Abstract
The Gene Ontology (GO) is an important component of modern biological knowledge representation with great utility for computational analysis of genomic and genetic data. The Gene Ontology Consortium (GOC) consists of a large team of contributors including curation teams from most model organism database groups as well as curation teams focused on representation of data relevant to specific human diseases. Key to the generation of consistent and comprehensive annotations is the development and use of shared standards and measures of curation quality. The GOC engages all contributors to work to a defined standard of curation that is presented here in the context of annotation of genes in the laboratory mouse. Comprehensive understanding of the origin, epistemology, and coverage of GO annotations is essential for most effective use of GO resources. Here the application of comparative approaches to capturing functional data in the mouse system is described.
Introduction
The Gene Ontology (GO, The Gene Ontology Consortium 2000, 2015) provides a structured, controlled vocabulary used by a wide range of biological knowledge bases to create annotations that describe a gene product’s function, the overall biological objective of the function, and the cellular location where the function occurs. GO is a widely used biomedical ontology, utilized extensively in data analysis pipelines especially for functional analysis of large datasets. Core methods for providing GO annotations for gene products include curating data from the biomedical literature, inferring information from structural parameters of the gene product, and inferring information based on data mined from homology and phylogenetic assertions to other gene products. Understanding the structure, scope, and origin of GO annotations that summarize current knowledge ensures the best use of GO resources by the research community. Here we focus primarily on the cross-species approach to generating GO annotations, using the Mouse Genome Database-GO curation workflow as an illustrative example. We then provide context for use of cross-species annotations in data analysis applications.
The key elements of a GO annotation are (1) the association of a gene product with a GO term, (2) a statement of the kind of evidence used to make the association (typically an evidence code), and (3) an authority from which the association is made (typically a publication). Here the term ‘gene product’ is used to capture all types of functional entities encoded by genome features including proteins, functional RNAs, and protein complexes. General information about the generation and quality control for GO annotations are discussed in Balakrishnan et al. (2013) and can be found at the GOC website (www.geneontology.org).
The Mouse Genome Database (MGD), the model organism database component of the Mouse Genome Informatics system (MGI; www.informatics.jax.org) (Eppig et al. 2015), makes use of GO terminology to provide functional information about mouse gene products. The MGD curation pipeline proceeds in the environment of curation paradigms developed by the GO Consortium (GOC). These paradigms are employed by all GO annotation providers ensuring consistency in generation and reporting of annotations (http://geneontology.org/page/annotation).
Recently, the GO curation workflow has expanded, so that the curation record can include more details about the context in which the gene product is functioning. This capture of contextual information includes the ability to provide information relative to precise protein forms including isoforms generated by alternative splicing and/or start/stop sites, as well as to protein forms having various post-translational modifications. The capture of contextual information includes describing cell type, anatomical location, time, and other aspects of the functioning of the gene product.
MGD is the authority for providing the comprehensive set of GO annotations for the laboratory mouse to the bioinformatics community. While the majority of mouse annotations are generated within the MGD project, other projects such as the GOA project at UniProt (Huntley et al. 2015) and the PAINT project within the GOC (Gaudet et al. 2011) also generate mouse annotations. These literature-based and sequence similarity-based annotations are imported and integrated into the MGD-authoritative mouse GO annotation file on a weekly basis (ftp://informatics.jax.org/pub/reports/index.html#go). These mouse annotations are then incorporated into the GO data resources such as AmiGO (http://amigo.geneontology.org/amigo, Carbon et al. 2009) and NCBI (NCBI Resource Coordinators 2015), and into other data resources representing current knowledge about mouse genes.
GO annotation via literature curation
Literature curation remains the primary source for experimentally based knowledge about molecular functions of genes and gene products. Currently, MGD has more than 112,500 curated, literature-based annotations to over 12,300 mouse genes obtained from over 23,200 publications. The data and information captured from the primary literature forms the basis for generation of annotation based on comparative inference. The workflow for obtaining literature and prioritizing papers to curate have been described in detail previously (Drabkin and Blake 2012). In brief, biologist curators with experience in specialized biomedical research fields identify appropriate literature with the assistance of data mining tools, confirm specific entities (such as genes or proteins), and select appropriate GO terms to represent the experimental results reported about these entities. Within MGD, the highest priority for literature curation is given to papers that describe new knowledge about genes for which the GO knowledge capture system has no information. Priority is also given to literature with functional information about genes implicated in important disease processes and to literature for genes whose functional annotations consist only of those inferred through automated processes.
GO annotation via sequence similarity
Experiment-based annotations form the basis for functional characterization of a gene product. In addition, the use of orthology to infer knowledge about a gene product from experiment-based annotations to a related gene product makes use of the expert knowledge captured and integrated into other model organism databases and resources such as GOA (human and other species, Huntley et al. 2015), RGD (Rattus; Shimoyama et al. 2015), FlyBase (Drosophilia melanogaster; dos Santos et al. 2015), SGD (Saccharomyces cerevisiae; Costanzo et al. 2014), Pombase (Schizosaccharomyces pombe; McDowall et al. 2015) WormBase (Caenorhabditis elegans; Harris et al. 2014), and DictyBase (Dictyostelium discoideum; Basu et al. 2013). Since the development of model organism research systems is a consequence of the utility of different organisms for different types of biological investigation, these similarity-based methods for obtaining functional annotations for mouse gene products bring into the mouse system more global information than has been generated by the mouse experimental system alone. Different assays are employed in different systems, each utilizing the strength of that particular system (e.g., many genes associated with human disease are often studied using cloned cDNA encoding a human protein in cell culture systems). GO curation guidelines provide several sequence similarity-based evidence codes to support the variety of cross-species annotations (see Table 1). This type of comparative inference is used across all of the model organism databases that use GO for functional annotation.
Table 1.
Sequence-based evidence codes
| Inferred from sequence or structural similarity (ISS) |
| Inferred from sequence orthology (ISO) |
| Inferred from sequence alignment (ISA) |
| Inferred from sequence model (ISM) |
| Inferred from genomic context (IGC) |
| Inferred from biological aspect of ancestor (IBA) |
| Inferred from biological aspect of descendant (IBD) |
| Inferred from key residues (IKR) |
| Inferred from rapid divergence (IRD) |
A complete list of all evidence codes used by GO can be found at http://geneontology.org/page/guide-go-evidence-codes
Within MGD specifically, orthology-based annotations are either captured by MGD curators or generated via semi-automated pipelines. In all cases, only annotations based on experimental characterization are propagated from one species to another, preventing circular annotations between the contributing and receiving resources. Since all GO groups are generating GO annotations via the same paradigm, experimental annotations between these groups are concordant. The standards for generation of orthology data representations between mouse and other organisms is a key to the process. Within vertebrate systems, as with other specific taxonomic groups, assertions of orthology are complicated by gene duplication and paralog divergence events (Sonnhammer et al. 2014).
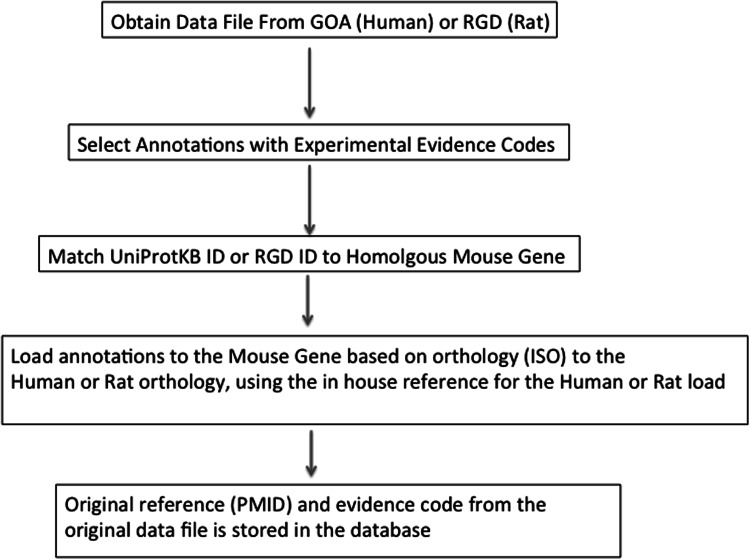
Rather than the MGD-vetted one-to-one orthology assertions that had been used previously, in 2013, MGD moved to a many-to-many orthology paradigm (see Dolan et al., Mammalian Genome this issue) through the use of an external resource, HomoloGene (NCBI Resource Coordinators 2015). Although one-to-one orthology assertions between mouse-human and rat genes still holds for over 90 % of protein-coding genes, MGD can now more clearly represent loci that include a more complex sequence of speciation and gene duplication events. In order to maximize the use of human-mouse orthology sets for comparative genomics in the context of phenotypes or disease, the May 2015 release of MGI also includes the use of HUGO Gene Nomenclature Committee (HGNC) (Gray et al. 2015) mouse–human orthology data. However, currently, HGNC orthology assertions are not used to transfer GO annotation from human to mouse genes. Figure 1 outlines the overall workflow for importing annotations from GOA or RGD based on orthology. Functional annotation of human and rat gene products coming into the MGD system are provided by GOA and by RGD, respectively. As mentioned above, these resources utilize the same GOC annotation guidelines in regard to literature curation of the experimental literature. However, because each species has unique aspects, a variety of rule-based systems have been developed in the MGD system to ensure the assertions result in reasonable predictions. For example, annotations to protein binding or using the NOT qualifier are excluded. Protein binding annotations are excluded because they are created in the context of a specific protein-binding event, something that cannot be reliably transferred between systems. The NOT qualifier is part of annotations where a protein has been demonstrated experimentally to NOT have some property. These cannot be reliably inferred in a cross-species manner. The change to a many-to-many orthology paradigm required careful attention to the development of rules appropriate for the transfer of functional annotation from human or rat experiments to mouse genes, especially in cases of paralogs. Specifically, for any case in which more than one gene per species is in a HomoloGene class, only experimental molecular function and cellular component annotations are transferred as ISOs. In addition, if any member of the class has a ‘NOT’ annotation, annotation to that term is not transferred to any member of the class. At present, the majority of GO annotations in MGD based on orthology/sequence similarity are based on orthology with rat and human genes. A summary of GO annotations based on orthology in MGD is found in Table 2.
Fig. 1.
Importing mouse annotations from rat or human genes based on orthology to mouse genes. Each specific load is assigned a specific MGD reference. Since the evidence code is assertion by orthology as determined by MGD, the provider of the annotations is MGD. Annotations are obtained from the designated authorities for GO annotation for human (GOA) or rat (RGD) genes
Table 2.
Summary of GO annotations in MGD from literature curation, orthology or electronic pipelines
| Annotation method | Total | Genes annotated only by orthology, phylogenetic, or electronic method | ||
|---|---|---|---|---|
| # Genes | # Annotations | # Genes | # Annotations | |
| Manual curation of experimental literature | 11,123 | 98,944 | NA | NA |
| Orthology transfer methods | 11,728a | 98,987 | 3728 | 18,012 |
| Transferred from human (GOA) via orthology | 10,515 | 65,988 | 3379 | 14,104 |
| Transferred from rat (RGD) via orthology | 4631 | 29,861 | 816 | 3271 |
| Curated by MGI curators | 1322 | 3138 | 268 | 637 |
| Phylogenetic methods | ||||
| PAINT | 4356 | 19,703 | 2285 | 10,841 |
| Electronic pipelines (IEA) | 14,653b | 98,980 | 5308 | 35,276 |
| Enzyme Commission (EC) | 1690 | 18,549 | 692 | 8848 |
| Swiss-Prot keywords | 14,270 | 55,754 | 5107 | 18,369 |
| InterPro | 9970 | 24,677 | 3346 | 8060 |
| All annotation methods | 24,179 | 357,251 | 7219 | 64,129 |
Numbers are as of May 5, 2015
aGenes can be annotated by multiple orthology methods, so this represents total number of genes annotated by any orthology method
bGenes can be annotated by multiple electronic pipelines, so this represents total number of genes annotated by any of them
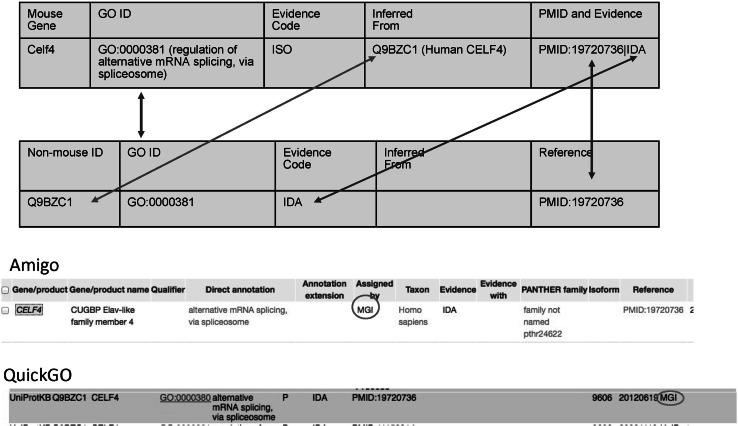
In addition to obtaining annotations for mouse genes from other species via orthology, MGD also generates experimentally supported orthology-based GO annotations for other species during curation of mouse genes. When appropriate, MGD curators may create annotations for the other species when the literature we are curating provides evidence for conservation of function between species. Annotations made by MGD curators using sequence similarity evidence codes (ISO/ISA/ISS) are converted by MGI to annotations to the non-mouse gene based on direct experimental evidence are supplied in GAF format to the GO Annotation (GOA) group at the EBI (European Bioinformatics Institute). For example, as shown in Fig. 2, an annotation for Celf4 was made by MGI based on orthology. The reference shown (J:73065, GO_REF:0000008) denotes that the annotation is made by orthology. The experimental evidence to base this on is obtained from a publication, which is stored at MGI. The experimentally based annotation for the human gene (CELF4) using that publication and the appropriate evidence code is then output to the GAF file given to GOA. Currently, MGI generates a file of 4877 annotations for over 30 non-mouse species from the ISO annotations MGD provides to the GOA resource. These include data from human, rat, cow, dog, hamster, rabbit, pig, macaque, zebra fish, chicken, and frog.
Fig. 2.
Exporting mouse annotations to non-mouse genes based on orthology. The orthologous non-mouse gene becomes the gene that is annotated by an experimental method described in the publication. The bottom two panels depict the non-mouse annotation at either the GOC site (Amigo browser) or GOA (QuickGO)
Phylogenetically based annotations (PAINT)
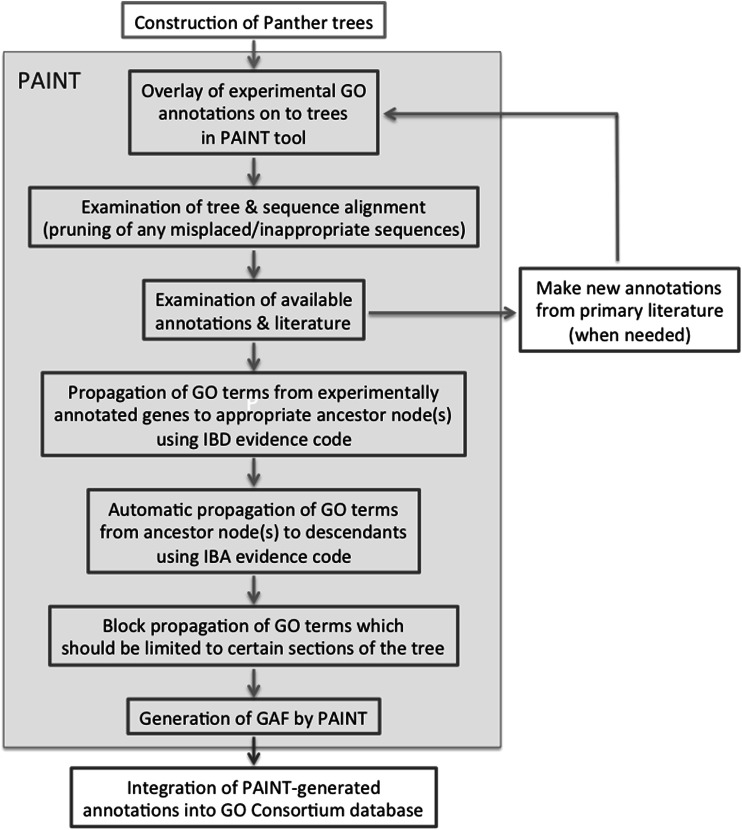
In order to facilitate the use of data from mouse and other species in the study of human biology and disease, the GOC has developed a pipeline for generation of cross-species annotations specifically focused on phylogenetic relationships extending across all taxa. These GO annotations are generated within the context of a given protein family as provided by the Panther system (Mi et al. 2009) and are based on the structure of the phylogenetic tree as well as the experimental evidence for characterized members of the family (Gaudet et al. 2011) Annotations asserted by direct experimental evidence, primarily not only from the twelve “reference” model organisms (see Table 3) but also from other experimentally characterized species when available, are overlaid upon a sequence-based evolutionary tree of all proteins in the Panther Tree database. Using the Phylogenetic Annotation and INference Tool (PAINT, Gaudet et al. 2011), curators determine which annotations can be propagated to a common ancestor node of the tree, indicating an ancient conserved function, where those ancestral annotations can be propagated to all descendent members of the tree unless there is evidence that a function has been lost within a branch of the tree (see Fig. 3).
Table 3.
Twelve model organisms (MODs) used for GO annotations backed by experimental evidence
| Arabidopsis thaliana (The Arabidopsis Information Resource (TAIR)) |
| Caenorhabditis elegans (WormBase) |
| Danio rerio (zebrafish; Zebrafish Model Organism Database (ZFIN)) |
| Dictyostelium discoideum (dictyBase) |
| Drosophila melanogaster (FlyBase) |
| Escherichia coli (PortEco) |
| Gallus gallus (AgBase) |
| Homo sapiens (human UniProtKB-Gene Ontology Annotation [UniProtKB-GOA] @ EBI) |
| Mus musculus (Mouse Genome Informatics) |
| Rattus norvegicus (Rat Genome Database (RGD)) |
| Saccharomyces cerevisiae (Saccharomyces Genome Database (SGD)) |
| Schizosaccharomyces pombe (Pombase) |
Fig. 3.
The PAINT tool overlays experimental GO annotations onto externally constructed Panther phylogenetic trees and allows curators to remove any inappropriate or misplaced sequences before propagating annotations. When needed, new annotations can be made which will be included in PAINT once they have been added to the GO Consortium annotation database. The curator can then determine which annotations represent ancestral functions which should be propagated to an ancestral sequence node. PAINT automatically propagates GO terms from the ancestor node to all descendant sequences that are not already annotated to that term experimentally, except where the curator blocks propagation due to divergence in function. The annotations are exported from PAINT and incorporated into the GO Consortium annotation database
The PAINT process is a powerful tool for cross-species annotation. Frequently, experimental work performed in one, or a few, experimental organism(s) is not going to be duplicated in others, and PAINT provides a mechanism to annotate genes from uncharacterized species based on the experimental work that has been done, wherever it may fall within the phylogenetic tree, often allowing use of more specific GO terms than are generated using some of the other annotation transfer pipelines.
How cross-species annotations aid mouse functional annotation
In a specific example, the nuclear RNA polymerase enzymes have been extensively characterized, supported by experimental data from human and yeast (S. cerevisiae). The yeast gene RPO26, and also the orthologous human gene POLR2F, is well characterized as a core subunit of three nuclear RNA polymerases, RNAP I, RNAP II, and RNAP III (Cramer et al. 2008; Thomas and Chiang 2006). However, the mouse gene Polr2f is not annotated with experimental evidence. PAINT allows the annotation of Polr2f with the GO terms specific to all three of these nuclear RNA polymerases. In contrast, the annotation based on the InterPRO domain provides only a general term without the specificity of which nuclear RNA polymerases Polr2f is part of, and the annotations generated by sequence orthology with human or rat are incomplete providing only the annotations specific to RNAP II but lacking the RNAP I and RNAP III annotations. Similarly, the axonemal dyneins are well characterized biochemically in Chlamydomonas reinhardtii as ATP-dependent microtubule motors, present either in the inner or outer dynein arms (King and Kamiya 2009); comparable biochemical work has not been done in mouse or human. PAINT annotation allows the detailed knowledge of which dyneins are present in which parts of the axoneme to be transferred to many other species based on the phylogenetic relationships. In cases like these, mouse genes receive detailed annotations more specific than that provided by the InterPRO domains, based on the experimental work from other species.
How mouse annotation helps cross-species annotations
In other cases, detailed work has been done in the mouse that allows transfer of information from mouse to other species. For example, the Doublesex AND MAB-3 Related Transcription (Dmrt) factor family is named partly for the Drosophila Doublesex gene, where it is involved in sex determination. In the vertebrates, there have been numerous duplications within this family, and some family members have acquired additional functions. Dmrt3 is involved in the regulation of odontogenesis and specification of ventral spinal cord interneurons (Ahituv et al. 2007; Andersson et al. 2012), Dmrt2 has been shown to be involved in the regulation of somitogenesis but does not appear to involved in sex determination (Seo et al. 2006; Seo 2007), and Dmrtb1 appears to have lost DNA binding ability (Murphy et al. 2007), as demonstrated by experimental work in the mouse.
Thus, transfer of experimental annotations via the PAINT tool can increase the annotation coverage across many species, from providing annotations for organisms that lack any experimental work in that area of biology, to filling in a few “missing” annotations for a relatively well-annotated species based on experimental work in a closely related species, e.g., where an enzymatic function has been characterized for a rat gene, but not for the human or mouse orthologs. The PAINT annotation process may also improve the specificity of the GO terms used, allowing more detailed knowledge to be represented in the GO annotations. This level of detailed annotation can provide important information. For example, knowing whether a gene is found in the inner versus the outer dynein arms of the axoneme may allow more accurate assessment of the expected phenotype within the spectrum of primary ciliary dyskinesia (PCD). For the Dmrt family of transcription factors, PAINT allows transfer of the different roles of various subclades of the family, including the fact that not all members of this family retain activity in regulation of sex determination.
MGD curators are active members of the PAINT annotation team. Besides making annotations to mouse genes within the MGI system, MGD curators have recently begun to annotate other species directly in UniProt’s GO annotation tool, Protein2GO (Huntley et al. 2015) when such annotations are needed for phylogenetic annotation via PAINT, even when there is not a direct comparison to mouse within the primary reference being annotated that would allow us to use one of our long-standing orthology transfer methods. Annotations to mouse genes made via the PAINT phylogenetic method are imported into the MGD on a weekly basis. A summary of GO annotations from PAINT in MGI is found in Table 2.
GO annotation via electronic pipelines
Additional MGD automated annotation strategies include data obtained from UniProtKB entries assigned to MGI Genes. These mappings include the Enzyme Commission number assignment, Swiss-Prot keywords, and InterPro. Currently MGI has approximately 99,183 the so-called electronic annotations (IEA) to over 14,650 genes. A summary of these annotations is found in Table 2. Note that because the InterPro mapping entries are manually annotated with terms from the GO (Burge et al. 2012), the annotations based on the mappings are considered of high quality. Mapping files can be found at http://geneontology.org/page/download-mappings#dir.
Use of GO cross-species/global annotations
Clearly, the generation of orthology- and phylogeny-based annotations brings significant added value to the comprehensive set of GO annotations available for mouse or for any organism. For research groups, including computational biologists and bioinformaticians who incorporate GO annotations in their data analysis streams, understanding the complexities and sources of GO annotations is an important element of effective data analysis (Blake 2013). The primary element in evaluating annotations in a cross-species manner is to review the origination of the knowledge assertion made by the annotation.
Uses of GO in complex queries
MGD is a component of the larger Mouse Genome Informatics (MGI) resource. The MGI system is made up of several resources in addition to MGD, such as the Gene Expression Database (GXD) and the Mouse Tumor Database (MTD). MGD curates not just functional information (GO) but also data about mutant mouse alleles, human diseases, and genome structure. GXD curates data on the expression of mouse genes during embryonic development. MTD curates data on the use of mouse models for hereditary cancer. The key paradigm linking these semi-independent curation efforts is achieved by data integration and specifically the fact that all the different types of data are linked to the same gene objects within the database. Thus, GO annotation can be used within the MGD/MGI system for complex queries, such as “show me all genes located on Chromosome 3 that have been annotated to ‘protein tyrosine kinase’ and are associated with Diabetes” (see Fig. 4), or “show me genes annotated to tyrosine kinase that are expressed in metanephric meschyme at Theiler Stage 17” (see Fig. 5).
Fig. 4.
Complex query for mouse genes located on chromosome 3 that are annotated to protein tyrosine kinase activity and are associated with diabetes
Fig. 5.
Complex GXD query for mouse genes annotated to protein tyrosine kinase activity and are expressed in Tyler Stages 17–19 metanephric mesenchyme
Available resources at MGD
Annotations for specific genes can be viewed starting at the Gene Detail page for any one gene. Links provided lead to summaries in tabular, graphical, and textual forms. MGD also provides links to FuncBase for mouse, where one can view computationally predicted GO annotations based on several methods including mutant phenotypes and ‘guilt-by association’ correlations (Beaver et al. 2010). The GO browser can be used to find GO terms as well as a summary of all mouse genes annotated to the term.
All GO annotations in the MGI database, updated nightly, in GAF file format can be found in gene_association.mgi, as well as go_terms.mgi, a file containing a list of all GO terms used at MGI. Annotations in Gene Product Association Data (GPAD, http://geneontology.org/page/gene-product-association-data-gpad-format) will soon be available. The GPAD format is designed to separate annotation object data (synonyms, ids, etc.) from annotation data to reduce redundancy and annotation file size.
There are also several tools available at MGD for use in analyzing GO data, summarized in Table 4.
Table 4.
Tools available at MGD for GO analysis
| Tool | Use | Comments | URL |
|---|---|---|---|
| GO Term Mapper | A tool for analyzing a mouse gene set based on mouse annotations using a method based on the GO Term Finder (Boyle et al. 2004) | Can exclude IEA annotations if desired | http://www.informatics.jax.org/gotools/MGI_Term_Finder.html |
| GO Slim Chart Tool: | A tool for categorizing a gene set according to a set of high-level GO terms, a ‘GO slim’ | Can exclude IEA annotations if desired | http://www.informatics.jax.org/gotools/MGI_GO_Slim_Chart.html |
| Vlad | A GO Term Finder type tool with a graphical output | Can select annotation set (MGI GO, or user supplied). Can supply reference set and filter on several evidence codes. Output can be graphical, or tabular | http://proto.informatics.jax.org/prototypes/vlad/ |
| MouseMine | An InterMine tool (Kalderimis et al. 2014) that provides access to mouse data for customized queries where the results can be downloaded or reused in subsequent queries | Can use the premade template queries in the FUNCTION section to access GO data in a variety of ways. Results can be further filtered to increase specificity of the query | http://www.mousemine.org/mousemine/begin.do |
Summary
MGD, as a representative member of the GOC, uses a variety of annotation strategies to provide the best possible annotation set for mouse genes and to contribute to the annotation of the other reference genomes. When genes are experimentally characterized in the mouse, we strive to represent this work with experimental GO annotations based on the published literature. However, some genes have not been experimentally characterized in the mouse. Some of these genes may never be fully experimentally characterized in the mouse, but highly conserved, homologous genes have been well characterized in another experimental system, and the findings may be applicable to mouse, e.g., RNA polymerase genes have been extensively characterized with human constructs and in S. cerevisiae, and axonemal dyneins have been experimentally characterized primarily in Chlamydomonas reinhardtii. For genes where there is experimental work on the orthologous gene in a closely related vertebrate such as rat or human, we are able to use our orthology-based sequence similarity annotation pipelines to provide informative GO annotations about the mouse genes. In other cases, where the experimental work has been done in an organism that is more distantly related and may not have a clear orthology with mouse, being able to make experimental annotations directly for the experimentally characterized organism allows us to use the PAINT tool to utilize the phylogenetic relationships to make informative annotations for evolutionarily related genes, from mouse and many other species. Thus, using direct experimental annotations, as well as a variety of orthology- and phylogeny-based tools to utilize experimental work from many species, MGD strives to provide a comprehensive set of annotations for all mouse genes and also contributes to the improvement in the annotations of genes from other species.
Acknowledgments
This work was funded through NIH Grant HG 002273.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin C-J, Patra K, Arnason T, Wellbring L, Hjalm G, et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature. 2012;488:642–646. doi: 10.1038/nature11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R, Harris MA, Huntley R, Van Auken K, Cherry JM. A guide to best practices for gene ontology (GO) manual annotation. Database. 2013;2013:bat054. doi: 10.1093/database/bat054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Fey P, Pandit Y, Dodson R, Kibbe WA, Chisholm RL. dictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 2013;41:D676–D683. doi: 10.1093/nar/gks1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver JE, Taşan M, Gibbons FD, Tian W, Hughes TR, Roth FP. FuncBase : a resource for quantitative gene function annotation. Bioinformatics. 2010;26:1806–1807. doi: 10.1093/bioinformatics/btq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA. Ten quick tips for using the gene ontology. PLoS Comput Biol. 2013;9:e1003343. doi: 10.1371/journal.pcbi.1003343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Kelly E, Lonsdale D, Mutowo-Muellenet P, McAnulla C, Mitchell A, Sangrador-Vegas A, Yong SY, Mulder N, Hunter S (2012) Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation. Database. doi:10.1093/database/bar068 [DOI] [PMC free article] [PubMed]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, the AmiGO Hub, and the Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Engel SR, Wong ED, Lloyd P, Karra K, Chan ET, Weng S, Paskov KM, Roe GR, Binkley G, et al. Saccharomyces genome database provides new regulation data. Nucleic Acids Res. 2014;42:D717–D725. doi: 10.1093/nar/gkt1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Armache K-J, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- Dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase Consortium FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H, Blake J. Manual gene ontology annotation workflow at the Mouse Genome Informatics Database. Database Oxf. 2012;2012:bas045. doi: 10.1093/database/bas045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, The Mouse Genome Database Group The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinf. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O’Donovan C. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:D1057–D1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J, Hu F, Smith R, Štěpán R, Sullivan J, et al. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42:W468–W472. doi: 10.1093/nar/gku301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S, Kamiya R. The Chlamydomonas Sourcebook. Amsterdam: Elsevier; 2009. Axonemal dyneins: assembly, structure, and force generation; pp. 131–208. [Google Scholar]
- McDowall MD, Harris MA, Lock A, Rutherford K, Staines DM, Bähler J, Kersey PJ, Oliver SG, Wood V. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 2015;43:D656–D661. doi: 10.1093/nar/gku1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucl Acids Res. 2009;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Zarkower D, Bardwell V. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KW. Double-knockout mice show severe defects in embryonic myogenesis. Comp Med. 2007;57:460–468. [PubMed] [Google Scholar]
- Seo KW, Wang Y, Kokubo H, Kettlewell JR, Zarkower DA, Johnson RL. Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev Biol. 2006;290:200–210. doi: 10.1016/j.ydbio.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Shimoyama M, De Pons J, Hayman GT, Laulederkind SJF, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang S-J, et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Gabaldón T, Sousa da Silva AW, Martin M, Robinson-Rechavi M, Boeckmann B, Thomas PD, Dessimoz C, The Quest for Orthologs consortium Big data and other challenges in the quest for orthologs. Bioinformatics. 2014;30:2993–2998. doi: 10.1093/bioinformatics/btu492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang C-M. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]