide RNA
- WT
- wild type
References
- Abrami L., Brandi L., Moayeri M., Brown M.J., Krantz B.A., Leppla S.H., and van der Goot F.G.. 2013. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Reports. 5:986–996. 10.1016/j.celrep.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti S.T., Chan E., and Nance J.. 2014. Polarized exocyst-mediated vesicle fusion directs intracellular lumenogenesis within the C. elegans excretory cell. Dev. Biol. 394:110–121. 10.1016/j.ydbio.2014.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., et al. 2012. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14:677–685. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., and Grant B.D.. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Bhattacharya M., Babwah A.V., Godin C., Anborgh P.H., Dale L.B., Poulter M.O., and Ferguson S.S.. 2004. Ral and phospholipase D2-dependent pathway for constitutive metabotropic glutamate receptor endocytosis. J. Neurosci. 24:8752–8761. 10.1523/JNEUROSCI.3155-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A., Krumeich S., Reyal F., Recchi C., Moita L.F., Seabra M.C., Ostrowski M., and Théry C.. 2012. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 72:4920–4930. 10.1158/0008-5472.CAN-12-0925 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brymora A., Valova V.A., Larsen M.R., Roufogalis B.D., and Robinson P.J.. 2001. The brain exocyst complex interacts with RalA in a GTP-dependent manner: identification of a novel mammalian Sec3 gene and a second Sec15 gene. J. Biol. Chem. 276:29792–29797. 10.1074/jbc.C100320200 [DOI] [PubMed] [Google Scholar]
- Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L.F., Théry C., and Raposo G.. 2013. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126:5553–5565. 10.1242/jcs.128868 [DOI] [PubMed] [Google Scholar]
- Corrotte M., Nyguyen A.P., Harlay M.L., Vitale N., Bader M.F., and Grant N.J.. 2010. Ral isoforms are implicated in Fc gamma R-mediated phagocytosis: activation of phospholipase D by RalA. J. Immunol. 185:2942–2950. 10.4049/jimmunol.0903138 [DOI] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., and Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 10:1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D.G., Mestre M.B., and Colombo M.I.. 2009. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta. 1793:1901–1916. 10.1016/j.bbamcr.2009.09.011 [DOI] [PubMed] [Google Scholar]
- Frand A.R., Russel S., and Ruvkun G.. 2005. Functional genomic analysis of C. elegans molting. PLoS Biol. 3:e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry L.R., Martin T.D., Reiner D.J., and Der C.J.. 2014. Ral small GTPase signaling and oncogenesis: More than just 15 minutes of fame. Biochim. Biophys. Acta. 1843:2976–2988. 10.1016/j.bbamcr.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R., Lembo F., Rubio A., Gaillard C.B., Bouchet J., Vitale N., Slavík J., Machala M., and Zimmermann P.. 2014. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 5:3477 10.1038/ncomms4477 [DOI] [PubMed] [Google Scholar]
- Gilleard J.S., Barry J.D., and Johnstone I.L.. 1997. cis Regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17:2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goi T., Shipitsin M., Lu Z., Foster D.A., Klinz S.G., and Feig L.A.. 2000. An EGF receptor/Ral-GTPase signaling cascade regulates c-Src activity and substrate specificity. EMBO J. 19:623–630. 10.1093/emboj/19.4.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J.C., Chaudhary V., Bartscherer K., and Boutros M.. 2012. Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 14:1036–1045. 10.1038/ncb2574 [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., and Pelham H.R.. 1992. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J. Cell Biol. 119:513–521. 10.1083/jcb.119.3.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K., Kimura S., Takatsu H., Ohmae M., Kawano S., Kitamura H., Ito M., Watarai H., Hazelett C.C., Yeaman C., and Ohno H.. 2009. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol. 11:1427–1432. 10.1038/ncb1990 [DOI] [PubMed] [Google Scholar]
- Hernandez J.M., Kreutzberger A.J., Kiessling V., Tamm L.K., and Jahn R.. 2014. Variable cooperativity in SNARE-mediated membrane fusion. Proc. Natl. Acad. Sci. USA. 111:12037–12042. 10.1073/pnas.1407435111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., and Lev S.. 2014. Tethering the assembly of SNARE complexes. Trends Cell Biol. 24:35–43. 10.1016/j.tcb.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Hoshino D., Kirkbride K.C., Costello K., Clark E.S., Sinha S., Grega-Larson N., Tyska M.J., and Weaver A.M.. 2013. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Reports. 5:1159–1168. 10.1016/j.celrep.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C., Morohashi Y., Yoshimura S., Manrique-Hoyos N., Jung S., Lauterbach M.A., Bakhti M., Grønborg M., Möbius W., Rhee J., et al. 2010. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 189:223–232. 10.1083/jcb.200911018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne V., Tremblay-Boudreault T., Velmurugan R., Grant B.D., Loerke D., and Labbé J.C.. 2012. RAB-5 controls the cortical organization and dynamics of PAR proteins to maintain C. elegans early embryonic polarity. PLoS ONE. 7:e35286 10.1371/journal.pone.0035286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Luo J.Q., Urano T., Frankel P., Lu Z., Foster D.A., and Feig L.A.. 1995. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 378:409–412. 10.1038/378409a0 [DOI] [PubMed] [Google Scholar]
- Jiu Y., Jin C., Liu Y., Holmberg C.I., and Jäntti J.. 2012. Exocyst subunits Exo70 and Exo84 cooperate with small GTPases to regulate behavior and endocytic trafficking in C. elegans. PLoS ONE. 7:e32077 10.1371/journal.pone.0032077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H., Simpson R.J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V.C., Borràs F.E., Breakefield X., Budnik V., et al. 2012. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10:e1001450 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kawato M., Shirakawa R., Kondo H., Higashi T., Ikeda T., Okawa K., Fukai S., Nureki O., Kita T., and Horiuchi H.. 2008. Regulation of platelet dense granule secretion by the Ral GTPase-exocyst pathway. J. Biol. Chem. 283:166–174. 10.1074/jbc.M705340200 [DOI] [PubMed] [Google Scholar]
- Kim D.K., Kang B., Kim O.Y., Choi D.S., Lee J., Kim S.R., Go G., Yoon Y.J., Kim J.H., Jang S.C., et al. 2013. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2 10.3402/jev.v2i0.20384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J.M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C.D., Hengartner M.O., and Ravichandran K.S.. 2008. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10:556–566. 10.1038/ncb1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., and Raposo G.. 2014. The complex ultrastructure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 6:a016857 10.1101/cshperspect.a016857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotuev I., Schwab Y., and Labouesse M.. 2010. A precise and rapid mapping protocol for correlative light and electron microscopy of small invertebrate organisms. Biol. Cell. 102:121–132. 10.1042/BC20090096 [DOI] [PubMed] [Google Scholar]
- Kowal J., Tkach M., and Théry C.. 2014. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29:116–125. 10.1016/j.ceb.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Laulagnier K., Grand D., Dujardin A., Hamdi S., Vincent-Schneider H., Lankar D., Salles J.P., Bonnerot C., Perret B., and Record M.. 2004. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 572:11–14. 10.1016/j.febslet.2004.06.082 [DOI] [PubMed] [Google Scholar]
- Liégeois S., Benedetto A., Garnier J.M., Schwab Y., and Labouesse M.. 2006. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 173:949–961. 10.1083/jcb.200511072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.A., Kwan E.P., Xie L., He Y., James D.E., and Gaisano H.Y.. 2008. The RalA GTPase is a central regulator of insulin exocytosis from pancreatic islet beta cells. J. Biol. Chem. 283:17939–17945. 10.1074/jbc.M800321200 [DOI] [PubMed] [Google Scholar]
- Luo J.Q., Liu X., Frankel P., Rotunda T., Ramos M., Flom J., Jiang H., Feig L.A., Morris A.J., Kahn R.A., and Foster D.A.. 1998. Functional association between Arf and RalA in active phospholipase D complex. Proc. Natl. Acad. Sci. USA. 95:3632–3637. 10.1073/pnas.95.7.3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J., and Söllner T.H.. 2011. Organization of SNAREs within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3:a005249 10.1101/cshperspect.a005249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., and Ting A.Y.. 2012. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30:1143–1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S., Henry D.O., Rosse C., Mirey G., Camonis J.H., and White M.A.. 2002. The exocyst is a Ral effector complex. Nat. Cell Biol. 4:66–72. 10.1038/ncb728 [DOI] [PubMed] [Google Scholar]
- Ohta Y., Suzuki N., Nakamura S., Hartwig J.H., and Stossel T.P.. 1999. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. USA. 96:2122–2128. 10.1073/pnas.96.5.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., et al. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12:19–30, 1–13. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- Page A.P., and Johnstone I.L.. 2007. The cuticle. WormBook. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., and Stoorvogel W.. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200:373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risselada H.J., Bubnis G., and Grubmüller H.. 2014. Expansion of the fusion stalk and its implication for biological membrane fusion. Proc. Natl. Acad. Sci. USA. 111:11043–11048. 10.1073/pnas.1323221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A., Fader C.M., Damiani M.T., and Colombo M.I.. 2005. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 6:131–143. 10.1111/j.1600-0854.2004.00257.x [DOI] [PubMed] [Google Scholar]
- Strauss K., Goebel C., Runz H., Möbius W., Weiss S., Feussner I., Simons M., and Schneider A.. 2010. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J. Biol. Chem. 285:26279–26288. 10.1074/jbc.M110.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K., Asano S., Tanaka K., Iwamatsu A., Okawa K., and Ohta Y.. 2002. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol. 4:73–78. 10.1038/ncb720 [DOI] [PubMed] [Google Scholar]
- Tamai K., Tanaka N., Nakano T., Kakazu E., Kondo Y., Inoue J., Shiina M., Fukushima K., Hoshino T., Sano K., et al. 2010. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 399:384–390. 10.1016/j.bbrc.2010.07.083 [DOI] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., and Simons M.. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 319:1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Vitale N., Mawet J., Camonis J., Regazzi R., Bader M.F., and Chasserot-Golaz S.. 2005. The Small GTPase RalA controls exocytosis of large dense core secretory granules by interacting with ARF6-dependent phospholipase D1. J. Biol. Chem. 280:29921–29928. 10.1074/jbc.M413748200 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhang F., Su Z., McNew J.A., and Shin Y.K.. 2005. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 12:417–422. 10.1038/nsmb921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.