Supplemental Digital Content is available in the text
Abstract
Acute respiratory infections (ARIs) cause large disease burden each year. The codetection of viral and bacterial pathogens is quite common; however, the significance for clinical severity remains controversial. We aimed to identify viruses and bacteria in hospitalized children with ARI and the impact of mixed detections.
Hospitalized children with ARI aged ≤16 were recruited from 2009 to 2013 at the Children's Hospital of Chongqing Medical University, Chongqing, China. Nasopharyngeal aspirates (NPAs) were collected for detection of common respiratory viruses by reverse transcription polymerase chain reaction (RT-PCR) or PCR. Bacteria were isolated from NPAs by routine culture methods. Detection and codetection frequencies and clinical features and severity were compared.
Of the 3181 hospitalized children, 2375 (74.7%) were detected with ≥1 virus and 707 (22.2%) with ≥1 bacteria, 901 (28.3%) with ≥2 viruses, 57 (1.8%) with ≥2 bacteria, and 542 (17.0%) with both virus and bacteria. The most frequently detected were Streptococcus pneumoniae, respiratory syncytial virus, parainfluenza virus, and influenza virus. Clinical characteristics were similar among different pathogen infections for older group (≥6 years old), with some significant difference for the younger. Cases with any codetection were more likely to present with fever; those with ≥2 virus detections had higher prevalence of cough; cases with virus and bacteria codetection were more likely to have cough and sputum. No significant difference in the risk of pneumonia, severe pneumonia, and intensive care unit admission were found for any codetection than monodetection.
There was a high codetection rate of common respiratory pathogens among hospitalized pediatric ARI cases, with fever as a significant predictor. Cases with codetection showed no significant difference in severity than those with single pathogens.
INTRODUCTION
Acute respiratory infection (ARI), which could lead to serious complications including pneumonia, acute bronchitis, otitis media, and others, are thought to cause 4.25 million deaths each year.1 The populations most susceptible to fatal respiratory infection are those very young, the elderly, and the immunocompromised.2 As the leading cause of illness in children, ARI causes approximately 1.8 million fatal cases of pneumonia per year among children <5 years of age.3,4 ARIs are classified as upper respiratory tract infections (URTIs) or lower respiratory tract infections (LRTIs). Most URTIs are caused by viruses, including influenza virus A or B (IFVA/B), respiratory syncytial virus (RSV), human rhinovirus (RV), parainfluenza virus (PIV), and human coronavirus (CoV). URTIs are common but rarely life threatening, whereas LRTIs can lead to more severe illnesses, such as pneumonia and bronchiolitis. Both of these infections can also be caused by bacterial pathogens, such as Streptococcus pneumoniae, Haemophilus influenzae, Haemophilus parainfluenzae, Klebsiella pneumoniae, and others. With the use of molecular detection techniques, simultaneous detection of multiple respiratory pathogens in patients with ARIs has been reported more frequently.5,6 The codetection of viral and bacterial pathogens is also quite common, especially in those less-developed countries.7,8 The significance of mixed detections for clinical severity is controversial with inconsistent results reported by the previous studies. The difficulties in establishing precise bacteriological or viral etiology and the frequent occurrence of mixed infections pose a challenge to medical treatment and clinical management. In the present study, we aimed to describe the detection of viruses and bacteria in hospitalized children with ARI in a subtropical city of mainland China, investigate the simultaneous detection pattern of multiple viruses and bacteria, and assess their association with clinical outcomes and severity.
MATERIALS AND METHODS
Sources of Data
We enrolled hospitalized children aged ≤16 years with ARI (determined based on syndromes of cough, rhinorrhea, and/or dyspnea, and/or fever of >37.5°C) admitted from June 2009 through October 2013 to the Children's Hospital of Chongqing Medical University, Chongqing, China. Fever caused by known chronic medical conditions was excluded. For recruited patients, nasopharyngeal aspirates (NPAs) samples were collected on hospital admission after obtaining informed consent. We used a standardized Respiratory Syndrome Cases Information Questionnaire to collect data on demographics, underlying medical conditions, selected laboratory tests, radiographic findings, clinical manifestations, management, and outcomes. Pneumonia was defined by the presence of patchy alveolar opacities on chest radiographs, in addition to symptoms of cough, dyspnea (lower chest wall indrawing), or tachypnea (in infants, >50–60 breaths/min; in older children, >40 breaths/min). Severe pneumonia was defined as pneumonia with hypoxemia (maintained Sao2 < 92% in air) or rising respiratory and pulse rates with clinical evidence of respiratory distress and exhaustion with or without raised Paco2.
This study was performed with the approval of the Ethical Committees of Children's Hospital of Chongqing Medical University and the Beijing Institute of Microbiology and Epidemiology. At recruitment, written informed consent was obtained from all parents or legal guardians of the participants.
Laboratory Detection
NPAs were preserved in virus transport medium (Hainan Xingnanfeng Medical equipment Enterprise Co, Ltd, Hainan, China) immediately after collection, and stored at −80°C prior to testing. Viral DNA and RNA were extracted from 200 μL of the NPA and eluted in 62 μL of AE Buffer by using QIAamp MinElute Virus Spin Kits (QIAGEN, Hilden, Germany). The complementary DNA sample was synthesized by using SuperScript First-Strand Synthesis System for reverse transcription polymerase chain reaction (RT-PCR) (Invitrogen, Camarillo, CA). All samples were screened by RT-PCR or PCR for common respiratory viruses: IFVA, IFVB, RSV (A and B subtypes), PIV (types 1, 2, 3, and 4), RV, CoV (229E and OC43), human metapneumovirus (MPV), adenovirus (AdV), and bocavirus (BoV) using standard methods as described elsewhere.9,10 As not all the samples were tested for subtypes of IFVA such as sH1N1, sH3N2, and pH1N1, here we did not evaluate by subtype. Qualitative and semiquantitative cultures for bacteria were performed immediately using standard microbiological methods.11,12 Macroscopically distinct colonies of the samples were isolated in pure culture, and standard methods were used for identification, typing with sensitivity patterns.
Codetection was defined as a positive detection of >1 microbe from the sample, and it was used instead of coinfection since coinfection means all the detected microbes must contribute to the pathogenic effect, whereas codetection may not indicate the necessary causative role.
Statistical Analysis
Detection and codetection frequencies of each pathogen in children were compared, and the difference of the clinical symptoms of various viral/bacterial infections was tested by Pearson's χ2 test, Fisher's exact test for dichotomous variables, or Kruskal–Wallis test (a nonparametric test that does not assume a normal distribution of the residuals) for continuous variables. To investigate the association of clinical symptoms, outcomes, and severity with different mono or codetection patterns, logistic regression was used for estimation of the odds ratio (OR) adjusting for age and sex. Statistical analyses were conducted in R software (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Detections and Codetections
In the study period, 3181 (77.0%) of the 4130 eligible patients were enrolled in our study. Among those enrolled hospitalized children, there were 2375 (74.7%) cases detected with ≥1 virus and 707 (22.2%) detected with ≥1 bacteria; 901 (28.3%) children detected with ≥2 viruses; 57 (1.8%) detected with ≥2 bacteria; whereas 542 (17.0%) were codetected with both virus and bacteria. Detection of viruses was more common among children aged <4 compared with those >4 years of age, especially, highest (76.7%) among those aged <1 (Table 1). However, the detection rate of bacteria was highest among those aged 1 to 4 years, then less among age group 0–1 and 4–6 years, and least among the older children aged >6. Virus and bacteria codetections were highest in the age group 1–4 years of age. Notably, there were 38.6% of the cases aged 6 to 16 who had negative detections for the pathogen of interest. Males had more frequent detections of viruses, bacteria, and codetection of both than females, and with less negative detections. Generally, detection rates of the viruses of interest across each year in this study (from June 2009 to October 2013) were significantly different, lower in 2013 than among the other years. However, the bacteria detection rates and virus and bacteria codetection rates were not significantly different over the years.
TABLE 1.
Pathogen Detection Rate by Sex, Age Group, and Year
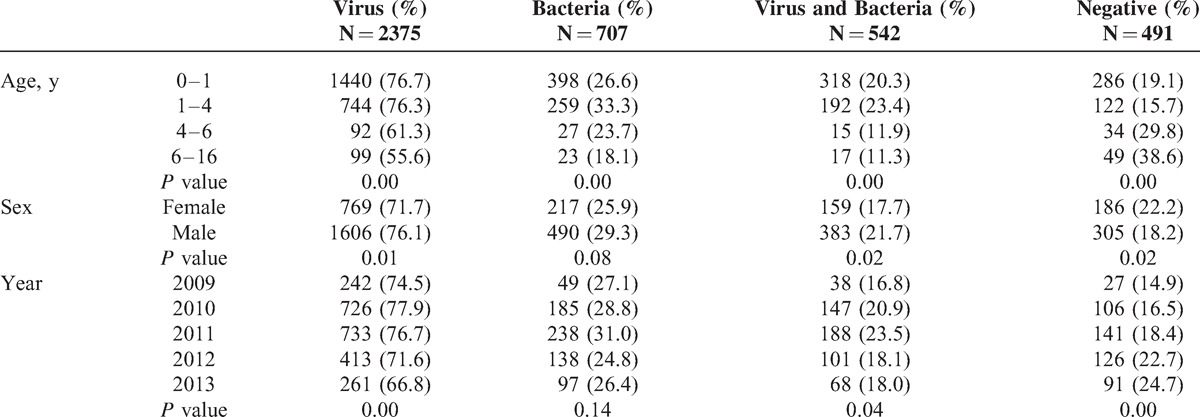
Generally, RSV (35.0%), PIV (24.9%), IFVA (14.9%), and BoV (12.3%) were more frequently detected among the 2375 case with virus detections; S pneumoniae (14.4%) was the most frequently detected bacteria among the 603 bacteria detections (Figure 1). For codetections between bacteria, H parainfluenzae had higher percentage (17.8%) of codetection with other bacteria. For codetection among different viruses, most of them had high percentage as codetected with other viruses, varying from 46.8% to 75.4%; BoV, IFVB, CoV, and MPV had higher percentages than the other virus. However, referring to codetections of both virus and bacteria, all the four kinds of bacteria-detected cases had high percentage of codetections with virus (74.4% to 78.0%); the samples detected with viruses have low proportion of codetection with bacteria, which varied from 21.6% for IFVA to 39.8% for MPV.
FIGURE 1.
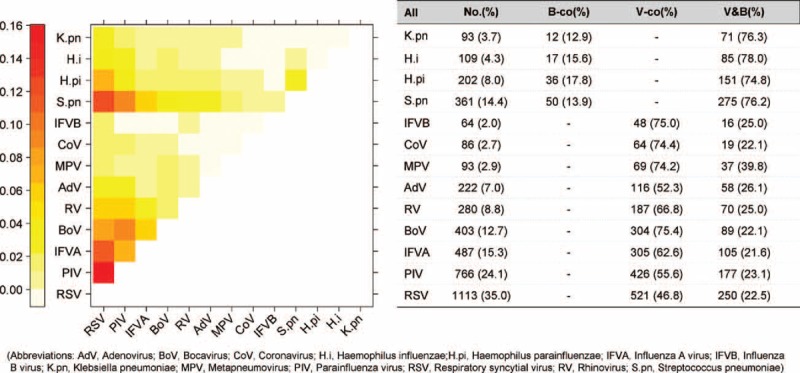
Codetection number and patterns between pathogens. AdV = adenovirus, ARI = acute respiratory infection, BoV = bocavirus, CoV = coronavirus, H.i = Haemophilus influenzae, H.pi = Haemophilus parainfluenzae, IFVA = influenza A virus, IFVB = influenza B virus, K.pn = Klebsiella pneumoniae, MPV = metapneumovirus, PIV = parainfluenza virus, RSV = respiratory syncytial virus, RV = rhinovirus, S.pn = Streptococcus pneumoniae.
In total, 22.8% of the children with virus detections were codetected with bacteria, and the most frequently codetected were S pneumoniae; and 76.7% of those with bacteria detections were codetected with virus, among which the noninfluenza virus were more frequent than influenza, specifically, with RSV (35.4%) and PIV (25%) more frequently codetected (Supplemental Figure, http://links.lww.com/MD/A248).
The heatmap in Figure 1 indicates the specific pattern of virus and bacteria codetection pairs. Among codetection within bacteria, S pneumoniae was more often codetected with H influenzae and H parainfluenzae, whereas there were very few codetections between others. For virus codetection, RSV, PIV, INFA, BoV, and RV were more frequently codetected with each other, especially, RSV and PIV. For virus and bacteria codetection, we found that S pneumoniae and H parainfluenzae were more frequently codetected with RSV, PIV, IFVA, and BoV.
Clinical Characteristics
Generally, there were no significant differences in clinical characteristics between patients with the virus and bacteria detections (data not shown). Among those detections with bacteria, most clinical characteristics were similar between different pathogen detection except for fever, cough, and mean age, which differed significantly (Supplemental Table 1-a, http://links.lww.com/MD/A248) (P < 0.05). S pneumoniae-detected cases had higher proportion of fever and older mean age than other viruses, whereas H influenzae and S pneumoniae-detected cases reported higher proportion of cough. For virus-detected cases, the clinical characteristics were quite different among different viruses (Supplemental Table 1-b, http://links.lww.com/MD/A248) (P < 0.05). AdV, IFVA, and IFVB had higher proportion of fever and greater mean age; RSV, PIV, and BoV-detected cases have higher proportion of cough than the others. IFVB and RSV had higher proportion of sputum; AdV, IFVB, RV, and RSV showed higher proportion of dyspnea; and PIV and RSV-detected cases had more presentation of diarrhea. When the clinical characteristics were compared by age groups, most of the differences were significant among younger age groups and fewer were significant for the older children aged >6 years (data not shown).
To assess the influence of virus and bacteria detection patterns (mono or codetection) on the clinical characteristics, we compared the clinical characteristics among the different detection patterns (Supplemental Table 2, http://links.lww.com/MD/A248). There were significant difference for fever, cough mean age (P < 0.05), and diarrhea (P < 0.1). Those cases detected with ≥2 bacteria had highest proportion of fever. Bacteria and virus codetected more frequently presented with cough. Cases codetected with ≥2 viruses or both virus and bacteria had a higher proportion presenting with diarrhea. The mean age was higher among cases with >2 bacteria. By further regressing the detection results against those clinical characteristic with age and sex adjusted in the logistic regression, we found that all cases with codetection of ≥2 virus (OR, 1.2; CI, 1.0–1.5), ≥2 bacteria (OR, 2.2; CI, 1.1–4.5), and virus and bacteria (OR, 1.4; CI, 1.1–1.7) were significantly more likely to present fever than those with monodetection (Table 2). Those with ≥2 virus codetection were 1.7 times more likely to have cough than monovirus-detected cases (OR, 1.7; CI, 1.0–2.8). Furthermore, those with both virus and bacteria detections were also more likely to present with cough (OR, 2.1; CI, 1.1–4.1) and sputum (OR, 1.3; CI, 1.0–1.7) than those with only 1 virus or bacteria (Supplemental Table 3, http://links.lww.com/MD/A248).
TABLE 2.
Clinical Characteristics Comparison Among Different Mono and Codetections

Clinical Outcomes and Severity
Among all those children with different pathogen detections, 82.8% to 95.3% had pneumonia, 8.6% to 28.8% had severe pneumonia based on the definition mentioned above, and <5% were transferred to the intensive care unit (ICU) (Figure 2). Specifically, cases detected with CoV and K pneumoniae had higher rate (95.3% and 94.6%) of pneumonia among all pathogens detections. Those with detections of AdV and K pneumoniae had higher risk (28.8% and 28.0%) of severe pneumonia than others, whereas IFVB and K pneumoniae-detected cases had a higher risk (4.7% and 3.2%) of being transferred to the ICU. To further investigate the impact of codetection on the clinical outcomes as compared with the single detection, we applied logistic regression to estimate the OR with adjustment for age. Generally, the risk of developing pneumonia, severe pneumonia, and transfer to the ICU were not significantly different between those cases with codetections and single detection of either virus or bacteria (data not shown).
FIGURE 2.
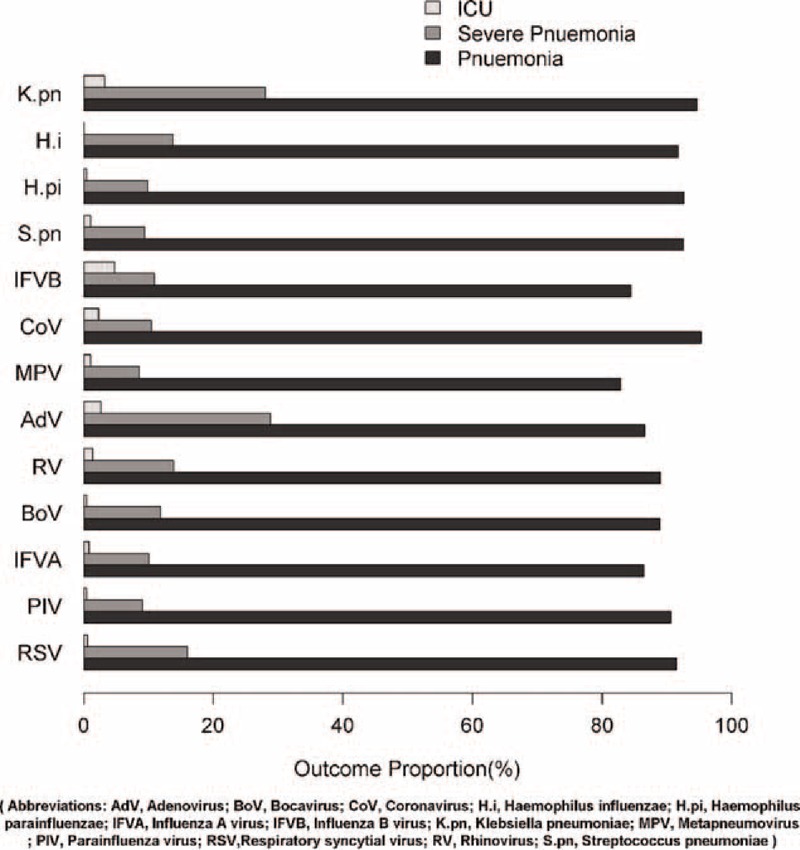
Clinical outcomes of the Acute Respiratory Infections cases by each pathogen. AdV = adenovirus, ARI = acute respiratory infection, BoV = bocavirus, CoV = coronavirus, H.i = Haemophilus influenzae, H.pi = Haemophilus parainfluenzae, ICU = intensive care unit, IFVA = influenza A virus, IFVB = influenza B virus, K.pn = Klebsiella pneumoniae, MPV = metapneumovirus, PIV = parainfluenza virus, RSV = respiratory syncytial virus, RV = rhinovirus, S.pn = Streptococcus pneumoniae.
DISCUSSION
In this study, by performing prolonged and systematic surveillance, we obtained a complete spectrum of viral and bacterial etiological agents among hospitalized children with ARIs. High codetection rate of respiratory microbial among those hospitalized pediatric ARI cases were demonstrated. Epidemiological and clinical features from single and mixed detections were described. Fever was found to be a significant predictor for codetection, whereas cases with codetection showed no significant increased severity of outcomes than monodetection in this study.
This is the first study to investigate the detection and codetection pattern based on large sample size in China. The overall positive detection for any respiratory virus was 74.7%, and 22.2% for bacteria, with S pneumoniae most commonly detected in bacteria, and RSV, PIV, IFVA, and BoV among viruses, which was comparable to some other etiological studies within children or whole populations.13–16S pneumoniae was more often codetected with H parainfluenzae and H influenzae, and previous literature has reported the interaction between these common bacteria and suggested the mechanism like synergism or competition.17,18 Margolis et al19 reported an increase of H influenzae when either S pneumoniae or Staphylococcus aureus was present, indicating synergism between these bacterial species. Previous experiments by Lysenko et al20 observed competition between S pneumoniae and H influenzae in a mouse model of colonization. For H parainfluenzae, it is often been seen as an upper respiratory tract contaminant with other pathogens, with potential pathogenic role in lower respiratory tract in patients with severely impaired host defense.21 In this study, it had a high chance of being codetected with other bacteria (17.8) and viruses (74.8%), which may account for the high pneumonia rate in those detections with H parainfluenzae.
For codetection within viruses, IFVA/B, CoV, BoV, MPV, and RV were more often codetected with other viruses. Some other studies indicated that viruses more frequently involving dual infection were RSV, INF, PIV, AdV, CoV, and RV.22 The mixed respiratory virus detection pattern varies widely in studies, with various of combinations like RSV and IFV, RSV and PIV, IFVA and IFVB, IFVA and AdV, RSV and RV, PIV and RV, MPV and RSV, and others.22–25 Although codetection may not always result in interactions for all virus species, virus–virus interaction is quite common and have 3 potential categories (genetic, host environment, and immunological mechanisms), which is critical for understanding of viral etiology and pathogenesis.26
The pathogens more frequently involving mixed detection of both virus and bacteria were S pneumoniae, RSV, PIV, IFVA, BoV, and RV. There were lots of studies that found that IFVA and RSV infections result in a predisposition to S pneumoniae, and H influenzae, which accounted for a substantial burden of morbidity and mortality.27–29 Some of the literature which reported codetection of RSV and S pneumoniae indicated that the underlying mechanism was RSV infection that could enhance adherence of S pneumoniae to human epithelial cells.30,31 In addition, IFVA and S pneumoniae codetection was also frequently reported, with S pneumoniae playing a role as complication during the previous influenza pandemics and contribute substantially to morbidity and mortality.32–34 Other studies have reported the concomitant presence of S pneumoniae with other respiratory viruses, such as RV, MPV, and ADV, with similar interaction.35–38 The mechanism by which viruses affect bacterial colonization and invasion is quite diverse in the previous literature, as it is multidimensional and involves a complex interplay among various hosts and pathogens.18
The large sample size allows us to perform the multivariate analysis, thus to acquire epidemiological and clinical features of the viruses/bacteria codetection. We found that the virus detection rate was higher among children aged <4, especially, those aged <1, which is consistent with some other studies on different population.13,22,39 However, bacteria detection and codetection of virus and bacteria were highest among those aged 1 to 4 years, whereas another study found that the mixed virus–bacterial infection was most common in children aged <2 years.40 Our data also indicated that males had higher detection and codetection rate of virus and bacteria than females, and this sex difference was similarly reported in the previous literature.39,41 The sex difference might be due to the effect of health-seeking behaviors or sex-specific risk for respiratory infection, which needs further study.
Our study found that the fever and cough were 2 main significant predictors for virus codetection; fever alone was a significant predictor for bacteria codetection; fever, cough, and sputum were significantly more frequent in virus and bacteria codetected cases than monodetected. This finding is partially in line with some other studies even with different population.42,43 This could be helpful for differentiating mono and codetection by preliminary symptoms, which could be useful for early treatment.
Regarding the clinical severity and impact of respiratory microbial codetections, the literature showed conflicting results. Our study found that there was no significant higher risk of pneumonia, severe pneumonia, and ICU admission for all codetections compared with monodetections based on the logistic regression adjusted for age and sex. However, some other studies revealed that some codetection did increase the severity of the respiratory illnesses, such as higher risk of sever pneumonia, longer hospital stays, and higher ICU admission.44–47 Some historical influenza pandemics had bacterial superinfections with increased mortality, most of which involving secondary pneumococcal pneumonia.33,48 Another study indicated the viral–bacterial coinfection, especially, S pneumoniae may cause most severe human MPV infection processing to pneumonia.35 Some other studies found that the outcomes including mortality that associated with bacterial–viral coinfection were not significantly different.49,50 Furthermore, there were a few studies that found the opposite result, such as RSV coinfections were inversely associated with hospitalization and RV-coinfected patients had less-severe disease.51,52
Conflicting results may be due to several factors, such as the study periods, target population, and environment factors. What is more, the presence of pathogens tested by molecular techniques or conventional culture may not confirm significant pathogenic role at that time. As viral–bacterial coinfection may lead to failure of the antibiotic therapy, this poses a challenge to efficient and effective treatments for those patients. Better understanding the pattern of mixed infection and their interaction, and assessing the clinical severity will be helpful for designing successful therapeutic strategies. Better and more sensitive etiologic diagnostic tool for respiratory microbial infection can reduce overuse of antibiotic in primary care.
There are some limitations of this study. First, the database of this study included limited information on the disease outcomes (eg, the death records), thus may underestimate the severity and disease burden. Second, the use of antibiotics might obscure the real association between these viral and bacterial detections in this study. Third, for the sputum as we had no information on the bacterial load of these positive detected cases, quantitative comparison could not be implemented in this study. Fourth, 79% of the cases had sputum culture results, with 667 cases undetected for bacteria, which may lead to underestimation of the bacterial burden. On the other hand, sputum might be contaminated by upper airway tract secretions, leading to overestimation. Last, the detection of the pathogens does not mean that it is the cause of the illness, thus it is difficult to determine the real cause of the patients’ illness.
We also acknowledge that these findings are based on pediatric patients from a subtropical city, and may not be generalized to other population or regions. As specific bacterial diagnosis based on the traditional methods is difficult, advanced bacteriological methods are needed in future. Further identification of predictive clinical parameters combined with accurate diagnosis is warrant for optimizing treatment including reasonable use of antibiotics.
Footnotes
Abbreviations: AdV = adenovirus, ARI = acute respiratory infection, BoV = bocavirus, CoV = coronavirus, ICU = intensive care unit, IFVA = influenza A virus, IFVB = influenza B virus, MPV = metapneumovirus, PIV = parainfluenza virus, RSV = respiratory syncytial virus, RV = rhinovirus.
This study was supported by grants from the China Mega-Project on Infectious Disease Prevention (No. 2013ZX10004202-002) and National Science Fund for Young Scholars (No. 81222037). BJC received research funding from MedImmune Inc. and Sanofi Pasteur.
BJC consults for Crucell NV. The other authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.The Acute Respiratory Infections Atlas. 2010. http://www.ariatlas.org/understanding_aris Accessed September 15, 2014. [Google Scholar]
- 2.Schaffer K, La Rosa AM, Whimbey E. Cohen J, Powderly WG, Opal SM. Respiratory viruses. Infectious Diseases 3rd ed. Philadelphia, PA: Mosby/Elsevier Medicine, 2010. [Google Scholar]
- 3.Black RE, Cousens S, Johnson HL, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 2010; 375:1969–1987. [DOI] [PubMed] [Google Scholar]
- 4.WHO. The global burden of disease: 2004 update. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 5.Calvo C, Garcia-Garcia ML, Blanco C, et al. Multiple simultaneous viral infections in infants with acute respiratory tract infections in Spain. J Clin Virol 2008; 42:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paranhos-Baccala G, Komurian-Pradel F, Richard N, et al. Mixed respiratory virus infections. J Clin Virol 2008; 43:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng D, Zhao D, Liu J, et al. Multipathogen infections in hospitalized children with acute respiratory infections. Virol J 2009; 6:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shann F, Gratten M, Germer S, et al. Aetiology of pneumonia in children in Goroka Hospital, Papua New Guinea. Lancet 1984; 2:537–541. [DOI] [PubMed] [Google Scholar]
- 9.Xu W, McDonough MC, Erdman DD. Species-specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol 2000; 38:4114–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiveljung-Lindell A, Rotzen-Ostlund M, Gupta S, et al. Development and implementation of a molecular diagnostic platform for daily rapid detection of 15 respiratory viruses. J Med Virol 2009; 81:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray PR, Washington JA. Microscopic and bacteriologic analysis of expectorated sputum. Mayo Clinic Proc 1975; 50:339–344. [PubMed] [Google Scholar]
- 12.Murray PR. Manual of Clinical Microbiology. 6th ed.Washington, DC: American Society for Microbiology Press; 1995. [Google Scholar]
- 13.Cilla G, Onate E, Perez-Yarza EG, et al. Viruses in community-acquired pneumonia in children aged less than 3 years old: high rate of viral coinfection. J Med Virol 2008; 80:1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan R, Rhodes J, Thamthitiwat S, et al. Incidence and etiology of acute lower respiratory tract infections in hospitalized children younger than 5 years in rural Thailand. Pediatr Infect Dis J 2014; 33:E45–E52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khor CS, Sam IC, Hooi PS, et al. Epidemiology and seasonality of respiratory viral infections in hospitalized children in Kuala Lumpur, Malaysia: a retrospective study of 27 years. BMC Pediatr 2012; 12:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falsey AR, Becker KL, Swinburne AJ, et al. Bacterial complications of respiratory tract viral illness: a comprehensive evaluation. J Infect Dis 2013; 208:432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby P, Watson K, Bowman J, et al. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine 2007; 25:2458–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosch AA, Biesbroek G, Trzcinski K, et al. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathogens 2013; 9:e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolis E, Yates A, Levin BR. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host's immune response. BMC Microbiol 2010; 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lysenko ES, Ratner AJ, Nelson AL, et al. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathogens 2005; 1:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Middleton AM, Dowling RB, Mitchell JL, et al. Haemophilus parainfluenzae infection of respiratory mucosa. Respir Med 2003; 97:375–381. [DOI] [PubMed] [Google Scholar]
- 22.Drews AL, Atmar RL, Glezen WP, et al. Dual respiratory virus infections. Clin Infect Dis 1997; 25:1421–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caracciolo S, Minini C, Colombrita D, et al. Human metapneumovirus infection in young children hospitalized with acute respiratory tract disease: virologic and clinical features. Pediatr Infect Dis J 2008; 27:406–412. [DOI] [PubMed] [Google Scholar]
- 24.Kouni S, Karakitsos P, Chranioti A, et al. Evaluation of viral co-infections in hospitalized and non-hospitalized children with respiratory infections using microarrays. Clin Microbiol Infect 2013; 19:772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debiaggi M, Canducci F, Ceresola ER, et al. The role of infections and coinfections with newly identified and emerging respiratory viruses in children. Virol J 2012; 9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DaPalma T, Doonan BP, Trager NM, et al. A systematic approach to virus-virus interactions. Virus Res 2010; 149:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beadling C, Slifka MK. How do viral infections predispose patients to bacterial infections? Curr Opin Infect Dis 2004; 17:185–191. [DOI] [PubMed] [Google Scholar]
- 28.Korppi M, Leinonen M, Koskela M, et al. Bacterial coinfection in children hospitalized with respiratory syncytial virus infections. Pediatr Infect Dis J 1989; 8:687–692. [DOI] [PubMed] [Google Scholar]
- 29.Chertow DS, Memoli MJ. Bacterial coinfection in influenza: a grand rounds review. J Am Med Assoc 2013; 309:275–282. [DOI] [PubMed] [Google Scholar]
- 30.Avadhanula V, Rodriguez CA, Devincenzo JP, et al. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol 2006; 80:1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hament JM, Aerts PC, Fleer A, et al. Enhanced adherence of Streptococcus pneumoniae to human epithelial cells infected with respiratory syncytial virus. Pediatr Res 2004; 55:972–978. [DOI] [PubMed] [Google Scholar]
- 32.Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis 2006; 6:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chien YW, Klugman KP, Morens DM. Bacterial pathogens and death during the 1918 influenza pandemic. N Engl J Med 2009; 361:2582–2583. [DOI] [PubMed] [Google Scholar]
- 34.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008; 198:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhi SA, Ludewick H, Kuwanda L, et al. Pneumococcal coinfection with human metapneumovirus. J Infect Dis 2006; 193:1236–1243. [DOI] [PubMed] [Google Scholar]
- 36.Peltola V, Heikkinen T, Ruuskanen O, et al. Temporal association between rhinovirus circulation in the community and invasive pneumococcal disease in children. Pediatr Infect Dis J 2011; 30:456–461. [DOI] [PubMed] [Google Scholar]
- 37.Hakansson A, Kidd A, Wadell G, et al. Adenovirus infection enhances in vitro adherence of Streptococcus pneumoniae. Infect Immunity 1994; 62:2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishizuka S, Yamaya M, Suzuki T, et al. Effects of rhinovirus infection on the adherence of Streptococcus pneumoniae to cultured human airway epithelial cells. J Infect Dis 2003; 188:1928–1939. [DOI] [PubMed] [Google Scholar]
- 39.Chorazy ML, Lebeck MG, McCarthy TA, et al. Polymicrobial acute respiratory infections in a hospital-based pediatric population. Pediatr Infect Dis J 2013; 32:460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korppi M. Mixed microbial aetiology of community-acquired pneumonia in children. APMIS 2002; 110:515–522. [DOI] [PubMed] [Google Scholar]
- 41.Nair H, Simoes EA, Rudan I, et al. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet 2013; 381:1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huijskens EG, Koopmans M, Palmen FM, et al. The value of signs and symptoms in differentiating between bacterial, viral and mixed aetiology in patients with community-acquired pneumonia. J Med Microbiol 2014; 63 (pt 3):441–452. [DOI] [PubMed] [Google Scholar]
- 43.Michelow IC, Olsen K, Lozano J, et al. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics 2004; 113:701–707. [DOI] [PubMed] [Google Scholar]
- 44.Johansson N, Kalin M, Hedlund J. Clinical impact of combined viral and bacterial infection in patients with community-acquired pneumonia. Scand J Infect Dis 2011; 43:609–615. [DOI] [PubMed] [Google Scholar]
- 45.Templeton KE, Scheltinga SA, van den Eeden WC, et al. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis 2005; 41:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semple MG, Cowell A, Dove W, et al. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis 2005; 191:382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papadopoulos NG, Moustaki M, Tsolia M, et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med 2002; 165:1285–1289. [DOI] [PubMed] [Google Scholar]
- 48.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 2006; 19:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi SH, Hong SB, Ko GB, et al. Viral infection in patients with severe pneumonia requiring intensive care unit admission. Am J Respir Crit Care Med 2012; 186:325–332. [DOI] [PubMed] [Google Scholar]
- 50.De Paulis M, Gilio AE, Ferraro AA, et al. Severity of viral coinfection in hospitalized infants with respiratory syncytial virus infection. J Pediatr 2011; 87:307–313. [DOI] [PubMed] [Google Scholar]
- 51.Esper FP, Spahlinger T, Zhou L. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J Infect 2011; 63:260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papenburg J, Hamelin ME, Ouhoummane N, et al. Comparison of risk factors for human metapneumovirus and respiratory syncytial virus disease severity in young children. J Infect Dis 2012; 206:178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]


