The Cancer Genome Atlas (TCGA) project in bladder cancer reported the integrated genomic analysis of the first 131 patients in 2014 [1]. We analyzed chemotherapy-naive, muscle-invasive urothelial cancers for somatic mutations, DNA copy number variants, mRNA and microRNA (miRNA) expression, protein and phosphorylated protein expression (reverse-phase protein array [RPPA]), and DNA methylation, integrated with comprehensive clinical and pathologic data. This muscle-invasive cohort has one of the highest somatic mutation rates, with mean and median somatic mutation rates of 7.7 and 5.5 per megabase, respectively, similar to adenocarcinoma and squamous cell carcinoma of the lung and melanoma. We reported 32 significantly mutated genes (SMGs) involved in multiple pathways including cell cycle regulation (93% of tumors), chromatin remodeling (76%), DNA damage response, transcription factors, and RAS/RTK/PI3K (72%) signaling pathways. Four SMGs—ARID1a, MLL2, KDM6a, and EP300—are involved in epigenetic regulation and were mutated in up to one-quarter of the tumors. One-third of the tumors were characterized by cancer-specific DNA hypermethylation.
An additional 281 tumors are in the pipeline, and the complete data set will be analyzed beginning in early 2015. We have preliminary mutation data on 238 samples (including the first 131), and six additional SMGs have been identified. Several of the 38 SMGs have not been described previously in bladder cancer.
Combining copy number variation and somatic mutations, 69% of tumors harbor one potentially actionable target or more. Three mutation clusters were identified and characterized as focally amplified, which were enriched in focal copy number alterations (eg, 3p loss/PPARG) and MLL2 mutations; enriched for TP53 and RB1 mutations and E2F3 amplifications; or papillary histology, which were FGFR3-mutant CDKN2A deficient.
Unsupervised clustering of mRNA, miRNA, and RPPA suggests four expression clusters, three of which were fully characterized as follows. Cluster 1 showed papillary morphology and FGFR3 dysregulation. Clusters 1 and 2 expressed high HER2 (ERBB2) and estrogen receptor-β signaling signature, sharing features with luminal A breast cancer. Cluster 3 showed similarities to basal-like breast and squamous cell head and neck carcinomas. Cluster 3 had a high cancer stem cell content, similar to the basal phenotype reported earlier by Volkmer et al [2] and Ho and colleagues [3]. Cluster 3 was also enriched with tumors with mixed squamous histology, similar to the subtype described by Sjodahl et al [4], which had the worst prognosis compared with three other subtypes that correlate well with the four subtypes described in the TCGA data set. Cluster 3 also correlated with other TCGA subtypes including basal-like breast carcinoma, head and neck squamous cell carcinoma, and lung squamous cell carcinoma [1]. This has been confirmed with a recent cluster of cluster TGCA pan-cancer analysis, which stratified the bladder cancer cohort into three subtypes. Two were similar to lung adenocarcinoma and head and neck and lung squamous carcinoma, and the third, which contained the majority of tumors, was unique to bladder cancer and had the best prognosis compared with the two other subtypes [5].
We have now integrated the mutation subtypes and the expression clusters for 234 tumors. Expression cluster 1 is enriched with FGFR3 mutations, amplification, and expression and urothelial differentiation markers (UPK1A, 2; KRT 20). Cluster 3 is enriched with squamous morphology, basal markers (KRT 14, KRT 5, TP63), and expression of several immune response genes. We have also performed unsupervised clustering of miRNA in 310 tumors and provide an integrated analysis of anticorrelation of clusters of miRNA regulating many pathways including epithelial–mesenchymal transition, DNA methylation, and FGFR3 expression, among many others.
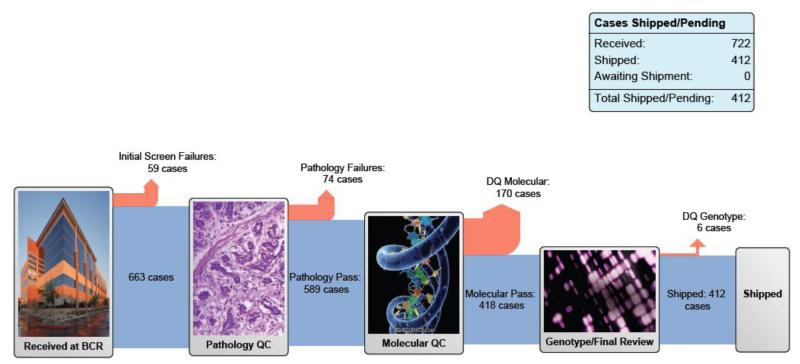
The total cohort now includes 412 tumors that have met the prespecified quality controls for pathology and RNA quality and have been distributed to the genome characterization centers [6] (Fig. 1). The complete data set will increase the power to detect additional low-frequency events [7], validate the cluster analyses, test hypotheses regarding chemotherapy resistance, and provide a host of translational opportunities for functional validation and targeted therapy trials. Outcome analyses were deliberately not included in the analysis of the first 131 tumors, as the follow-up data were not mature. We expect to be able to include this in the final analysis of the full cohort. The analysis working group will reconvene in early 2015 to begin the final analysis, with the expectation of publishing an updated comprehensive integrated analysis.
Fig. 1. The Cancer Genome Atlas muscle-invasive bladder cancer pipeline. Reproduced with permission from the National Cancer Institute [6].
There is a large unmet need for comprehensive genomic characterization of non–muscle-invasive bladder cancer. FGFR3 mutations characterize low-grade Ta tumors, and high-grade tumors share similar genomic alterations with muscle-invasive carcinomas. The National Cancer Institute is sponsoring a clinical trial planning meeting to be held in March 2015, with the goal of designing a targeted therapy trial. A more comprehensive understanding of the genomic landscape is a critical step in this process. A recent landmark study of aristolochic acid–induced upper tract tumors, using whole-exome sequencing, found a remarkably high somatic mutation rate and a unique mutation signature [8]. There are funding opportunities specifically for rare cancers (3–6 per 100 000), and this presents unique collaborative opportunities for important research on the biology and treatment of upper tract urothelial carcinoma [9].
Take-home message.
The Cancer Genome Atlas project in bladder cancer reported the integrated genomic analysis of the first 131 patients in 2014. This muscle-invasive cohort was found to have one of the highest somatic mutation rates. The analysis working group will reconvene in early 2015 to begin the final analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The author has no conflicts of interest relevant to this project. The author does receive compensation for consulting and advisor board participation from BioCancell, Theracoat, Nucleix, Vaxiion, OncoGeneX, Sitka, Taris and Telestra. The author receives support for clinical trials from FKD and Endo
References
- [1].Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Volkmer JP, Sahoo D, Chin RK, et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci U S A. 2012;109:2078–83. doi: 10.1073/pnas.1120605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ho PL, Kurtova A, Chan KS. Normal and neoplastic urothelial stem cells: getting to the root of the problem. Nat Rev Urol. 2012;9:583–94. doi: 10.1038/nrurol.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- [5].Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. [Accessed November 8, 2014];BCR pipeline report. The Cancer Genome Atlas Web site. https://tcga-data.nci.nih.gov/datareports/BCRPipelineReport.htm.
- [7].Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hoang ML, Chen CH, Sidorenko VS, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5:197ra02. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Participation in Trials of Rare Cancers National Cancer Institute Web site. http://cancercenters.cancer.gov/news/news-announ-comm.html.