Supplemental Digital Content is available in the text
Abstract
Aberrant DNA methylation that leads to the inactivation of tumor suppressor genes is known to play an important role in the development and progression of breast cancer. Methylation status of cancer-related genes is considered to be a promising biomarker for the early diagnosis and prognosis of tumors. This study investigated the methylation status of the Sox17 gene in breast cancer tissue and its corresponding plasma DNA to evaluate the association of methylation levels with clinicopathological parameters and prognosis.
The methylation status of the Sox17 gene promoter was evaluated with methylation-specific polymerase chain reaction (MSP) in 155 paired breast cancer tissue and plasma samples and in 60 paired normal breast tissue and plasma samples. Association of Sox17 methylation status with clinicopathological parameters was analyzed by χ2 tests. Overall and disease-free survival (DFS) curves were calculated using Kaplan–Meier analysis, and the differences between curves were analyzed by log-rank tests.
The frequency of Sox17 gene methylation was 72.9% (113/155) in breast cancer tissues and 58.1% (90/155) in plasma DNA. Sox17 gene methylation was not found in normal breast tissues or in their paired plasma DNA. There was a significant correlation of Sox17 methylation between corresponding tumor tissues and paired plasma DNA (r = 0.688, P < 0.001). Aberrant Sox17 methylation in cancer tissues and in plasma DNA was significantly associated with the tumor node metastasis stage (P = 0.035 and P = 0.001, respectively) and with lymph node metastasis (P < 0.001 and P = 0.001, respectively). Kaplan–Meier survival curves showed that aberrant Sox17 promoter methylation in cancer tissues and plasma DNA was associated with poor DFS (P < 0.005) and overall survival (OS) (P < 0.005). Multivariate analysis showed that Sox17 methylation in plasma DNA was an independent prognostic factor in breast cancer for both DFS (P = 0.020; hazard ratio [HR] = 2.142; 95% confidence interval [CI]: 1.128–4.067) and for OS (P = 0.001; HR = 4.737; 95% CI: 2.088–10.747).
Sox17 gene promoter methylation may play an important role in breast cancer progression and could be used as a prognostic biomarker to identify patients at risk of developing metastasis or recurrence after mastectomy.
INTRODUCTION
Breast cancer is the most prevalent cancer and is a major cause of cancer-related death in women worldwide.1 Over the past 3 decades, despite numerous advances having been made in breast cancer early detection and comprehensive therapies, a great many patients still finally die of cancer recurrence and metastasis. The detailed mechanisms underlying this malignancy remain largely unknown and current detection and treatment measures do not adequately improve the survival chances of women with this disease. Thus, the identification of markers for the early detection and effective therapeutic targets for breast cancer patients is urgent and necessary.
It is now recognized that solid malignant tumors can release a significant amount of genomic DNA into circulation in the blood,2 and such DNA can account for >90% of total circulating cell-free DNA3–6 and can be characteristic of the overall heterogeneity of the tumor from which this DNA was released.7 The presence of abnormally high DNA concentrations in plasma has been reported in breast cancer, and the association of changes in the levels of circulating DNA with tumor burden and progression has been repeatedly confirmed.8,9 To date, almost all of the markers associated with genetic alterations, including epigenetic alterations, have been described in circulating DNA.10 Epigenetic silencing due to hypermethylation of tumor-related genes is known to play critical roles in the initiation and progression of breast cancer; this has been demonstrated in DNA damage repair genes, cell cycle regulation genes, and cell signal transduction genes, among others.11–13 Increasing amounts of data strongly suggest that DNA methylation can be a useful biomarker in risk assessment,14 early diagnosis,15 prognosis,16–20 and treatment18,21 for breast cancer patients. Furthermore, some studies have also shown that methylation patterns found in circulating cell-free DNA were similar to the patterns in primary tumors,22–24 indicating the potential utility of minimally invasive blood-based methods for breast cancer detection.
Sox17, a member of the Sry-related high-mobility group box gene family, is a high-mobility group box transcription factor that is known to function as a key regulator in various developmental and disease contexts, including endoderm organ development,25,26 vascular development,27 oligodendrocyte development,28,29 and stem cell function regulation.30,31 In addition, Sox17 can act as a negative regulation factor of β-catenin/TCF transcription activity in the Wnt/β-catenin signal transduction pathway.25,29,32–36 Recently, growing evidence has indicated that Sox17 also plays an important role in human carcinogenesis. The downregulated expression of Sox17 has been detected in colorectal cancer, hepatocellular carcinoma, gastric cancer, and esophageal carcinoma, among other cancers.32–41 Further studies have revealed that Sox17 gene silencing is associated with hypermethylation of the Sox17 promoter. Hypermethylation of the Sox17 promoter is correlated with poor prognosis in several cancers.34,37–38,40 In a previous study, we demonstrated that Sox17 is often hypermethylated and provides important prognostic information in breast cancer patients.34 Recent studies have shown that Sox17 is also epigenetically silenced in circulating tumor cells isolated from the peripheral blood of patients with breast or gastric cancer,41–43 and that such silencing can be used as a molecular diagnostic marker in early-stage gastric cancer.43 Therefore, the aim of this study was to evaluate the prognostic significance of Sox17 promoter methylation in breast cancer patients.
MATERIALS AND METHODS
Patients and Samples
From January 2007 to June 2008, 155 patients with breast cancer, from Northern Jiangsu People's Hospital, Yangzhou, China, were enrolled in this study. All tissue specimens were flash frozen in liquid nitrogen and stored at −80°C for DNA extraction immediately after resectioning. Pathological information was obtained for the following: histological tumor type, primary tumor size, axillary lymph nodal status, histological grade, estrogen and progesterone receptors status, and HER2/neu status. The disease stage of the breast cancer cases was classified according to the American Joint Committee on Cancer-7 tumor node metastasis (TNM) staging system. Meanwhile, paired blood samples from all recruited individuals were collected before surgery. Sixty normal tissues adjacent to benign breast tumors and paired plasma samples were collected as controls. All patients gave written informed consent for the use of their samples in this research, and the study was approved by the Ethical Committee and Institutional Review Board of Northern Jiangsu People's Hospital.
Collection and Processing of Samples and DNA Preparation
Ten milliliters of blood samples were collected in BD Vacutainer® EDTA Tubes (Becton Dickinson, Franklin Lakes, NJ, USA) tubes before any invasive procedures or any treatment had been performed. Plasma was immediately separated from the cellular fraction by 2 rounds of centrifugation at 3000 rpm for 10 minutes at room temperature and then stored at −80°C for later use. Genomic DNA from breast cancer tissues was extracted with a DNeasy Tissue Kit (Qiagen, Hilden, Germany), and plasma DNA was isolated with a QIAamp DNA Blood Mini Kit (Qiagen) according to the manufacturer's instructions. The extracted DNA was quantified spectrophotometrically and stored at −20°C.
Sodium Bisulfite Modification and MSP
DNA was modified with an EZ DNA Methylation-Gold Kit (ZYMO Research Co., Orange, CA) as previously described.34 The methylation status of Sox17 in breast cancer tissues and in circulating cell-free DNA was detected with the methylation-specific polymerase chain reaction (MSP) method. The primer pairs for both the methylated and the unmethylated sequences and the thermocycling conditions of MSP were to those reported in our previous study.34 Each MSP reaction included 2 μL of DNA template, 0.18 μL of each primer, 0.45 μL of 10 mM dNTP Mix (Promega Corp., Madison, WI, USA) 1.5 μL 10× PCR buffer, and 0.12 μL of HotStart Taq DNA Polymerase (Sigma, Germany) in a final reaction volume of 15 μL. MSP products (4 μL) were loaded onto 2% agarose gels and visualized by ethidium bromide staining. SssI-methylated DNA was used as a positive control; whole-genome amplification DNA of normal peripheral lymphocytes was used as a negative control.
Follow-Up
Patients were tracked until September 30, 2014. For every patient enrolled, a complete diagnostic evaluation consisting of chest x-rays, mammography, ultrasounds of the liver, and a whole-body bone scan before surgery was performed to exclude the presence of distant metastasis. Patients were given a physical examination every 3 months for the first 2 years postoperatively and were subsequently examined every 6 months. Disease-free survival (DFS) was defined as the duration from the date of surgery to the date of first evidence of local recurrence, distant metastasis, or last contact. Overall survival (OS) was defined as the time from the date of surgery to the date of death or the date of last contact if the patient was still alive.
Statistical Analyses
Statistical analyses were performed with SPSS statistical software, version 16.0, for Windows (SPSS Inc., Chicago, IL). Categorical data were analyzed by χ2 or Fisher exact tests. Correlations between the methylation statuses of both plasma DNA and tumor tissues were analyzed with Spearman correlation coefficient analysis. DFS and OS curves were calculated using the Kaplan–Meier method and comparisons were performed using the log-rank tests. A univariate Cox regression analysis was used to determine identified prognostic factors, and multivariate Cox regression analysis was used to explore combined effects. All P values presented are 2-sided; a P value <0.05 was considered to indicate statistical significance.
RESULTS
Patient Characteristics
A total of 155 patients with breast cancer were enrolled in this study. The median age was 48.4 years (range 26–75 years); 117 patients were of stages I and II, and 38 patients were of stage III. Patient demographics and pathological features are summarized in Table 2. Control tissue samples adjacent to benign breast tumors, along with paired plasma samples were obtained from 60 patients; these samples incuded 6 intraductal papilloma, 24 mastopathies, 20 fibroadenoma, and 10 breast adipomas.
TABLE 2.
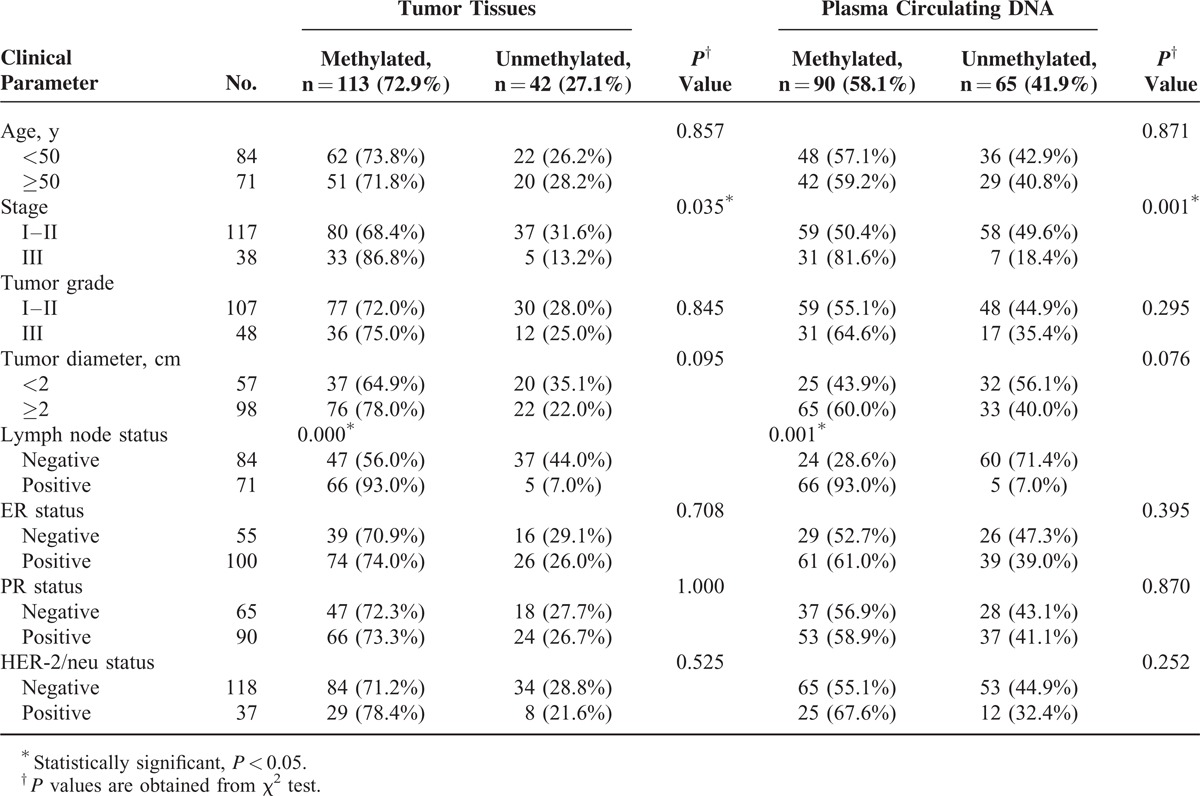
Clinical Characteristics of 155 Patients With and Without Sox17 Methylation in Cancer Tissues and/or Plasma DNA
Frequency of Sox17 Promoter Methylation in Breast Cancer Tissues and Paired Plasma DNA
Aberrant Sox17 promoter methylation was present in 113 of 155 (72.9%) primary tumors, whereas 0/60 (0%) of the normal breast specimens exhibited this. There was a significant difference in Sox17 promoter methylation between the breast cancer and the normal tissues (P < 0.001). Sox17 promoter methylation was found in 90 of 155 (58.1%) plasma DNA from patients with breast cancer; it was not found in any of the control plasma samples. There was a significant difference between the 2 groups (P < 0.001). Representative results of MSP assays for Sox17 methylation are shown in Figure 1.
FIGURE 1.
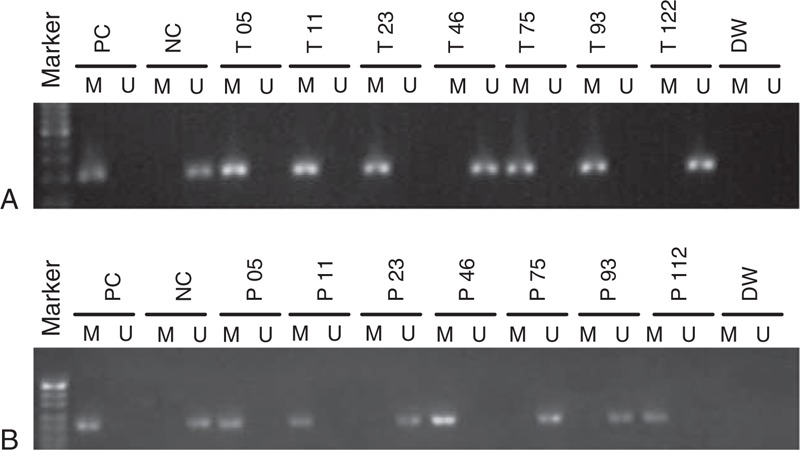
Methylation status of the Sox17 promoter in breast cancer tissues and paired plasma DNA. (A) Representative results of MSP assays of Sox17 methylation in primary breast cancer tissues; B. Representative results of MSP of Sox17 methylation in paired plasma DNA. DW = distilled water, M = methylated, MSP = methylation-specific polymerase chain reaction, NC = negative control, P = plasma DNA, PC = positive control, T = breast cancer tissue, U = unmethylated.
Correlation of Sox17 Methylation in Breast Cancer Tissues and Paired Plasma DNA
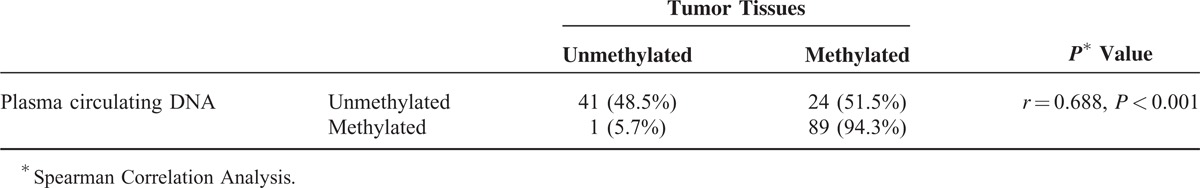
We further analyzed the relationship of the methylation status of Sox17 in breast cancer tissues with the methylation status of Sox17 in paired plasma DNA samples in these patients. Eighty-nine cases (57.42%) had Sox17 promoter methylation in both tumor tissues and in circulating DNA. Nonmethylated Sox17 in both tumor tissues and plasma DNA was found in 41 (33.3%) of the cases; 78.8% (89/113) methylated Sox17 in tumor tissues could be detected in plasma DNA, only 1 case showed Sox17 methylation in plasma DNA but not in tumor tissue. The consistency between tissues and plasma samples was 130 of 155 (83.9%), and methylation in plasma was closely correlated with the Sox17 methylation in tumor tissues (r = 0.688, P < 0.001; Table 1).
TABLE 1.
Correlation Analysis of Sox17 Gene Methylation in Breast Cancer Tissues and Paired Plasma DNA
Association of Sox17 Methylation Status in Breast Tissues and Plasma DNA With Clinicopathological Parameters of Breast Cancer Patients
Table 2 summarizes the association of Sox17 methylation status in tumors and in plasma DNA with various clinicopathological parameters of breast cancer patients. The aberrant methylation status of Sox17 in both cancer tissues and in plasma DNA was significantly associated with TNM stage III status and with lymph node metastasis (P < 0.05). There were no statistically significant associations between Sox17 promoter methylation status in both tumors and plasma samples with age, histological grade, tumor diameter, estrogen receptor status, progesterone receptor status, or HER2 status (P > 0.05).
Sox17 Promoter Methylation and Survival in Patients With Breast Cancer
After a median follow-up period of 66 months (range 3–91 months), 45 of 155 (29.0%) patients relapsed, and 32 of 155 (20.6%) died of the disease. Relapse was detected in 33.6% (38/113) of patients with Sox17 methylation in tumor tissues, and 24.8% (28/113) of patients with Sox17 methylation in tumor tissues died. Both of these rates were significantly higher than those of patients without Sox17 methylation in tumor tissues (P = 0.039 and 0.037, respectively) (Table 3). Patients with methylated Sox17 had worse DFS and OS than did those with nonmethylated Sox17 (log-rank test, P = 0.033 and 0.037, respectively) (Figure 2).
TABLE 3.
Incidence of Disease-Relapse and Disease-Related Death According to the Methylation Status of the Sox17 Promoter
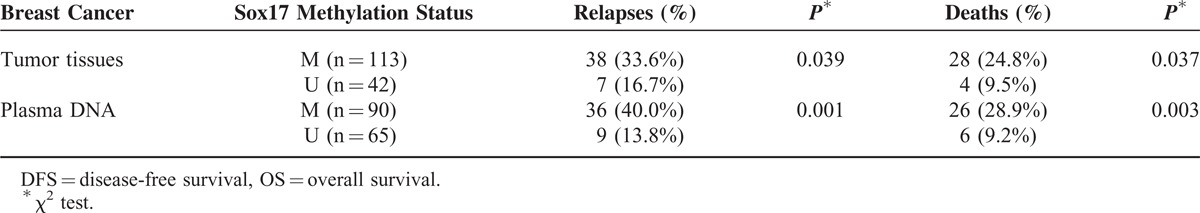
FIGURE 2.
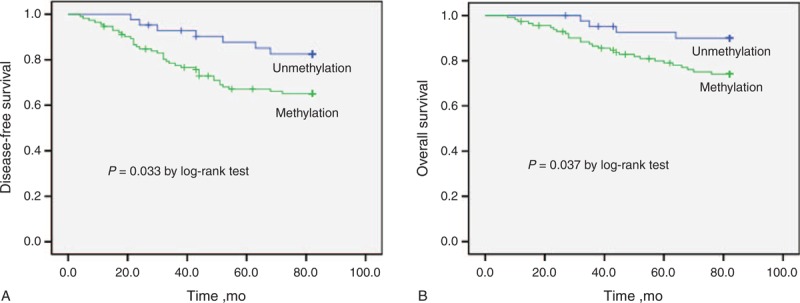
Kaplan–Meier estimates of (A) disease-free survival and (B) overall survival for breast cancer patients with or without Sox17 promoter methylation in tumor tissues.
In the paired plasma DNA, 40.0% (36/90) of patients with Sox17 methylation experienced relapse and 28.9% (26/90) of patients with Sox17 methylation died of breast cancer. In contrast, the rates of relapse and death were only 13.8% (9/65) and 9.2% (6/65), respectively, in those patients without Sox17 methylation. Both the rate of relapse and death differed significantly between the patients with or without Sox17 methylation in plasma DNA (P = 0.001 and 0.003, respectively) (Table 3). Worse DFS and OS were also observed for patients in the Sox17 methylation group (P < 0.001 and 0.002, respectively) (Figure 3).
FIGURE 3.
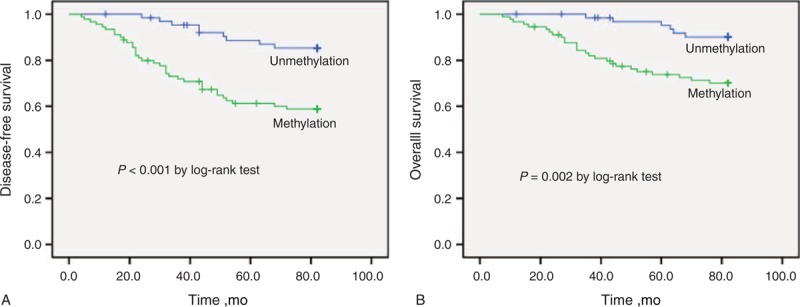
Kaplan–Meier estimates of (A) disease-free survival and (B) overall survival for breast cancer patients with or without Sox17 promoter methylation in plasma DNA.
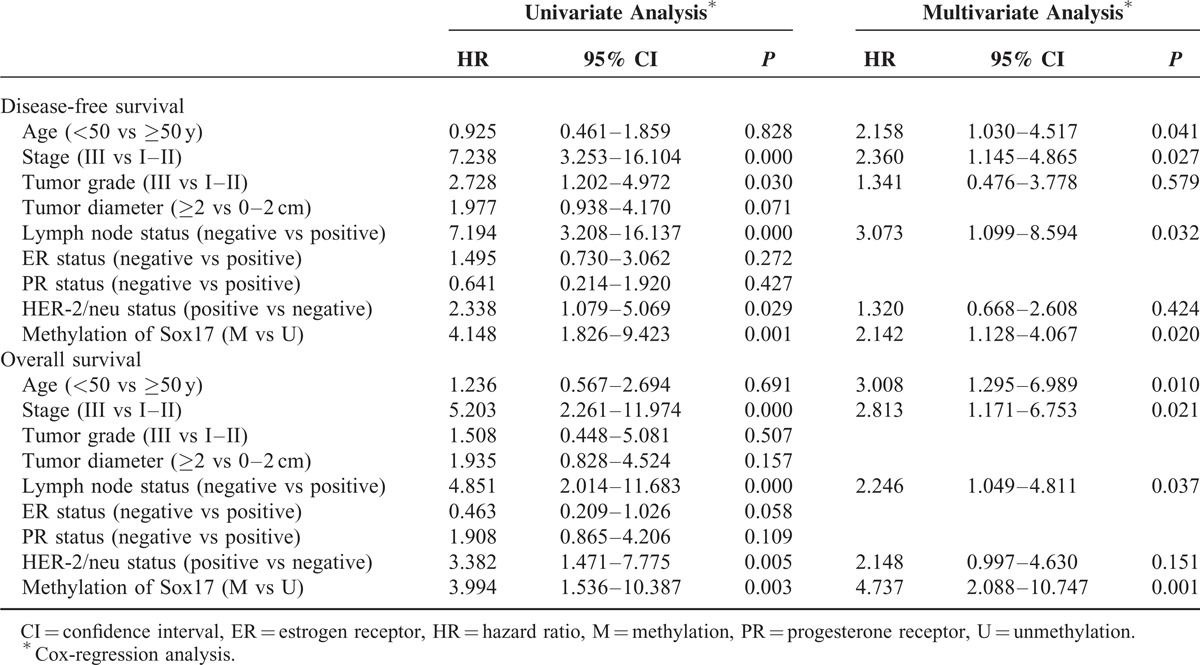
Univariate analysis indicated that TNM stage (P < 0.001), histological grade (P = 0.030), lymph node metastasis (P < 0.001), and HER-2/neu status (P = 0.029) were associated with a decreased DFS. TNM stage (P < 0.001), lymph node status (P < 0.001), and HER2 status (P = 0.005) were significantly associated with worse OS (Table 4). Using a Cox regression model, all factors that determined to be significant in the univariate analysis were tested with multivariate analysis for association with DFS and OS. The results of this analysis are shown in Table 4. Multivariate analysis demonstrated that Sox17 methylation (hazard ratio [HR] = 2.142; 95% confidence interval [CI]: 1.128–4.067; P = 0.020), lymph node status (HR = 3.073; 95% CI: 1.099–8.594; P = 0.032), and TNM stage (HR = 2.360; 95% CI: 1.145–4.865; P = 0.027) were independently associated with a decreased DFS. Sox17 methylation (HR = 4.737; 95% CI: 2.088–10.747; P = 0.001), lymph node status (HR = 2.246; 95% CI: 1.049–4.811; P = 0.037), and tumor stage (HR = 2.813; 95% CI: 1.145–4.865; P = 0.021) were independently associated with a shorter OS. Similarly, results were also observed from Sox17 promoter methylation in breast cancer tissues (Supplement Table 1, http://links.lww.com/MD/A230).
TABLE 4.
Sox17 Promoter Methylation in Plasma DNA of Patients With Breast Cancer: Univariate and Multivariate Analysis for DFS and OS
DISCUSSION
Although multiple epigenetic and genetic changes have been associated with breast cancer, the precise molecular mechanisms in breast cancer carcinogenesis and progression remain unknown. Therefore, sensitive and specific prognostic indicators that can reflect specific alterations in tumors are needed for use in clinical settings. Cell-free DNA in plasma is a type of blood-based biomarker; most of this DNA is released from cancer cells, and it can be used to glean important information about the tumor(s) that released it.44,45 Cell-free DNA in plasma is therefore considered to be a potentially useful noninvasive biomarker in breast cancer diagnosis and prognosis.
In a previous study, we found that the mRNA expression level of the Sox17 gene was significantly decreased in both breast cancer cell lines and in the majority of breast cancer tissues, and observed that the expression level of Sox17 was closely related to the methylation status of its promoter. Furthermore, Sox17 methylation was found to be significantly related to breast cancer staging and lymph node metastasis.34 In the present study, the Sox17 promoter was methylated in 72.9% of cancer tissues and was not methylated in any of the control samples. This high rate of positivity in cancer tissues indicates that Sox17 methylation may represent not only a frequent event in human breast cancer but may also be a useful marker in distinguishing malignant from nonmalignant breast lesions. The fact that Sox17 methylation correlated with TNM stage and lymph node metastasis suggested that epigenetic silencing of Sox17 may also accelerate the spread of cancer through influencing the development of an invasive and biologically aggressive phenotype and thus expedite the progression of breast cancer.
Compared with tumor tissue, plasma samples are near ideal clinical specimen that are readily available, convenient, noninvasive, and can be sampled repeatedly over time. This study shows that Sox17 promoter methylation was found in 58.1% of paired plasma samples, but not detected in any of the control plasma samples. Furthermore, we observed a high degree of consistency between Sox17 methylation in plasma DNA and tumor tissue, suggesting that Sox17 methylation in peripheral blood samples may be a good tumor marker for the diagnosis of breast cancer. Further analysis showed that aberrant Sox17 promoter methylation in plasma DNA was associated with poor DFS and shorter OS, and it was an independent prognostic factor in breast cancer for both DFS and OS by analyzing with multivariate analysis. Thus, Sox17 promoter methylation in plasma DNA is highly specific and can provide important prognostic information for patients with breast cancer.
Many studies have demonstrated that the Sox17 gene can perform tumor suppression functions; it is known to be an important antagonist in the Wnt/β-catenin signaling pathway.40–43 Thus, Sox17 gene silencing due to promoter methylation may deactivate its tumor suppressor role and thereby contribute to poorer outcomes in breast cancer patients. The tumor suppressing function of nonmethylated Sox17 may be partially maintained in patients without Sox17 methylation, and this may slow the progress of tumor development. Furthermore, the occurrence of methylation of Sox17 in plasma DNA provides additional information with clinical relevance. Namely, the situation is an indication that tumor cells may circulating in blood.46 In this light, detection of Sox17 methylation in plasma DNA may also be useful in detecting the development of distant metastasis. MSP of Sox17 methylation in plasma may therefore prove effective for the early detection of residual and/or recurrent tumors. Such detection could motivate successful salvage treatments for these patients in a timely manner. Moreover, Sox17 methylation can be detected in some early-stage patients, which implies that it may be useful in the clinical application of screening and diagnosing breast cancer.
There were some limitations in this study. First, the MSP method is only a qualitative method to identify the presence of methylation and is thus not highly informative. Quantitative MSP, by contrast, is a highly sensitive assay that can detect 1 copy of the methylated gene among 10 000 unmethylated copies. This technology has the potential to screen hundreds to thousands of samples rapidly.47 However, the main aim of this study was to identify whether or not Sox17 promoter methylation in plasma DNA could be a useful and noninvasive biomarker for breast cancer diagnosis and prognosis. Furthermore, MSP can be performed easily and economically in most clinical laboratories. Second, this was only a retrospective study and the number of cases in the study was limited. Further studies with larger sample sizes from multiple clinical centers will be needed to prove the clinical value of Sox17 promoter methylation in plasma DNA.
In summary, our results clearly indicate that methylation of the Sox17 promoter provides important prognostic information for breast cancer patients. Sox17 methylation in plasma DNA can serve as a valuable noninvasive biomarker in breast cancer diagnosis and prognosis. This promising finding deserves further evaluation and validation in a larger patient cohort.
ACKNOWLEDGMENT
The authors would like to thank the patients of the study for their willingness to cooperate with the research.
Footnotes
Abbreviations: AJCC = American Joint Committee on Cancer, CI = confidence interval, DFS = disease-free survival, HR = hazard ratio, MSP = methylation-specific polymerase chain reaction, OS = overall survival.
This study was supported by grants from the National Natural Science Foundation of China (No. 81172508), the Foundation of Social Development of Jiangsu Province (BE2012705), the Foundation of China Postdoctoral Studies (M2013541699), the Foundation of Jiangsu Province Postdoctoral Studies (1302149C), and the Foundation of Young Scholars in Yangzhou (YZ2014046). The funders had no role in the study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011; 61:69–90. [DOI] [PubMed] [Google Scholar]
- 2.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977; 37:646–650. [PubMed] [Google Scholar]
- 3.Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers: blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol 2011; 8:142–150. [DOI] [PubMed] [Google Scholar]
- 4.Kohler C, Barekati Z, Radpour R, et al. Cell-free DNA in the circulation as a potential cancer biomarker. Anticancer Res 2011; 31:2623–2628. [PubMed] [Google Scholar]
- 5.Ignatiadis M, Sotiriou C, Pantel K. Minimal residual disease and circulating tumor cells in breast cancer: open questions for research. Recent Results Cancer Res 2012; 195:3–9. [DOI] [PubMed] [Google Scholar]
- 6.Sharma VK, Vouros P, Glick J. Mass spectrometric based analysis, characterization and applications of circulating cell free DNA isolated from human body fluids. Int J Mass Spectrom 2011; 304:172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia JM, Silva JM, Dominguez G, et al. Heterogeneous tumor clones as an explanation of discordance between plasma DNA and tumor DNA alterations. Genes Chromosomes Cancer 2001; 31:300–301. [DOI] [PubMed] [Google Scholar]
- 8.Silva JM, Silva J, Sanchez A, et al. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin Cancer Res 2002; 8:3761–3766. [PubMed] [Google Scholar]
- 9.Huang ZH, Li LH, Hua D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett 2006; 243:64–70. [DOI] [PubMed] [Google Scholar]
- 10.Anker P, Mulcahy H, Chen XQ, et al. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 1999; 18:65–73. [DOI] [PubMed] [Google Scholar]
- 11.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol 2004; 22:4632–4642. [DOI] [PubMed] [Google Scholar]
- 12.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene 2002; 21:5462–5482. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal A, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol 2007; 20:711–721. [DOI] [PubMed] [Google Scholar]
- 14.Müller HM, Widschwendter A, Fiegl H, et al. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res 2003; 63:7641–7645. [PubMed] [Google Scholar]
- 15.Matuschek C, Bölke E, Lammering G, et al. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res 2010; 15:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma G, Mirza S, Parshad R, et al. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci 2010; 87:83–91. [DOI] [PubMed] [Google Scholar]
- 17.Zurita M, Lara PC, del Moral R, et al. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer 2010; 10:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Galán J, Torres B, Del Moral R, et al. Quantitative detection of methylated ESR1 and 14-3-3-sigma gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biol Ther 2008; 7:958–965. [DOI] [PubMed] [Google Scholar]
- 19.Göbel G, Auer D, Gaugg I, et al. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res Treat 2011; 130:109–117. [DOI] [PubMed] [Google Scholar]
- 20.Jing F, Jun L, Yong Z, et al. Multigene methylation in serum of sporadic Chinese female breast cancer patients as a prognostic biomarker. Oncology 2008; 75:60–66. [DOI] [PubMed] [Google Scholar]
- 21.Liggett TE, Melnikov AA, Marks JR, et al. Methylation patterns in cell-free plasma DNA reflect removal of the primary tumor and drug treatment of breast cancer patients. Int J Cancer 2011; 128:492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 2009; 10:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan PS, Venkataramu C, Ibrahim A, et al. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res 2006; 12:6626–6636. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Yao X. Use of DNA methylation for cancer detection: promises and challenges. Int J Biochem Cell Biol 2009; 41:147–154. [DOI] [PubMed] [Google Scholar]
- 25.Sinner D, Rankin S, Lee M, et al. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development 2004; 131:3069–3080. [DOI] [PubMed] [Google Scholar]
- 26.Engert S, Burtscher I, Liao WP, et al. Wnt/β-catenin signalling regulates Sox17 expression and is essential for organizer and endoderm formation in the mouse. Development 2013; 140:3128–3138. [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Lee S, Lee S, et al. Sox17 promotes tumor angiogenesis and destabilizes tumor vessels in mice. J Clin Invest 2013; 123:418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohn J, Natale J, Chew LJ, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci 2006; 26:9722–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li-Jin C, Weiping S, Xiaotian M, et al. Sox17 regulates the Wnt-beta-catenin signaling pathway in oligodendrocyte progenitor cells. J Neurosci 2011; 31:13921–13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 2007; 130:470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Asakura M, Inoue H, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci USA 2007; 104:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Glöckner SC, Guo M, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res 2008; 68:2764–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinner D, Kordich JJ, Spence JR, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol 2007; 27:7802–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu DY, Wang ZM, Li-Chen, et al. Sox17, the canonical Wnt antagonist, is epigenetically inactivated by promoter methylation in human breast cancer. Breast Cancer Res Treat 2010; 119:601–612. [DOI] [PubMed] [Google Scholar]
- 35.Jia Y, Yang Y, Liu S, et al. SOX17 antagonizes WNT/β-catenin signaling pathway in hepatocellular carcinoma. Epigenetics 2010; 5:743–749. [DOI] [PubMed] [Google Scholar]
- 36.Yin D, Jia Y, Yu Y, et al. SOX17 methylation inhibits its antagonism of Wnt signaling pathway in lung cancer. Discov Med 2012; 14:33–40. [PMC free article] [PubMed] [Google Scholar]
- 37.Ye YW, Wu JH, Wang CM, et al. Sox17 regulates proliferation and cell cycle during gastric cancer progression. Cancer Lett 2011; 307:124–131. [DOI] [PubMed] [Google Scholar]
- 38.Oishi Y, Watanabe Y, Yoshida Y, et al. Hypermethylation of Sox17 gene is useful as a molecular diagnostic application in early gastric cancer. Tumour Biol 2012; 33:383–393. [DOI] [PubMed] [Google Scholar]
- 39.Du YC, Oshima H, Oguma K, et al. Induction and down-regulation of Sox17 and its possible roles during the course of gastrointestinal tumorigenesis. Gastroenterology 2009; 137:1346–1357. [DOI] [PubMed] [Google Scholar]
- 40.Kuo IY, Wu CC, Chang JM, et al. Low SOX17 expression is a prognostic factor and drives transcriptional dysregulation and esophageal cancer progression. Int J Cancer 2014; 135:563–573. [DOI] [PubMed] [Google Scholar]
- 41.Chen HL, Chew LJ, Packer RJ, et al. Modulation of the Wnt/beta-catenin pathway in human oligodendroglioma cells by Sox17 regulates proliferation and differentiation. Cancer Lett 2013; 335:361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chimonidou M, Strati A, Malamos N, et al. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin Chem 2013; 59:270–279. [DOI] [PubMed] [Google Scholar]
- 43.Balgkouranidou I, Karayiannakis A, Matthaios D, et al. Assessment of SOX17 DNA methylation in cell free DNA from patients with operable gastric cancer. Association with prognostic variables and survival. Clin Chem Lab Med 2013; 51:1505–1510. [DOI] [PubMed] [Google Scholar]
- 44.Esposito A, Bardelli A, Criscitiello C, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev 2014; 40:648–655. [DOI] [PubMed] [Google Scholar]
- 45.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497:108–112. [DOI] [PubMed] [Google Scholar]
- 46.Van der Auwera I, Elst HJ, Van Laere SJ, et al. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br J Cancer 2009; 100:1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann U, Länger F, Feist H, et al. Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol 2002; 160:605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]


