The exon junction core complex associates with Balbiani ring (BR) premRNPs during transcription and in relation to splicing, whereas the export factor NXF1 is recruited in the interchromatin, and BR mRNPs become export competent only after passage through the interchromatin.
Abstract
Eukaryotic gene expression requires the ordered association of numerous factors with precursor messenger RNAs (premRNAs)/messenger RNAs (mRNAs) to achieve efficiency and regulation. Here, we use the Balbiani ring (BR) genes to demonstrate the temporal and spatial association of the exon junction complex (EJC) core with gene-specific endogenous premRNAs and mRNAs. The EJC core components bind cotranscriptionally to BR premRNAs during or very rapidly after splicing. The EJC core does not recruit the nonsense-mediated decay mediaters UPF2 and UPF3 until the BR messenger RNA protein complexes (mRNPs) enter the interchromatin. Even though several known adapters for the export factor NXF1 become part of BR mRNPs already at the gene, NXF1 binds to BR mRNPs only in the interchromatin. In steady state, a subset of the BR mRNPs in the interchromatin binds NXF1, UPF2, and UPF3. This binding appears to occur stochastically, and the efficiency approximately equals synthesis and export of the BR mRNPs. Our data provide unique in vivo information on how export competent eukaryotic mRNPs are formed.
Introduction
Expression of protein coding genes in eukaryotes requires that several multicomponent machineries and processes are coordinated in time and space, e.g., transcription, premRNA processing, and export are coupled (Maniatis and Reed, 2002; Perales and Bentley, 2009; Bentley, 2014). The nuclear events also influence the fate of the mRNAs in the cytoplasm, for example localization (Percipalle, 2014), translation efficiency (Nott et al., 2004), and quality control (Popp and Maquat, 2014). It is therefore essential to learn about the composition of the molecular machines associating with premRNA/mRNA, where and when they assemble, and how they interact with each other.
Splicing promotes gene expression by enhancing mRNP export (Valencia et al., 2008) and translation (Nott et al., 2003). Splicing changes the protein composition of the precursor messenger RNA protein complexes (premRNPs)/messenger RNA protein complexes (mRNPs), and as a consequence, the exon junction complex (EJC), assembles 20–24 nucleotides upstream the exon junction, in a sequence-independent manner (Le Hir et al., 2000; Kataoka et al., 2001; Bono and Gehring, 2011). Most exon junctions are marked by an EJC, but EJCs may assemble at noncanonical sites on mRNAs (Saulière et al., 2012; Singh et al., 2012). EJCs can be deposited on specific junctions, depending on cis-acting RNA sequences (Saulière et al., 2010). Together with SR proteins, a family of splicing factors (Long and Cáceres, 2009; Zhou and Fu, 2013), EJCs influence the overall folding of the mRNA (Singh et al., 2012). Based on in vitro studies, the spliceosomal component CWC22 recruits EJC (Alexandrov et al., 2012; Barbosa et al., 2012; Steckelberg et al., 2012) during the second step of splicing (Gehring et al., 2009), and the intron binding protein IBP160 is involved in recruiting the EJC (Ideue et al., 2007). Splicing is largely cotranscriptional (Brugiolo et al., 2013; Bentley, 2014), and in accordance, EJC components are recruited to sites of transcription (Custódio et al., 2004). The relationship between splicing and EJC formation is however not yet analyzed for defined endogenous premRNAs in vivo.
Four proteins form the core of the EJC, eIF4AIII, Y14, Mago, and MLN51/Barentsz (Btz). The structure of the EJC core has been described (Ballut et al., 2005; Andersen et al., 2006; Bono et al., 2006). The EJC core remains associated with the mRNP during export and cytoplasmic events, being removed from the mRNP by the translating ribosome and the ribosome-associated protein PYM (Gehring et al., 2009; Ghosh et al., 2014). The EJC core has specific functions, e.g., influencing splicing (Hayashi et al., 2014; Malone et al., 2014). In addition, eIF4AIII is essential for nonsense-mediated decay (NMD; Palacios et al., 2004; Shibuya et al., 2004). Y14 and Mago promote translation (Nott et al., 2004), are important for development in plants and animals (Gong et al., 2014), and influence cytoplasmic mRNP localization (Palacios, 2002). Btz interacts with eIF3, a translational initiation factor, and enhances translation (Chazal et al., 2013). It also influences P-body disassembly (Cougot et al., 2014).
The EJC core serves as a versatile platform that interacts with several proteins, thereby being important for feeding the mRNP into posttranscriptional pathways. The EJC-interacting proteins influence translational efficiency (Le Hir and Séraphin, 2008) and NMD (Chamieh et al., 2008). The UPF1, UPF2, and UPF3 proteins are conserved effectors of NMD (Mühlemann et al., 2008). UPF3 is the link between the EJC and the NMD machinery and is recruited to the EJC in the nucleus. UPF2 is believed to associate with UPF3 at the cytoplasmic side of the nuclear membrane (Lykke-Andersen et al., 2000; Serin et al., 2001), whereas UPF1 is added to form an UPF3-UPF2-UPF1 complex on triggering of NMD. As yet, binding of UPF proteins to specific endogenous mRNPs in vivo has not been analyzed.
An important aspect of mRNP maturation is the recruitment of export factors. NXF1 and its cofactor p15 is the main export factor for mRNPs (Natalizio and Wente, 2013). NXF1 binds to mRNPs via different adapters, and association of these adapters with mRNPs is likely to be influenced by different processes. Aly/REF, together with the THO component Toc5 is an adapter for NXF1 (Katahira et al., 2009; Viphakone et al., 2012). Aly/REF, the THO complex, UAP56, and Tex1 constitute the conserved TREX complex that couples transcription and mRNA export. In yeast, TREX is recruited to the RNA polymerase II CTD through its THO complex and increases along the active gene from 5′ to 3′ (Meinel et al., 2013). In humans, the THO complex is associated with spliced mRNAs (Masuda et al., 2005; Chi et al., 2013). In yeast, Yra1p (the Aly/REF equivalent) is recruited to the TREX complex during 3′ end processing by binding to Pcf11, and this interaction is conserved between their human homologoues (Johnson et al., 2009). It has also been reported that the human TREX complex is recruited in a splicing- and CBP80-dependent way to a position close to the 5′ end of the mRNA (Cheng et al., 2006).
In addition, the EJC has been reported to serve as a binding platform for Aly/REF and NXF1/p15 (Le Hir et al., 2001; Tange et al., 2004), and certain SR proteins can serve as NXF1 adapters (Huang and Steitz, 2001; Huang et al., 2003). To understand how an mRNP gets export competent, it is important to study when and where in the nucleus NXF1 binds to premRNPs/mRNPs in vivo.
Here we use unique experimental advantages of the Balbiani ring (BR) genes in the dipteran Chironomus tentans to analyze the spatial and temporal aspects of the association of the EJC core, the NMD effectors UPF2 and UPF3, and the export factor NXF1 with gene-specific premRNPs/mRNPs in vivo.
The BR1 and BR2 genes allow identification of gene-specific premRNPs and mRNPs in intact cell nuclei. The BR premRNAs assemble into premRNA-protein complexes rapidly (Daneholt, 2001). The BR1 and BR2 premRNAs contain four introns each, and these introns are excised essentially cotranscriptionally (Baurén and Wieslander, 1994). When the BR mRNPs are released from the genes, they move by diffusion until they dock at the nuclear pore complexes (NPCs; Singh et al., 1999) and are subsequently translocated into the cytoplasm (Siebrasse et al., 2008). In the EM, individual BR mRNPs are identified in the interchromatin as uniform 50-nm spherical structures, and they can be morphologically seen during NPC docking and translocation (Daneholt, 2001). Using immuno-EM, we show that the C. tentans EJC core binds to BR premRNPs cotranscriptionally in relation to splicing, whereas UPF2, UPF, and NXF1 bind to BR mRNPs in the interchromatin. Therefore, BR mRNPs become export competent first after entering the interchromatin space.
Results
Localization of the EJC core
Specific antibodies (Fig. S1, A and C) located the EJC core in diploid cells (Fig. 1). eIF4AIII, Y14, and Mago had a widespread distribution in the nucleus and cytoplasm. Btz was predominantly cytoplasmic, although significant labeling was also detected in the nucleus.
Figure 1.
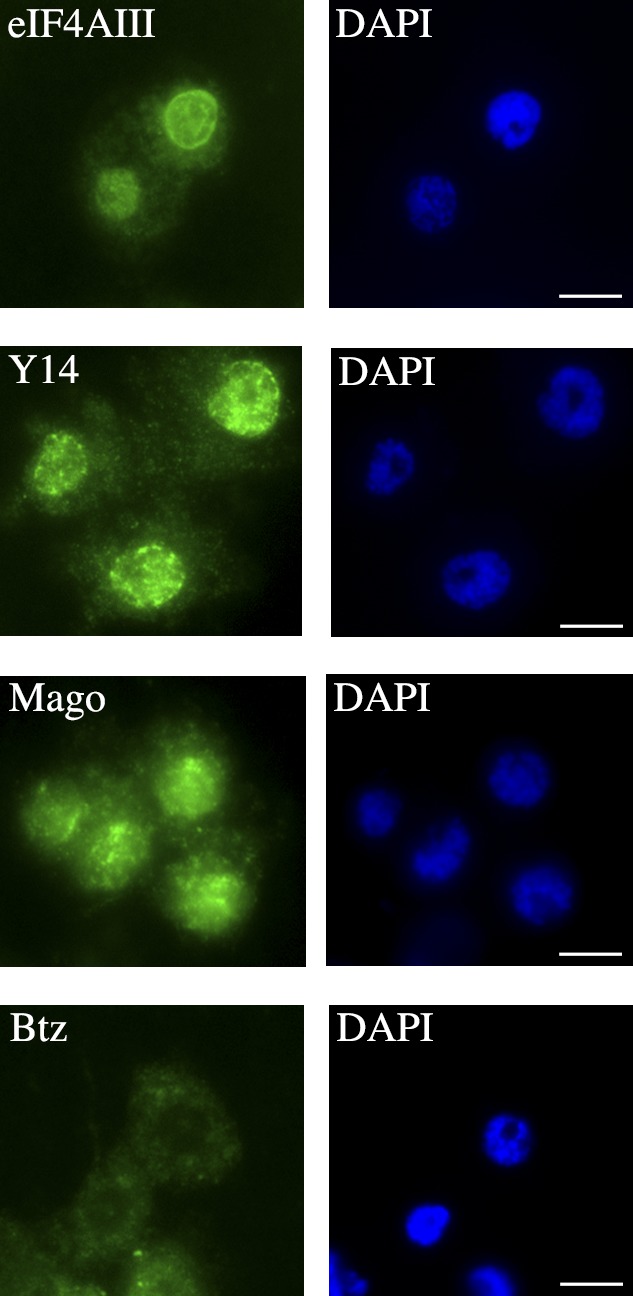
Localization of EJC core proteins in C. tentans diploid cells. Cells stained with anti-eIF4AIII, anti-Y14, anti-Mago, or anti-Btz antibodies (left). DAPI staining (right). Bars, 2 µm.
Immunostaining of sections of polytene cells provided a more detailed localization (Fig. S2). In these nuclei, the interchromatin forms large volumes surrounding the well-defined polytene chromosomes. eIF4AIII, Y14, and Mago were present throughout the interchromatin space, but no enrichment in distinct sites could be detected. Specific chromosome bands were stained, showing that these components were present in specific gene loci. No staining could be seen in the nucleoli.
In agreement with the localization in diploid cells, Btz was weakly detected in the polytene nuclei. The localization of Btz was similar to that of the other EJC core components; however, the signal intensity suggested that in comparison with the cytoplasm, less Btz was present in the nucleus.
To simplify interpretation, isolated polytene chromosomes were separately stained with each of the four anti-EJC core component antibodies. Chromosome IV has four active BR genes, BR1 in the BR1 locus, BR2.1 and BR2.2 in the BR2 locus, and BR3 in the BR3 locus. The BR1, BR2.1, and BR2.2 genes are closely related, including exon-intron organization (Wieslander, 1994). The morphology of their premRNPs/mRNPs is also very similar (Daneholt, 2001). Fig. 2 shows that the EJC core was present throughout the active BR gene loci. Immuno-EM (see Fig. 5) showed that the premRNPs in the BR1 and BR2 gene loci were labeled for each of the four EJC core components. In agreement, RNase treatment of chromosome IV before staining with anti-eIF4AIII antibodies drastically diminished the signal in the transcribing gene loops (Fig. S3). We conclude that the EJC core associates with the BR premRNPs cotranscriptionally.
Figure 2.
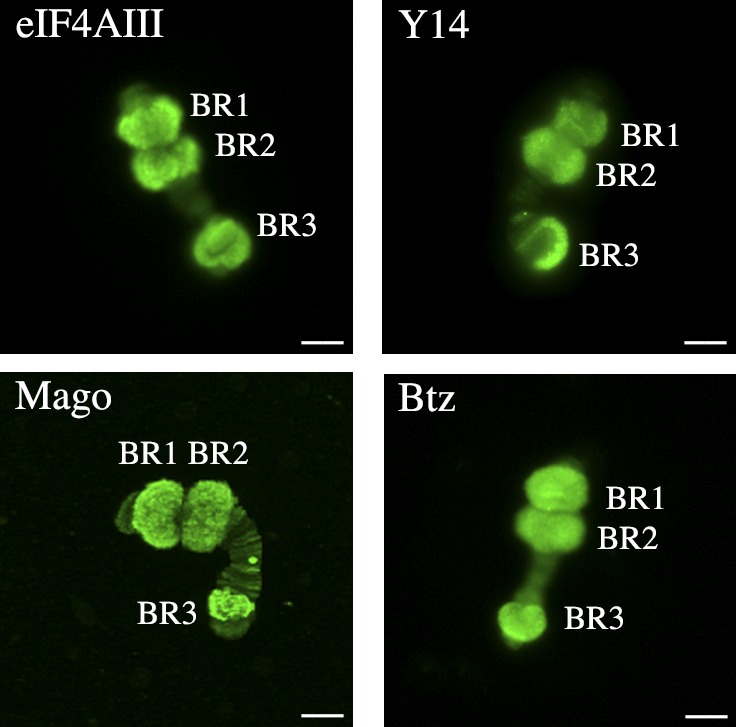
EJC core is present in BR gene loci. Chromosome IV stained with anti-eIF4AIII, anti-Y14, anti-Mago, or anti-Btz antibodies. The BR1, BR2, and BR3 loci are indicated. Bars, 10 µm.
Figure 5.
Immuno-EM analyses of EJC core in BR premRNPs. Chromosome IV was stained with anti-eIF4AIII, anti-Y14, anti-Mago, or anti-Btz antibodies, and secondary antibodies were conjugated with gold. (A) Section through part of chromosome IV with the BR1 and BR2 gene loci. (B) For orientation purposes, part of the BR1 locus is shown at a higher magnification (boxed in A). Transcribing BR1 gene loops are seen. Proximal (p), middle (m), and distal (d) segments of the active gene are indicated. (C) Schematic representation of an active BR1 or BR2 gene loop. The proximal (p), middle (m), and distal (d) segments are indicated. (D) Schematic representation of a BR1 or BR2 gene showing the four introns (i1–4). Under this, the lengths of the gene segments are shown (black lines labeled p, m, and d). The red lines indicate when spliceosome assembly and splicing take place. (E) Detailed views of immunogold-labeled BR premRNPs in the proximal, middle, and distal segments for eIF4AIII, Y14, Mago, and Btz. To the right of each image, a schematic drawing of selected immunolabeled BR premRNPs is shown. Bars: (A) 5 µm; (B and E) 100 nm.
eIF4AIII is distributed unevenly along nascent transcripts in relation to exon junctions
We analyzed the presence of eIF4AIII, as a representative of the core EJC, within premRNPs in relation to BrUTP incorporation that provided a measure of premRNA amounts. Polytene chromosomes were stained with anti-eIF4AIII and anti-BrUTP antibodies. BrUTP incorporation performed in this way does not affect splicing of BR premRNAs (Singh et al., 1999). Chromosome I provided an overview of a large number of active gene loci (Fig. 3 A). Although we cannot identify the genes, the staining pattern revealed that the relationship between BrUTP incorporation and the amount of eIF4AIII varied for different gene loci. Some loci contained relatively more eIF4AIII (Fig. 3 A, peaks 2 and 3) and others contained relatively more BrUTP (Fig. 3 A, peaks 1 and 4). Therefore, the amount of premRNA and the amount of eIF4AIII were not strictly correlated, suggesting that at many gene loci, eIF4AIII is not distributed equally along the transcripts. If eIF4AIII had been evenly distributed along the transcripts, we should have seen a closer correlation between the eIF4AIII and premRNA amounts. eIF4AIII binds preferentially at canonical sites upstream of most exon junctions, but also at other sites along an mRNA (Saulière et al., 2012; Singh et al., 2012). Because the exon-intron organization varies for different gene loci, a plausible interpretation of the staining patterns is that eIF4AIII preferentially associates with the premRNA in relation to exon junctions.
Figure 3.

eIF4AIII staining and BrUTP incorporation are not correlated. Salivary glands were incubated with BrUTP, chromosomes I (A) and IV (B) were isolated, and staining intensities for anti-eIF4AIII and anti-BrUTP antibodies were analyzed. In A, line scan analyses (right bottom panel) were performed on the merged image (along the white line, right top panel). Numbers 1–4 show selected peaks corresponding to the chromosomal loci indicated by the vertical white lines from left to right. In B, the BR1, BR2, and BR3 gene loci are indicated. Immunostaining intensities were measured in these loci and analyzed in Table 1. Bars, 10 µm. The data shown are from single representative experiments out of five repeats.
We analyzed the relationship between eIF4AIII, the amount of nascent transcripts, the exon-intron structure, and splicing more closely by analyzing the BR1, BR2, and BR3 loci, for which these parameters are known. For the BR1 and BR2 loci (Fig. 3 B), the ratio between eIF4AIII and BrUTP staining intensities was the same (Table 1), suggesting that the BR1 and BR2 premRNPs had a similar relationship between eIF4AIII binding and the amount of RNA. The BR3 locus contained considerably more eIF4AIII in relation to the amount of premRNA (Table 1). To extend our analyses to the relationship of eIF4AIII to spliceosome assembly and splicing, we analyzed the relationship between eIF4AIII and the spliceosomal U2 snRNP component U2B″, as a representative of the spliceosome. In chromosome I, we observed considerable overlap between staining for eIF4AIII and U2B″ (Fig. 4 A). There was also a variation in the staining of the gene loci, with relatively more eIF4AIII (Fig. 4 A, peaks 1 and 4) or relatively more U2B″ (Fig. 4 A, peaks 2 and 3). Therefore, in the nascent transcripts, there was not a simple relationship between the U2B″ and eIF4AIII content. In the BR1 and BR2 gene loci (Fig. 4 B), we observed similar ratios between eIF4AIII and U2B″ staining intensities (Table 2). In contrast, the eIF4AIII to U2B″ ratio was 2–3 times lower in the BR3 locus (Table 2), meaning that U2B″ was in excess of eIF4AIII in the BR3 locus compared with the BR1 and BR2 loci.
Table 1. Colocalization of eIF4AIII and nascent transcripts.
| BR gene | Ratio eIF4AIII/BrUTP |
|---|---|
| BR1 | 1.06 (0.93–1.19) |
| BR2 | 0.97 (0.77–1.22) |
| BR3 | 1.51 (1.17–1.94) |
The ratio of eIF4AIII to BrUTP staining intensities was calculated for each of the BR genes, respectively. The geometric means (BR1, BR2, and BR3 on five independently analyzed chromosomes) and the 95% confidence intervals are shown. The individual measurements are presented in Table S1. Analysis of variance on log-transformed ratios and subsequent pairwise Tukey test showed significant differences between BR1 and BR3 (PTukey = 0.02) and between BR2 and BR3 (PTukey = 0.006), but not between BR1 and BR2 (PTukey = 0.699).
Figure 4.
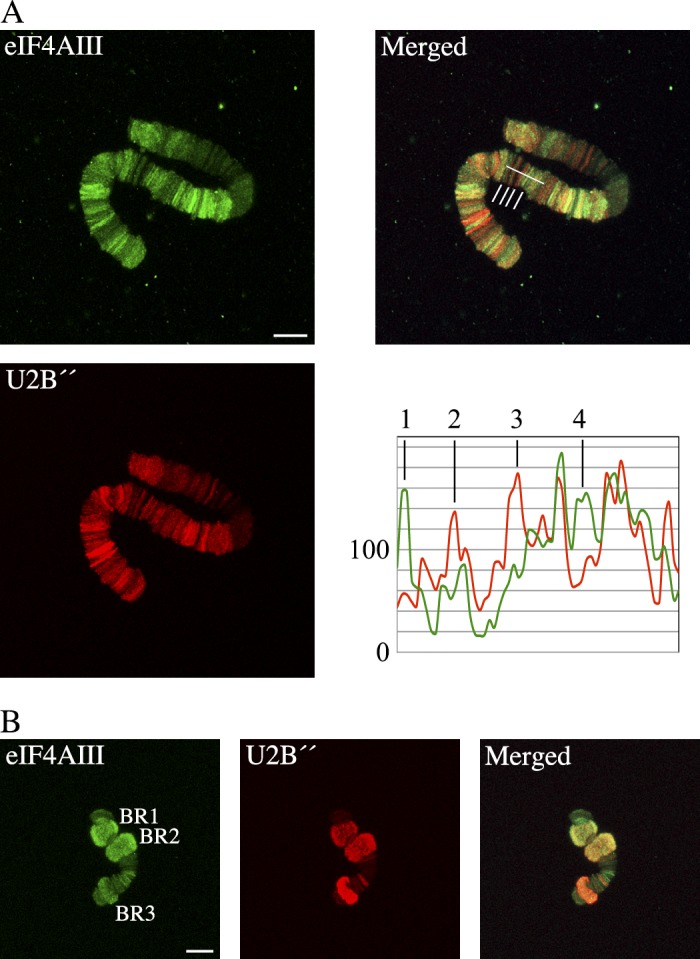
eIF4AIII and U2B″ staining intensities are not correlated. Chromosomes I and IV (A and B, respectively) were stained with anti-eIF4AIII and anti-U2B″ antibodies. (A) Line scan analyses (right bottom panel) were performed on the merged image (along the white line, right top panel). Numbers 1–4 show selected peaks corresponding to the chromosomal loci indicated by the vertical white lines from left to right. (B) The BR1, BR2, and BR3 gene loci are indicated. Immunostaining intensities were measured in these loci and analyzed in Table 2. Bars, 10 µm. The data shown are from single representative experiments out of three/four repeats.
Table 2. Colocalization of eIF4AIII and U2B″.
| BR gene | Ratio of eIF4AIII/U2B″ |
|---|---|
| BR1 | 1.53 (0.54–4.28) |
| BR2 | 1.21 (0.69–2.11) |
| BR3 | 0.51 (0.41–0.64) |
The ratio of eIF4AIII to U2B″ staining intensities was calculated for each of the BR genes, respectively. The geometric means (three/four independent experiments) and the 95% confidence intervals are shown. The individual measurements are presented in Table S2. Analysis of variance on log-transformed ratios and subsequent pairwise Tukey test showed significant differences between BR1 and BR3 (PTukey = 0.001) and between BR2 and BR3 (PTukey = 0.001), but not between BR1 and BR2 (PTukey = 0.145).
To interpret the differences in staining intensities for eIF4AIII, BrUTP, and U2B″ in the BR3 versus the BR1 and BR2 gene loci, it is necessary to consider the amount of nascent transcripts, the number of introns, and the cotranscriptional splicing characteristics for the three gene loci. These characteristics have been outlined in previous studies (Baurén and Wieslander, 1994; Baurén et al., 1998; Wetterberg et al., 2001). At a given moment, a BR1 or BR2 gene contains ∼100 nascent transcripts, and this population contains ∼65 introns and 240 exon junctions. The BR3 gene has ∼25 nascent transcripts, and this population contains ∼250 introns and 250 exon junctions. The BR3 locus has ∼1/5 of the amount of RNA compared with a BR1 or BR2 locus (Edström et al., 1978b). The lower amount of premRNA in the BR3 locus contains as many exon junctions and more introns as a larger amount of premRNA in a BR1 or BR2 locus, explaining the differences in staining intensity ratios for these gene loci (higher eIF4AIII to BrUTP staining intensitiy in BR3).
The higher staining intensity for U2B″ relative to eIF4AIII in the BR3 locus compared with the BR1 and BR2 genes is likely explained by the differences in splicing characteristics and exon junctions. Exon junctions outnumber ongoing splicing events in the BR1 and BR2 genes, whereas they are approximately equal in the BR3 locus. U2B″ is present in all BR3 nascent transcripts (Wetterberg et al., 2001), but it is present only in a minority of the BR1 and BR2 transcripts (Kiseleva et al., 1994). This agrees with a higher U2B″/eIF4AIII ratio in the BR3 locus, even if eIF4AIII is present during late splicing steps. The BR3 versus BR1 and BR2 staining results therefore show that eIF4AIII binding to premRNAs is dependent on the exon-intron structure and the splicing characteristics of the specific premRNAs. Because mRNPs are formed and released from the gene loci, the presence and locations of exon junctions will determine the extent and location of eIF4AIII binding to the mRNP.
Cotranscriptional assembly of the EJC core on BR premRNPs in vivo
Immuno-EM revealed the distribution of the EJC core within BR1 and BR2 premRNPs along the active genes. The morphology of the BR1 and BR2 premRNPs is characteristic and changes reproducibly along the transcribing genes, largely as a result of the addition of more RNA, recruitment of proteins, and assembly into premRNPs. It is therefore possible to determine the position along the gene for each individual premRNP, including the ones labeled with gold conjugated antibody. According to the morphology of the premRNPs, the transcribing gene is divided into three segments, proximal, middle, and distal segments, representing ∼20%, 60%, and 20% of the gene, respectively (see Materials and methods; Fig. 5). Each of the four anti-EJC core component antibodies stained the premRNPs along the genes, and the distributions are analyzed in Table 3 and Table S3 (A and B). The data have been normalized to represent gold particle/premRNP (see Materials and methods). Most labeling was detected in the proximal segment. The labeling intensity then decreased in the middle segment and increased again in the distal segment. Provided that the detectability of the epitopes was not different in the premRNPs present in the different segments, this shows that there is a higher concentration of core components in the premRNPs located in the proximal segment. Here, three introns are incorporated into the premRNA and subsequently removed before transcription has reached very far into the middle segment. In the middle segment, we recorded a significantly lower signal, showing that core components are not added continuously during synthesis of the premRNA. The distribution also suggests that less EJC core components are present in the premRNPs after splicing is completed, compared with ongoing splicing. Therefore, we hypothesize that a high concentration of EJC core components is present in the proximal segment when splicing is going on, but not all of these components associate stably with the premRNPs. In the distal segment, a fourth intron is present, and this is excised to a considerable extent at the gene locus (Baurén and Wieslander, 1994; Baurén et al., 1998). Increased labeling was recorded here, indicating that additional EJC core components become associated with the premRNP at this stage.
Table 3. Distribution of EJC core proteins in the proximal, middle, and distal segments of the BR1 and BR2 genes.
| Protein | Ratio proximal/middle | Ratio distal/middle |
|---|---|---|
| eIF4AIII | 9.37 (8.39–10.50) | 2.85 (2.48–3.27) |
| Y14 | 8.53 (7.63–9.54) | 2.87 (2.49–3.30) |
| Mago | 7.90 (7.09–8.79) | 2.63 (2.30–3.01) |
| Btz | 4.04 (3.48–4.69) | 2.65 (2.25–3.13) |
Gold particle counts, pooled over four experiments, were analyed using a generalized linear model with Poisson distribution and a log link. Offsets were used and correspond to the estimated segment proportion in the experiment (∼20% proximal, 60% middle, and 20% distal). The table shows gold particles/length unit ratios for the proximal and distal segments against the middle segment, together with 95% confidence intervals. The individual measurements are presented in Table S3. The difference in gold particles/length unit ratios between proteins (eIF4AIII, Y14, and Mago compared with Btz) is highly significant (likelihood ratio test, P < 0.00005) and is caused by the lower gold particles/length unit ratio for Btz in the proximal/middle segment.
It is not previously clear when Btz associates with the EJC core on endogenous transcripts. Our data show that Btz associates cotranscriptionally and in relation to splicing. We observed a slightly different distribution with relatively less labeling in the proximal segment compared with eIF4AIII, Y14, and Mago. This may reflect a difference in association characteristics for Btz compared with the other core components.
Association of UPF2, UPF3, and NXF1 with BR mRNPs
With the aim to reach a more comprehensive view on how export competent BR mRNPs form, we analyzed when and where three additional proteins become associated with the BR mRNPs. UPF2 and UPF3 associate with mRNPs via the EJC and promote translation and mediate NMD. UPF3 binds directly to the EJC and bridges the UPF2-EJC association. When and where UPF2 and UPF3 are recruited to EJCs on endogenous transcripts are still unclear.
NXF1 is the main mRNA export factor. It is unclear when and where NXF1 associates with mRNPs; however, it does so via one of several adapters.
Specific antibodies (Fig. S1, B and C) were used to localize UPF2, UPF3, and NXF1. We located UPF2 and UPF3 both in the cytoplasm and nucleus (Fig. S4), in agreement with previous studies (Lykke-Andersen et al., 2000; Serin et al., 2001). Staining of chromosome IV did not show significant amounts of either UPF2 or UPF3 in the BR gene loci (Fig. S5), showing that these proteins do not associate with the BR premRNPs. Immuno-EM confirmed that UPF2 and UPF3 did not bind to BR premRNPs cotranscriptionally (Fig. 6, E and F). Both proteins were however present in the interchromatin. This was supported by biochemical analyses of nuclear extracts (Fig. S4 B). We found that throughout the interchromatin, UPF2 and UPF3 are bound to ∼10% of the BR mRNPs. UPF2 and UPF3 association with BR mRNPs was enriched at the nuclear membrane, when the BR mRNPs were located in close proximity to the NPCs (Fig. 6 and Table 4).
Figure 6.
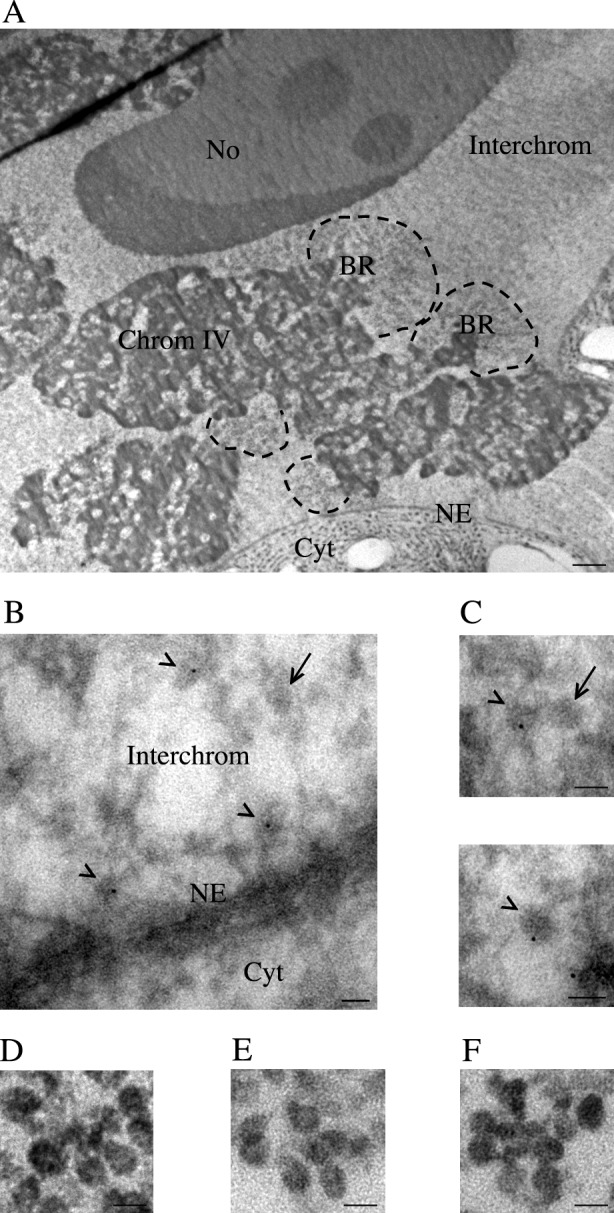
Immuno-EM analyses of NXF1, UPF2, and UPF3 in BR mRNPs in the interchromatin. (A) Section through a polytene cell. Part of the cytoplasm (Cyt) and the nuclear membrane (NE) are visible. The interchromatin (Interchrom) accounts for large volumes devoid of chromatin. Part of chromosome IV (Chrom IV) with its BR genes (BR) is seen. In the BR loci, the volumes occupied by transcriptionally active gene loops are delimited by broken lines. No., nucleolus. (B) BR mRNPs in the interchromatin. A BR mRNP containing NXF1 (gold particle) is marked (arrowhead). A BR mRNP, not labeled by the anti-NXF1 antibodies, is also observed (arrow). Parts of the cytoplasm (Cyt) and nuclear membrane (NE) are visible. Two docked BR mRNPs contain NXF1 (gold particles, arrowheads). (C) Three BR mRNPs in the interchromatin are detected. (top) Anti-UPF3 antibodies label one out of two BR mRNPs (gold particle, arrowhead). Unlabeled BR mRNP are indicated (arrow). (bottom) Anti-UPF2 antibodies label a BR mRNP (gold particle, arrowhead). (D) Anti-NXF1 antibodies do no stain BR premRNPs. (E) Anti-UPF2 antibodies do not stain BR mRNPs. (F) Anti-UPF3 antibodies do not stain BR mRNPs. Bars: (A) 2.5 µm; (B–F) 50 nm.
Table 4. Immunogold labeling of EJC-associated proteins in the BR mRNPs in the interchromatin and docked at the NPC.
| Protein | Labeled BR mRNPs: docked at NPC/interchromatin |
|---|---|
| NXF1 | 4.15 (2.52–6.81) |
| UPF2 | 2.56 (1.36–4.82) |
| UPF3 | 3.60 (2.25–5.76) |
Data, pooled over three experiments, were analyzed using a generalized linear model with binomial distribution and a logit link. The relative proportions, together with 95% confidence intervals, are shown. The individual measurements are presented in Table S4. There is no significant difference in relative proportion over proteins (likelihood ratio test, P = 0.38). In addition, we performed Fischer exact test for NXF1, UPF2, and UPF3 separately with pooled data for the three experiments to test the hypothesis that the percentage labeled BR mRNPs was the same in the interchromatin and docked at the NPCs. The hypothesis was rejected: NXF1, P < 0.0001; UPF2, P = 0.008; UPF3, P < 0.0001.
We conclude that UPF2 and UPF3 bind to BR mRNPs in the nucleus, most likely in the interchromatin.
Immunostaining of diploid C. tentans cells (Fig. S4 A) and Western blots of nuclear and cytoplasmic extracts (Fig. S4 B) showed that NXF1 was mainly present in the nucleus, with a substantial amount also in the cytoplasm. Frequently, a higher concentration of NXF1 was detected at the nuclear periphery. Staining of chromosome IV revealed no or very little NXF1 (Fig. S5). NXF1 could not be detected in any other gene loci in the polytene chromosomes (unpublished data). In agreement, immuno-EM did not show any significant labeling for NXF1 in BR premRNPs (Fig. 6 D). Analysis of chromatin-associated mRNPs and mRNPs in interchromatin agreed with these data (Fig. S4 B). It therefore appears that NXF1 does not associate with the BR premRNPs during transcription.
According to quantification of the immunogold labeling, NXF1 is spread relatively even throughout the interchromatin space. We could not determine if the labeling detected free NXF1 or NXF1 bound to various mRNPs (only BR mRNPs were identifiable in our experiment). We assume that we detected both. However, the concentration of NXF1 was approximately four times higher in a 50-nm zone close to the nuclear membrane (unpublished data). This is in agreement with staining of diploid C. tentans cells (Fig. S4 A). Examination of the nuclear membrane and 50-nm wide zones at the nuclear and cytoplasmic sides, respectively, showed that NXF1 was present at higher concentrations at both sides of the membrane compared with inside the NPC channels (Table 5). The same distribution was observed for UPF2 and UPF3 (Table 5). This situation may reflect a rapid translocation of NXF1, UPF2, and UPF3 through the NPCs, free or bound to mRNPs, and relative delayed residence times at both sides of the NPCs.
Table 5. Distribution of immunogold labeling of EJC-associated proteins at the nuclear membrane: nuclear side, middle, and cytoplasmic side.
| Protein | Ratio nuclear/middle | Ratio cytoplasmic/middle |
|---|---|---|
| NXF1 | 3.89 (1.87–8.09) | 3.89 (1.87–8.09) |
| UPF2 | 3.71 (1.61–8.56) | 3.43 (1.48–7.69) |
| UPF3 | 3.08 (1.65–5.75) | 2.08 (1.07–4.03) |
Gold particle counts, pooled over three experiments, were analyzed using a generalized linear model with Poisson distribution and a log link. The gold particle distribution ratios for the nuclear and cytoplasmic sides against the middle, together with 95% confidence intervals, are shown. The individual measurements are presented in Table S5. There is no significant difference in the gold particles/area unit ratios between proteins (likelihood ratio test, P < 0.68).
Immuno-EM demonstrated that BR mRNPs in the interchromatin contained NXF1 (Fig. 6 B). We analyzed different sites within the interchromatin, including sites far from the BR genes and sites at different distances from the nuclear membrane. In all sites, ∼10% of the BR mRNPs contained NXF1. Only one location differed. In a 50-nm zone close to the nuclear membrane, ∼40% of the BR mRNPs contained NXF1 (Fig. 6 B and Table 4). Most of these BR mRNPs appeared to be docked at the basket of the NPCs.
Discussion
BR premRNPs are equipped with core EJCs
In C. tentans, eIF4AIII binds to premRNAs at a large number of endogenous genes, in agreement with the presence of eIF4AIII at transcription sites in mammalian cells (Custódio et al., 2004). All four EJC core components bind to the nascent BR1 and BR2 gene transcripts in vivo. The amount of eIF4AIII associated with BR premRNAs was correlated to the number of introns and exon junctions and not to the amount of premRNA (Figs. 3 and 4). Therefore, eIF4AIII is not evenly positioned along the premRNAs. Instead, the finding suggests that eIF4AIII is mainly bound to the nascent RNA close to exon junctions, as observed in large-scale studies of eIF4AIII binding sites on mRNAs (Saulière et al., 2012; Singh et al., 2012). We show that binding of the EJC core to the BR premRNAs occurs in relation to spliceosome formation and intron excision (Baurén and Wieslander, 1994; Wetterberg et al., 1996), in agreement with in vitro data showing that EJC formation is coupled to splicing.
The EJC core is enriched in the BR premRNPs where and when splicing takes place, suggesting that an excess of EJC core components is present during splicing before a stable EJC is formed at the exon junction. This finding may reflect that several splicing-related factors attract EJC components, possibly to ensure efficient, splicing-dependent delivery. It has also been reported that Y14 and Mago associate with nascent transcripts with a high on and off rate (Schmidt et al., 2009), which could contribute to such a situation.
It was so far unclear when Btz is recruited to the EJC core. We demonstrate that Btz binds to BR premRNPs, but possibly slightly later than the other EJC core components. In any case, our data show that all four EJC core components are associated with the BR premRNAs, a prerequisite for cotranscriptional interaction with each other as an EJC core complex.
EJC formation may be important for compaction of the BR premRNPs
The BR1 and BR2 premRNPs distinctly change morphology ∼8 kb from the transcription start site, corresponding to the beginning of the middle gene segment. The 5′ proximal part of the premRNP then folds into a globular structure (Daneholt, 2001), most likely as a result of a new type of packaging of the premRNP fiber. This morphological change occurs approximately when the 5′ located introns have been excised (Baurén and Wieslander, 1994), and according to our present results, when EJCs have assembled at the formed exon junctions. EJCs interact with SR proteins resulting in mRNP compaction (Singh et al., 2012). The BR premRNPs contain several different SR proteins (Björk et al., 2009), and if EJCs form at a relatively specific time after transcription initiation, such an EJC–SR interaction can be involved in the restructuring and compaction of the BR premRNPs.
UPF2 and UPF3 bind to BR mRNPs in the interchromatin
The EJC is important for translation and for binding the effectors of NMD: UPF1, UPF2, and UPF3 (Mühlemann et al., 2008). UPF3 binds to a surface formed by eIF4AIII, Mago, and Y14 (Buchwald et al., 2010). Previous data suggest binding of UPF3 to the EJC core in the nucleus (Lykke-Andersen et al., 2000; Serin et al., 2001). We show that UPF3 is associated with a fraction of the BR mRNPs in the interchromatin, but not with BR premRNPs. As for NXF1 (seeFig. 6), BR mRNPs containing UPF3 are enriched at the NPCs, presumably because the BR mRNPs that will be exported are retained at the basket of the NPC.
UPF2 is believed to associate with UPF3 at the cytoplasmic side of the nuclear membrane (Lykke-Andersen et al., 2000; Serin et al., 2001). In the interchromatin, as many BR mRNPs contain UPF2 as UPF3, arguing that UPF2 can bind to the EJC in the nucleus and that this takes place rapidly after binding of UPF3.
BR mRNPs emerge into the cytoplasm with their 5′ end leading the way (Mehlin et al., 1995). Preliminary immuno-EM analyses suggest that both UPF2 and UPF3 are associated with the 5′ end of the BR mRNPs during translocation through the NPCs (unpublished data). This would be compatible with EJCs binding just upstream the exon junctions at the 5′ end of the BR mRNAs (Fig. 5 and Table 3). The BR mRNPs rapidly bind eIF4H in the cytoplasmic perinuclear region (Björk et al., 2003), and according to morphological observations, ribosomes can rapidly initiate translation there (Mehlin et al., 1992). Because UPF3 and UPF2 are likely to be part of the BR mRNPs during translocation, NMD involving BR mRNPs could take place in the cytoplasmic perinuclear region.
NXF1 and BR mRNP export competence
It is unclear when and where the export factor NXF1 binds to mRNPs. In mammalian cells, NXF1 was not detected at transcription sites (Custódio et al., 2004). In HeLa cells, NXF1 is present diffusely throughout the nucleus, and interaction with Aly/REF, an NXF1 adapter, was detected in the interchromatin outside speckles (Teng and Wilson, 2013). In our study, we can directly visualize the presence of NXF1 in specific endogenous premRNPs/mRNPs. NXF1 does not bind to BR premRNPs (at least below our detection level). NXF1 is bound to a fraction of BR mRNPs throughout the interchromatin and to a four times larger fraction of BR mRNPs docking at the NPCs. Preliminary immuno-EM observations suggest that NXF1 is associated with the 5′ end of the BR mRNPs during translocation through the NPCs (unpublished data). This is compatible with the hypothesis that NXF1 binds to the 5′ end of mRNPs in a CBP80-dependent way (Cheng et al., 2006). A detailed analysis of NXF1 binding during BR mRNP translocation through NPCs remains to be performed.
Several of the identified NXF1 adapters associate with BR premRNPs already at the gene. The BR mRNPs have a cap binding complex (Visa et al., 1996), they are spliced (Baurén and Wieslander, 1994), they are polyadenylated (Baurén et al., 1998) and equipped with Aly/REF (Yra1; Kiesler et al., 2002), they have SR proteins (Björk et al., 2009), and they have the EJC core (this study) at the gene. In spite of this, no NXF1 could be detected in BR premRNPs. The BR mRNPs are therefore not fully export competent as they leave the gene. Because only a fraction of the BR mRNPs in the interchromatin contained NXF1, it is unlikely that the presence of NXF1 is required for release of the BR mRNPs from their genes.
After synthesis, the BR mRNPs move away from the BR genes in all directions, forming a nuclear pool of BR mRNPs (Singh et al., 1999; Fig. 7). The mobility of BR mRNPs is complex, and a single diffusion coefficient does not exist. Instead, the BR mRNPs repeatedly and transiently interact with nonchromatin structures (Miralles et al., 2000; Veith et al., 2010). As the BR mRNPs move around, they can contact the NPCs, but only ∼25% of these NPC interactions result in export (Siebrasse et al., 2012). BR mRNPs therefore often return into the interchromatin. Nonproductive interactions between mRNPs and NPCs have also been observed in diploid nuclei (Ma et al., 2013). When export of BR mRNPs occurred, a rate-limiting step was observed at the basket of the NPC.
Figure 7.
Model of BR mRNP acquisition of EJC core, NXF1, UPF2, and UPF3 in relation to transcription, processing, and export. (a) At the BR gene (gray line), premRNPs (light blue) are assembled, and the premRNAs are largely spliced. In connection with splicing, the EJC core (purple circles) becomes associated with the premRNP. The EJC core remains associated with the mRNP in the interchromatin and during export. (b) New BR mRNPs (light blue) are released into the interchromatin and move in all directions. (c) They become part of an interchromatin pool of BR mRNPs. In the interchromatin, the BR mRNPs bind NXF1, UPF2, and UPF3 (yellow and orange crosses). At a given moment 10–25% of the BR mRNPs contain these proteins. The pool contains old BR mRNPs (dark blue, boxed by dark blue line) and new BR mRNPs (light blue, boxed by light blue line). NXF1, UPF2, and UPF3 bind to both old and new BR mRNPs. Some BR mRNPs have recruited the proteins at an earlier time point (orange crosses), and some BR mRNPs have just recruited the proteins (yellow crosses). In steady state, the pool receives newly synthesized BR mRNPs and loses an approximately equal number through export to the cytoplasm. Both new and old BR mRNPs are exported if they contain NXF1, UPF, and UPF3 (boxed by black line). (d) BR mRNPs that are exported are a mixture of those that have contained the proteins for some time and those that have just obtained them. (e) BR mRNPs that do not yet contain NXF1, UPF2, and UPF3 can diffuse and interact with the basket of the NPC, but they are rejected and return into the interchromatin.
We found two BR mRNP populations in the interchromatin, one containing NXF1 (10%) and the other without NXF1. Both are likely to encounter NPCs. Because interactions between BR mRNPs and NPCs can lead both to export and nonexport (Siebrasse et al., 2012), it is possible that BR mRNPs that cannot be exported do not yet contain NXF1.
We do not know the efficiency of detection in the immuno-EM experiments, but the measured 10% is likely a minimum value. Of the BR mRNPs, 40% were docked at the NPCs contained NXF1. These BR mRNPs initially have the same morphology as in the interchromatin. This detection of 40% indicates that the 10% value for BR mRNPs in the interchromatin does not represent the maximum methodological detection level. We therefore propose that the fraction of BR mRNPs that contain NXF1 in the interchromatin is considerably smaller than the fraction lacking NXF1.
Our results do not conclusively demonstrate where NXF1 binds to BR mRNPs. One possibility is that NXF1 binds to BR mRNPs at the NPCs. If so, a considerable number of BR mRNPs with NXF1 must then be released from the NPCs and diffused into the interchromatin to explain the pool of NXF1 containing BR mRNPs there. In view of the finding that many BR mRNPs interact with the NPCs transiently (Siebrasse et al., 2012), this possibility cannot be ruled out.
An alternative, and more plausible possibility, is that NXF1 binds to the BR mRNPs at many locations throughout the interchromatin. This model is consistent with our finding that throughout the interchromatin space, approximately the same fraction of BR mRNPs contains NXF1. The recorded higher fraction of NXF1 containing BR mRNPs at the NPCs then reflects enrichment there. Because a rate-limiting step takes place at the NPC basket (Siebrasse et al., 2012), an enrichment of export competent BR mRNPs there is expected. In both models, export competence in terms of acquisition of NXF1 is not obtained until the BR mRNPs have moved around in the interchromatin.
Why is NXF1 binding to BR mRNPs a late step in acquiring export competence? One possibility is that some so far unknown factor is required for NXF1 binding and that this factor may be located in the interchromatin. A second possibility is that structural reorganizations in the mRNPs, involving the adapters, have to take place. This may only occur in fully processed mRNPs equipped with all the processing associated factors and therefore only after exit from the gene.
The events for BR premRNPs/mRNPs are identified in the polytene nuclei because of their large volumes of interchromatin devoid of chromatin and the presence of morphologically identifiable BR mRNPs. The lack of defined and identifiable mRNPs in diploid nuclei and their more complex chromatin/interchromatin organization make it more difficult to quantitatively analyze the events in these cells. We propose that a similar scenario excist in diploid nuclei. However, the different chromatin/interchromatin organization in these cells may very well influence the characteristics of the events.
Materials and methods
Animals and cells
C. tentans was cultured as described previously (Meyer et al., 1983). A C. tentans embryonic epithelial cell line was cultured as described previously (Wyss, 1982).
Identification of C. tentans proteins
The C. tentans and Drosophila melanogaster proteins represent orthologues according to extensive analyses (Kutsenko et al., 2014). The EMBOSS Needle software was used (European Bioinformatics Institute).
eIF4AIII, 92.4% identity, 94,9% similarity; Y14, 76.5% identity, 89,2% similarity; Mago, 85.9% identity, 91.9% similarity; Btz (PA), 30.0% identity, 38.8% similarity; NXF1 (small bristles), 41.7% identity, 58.2% similarity; UPF2, 49.1%, identity, 65.6% similarity; and UPF3, 26.8% identity, 43.2% similarity (see Table S6).
For some of the proteins, antibodies were raised against specific peptides present in the C. tentans proteins. In these cases, the peptide sequences are underlined.
Cloning and expression of C. tentans proteins
C. tentans Y14 cDNA was isolated by RT-PCR using degenerate primers corresponding to sequences in the homologous gene in D. melanogaster. Poly(A) + RNA was isolated from C. tentans epithelial tissue culture cells and reverse transcribed using oligo dT priming. The degenerate primers were used for PCR. The obtained fragment was sequenced. The full-length cDNA sequence was isolated from a C. tentans lambda Zap cDNA library. This library was made using the Zap-cDNA Synthesis kit (Agilent Technologies) and the Uni-Zap XR Cloning kit (Agilent Technologies). Programs in the Genetics Computer Group package (Devereux et al., 1984) and the Biology Workbench package (San Diego Supercomputer Center) were used to analyze DNA and protein sequences. Y14 was expressed as a His-tagged fusion protein from the vector pET15B (Novagen) in BL21 bacteria (Agilent Technologies) and purified on Ni-NTA (QIAGEN).
The protein coding sequences of C. tentans eIF4AIII, Btz, NXF1, and UPF3 were obtained by PCR using cDNA synthesized from mRNA as template and cloned into the expression vector pET-46Ek/LIC (Novagen). Proteins were expressed in BL21 bacteria and purified on Ni-NTA Agarose (QIAGEN).
Antibodies
Polyclonal antibodies against Y14, Mago, NXF1, UPF2, and UPF3 were raised in rabbits (Agrisera). The cDNA encoding Y14 was cloned and expressed in Escherichia coli. The purified Y14 was immunized into rabbits. Synthetic peptides spanning amino acids (in all cases referring to the C. tentans proteins) 73–88 (CMAEDDSLWPPADRVGR) of Mago, amino acids 43–56 (CKGRFRNRKGSPIPN) of NXF1, amino acids 1092–1105 (CKVQKFKHQRGVPEI) of UPF2, and amino acids 177–190 (CNKDRPTIQIYRPKR) of UPF3 were conjugated to keyhole limpet hemocyanin and immunized into rabbits. All antibodies were purified by affinity chromatography. The D. melanogaster anti-Dm-eIF4AIII polyclonal rabbit antibodies (Palacios et al., 2004) and the anti-Dm-Barentsz polyclonal rabbit antibodies (van Eeden et al., 2001) were a gift from I. Palacios (University of Cambridge, Cambridge, UK). The antibodies specifically detected single proteins of the expected relative molecular mass in Western blots of C. tentans cell extracts (Fig. S1).
Monoclonal mouse antibodies against hrp45 and hrp36 were a gift from B. Daneholt (Karolinska Institutet, Stockholm, Sweden). The PKC rabbit polyclonal antibodies, the anti-BrUTP mouse monoclonal antibody, the anti-His (H1029) mouse monoclonal antibody, and the anti-U2B″ protein mouse monoclonal antibody were purchased (Santa Cruz Biotechnology Inc., Roche, Sigma-Aldrich, and ICN/Cappel, respectively). The secondary antibodies used for Western blots were swine antirabbit Ig HRP and goat antimouse Ig HRP (Dako) diluted 1:3,000. The secondary antibodies used in immunofluorescence were swine antirabbit Ig FITC (Dako) diluted 1:100 and goat antimouse IgG Texas red (Jackson ImmunoResearch Laboratories Inc.) diluted 1:100. The secondary antibodies used in immuno-EM were goat antirabbit IgG conjugated with 6-nm colloidal gold (Jackson ImmunoResearch Laboratories Inc).
Protein preparation and Western blotting
C. tentans epithelial tissue culture cells were boiled in SDS-PAGE sample buffer. Nuclear and cytoplasmic extracts were prepared essentially as previously described (Wurtz et al., 1996). The protein samples were separated on 12% SDS–polyacrylamide gels and transferred to Immobilon-P polyvinylidene filters (EMD Millipore) by semidry electrophoresis. HRP-labeled secondary antibodies were detected by the enhanced chemiluminescence method (GE Healthcare).
Immunofluorescence analyses
Cultured C. tentans diploid cells were prepared and immunostained essentially as previously described (Baurén et al., 1996), with the modification of using 0.2% Triton X-100 instead of SDS. Polytene chromosomes were isolated from C. tentans salivary gland cells and processed for immunofluorescence, and sections of polytene nuclei were prepared and immunostained, as described previously (Björk and Wieslander, 2009). For BrUTP incorporation, C. tentans salivary glands were incubated in hemolymph containing 4 mM BrUTP for 45 min at room temperature before isolation of chromosomes.
Immuno-EM on isolated chromosomes and cryosections
Polytene chromosomes were isolated from C. tentans salivary gland cells and processed for EM as described previously (Sjölinder et al., 2005; Björk and Wieslander, 2009). In brief, the glands were fixed in 2% paraformaldehyde in 10 mM triethanolamine-HCl, pH 7.0, 100 mM KCl, and 1 mM MgCl2 (TKM), and polytene chromosomes were isolated by pipetting. Individual polytene chromosomes were fixed in 4% paraformaldehyde in TKM. The chromosomes were treated with 2% BSA and incubated with antibodies diluted in TKM containing 0.5% BSA. After washing, the chromosomes were fixed in TKM containing 2% glutaraldehyde, dehydrated in ethanol, and finally embedded in R1031 Agar 100 resin (Agar Scientific). The chromosomes were sectioned and placed on EM grids. For increased contrast, the sectiones were incubated in 2% UAc in 50% ethanol.
Classification of BR gene segments according to BR premRNP morphology
In sections through active BR1 and BR2 gene loci, proximal, middle, and distal segments of these BR genes can be classified based on the morphology of the premRNPs (Fig. 5; Daneholt et al., 1982; Daneholt, 2001). In the proximal segment, the premRNPs form fiber-like structures lacking distinct granules at their ends. In the middle segment, the premRNPs have stalks with distinct granules at their ends. The granules increase in size along the segment, reaching a diameter of 35 nm. In the distal segment, the granules grow until they reach a size of 50 nm, and the premRNPs are dominated by these more compact granular structures. In reconstructions of complete BR gene loops from serial sections, the proximal, middle, and distal segments constitute 20%, 60%, and 20% of the gene, respectively. These morphological criteria are reproducible. For each immuno-EM experiment, we calculated the area occupied by the three gene segments based on the morphology of the BR premRNPs. In all experiments, the three gene segments amounted to the 20% (proximal), 60% (middle), and 20% (distal) proportions with very little variation (Table S3 B).
Sections of polytene chromosomes were immunostained for each of the four components of the EJC core. The distribution of each analyzed protein along the BR1 and BR2 genes was determined by assigning each individual immunogold particle to one of the three gene segments, identified by the morphology of the BR premRNPs. Four independent experiments were performed for each antibody. The number of counted gold particles was 2097 for eIF4AIII, 973 for Btz, 1957 for Y14, and 2132 for Mago. The distribution of gold labeling for each antibody was evaluated statistically (see legend to Table 3).
The distribution of the EJC-associated proteins in BR mRNPs, in the interchromatin, and at the nuclear membrane, was analyzed by immunostaining of cryosections of polytene nuclei. A BR mRNP was considered labeled when an immunogold particle was on top of or within 20 nm from a BR mRNP (estimated length of primary plus secondary antibody). The ratio of immunogold-labeled BR mRNPs to unlabeled BR mRNPs, was calculated in the interchromatin and at the nuclear membrane. The total number of counted gold particles labeling BR mRNPs in the interchromatin was 44 for NXF1, 35 for UPF2, and 65 for UPF3. Three independent experiments were performed for each antibody. At the nuclear membrane, the ratio of immunogold-labeled BR mRNPs docked at the NPCs (within 50 nm from the nuclear membrane and morphologically at the position of an NPC) to docked unlabeled BR mRNPs was calculated. The total number of labeled/unlabeled docked BR mRNPs was 14/36 for NXF1, 10/46 for UPF2, and 14/36 for UPF3. Three independent experiments were performed for each antibody. Statistical analyses were performed in all cases (see legend to Table 4). Background labeling was checked after staining with only secondary antibody and was found to be essentially zero.
The number of BR mRNPs synthesized per time unit was calculated based on the measured transcription time of 20 min for a BR1 or BR2 gene (Egyházi, 1975; Edström et al., 1978a), the number of premRNPs (100) simultaneously present on an active BR1 or BR2 gene (Lamb and Daneholt, 1979), and the number of BR1 or BR2 genes (8,000) in a polytene chromosome IV.
Microscopy
Preparations for immunofluorescence analyses were mounted in Vectashield mounting medium (Vector Laboratories). The specimens were viewed in either an Axioplan 2 microscope equipped with a Hamamatsu ORCA-ER camera or an LSM 510 META confocal microscope system equipped with an Axiovert 200M microscope (Carl Zeiss). Images were collected using either a 40x oil immersion 1.3 NA Plan-NeoFluar lens, at room temperature, and the AxioVision 4.6 software or a 40x oil immersion 1.0 NA Plan-Apochromat lens, at room temperature, and the LSM 510 software (Carl Zeiss). Staining intensities were analyzed using ImageJ software (National Institutes of Health).
EM preparations were viewed and photographed at 80 kV in a 120-kV electron microscope (Tecnai; FEI) using a charged-coupled device camera (1000P; Gatan) and the DigitalMicrograph acquisition software (Gatan). Images were processed in Photoshop version CS4 (Adobe).
Online supplemental material
Fig. S1 shows Western blot analyses of C. tentans cell extracts and E. coli expressed proteins probed with the indicated antibodies. Fig. S2 shows localization of the four EJC core components in sections of polytene C. tentans cells. Fig. S3 shows immunostaining of chromosome IV for eIF4AIII after RNase treatment. Fig. S4 A shows the location of NXF1, UPF2, and UPF3 in diploid C. tentans cells. Fig. S4 B shows Western blot analyses of NXF1, UPF2, and UPF3 in nuclear and cytoplasmic extracts. PKC and hrp45 are markers for the cytoplasm and nucleus, respectively. It also shows presence of indicated proteins in interchromatin and proteins associated with chromatin. Fig. S5 shows immunostaining of isolated chromosome IV with antibodies against NXF1, UPF2, and UPF3. Staining for hrp36 was a positive control, and staining with only the secondary antibody was a negative control. Table S1 shows colocalization of eIF4AIII and nascent transcripts. Table S2 shows colocalization of eIF4AIII and U2B″. Table S3 shows distributions of EJC core proteins in the proximal, middle, and distal segments of the BR1 and BR2 genes. Table S4 shows immunogold labeling of proteins associated with BR mRNPs in the interchromatin and docked at the NPC. Table S5 shows distribution of immunogold particles at the nuclear membrane: nuclear side, middle, and cytoplasmic side. Table S6 shows alignments of C. tentans and D. melanogaster proteins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412017/DC1.
Supplementary Material
Acknowledgments
We thank I. Palacios and B. Daneholt for gifts of antibodies. We are grateful to N. Visa for valuable comments.
The Swedish Research Council supported this study. The Imaging Facility at Stockholm University is acknowledged.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BR
- Balbiani ring
- Btz
- Barentsz
- EJC
- exon junction complex
- mRNP
- messenger RNA protein complex
- NMD
- nonsense-mediated decay
- NPC
- nuclear pore complex
- premRNP
- precursor mRNP
References
- Alexandrov A., Colognori D., Shu M.D., and Steitz J.A.. 2012. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc. Natl. Acad. Sci. USA. 109:21313–21318. 10.1073/pnas.1219725110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C.B., Ballut L., Johansen J.S., Chamieh H., Nielsen K.H., Oliveira C.L., Pedersen J.S., Séraphin B., Le Hir H., and Andersen G.R.. 2006. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 313:1968–1972. 10.1126/science.1131981 [DOI] [PubMed] [Google Scholar]
- Ballut L., Marchadier B., Baguet A., Tomasetto C., Séraphin B., and Le Hir H.. 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12:861–869. 10.1038/nsmb990 [DOI] [PubMed] [Google Scholar]
- Barbosa I., Haque N., Fiorini F., Barrandon C., Tomasetto C., Blanchette M., and Le Hir H.. 2012. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol. 19:983–990. 10.1038/nsmb.2380 [DOI] [PubMed] [Google Scholar]
- Baurén G., and Wieslander L.. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 76:183–192. 10.1016/0092-8674(94)90182-1 [DOI] [PubMed] [Google Scholar]
- Baurén G., Jiang W.Q., Bernholm K., Gu F., and Wieslander L.. 1996. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J. Cell Biol. 133:929–941. 10.1083/jcb.133.5.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G., Belikov S., and Wieslander L.. 1998. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 12:2759–2769. 10.1101/gad.12.17.2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. 2014. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15:163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk P., and Wieslander L.. 2009. Gene expression in polytene nuclei. Methods Mol. Biol. 464:29–54. 10.1007/978-1-60327-461-6_3 [DOI] [PubMed] [Google Scholar]
- Björk P., Baurén G., Gelius B., Wrange O., and Wieslander L.. 2003. The Chironomus tentans translation initiation factor eIF4H is present in the nucleus but does not bind to mRNA until the mRNA reaches the cytoplasmic perinuclear region. J. Cell Sci. 116:4521–4532. 10.1242/jcs.00766 [DOI] [PubMed] [Google Scholar]
- Björk P., Jin S., Zhao J., Singh O.P., Persson J.O., Hellman U., and Wieslander L.. 2009. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J. Cell Biol. 184:555–568. 10.1083/jcb.200806156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F., and Gehring N.H.. 2011. Assembly, disassembly and recycling: The dynamics of exon junction complexes. RNA Biol. 8:24–29. 10.4161/rna.8.1.13618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F., Ebert J., Lorentzen E., and Conti E.. 2006. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 126:713–725. 10.1016/j.cell.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Brugiolo M., Herzel L., and Neugebauer K.M.. 2013. Counting on co-transcriptional splicing. F1000Prime Rep. 5:9 10.12703/P5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G., Ebert J., Basquin C., Saulière J., Jayachandran U., Bono F., Le Hir H., and Conti E.. 2010. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc. Natl. Acad. Sci. USA. 107:10050–10055. 10.1073/pnas.1000993107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H., Ballut L., Bonneau F., and Le Hir H.. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 15:85–93. 10.1038/nsmb1330 [DOI] [PubMed] [Google Scholar]
- Chazal P.E., Daguenet E., Wendling C., Ulryck N., Tomasetto C., Sargueil B., and Le Hir H.. 2013. EJC core component MLN51 interacts with eIF3 and activates translation. Proc. Natl. Acad. Sci. USA. 110:5903–5908. 10.1073/pnas.1218732110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.-S., Hsu J.L., Dias A., and Reed R.. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 127:1389–1400. 10.1016/j.cell.2006.10.044 [DOI] [PubMed] [Google Scholar]
- Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., and Cheng H.. 2013. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 41:1294–1306. 10.1093/nar/gks1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Daguenet E., Baguet A., Cavalier A., Thomas D., Bellaud P., Fautrel A., Godey F., Bertrand E., Tomasetto C., and Gillet R.. 2014. Overexpression of MLN51 triggers P-body disassembly and formation of a new type of RNA granules. J. Cell Sci. 127:4692–4701. 10.1242/jcs.154500 [DOI] [PubMed] [Google Scholar]
- Custódio N., Carvalho C., Condado I., Antoniou M., Blencowe B.J., and Carmo-Fonseca M.. 2004. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 10:622–633. 10.1261/rna.5258504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B. 2001. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. USA. 98:7012–7017. 10.1073/pnas.111145498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B., Andersson K., Björkroth B., and Lamb M.M.. 1982. Visualization of active 75 S RNA genes in the Balbiani rings of Chironomus tentans. Eur. J. Cell Biol. 26:325–332. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., and Smithies O.. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387–395. 10.1093/nar/12.1Part1.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edström J.E., Ericson E., Lindgren S., Lönn U., and Rydlander L.. 1978a Fate of Balbiani-ring RNA in vivo. Cold Spring Harb. Symp. Quant. Biol. 42:877–884. 10.1101/SQB.1978.042.01.089 [DOI] [PubMed] [Google Scholar]
- Edström J.E., Lindgren S., Lönn U., and Rydlander L.. 1978b Balbiani ring RNA content and half life in nucleus and cytoplasm of Chironomus tentans salivary gland cells. Chromosoma. 66:33–44. 10.1007/BF00285814 [DOI] [Google Scholar]
- Egyházi E. 1975. Inhibition of Balbiani ring RNA synthesis at the initiation level. Proc. Natl. Acad. Sci. USA. 72:947–950. 10.1073/pnas.72.3.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H., Lamprinaki S., Kulozik A.E., and Hentze M.W.. 2009. Disassembly of exon junction complexes by PYM. Cell. 137:536–548. 10.1016/j.cell.2009.02.042 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Obrdlik A., Marchand V., and Ephrussi A.. 2014. The EJC binding and dissociating activity of PYM is regulated in Drosophila. PLoS Genet. 10:e1004455 10.1371/journal.pgen.1004455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Zhao M., and He C.. 2014. Slow co-evolution of the MAGO and Y14 protein families is required for the maintenance of their obligate heterodimerization mode. PLoS One. 9:e84842 10.1371/journal.pone.0084842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R., Handler D., Ish-Horowicz D., and Brennecke J.. 2014. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes Dev. 28:1772–1785. 10.1101/gad.245738.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., and Steitz J.A.. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 7:899–905. 10.1016/S1097-2765(01)00233-7 [DOI] [PubMed] [Google Scholar]
- Huang Y., Gattoni R., Stévenin J., and Steitz J.A.. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 11:837–843. 10.1016/S1097-2765(03)00089-3 [DOI] [PubMed] [Google Scholar]
- Ideue T., Sasaki Y.T.F., Hagiwara M., and Hirose T.. 2007. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 21:1993–1998. 10.1101/gad.1557907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.A., Cubberley G., and Bentley D.L.. 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell. 33:215–226. 10.1016/j.molcel.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Inoue H., Hurt E., and Yoneda Y.. 2009. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 28:556–567. 10.1038/emboj.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Diem M.D., Kim V.N., Yong J., and Dreyfuss G.. 2001. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 20:6424–6433. 10.1093/emboj/20.22.6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler E., Miralles F., and Visa N.. 2002. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr. Biol. 12:859–862. 10.1016/S0960-9822(02)00840-0 [DOI] [PubMed] [Google Scholar]
- Kiseleva E., Wurtz T., Visa N., and Daneholt B.. 1994. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J. 13:6052–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsenko A., Svensson T., Nystedt B., Lundeberg J., Björk P., Sonnhammer E., Giacomello S., Visa N., and Wieslander L.. 2014. The Chironomus tentans genome sequence and the organization of the Balbiani ring genes. BMC Genomics. 15:819 10.1186/1471-2164-15-819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb M.M., and Daneholt B.. 1979. Characterization of active transcription units in Balbiani rings of Chironomus tentans. Cell. 17:835–848. 10.1016/0092-8674(79)90324-6 [DOI] [PubMed] [Google Scholar]
- Le Hir H., and Séraphin B.. 2008. EJCs at the heart of translational control. Cell. 133:213–216. 10.1016/j.cell.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Le Hir H., Moore M.J., and Maquat L.E.. 2000. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon-exon junctions. Genes Dev. 14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., and Moore M.J.. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987–4997. 10.1093/emboj/20.17.4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., and Cáceres J.F.. 2009. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 417:15–27. 10.1042/BJ20081501 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.-D., and Steitz J.A.. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 103:1121–1131. 10.1016/S0092-8674(00)00214-2 [DOI] [PubMed] [Google Scholar]
- Ma J., Liu Z., Michelotti N., Pitchiaya S., Veerapaneni R., Androsavich J.R., Walter N.G., and Yang W.. 2013. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat. Commun. 4:2414 10.1038/ncomms3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.D., Mestdagh C., Akhtar J., Kreim N., Deinhard P., Sachidanandam R., Treisman J., and Roignant J.-Y.. 2014. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes Dev. 28:1786–1799. 10.1101/gad.245829.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., and Reed R.. 2002. An extensive network of coupling among gene expression machines. Nature. 416:499–506. 10.1038/416499a [DOI] [PubMed] [Google Scholar]
- Masuda S., Das R., Cheng H., Hurt E., Dorman N., and Reed R.. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512–1517. 10.1101/gad.1302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin H., Daneholt B., and Skoglund U.. 1992. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 69:605–613. 10.1016/0092-8674(92)90224-Z [DOI] [PubMed] [Google Scholar]
- Mehlin H., Daneholt B., and Skoglund U.. 1995. Structural interaction between the nuclear pore complex and a specific translocating RNP particle. J. Cell Biol. 129:1205–1216. 10.1083/jcb.129.5.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinel D.M., Burkert-Kautzsch C., Kieser A., O’Duibhir E., Siebert M., Mayer A., Cramer P., Söding J., Holstege F.C., and Sträßer K.. 2013. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet. 9:e1003914 10.1371/journal.pgen.1003914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Mähr R., Eppenberger H.-M., and Lezzi M.. 1983. The activity of Balbiani rings 1 and 2 in salivary glands of Chironomus tentans larvae under different modes of development and after pilocarpine treatment. Dev. Biol. 98:265–277. 10.1016/0012-1606(83)90357-3 [DOI] [PubMed] [Google Scholar]
- Miralles F., Öfverstedt L.G., Sabri N., Aissouni Y., Hellman U., Skoglund U., and Visa N.. 2000. Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J. Cell Biol. 148:271–282. 10.1083/jcb.148.2.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O., Eberle A.B., Stalder L., and Zamudio Orozco R.. 2008. Recognition and elimination of nonsense mRNA. Biochim. Biophys. Acta. 1779:538–549. 10.1016/j.bbagrm.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Natalizio B.J., and Wente S.R.. 2013. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 23:365–373. 10.1016/j.tcb.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Meislin S.H., and Moore M.J.. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA. 9:607–617. 10.1261/rna.5250403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Le Hir H., and Moore M.J.. 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes Dev. 18:210–222. 10.1101/gad.1163204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I.M. 2002. RNA processing: Splicing and the cytoplasmic localisation of mRNA. Curr. Biol. 12:R50–R52. 10.1016/S0960-9822(01)00671-6 [DOI] [PubMed] [Google Scholar]
- Palacios I.M., Gatfield D., St Johnston D., and Izaurralde E.. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 427:753–757. 10.1038/nature02351 [DOI] [PubMed] [Google Scholar]
- Perales R., and Bentley D.. 2009. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 36:178–191. 10.1016/j.molcel.2009.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P. 2014. New insights into co-transcriptional sorting of mRNA for cytoplasmic transport during development. Semin. Cell Dev. Biol. 32:55–62. 10.1016/j.semcdb.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Popp M.W., and Maquat L.E.. 2014. The dharma of nonsense-mediated mRNA decay in mammalian cells. Mol. Cells. 37:1–8. 10.14348/molcells.2014.2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulière J., Haque N., Harms S., Barbosa I., Blanchette M., and Le Hir H.. 2010. The exon junction complex differentially marks spliced junctions. Nat. Struct. Mol. Biol. 17:1269–1271. 10.1038/nsmb.1890 [DOI] [PubMed] [Google Scholar]
- Saulière J., Murigneux V., Wang Z., Marquenet E., Barbosa I., Le Tonquèze O., Audic Y., Paillard L., Roest Crollius H., and Le Hir H.. 2012. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol. 19:1124–1131. 10.1038/nsmb.2420 [DOI] [PubMed] [Google Scholar]
- Schmidt U., Im K.-B., Benzing C., Janjetovic S., Rippe K., Lichter P., and Wachsmuth M.. 2009. Assembly and mobility of exon-exon junction complexes in living cells. RNA. 15:862–876. 10.1261/rna.1387009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G., Gersappe A., Black J.D., Aronoff R., and Maquat L.E.. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21:209–223. 10.1128/MCB.21.1.209-223.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T., Tange T.Ø., Sonenberg N., and Moore M.J.. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11:346–351. 10.1038/nsmb750 [DOI] [PubMed] [Google Scholar]
- Siebrasse J.P., Veith R., Dobay A., Leonhardt H., Daneholt B., and Kubitscheck U.. 2008. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc. Natl. Acad. Sci. USA. 105:20291–20296. 10.1073/pnas.0810692105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebrasse J.P., Kaminski T., and Kubitscheck U.. 2012. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 109:9426–9431. 10.1073/pnas.1201781109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., and Moore M.J.. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 151:750–764. 10.1016/j.cell.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh O.P., Björkroth B., Masich S., Wieslander L., and Daneholt B.. 1999. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp. Cell Res. 251:135–146. 10.1006/excr.1999.4490 [DOI] [PubMed] [Google Scholar]
- Sjölinder M., Björk P., Söderberg E., Sabri N., Farrants A.K., and Visa N.. 2005. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 19:1871–1884. 10.1101/gad.339405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckelberg A.L., Boehm V., Gromadzka A.M., and Gehring N.H.. 2012. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Reports. 2:454–461. 10.1016/j.celrep.2012.08.017 [DOI] [PubMed] [Google Scholar]
- Tange T.Ø., Nott A., and Moore M.J.. 2004. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16:279–284. 10.1016/j.ceb.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Teng I.F., and Wilson S.A.. 2013. Mapping interactions between mRNA export factors in living cells. PLoS One. 8:e67676 10.1371/journal.pone.0067676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia P., Dias A.P., and Reed R.. 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA. 105:3386–3391. 10.1073/pnas.0800250105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden F.J.M., Palacios I.M., Petronczki M., Weston M.J.D., and St Johnston D.. 2001. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J. Cell Biol. 154:511–524. 10.1083/jcb.200105056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith R., Sorkalla T., Baumgart E., Anzt J., Häberlein H., Tyagi S., Siebrasse J.P., and Kubitscheck U.. 2010. Balbiani ring mRNPs diffuse through and bind to clusters of large intranuclear molecular structures. Biophys. J. 99:2676–2685. 10.1016/j.bpj.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N., Hautbergue G.M., Walsh M., Chang C.-T., Holland A., Folco E.G., Reed R., and Wilson S.A.. 2012. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 3:1006 10.1038/ncomms2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Izaurralde E., Ferreira J., Daneholt B., and Mattaj I.W.. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 133:5–14. 10.1083/jcb.133.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I., Baurén G., and Wieslander L.. 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA. 2:641–651. [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I., Zhao J., Masich S., Wieslander L., and Skoglund U.. 2001. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 20:2564–2574. 10.1093/emboj/20.10.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. 1994. The Balbiani ring multigene family: coding repetitive sequences and evolution of a tissue-specific cell function. Prog. Nucleic Acid Res. Mol. Biol. 48:275–313. 10.1016/S0079-6603(08)60858-2 [DOI] [PubMed] [Google Scholar]
- Wurtz T., Kiseleva E., Nacheva G., Alzhanova-Ericcson A., Rosén A., and Daneholt B.. 1996. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 16:1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. 1982. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp. Cell Res. 139:309–319. 10.1016/0014-4827(82)90255-5 [DOI] [PubMed] [Google Scholar]
- Zhou Z., and Fu X.D.. 2013. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 122:191–207. 10.1007/s00412-013-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




