Supplemental Digital Content is available in the text
Abstract
Papillary renal cell carcinoma (pRCC) is the second most prevalent subtype of kidney cancers. In the current study, we analyzed the global microRNA (miRNA) expression profiles in pRCC, with the aim to evaluate the relationship of miRNA expression with the progression and prognosis of pRCC.
A total of 163 treatment-naïve primary pRCC patients were identified from the Cancer Genome Atlas dataset and included in this retrospective observational study. The miRNA expression profiles were graded by tumor-node-metastasis information, and compared between histologic subtypes. Furthermore, the training-validation approach was applied to identify miRNAs of prognostic values, with the aid of Kaplan–Meier survival, and univariate and multivariate Cox regression analyses. Finally, the online DAVID (Database for Annotation, Visualization, and Integrated Discover) program was applied for the pathway enrichment analysis with the target genes of prognosis-associated miRNAs, which were predicted by 3 computational algorithms (PicTar, TargetScan, and Miranda).
In the progression-related miRNA profiles, 26 miRNAs were selected for pathologic stage, 28 for pathologic T, 16 for lymph node status, 3 for metastasis status, and 32 for histologic types, respectively. In the training stage, the expression levels of 12 miRNAs (mir-134, mir-379, mir-127, mir-452, mir-199a, mir-200c, mir-141, mir-3074, mir-1468, mir-181c, mir-1180, and mir-34a) were significantly associated with patient survival, whereas mir-200c, mir-127, mir-34a, and mir-181c were identified by multivariate Cox regression analyses as potential independent prognostic factors in pRCC. Subsequently, mir-200c, mir-127, and mir-34a were confirmed to be significantly correlated with patient survival in the validation stage. Finally, target gene prediction analysis identified a total of 113 target genes for mir-200c, 37 for mir-127, and 180 for mir-34a, which further generated 15 molecular pathways.
Our results identified the specific miRNAs associated with the progression and aggressiveness of pRCC, and 3 miRNAs (mir-200c, mir-127, and mir-34a) as promising prognostic factors of pRCC.
INTRODUCTION
Renal cell carcinoma (RCC) accounts for approximately 90% of all kidney tumors and 2% to 3% of all human malignancies, with an estimated 63,920 newly diagnosed cases and 13,860 RCC-related deaths for 2014 in the United States.1 As the second most common histologic subtype after clear cell RCC (ccRCC), papillary RCC (pRCC) represents 10% to 15% of all RCC cases.2 In the current clinical practice, the histopathologic parameters including tumor-node-metastasis (TNM) stage, histologic subtype (types 1 and 2), tumor necrosis and microvascular invasion, and preexisting health problems such as type 2 diabetes mellitus have been established and validated as potent prognostic factors; however, they are of limited values as the clinical behavior and long-term outcomes of pRCC are highly variable.3–6 Hence, the identification of novel molecular biomarkers that are predictive of pRCC aggressiveness and patient outcome is of great importance, which could help identify the molecular mechanisms and improve the ability to manage pRCC patients.
MicroRNAs (miRNAs) are short (about 19–25 nucleotides in length), noncoding, and single-stranded RNAs that regulate gene expression post-transcriptionally through the epigenetic mechanism of RNA interference.7 miRNAs are implied in a wide range of biological processes including cellular proliferation, differentiation, and apoptosis, via regulating the expression of hundreds of target genes.8 Furthermore, miRNAs are aberrantly expressed or mutated in human cancers, suggesting that they may exert critical functions as a novel class of tumor suppressive or carcinogenic factors.9–13 The diagnostic and prognostic capabilities of miRNAs have been thoroughly explored in ccRCC, and the cancer-specific miRNA expression profiles have been identified, which were significantly associated with patient survival.14–16 However, the clinical utility of miRNAs in pRCC remains to be elucidated.
Hence, we stringently designed a stepwise study with the data from The Cancer Genome Atlas (TCGA) project, which provides a collection of clinicopathological data and the global miRNA expression profiles.17,18 In the current study, we explored the miRNAs associated with the progression and prognosis of pRCC, with the hope to identify the specific miRNAs that could predict the clinical phenotypes and prognosis in pRCC.
MATERIALS AND METHODS
Patients and Samples
This retrospective observational study was conducted and reported in accordance with the STROBE (Strengthening the Reporting of Observational studies in Epidemiology) guidelines.19–22 All pRCC patients were identified from the multi-institutional TCGA project (http://cancergenome.nih.gov/) that underwent nephrectomy from 1996 to 2013 for sporadic pRCC.23 The full clinical data (Level 1 and Level 2) were downloaded from the TCGA data portal (up to October 1, 2014) and double checked for the further assessment of the eligibility. The subjects with history of other malignancies or neoadjuvant therapy (chemotherapy or radiation therapy) were excluded. Furthermore, the pathological stage was reevaluated and confirmed by 2 experienced pathologists according to the 7th edition of TNM classification of the American Joint Committee on Cancer (AJCC). Overall, a total of 163 pRCC patients were enrolled with full annotation of the corresponding clinicopathological data including age, sex, race, histologic subtype, and AJCC TNM information (Table 1). In the current study, the 2-stage training-validation approach was adopted to identify miRNAs predictive of patient survival. Among the 163 pRCC patients routinely followed up at the corresponding centers, 129 subjects (79.1%) were followed up for >30 days and included in the training stage. In the subsequent validation stage, all subjects (n = 163) were included with limited follow-up time (2 year, 730 days) to confirm the prognostic power of the miRNAs selected in the training stage. All data collection and procession (including the consenting process) were conducted with approvals by the corresponding institutional review boards and in agreement with the human subject protection and data access policies of TCGA.
TABLE 1.
Clinicopathological Characteristics of Patients With Papillary Renal Cell Carcinoma

Microarray Data Procession
The miRNA expression profiling was performed with the Illumina HiSeq platform (Illumina Inc, San Diego, CA) and quantified by relative miRNA read counts to the total miRNAs read counts and presented as reads per million (RPM) counts. After downloading from TCGA data portal (up to October 15, 2014), the summary miRNA data was processed using BRB-Array tools (version 4.4.0; National Cancer Institute, Bethesda, MD) that were developed by Dr Richard Simon and BRB-Array Tools Development Team (http://brb.nci.nih.gov/BRB-ArrayTools.html).24 Briefly, the miRNAs were retained when they were >1 RPM in at least 10% of all samples and had changes of >1.5 fold from the median value in at least 20% of samples. Subsequently, the expression level of each individual miRNA was log2 transformed for further analysis.
Target Gene Prediction and Pathway Analysis
Three computational algorithms, PicTar (http://pictar.mdc-berlin.de/),25 TargetScan (http://www.targetscan.org/),7 and Miranda (http://www.microrna.org),26 were employed to predict miRNA targets, and the genes predicted by at least 2 independent tools were taken into consideration. The selected genes of each individual miRNA were uploaded to the online Database for Annotation, Visualization, and Integrated Discovery (DAVID) program (http://david.abcc.ncifcrf.gov/)27 for the analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.28 In the functional annotation analysis, the minimum number of genes for the corresponding term was set as 2 to identify appropriate functional categories. To examine the significance of gene-term enrichment, a modified Fisher test was conducted and the significant P value was set as 0.1.27
Statistical Analysis
The continuous variables were presented as mean ± standard deviation (SD), after the normality status was explored with Kolmogorov–Smirnov and Shapiro–Wilk tests. The miRNA expression levels between the 2 different groups (stages III + IV vs stages I + II, T3 + T4 vs T1 + T2, N1 + N2 vs N0, M1 vs M0, and type 2 vs type 1) were ascertained with Student t test (significance level was set as 0.001). To generate a tree cluster showing the separation of different classes, the hierarchical cluster analysis was performed by Euclidian distance and average linkage with BRB-Array tools.
Kaplan–Meier survival and univariate Cox proportional hazards regression analyses were conducted to explore the effects of age, sex, race, AJCC stage (stages III + IV vs stages I + II), tumor size (T3 + T4 vs T1 + T2), and miRNA expression levels (cutoff point: median value) on patient survival. Furthermore, the multivariate Cox proportional hazards regression analysis was performed by combining all potential prognostic factors, with the stepwise backward likelihood ratio method. In the Cox proportional hazards regression analyses, the results were expressed as hazard ratio (HR) with the corresponding 95% confidential interval (CI). Statistical significance was taken as a 2-sided P value <0.05 unless specifically indicated. The statistical analyses were performed with the use of BRB-Array Tools and SPSS (version 21.0; SPSS Institute Inc, Chicago, IL), as appropriate.
RESULTS
Patient Characteristics
All 163 patients enrolled in the current study were clinically and pathologically diagnosed with pRCC and underwent nephrectomy without history of other malignancies and neoadjuvant therapy (chemotherapy or radiation therapy). The mean age for all 163 patients was 59.9 years (SD: 12.6 years), and the detailed demographic and clinicopathologic information was summarized in Table 1.
MiRNAs in Relation to Tumor Progression of pRCC
To identify miRNAs related to progression for each clinical feature, the class comparison analyses were conducted. A summary of 26 miRNAs were selected for AJCC stage, 28 for pathologic T, 16 for lymph node involvement status, 3 for distant metastasis status, and 32 for histologic types (see Supplementary Table S1, http://links.lww.com/MD/A250, Supplementary Digital Content 1, which illustrates miRNAs associated with the histopathological characteristics of pRCC). Additionally, the unsupervised hierarchical clustering with the miRNAs expression data could clearly separate different classes according to the stage, pathologic T, lymph node involvement, and histologic type (Figure 1).
FIGURE 1.
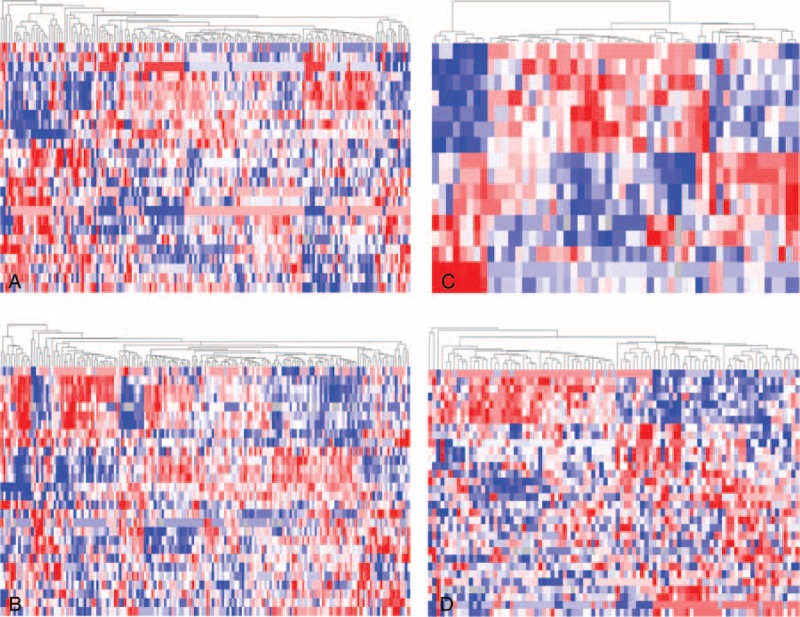
Unsupervised hierarchical cluster analysis of the differentially expressed microRNAs. The papillary renal cell carcinoma patients were compared between 2 groups according to (A) stage (stages III + IV vs stages I + II); (B) pathologic T (T3 + T4 vs T1 + T2); (C) pathologic N (N1–2 vs N0); and (D) histologic subtype (type 2 vs type 1).
MiRNA Expression Profiles Associated With pRCC Prognosis
In the training stage, among the 129 pRCC patients followed up for >30 days, 18 (14.0%) participants had died during the median follow-up time of 12.1 months. As indicated in the univariate Cox regression analyses (Table 2), a total of 12 miRNAs (mir-134, mir-379, mir-127, mir-452, mir-199a, mir-200c, mir-141, mir-3074, mir-1468, mir-181c, mir-1180, and mir-34a) were significantly associated with patient survival in pRCC, which were further confirmed by Kaplan–Meier survival analyses (Figures 2 and 3). Furthermore, the multivariate Cox regression analyses demonstrated that AJCC stage (P = 0.019), mir-200c (P = 0.007), mir-127 (P = 0.001), mir-34a (P = 0.010), and mir-181c (P = 0.001) were potential independent prognostic factors (Table 2 and Figure 3). Subsequently, the prognostic probabilities of mir-200c, mir-127, mir-34a, and mir-181c were explored in the validation stage with all subjects (n = 163) followed up for 2 years. As shown in Figure 4, mir-200c (P = 0.010), mir-127 (P = 0.016), and mir-34a (P = 0.001) except mir-181c (P = 0.638) were significantly associated with patient survival of pRCC patients, which were further confirmed by univariate Cox regression analyses: mir-200c (HR = 5.894, 95% CI: 1.271–27.33; P = 0.023), mir-127 (HR = 8.336, 95% CI: 1.066–65.21; P = 0.043), mir-34a (HR = 0.072, 95% CI: 0.009–0.561; P = 0.012), and mir-181c (HR = 1.329, 95% CI: 0.405–4.355; P = 0.639). Overall, mir-200c, mir-127, and mir-34a could serve as promising prognostic biomarkers in pRCC, even though only mir-34a (HR = 0.046, 95% CI: 0.004–0.485; P = 0.010) passed the multivariate Cox regression analysis in the validation stage.
TABLE 2.
Univariate and Multivariate Cox Regression Analysis of Overall Survival in Papillary Renal Cell Carcinoma Patients
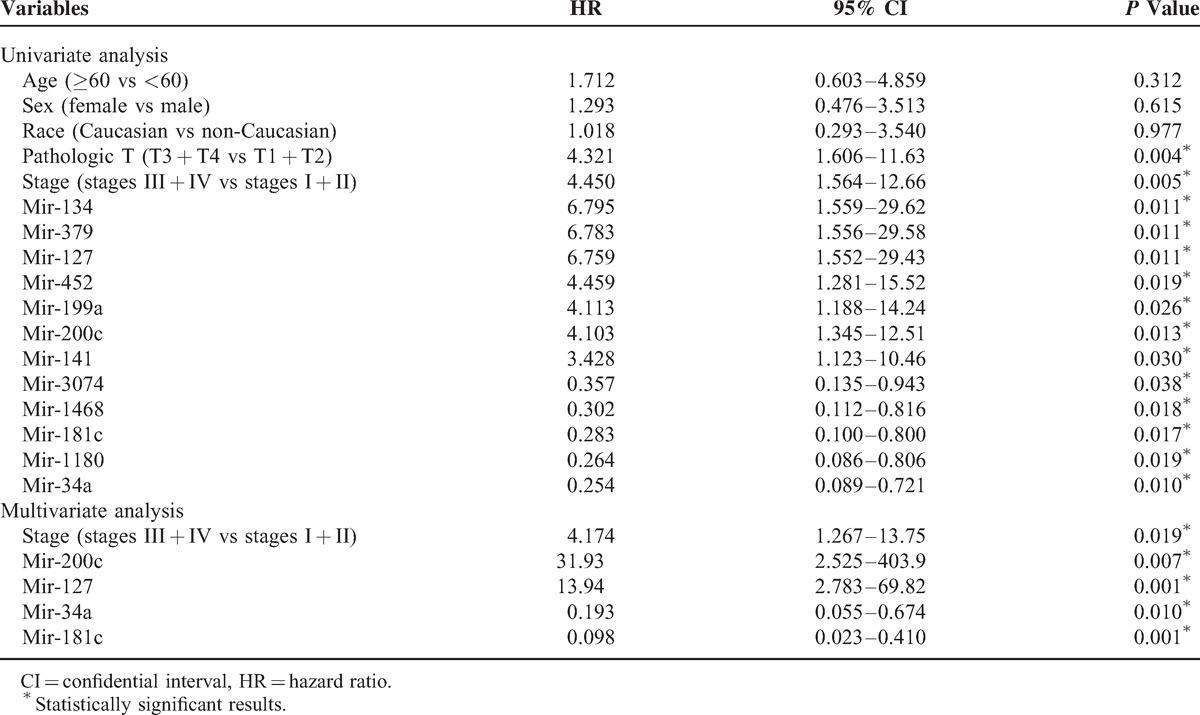
FIGURE 2.

Kaplan–Meier curve analysis of microRNAs for the survival in papillary renal cell carcinoma patients. A total of 8 microRNAs are presented, including mir-134, mir-379, mir-452, mir-199a, mir-141, mir-3074, mir-1468, and mir-1180.
FIGURE 3.
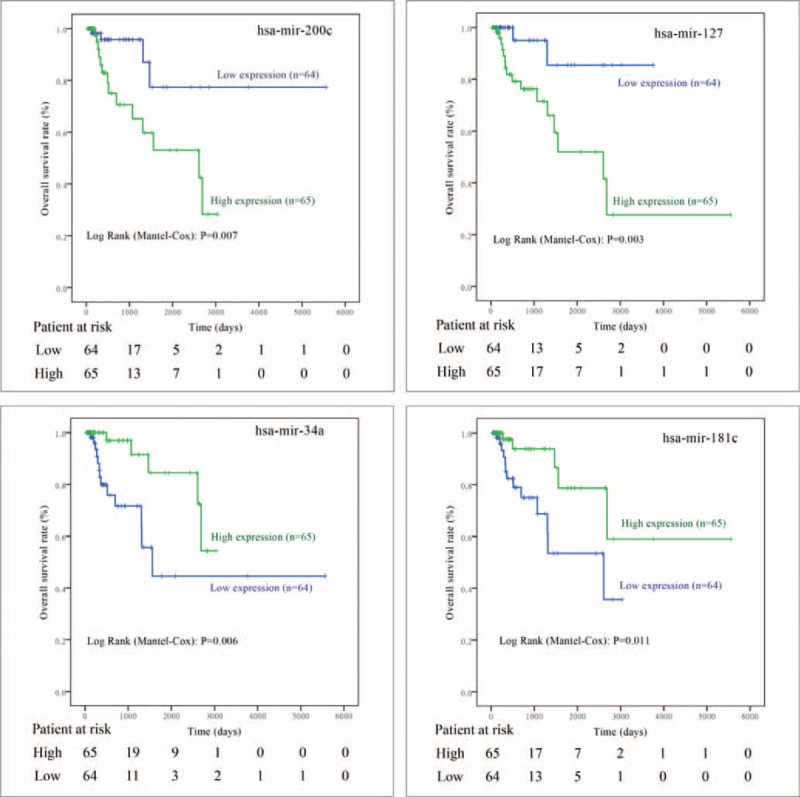
Kaplan–Meier curve analysis of microRNAs for the survival in papillary renal cell carcinoma patients. The 4 microRNAs (mir-200c, mir-127, mir-34a, and mir-181c) are potent prognostic factors confirmed by multivariate Cox regression analyses in the training stage.
FIGURE 4.
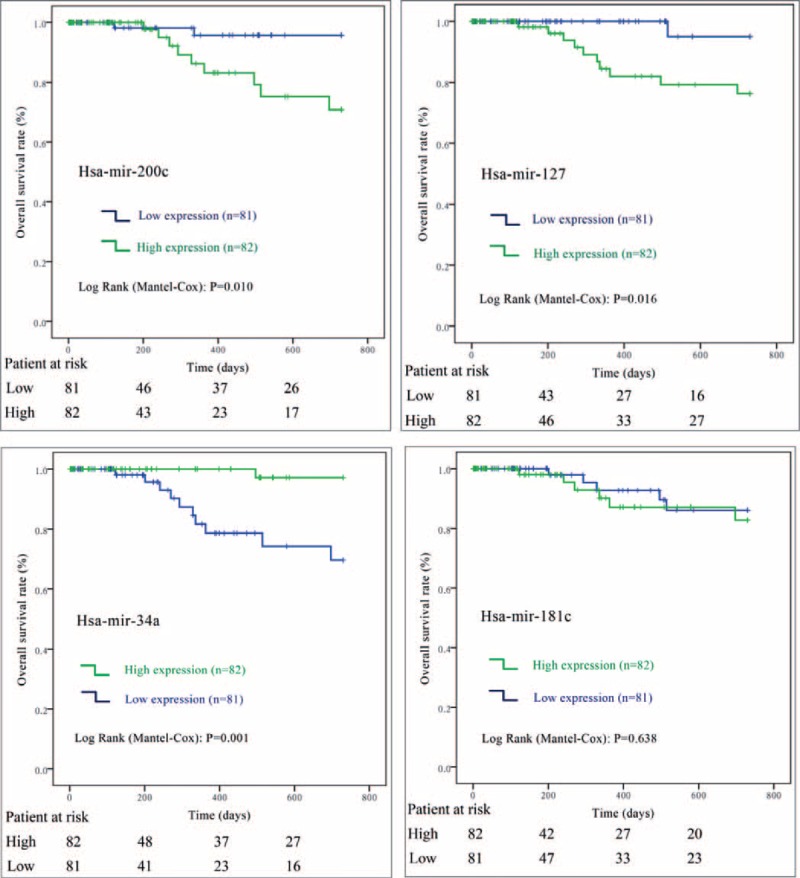
Kaplan–Meier curve analysis of microRNAs for the survival in papillary renal cell carcinoma patients (the validation stage). The 3 microRNAs (mir-200c, mir-127, and mir-34a) except mir-181c are significantly associated with patient survival.
Target Gene Prediction and Pathway Mapping Analysis
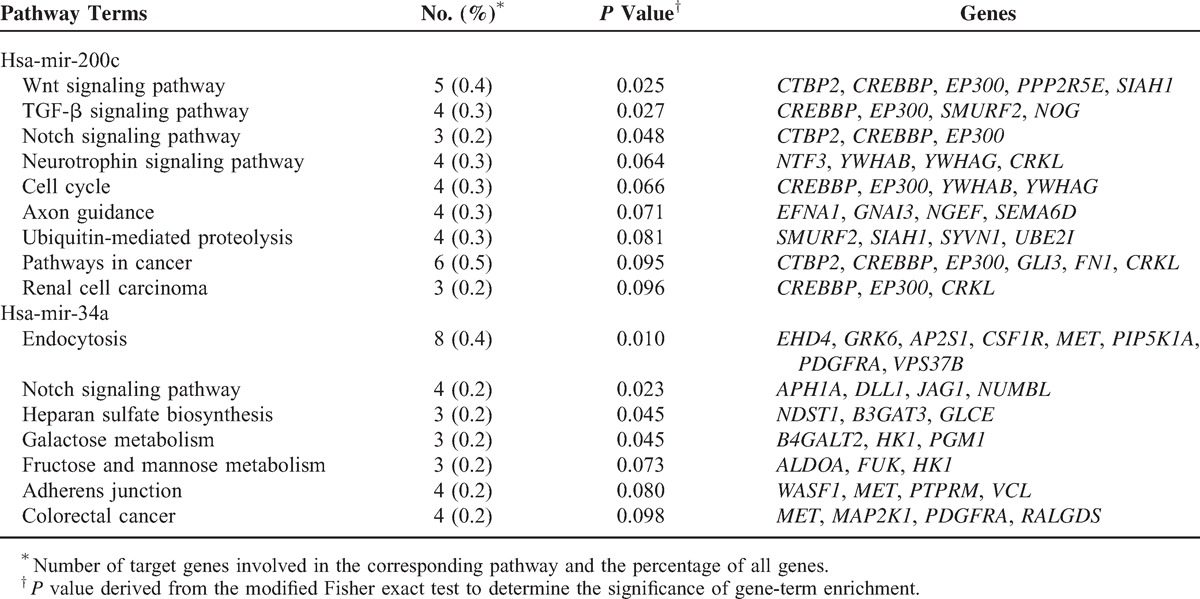
To explore the biological functions of the prognosis-related miRNAs, 3 computational algorithms (TargentScan, PicTar, and Miranda) were applied to predict the potential targets. Overall, a total of 113 target genes were selected for mir-200c, 37 for mir-127, and 180 for mir-34a by at least 2 of the 3 independent tools (see Supplementary Table S2, http://links.lww.com/MD/A250, Supplementary Digital Content 2, which illustrates the predicted target genes of mir-200c, mir-127, and mir-34a). Furthermore, the putative targets were assigned into KEGG pathway enrichment analysis via the online DAVID program. As summarized in Table 3, a total of 15 pathways were enriched, among which 9 were associated with mir-200c and 7 with mir-34a. Of note, the results provided evidence that mir-200c was involved in the pathway “renal cell carcinoma” through modulating 3 target genes (CREBBP, EP300, and CRKL), and 1 pathway could gather the putative targets for 2 miRNAs, such as Notch signaling pathway for mir-200c and mir-34a.
TABLE 3.
Kyoto Encyclopedia of Genes and Genomes Terms of the Predicted Target Genes for Mir-200c and Mir-34a
DISCUSSION
pRCC represents the second most common subtype of RCC, with highly variable clinical behaviors and long-term outcomes. However, the currently established prognostic models are based merely on clinicopathological and imaging data, which cannot compensate for the inherent limited information.4,5,29 In the present study, we explored the global miRNA expression profiles in pRCC, with the aim to identify the specific miRNAs associated with the progression and prognosis of pRCC, which could fulfill the demands for improving risk stratification and prognosis prediction for pRCC patients.
As an endogenous class of small, noncoding RNAs, miRNAs have been documented to be involved in a wide range of pathophysiological processes including carcinogenesis and metastasis.30,31 To gain more insight into the molecular mechanisms underlying carcinogenesis, the miRNA expression profiling has emerged as a potent technique approach and identified specific miRNA signatures associated with the aggressiveness and prognosis in human malignancies.32,33 In the current study, we explored the genome-wide miRNA expression profiles in pRCC, after identifying 163 treatment-naïve primary pRCC patients from the multi-institutional TCGA project.
Overall, 26 miRNAs were selected for AJCC stage, 28 for pathologic T, 16 for lymph node involvement status, and 3 for metastasis status. Of note, as pRCC is relatively indolent with low incidences of metastasis,4 and the patients with distant metastasis are rarely recommended to undergo surgery, the number of M1 patients was only 9, which could explain the result of 3 metastasis-related miRNAs to some extent. Furthermore, as pRCC can be divided into 2 distinct subtypes (type 1 and type 2), and type 2 pRCC appears to have a worse prognosis than type 1,4,34 we compared the miRNA expression profiles according to the histologic subtypes and identified 32 differentially expressed miRNAs. In the training stage of survival analysis, a summary of 12 miRNAs (mir-134, mir-379, mir-127, mir-452, mir-199a, mir-200c, mir-141, mir-3074, mir-1468, mir-181c, mir-1180, and mir-34a) were demonstrated to be significantly associated with prognosis by univariate Cox regression and Kaplan–Meier survival analyses. After final stepwise multivariate Cox regression analysis, 4 miRNAs (mir-200c, mir-127, mir-34a, and mir-181c) were identified as potential independent prognostic parameters, and the high expression of mir-200c and mir-127 predicts poor outcomes in pRCC. In the subsequent validation stage, mir-200c, mir-127, and mir-34a were confirmed to be significantly correlated with overall survival of pRCC patients.
Of all the 3 prognosis-related miRNAs (mir-200c, mir-127, and mir-34a), none has been explored in pRCC. As 1 member of the tumor suppressive mir-200 family, mir-200c has been involved in the process of mesenchymal-epithelial transition.35 However, the high expression of mir-200c was proved to be associated with a poor prognosis in pRCC, which were in consistent with the reports in other malignancies such as breast cancer,36 colorectal cancer,37 and ovarian cancer.38 The computational studies demonstrated that mir-200c was targeting 113 genes and implied in 9 pathways including “renal cell carcinoma” by KEGG enrichment analysis. Mir-127 is located on chromosome region 14q32.2 with 37 putative target genes. Even though no pathway was identified because of the small number of target genes, mir-127 has been proven to promote glioblastoma cell migration and invasion via targeting the tumor suppressor genes.39 In pRCC, mir-127 was upregulated in pathologic T3+T4 tumors and associated with a poor prognosis, which totally differed from ccRCC, as the high expression of mir-127 was significantly associated with longer relapse-free survival time in ccRCC.40 The remaining 1 miRNA, mir-34a, was a protective miRNA, as the higher expression levels correlated with a better prognosis in pRCC. In agreement with the current study, mir-34a acts as a tumor suppressor in ccRCC via inhibiting cell proliferation and metastasis.41,42 Furthermore, the bioinformatics analysis identified a total 180 target genes and 7 pathways including “endocytosis,” “Notch signaling pathway,” “heparan sulfate biosynthesis,” and others for mir-34a, which could deepen the understanding of mir-34a in RCC (including ccRCC and pRCC). Overall, all these 3 miRNAs exert important functions in the carcinogenesis of pRCC through targeting a wide range of genes, regulating various pathways and complicated cross-talk with each other, which warrant further functional analysis.
Some limitations should be acknowledged in interpreting the results. First, as the status of lymph node involvement, distant metastasis, and histologic subtype remains unclear in a substantial group of pRCC patients (65.9%, 42.6%, and 44.2%, respectively), these histopathological parameters were excluded from the survival analysis, which could reduce the statistical power of the survival analysis. Second, the participants were majorly recruited from Caucasians, which limited the application of the miRNA profiles in the whole population, even though ethnicity has been disproved as an independent prognostic factor.43,44 Third, the limited number of pRCC patients enrolled in the current study and relatively short follow-up time, which could reduce the statistical power to identify more significant miRNAs. Fourth, even though we strictly selected the subjects to control the potential heterogeneity, the false-positive results do potentially exist, which need an external validation cohort to validate the results with different methods such as quantificational real-time polymerase chain reaction.
CONCLUSION
In summary, by analyzing the global miRNA expression profiles in an independent pRCC patient cohort, our study identified specific miRNAs in relation with the progression and aggressiveness of pRCC and 3 miRNAs (mir-200c, mir-127, and mir-34a) as the promising prognostic factors of pRCC. Additionally, further well-designed and unbiased studies with larger sample size and longer follow-up time should be conducted to verify our findings. Furthermore, the functional studies of miRNAs in pRCC are warranted that will help to understand the underlying molecular mechanisms.
Acknowledgments
The authors would like to sincerely thank The Cancer Genome Atlas (TCGA) project for generating, curating, and providing high-quality biological and clinical data about papillary renal cell carcinoma.
Footnotes
Abbreviations: AJCC = American Joint Committee on Cancer, ccRCC = clear cell renal cell carcinoma, CI = confidential interval, DAVID = Database for Annotation, Visualization, and Integrated Discover, HR = hazard ratio, KEGG = Kyoto Encyclopedia of Genes and Genomes, miRNA = microRNA, pRCC = papillary renal cell carcinoma, RCC = renal cell carcinoma, RPM = reads per million, STROBE = Strengthening the Reporting of Observational studies in Epidemiology, TCGA = The Cancer Genome Atlas, TNM = tumor-node-metastasis.
This study was supported by grants from the National Natural Science Foundation of China (81070597 and 81370853), Science and Education Development Program of the Jiangsu Province Health Board (LJ201107), Six Talent Peaks of the Jiangsu Province Health Bureau (2011-WS-093), and the Research and Innovation Program for Graduates of Jiangsu Province (CXZZ13_0583). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA: Cancer J Clin 2014; 64:9–29. [DOI] [PubMed] [Google Scholar]
- 2.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009; 373:1119–1132. [DOI] [PubMed] [Google Scholar]
- 3.Cronin RE, Kaehny WD, Miller PD, et al. Renal cell carcinoma: unusual systemic manifestations. Medicine 1976; 55:291–311. [DOI] [PubMed] [Google Scholar]
- 4.Warrick JI, Tsodikov A, Kunju LP, et al. Papillary renal cell carcinoma revisited: a comprehensive histomorphologic study with outcome correlations. Hum Pathol 2014; 45:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Twardowski PW, Mack PC, Lara PN., Jr Papillary renal cell carcinoma: current progress and future directions. Clin Genitourinary Cancer 2014; 12:74–79. [DOI] [PubMed] [Google Scholar]
- 6.Vavallo A, Simone S, Lucarelli G, et al. Pre-existing type 2 diabetes mellitus is an independent risk factor for mortality and progression in patients with renal cell carcinoma. Medicine 2014; 93:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 8.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell 2013; 153:516–519. [DOI] [PubMed] [Google Scholar]
- 9.Monzo M, Martinez-Rodenas F, Moreno I, et al. Differential MIR-21 expression in plasma from mesenteric versus peripheral veins: an observational study of disease-free survival in surgically resected colon cancer patients. Medicine 2015; 94:e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Qiang F, Gao Y, et al. Evaluation of a novel functional single-nucleotide polymorphism (rs35010275 G>C) in MIR196A2 promoter region as a risk factor of gastric cancer in a Chinese population. Medicine 2014; 93:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Q, Li S, Lao X, et al. The association of common functional polymorphisms in mir-146a and mir-196a2 and hepatocellular carcinoma risk: evidence from a meta-analysis. Medicine 2014; 93:e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maher ER. Genomics and epigenomics of renal cell carcinoma. Semin Cancer Biol 2013; 23:10–17. [DOI] [PubMed] [Google Scholar]
- 13.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012; 6:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juan D, Alexe G, Antes T, et al. Identification of a microRNA panel for clear-cell kidney cancer. Urology 2010; 75:835–841. [DOI] [PubMed] [Google Scholar]
- 15.Osanto S, Qin Y, Buermans HP, et al. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PloS One 2012; 7:e38298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Weng L, Li X, et al. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PloS One 2012; 7:e35661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014; 26:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet 2007; 370:1453–1457. [DOI] [PubMed] [Google Scholar]
- 20.Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. PLoS Med 2007; 4:e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Ann Int Med 2007; 147:W163–W194. [DOI] [PubMed] [Google Scholar]
- 22.Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Epidemiology 2007; 18:805–835. [DOI] [PubMed] [Google Scholar]
- 23.Zovoilis A, Mungall AJ, Moore R, et al. The expression level of small non-coding RNAs derived from the first exon of protein-coding genes is predictive of cancer status. EMBO Rep 2014; 15:402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Inform 2008; 6:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37:495–500. [DOI] [PubMed] [Google Scholar]
- 26.Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008; 36 (database issue):D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2010; 38 (database issue):D355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun M, Shariat SF, Cheng C, et al. Prognostic factors and predictive models in renal cell carcinoma: a contemporary review. Eur Urol 2011; 60:644–661. [DOI] [PubMed] [Google Scholar]
- 30.Iorio MV, Croce CM. MicroRNA involvement in human cancer. Carcinogenesis 2012; 33:1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L, Qu L. The function of microRNAs in renal development and pathophysiology. J Genet Genomics 2013; 40:143–152. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834–838. [DOI] [PubMed] [Google Scholar]
- 33.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–866. [DOI] [PubMed] [Google Scholar]
- 34.Kim KH, You D, Jeong IG, et al. Type II papillary histology predicts poor outcome in patients with renal cell carcinoma and vena cava thrombus. BJU Int 2012; 110 (11 pt B):E673–E678. [DOI] [PubMed] [Google Scholar]
- 35.Feng X, Wang Z, Fillmore R, et al. MiR-200, a new star miRNA in human cancer. Cancer Lett 2014; 344:166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuomarila M, Luostari K, Soini Y, et al. Overexpression of microRNA-200c predicts poor outcome in patients with PR-negative breast cancer. PloS One 2014; 9:e109508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun 2014; 455:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilming Elgaaen B, Olstad OK, Haug KB, et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer 2014; 14:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang H, Hua D, Zhang J, et al. MicroRNA-127-3p promotes glioblastoma cell migration and invasion by targeting the tumor-suppressor gene SEPT7. Oncol Rep 2014; 31:2261–2269. [DOI] [PubMed] [Google Scholar]
- 40.Slaby O, Redova M, Poprach A, et al. Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer 2012; 51:707–716. [DOI] [PubMed] [Google Scholar]
- 41.Yamamura S, Saini S, Majid S, et al. MicroRNA-34a suppresses malignant transformation by targeting c-Myc transcriptional complexes in human renal cell carcinoma. Carcinogenesis 2012; 33:294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Li H, Wang J, et al. miRNA-34a suppresses cell proliferation and metastasis by targeting CD44 in human renal carcinoma cells. J Urol 2014; 192:1229–1237. [DOI] [PubMed] [Google Scholar]
- 43.Motzer RJ, Escudier B, Bukowski R, et al. Prognostic factors for survival in 1059 patients treated with sunitinib for metastatic renal cell carcinoma. Br J Cancer 2013; 108:2470–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol 2015; 67:85–97. [DOI] [PubMed] [Google Scholar]


