Supplemental Digital Content is available in the text
Abstract
Epidermal growth factor receptor (EGFR) activating mutations are a predictor of tyrosine kinase inhibitor effectiveness in the treatment of non–small-cell lung cancer (NSCLC). The objective of this study is to build a model for predicting the EGFR mutation status of brain metastasis in patients with NSCLC.
Observation and model set-up.
This study was conducted between January 2003 and December 2011 in 6 medical centers in Southwest China.
The study included 31 NSCLC patients with brain metastases.
Eligibility requirements were histological proof of NSCLC, as well as sufficient quantity of paraffin-embedded lung and brain metastases specimens for EGFR mutation detection. The linear discriminant analysis (LDA) method was used for analyzing the dimensional reduction of clinical features, and a support vector machine (SVM) algorithm was employed to generate an EGFR mutation model for NSCLC brain metastases. Training-testing-validation (3 : 1 : 1) processes were applied to find the best fit in 12 patients (validation test set) with NSCLC and brain metastases treated with a tyrosine kinase inhibitor and whole-brain radiotherapy.
Primary and secondary outcome measures: EGFR mutation analysis in patients with NSCLC and brain metastases and the development of a LDA-SVM-based EGFR mutation model for NSCLC brain metastases patients.
EGFR mutation discordance between the primary lung tumor and brain metastases was found in 5 patients. Using LDA, 13 clinical features were transformed into 9 characteristics, and 3 were selected as primary vectors. The EGFR mutation model constructed with SVM algorithms had an accuracy, sensitivity, and specificity for determining the mutation status of brain metastases of 0.879, 0.886, and 0.875, respectively. Furthermore, the replicability of our model was confirmed by testing 100 random combinations of input values.
The LDA-SVM-based model developed in this study could predict the EGFR status of brain metastases in this small cohort of patients with NSCLC. Further studies with larger cohorts should be carried out to validate our findings in the clinical setting.
INTRODUCTION
Lung cancer is the leading cause of cancer-related death worldwide, and non-small cell lung cancer (NSCLC) accounts for about 80% of all lung cancers.1,2 Autopsy data have shown that 44% of patients with NSCLC have brain metastases,3 and most patients have multiple metastases.4 The prognosis for patients with brain metastases is poor, with a median survival time of 1 to 2 months with corticosteroids,5 and 6 months for those who receive whole-brain radiation therapy (WBRT).6,7
Epidermal growth factor receptor (EGFR) activating mutations occur more frequently in nonsmokers, females, and people of Asian ethnicity, as well as in those with adenocarcinomas.8,9 Tyrosine kinase inhibitors (TKIs) have been shown to be useful for the treatment of patients with NSCLC, and tumors with EGFR-activating mutations demonstrate a better response to TKIs than those without mutations.10,11 For this reason, EGFR mutations are now recognized as a prognostic indicator in NSCLC patients treated with TKIs.10–12
TKIs, alone (eg, gefitinib and erlotinib) or combined with WBRT, represent a promising and effective strategy for treating NSCLC brain metastases.13–15 In vitro studies have shown that cells with EGFR mutations are more sensitive to radiation than those expressing wild-type EGFR.15 NSCLC with mutations in exons 19 and 21 are more susceptible to treatment with TKIs alone or with concurrent WBRT.10,11,16,17 A retrospective study has also shown that NSCLC brain metastases with EGFR mutations are more sensitive to the erlotinib monotherapy than metastases expressing wild-type EGFR.14 Furthermore, the presence of EGFR mutations in NSCLC patients with brain metastases is an independent predictor of the efficacy of WBRT.15 Patients with EGFR mutation-positive disease had significantly longer median progression free survival versus those with wild-type EGFR disease (15.2 months vs 4.4 months, respectively).18 Welsh et al19 reported that among NSCLC patients with brain metastases who received WBRT and erlotinib, those with EGFR mutations had better overall survival compared with EGFR wild-type patients. Interestingly, Shin et al20 reported that the risk of brain metastases is higher in patients with pulmonary adenocarcinoma when the primary tumor is positive for EGFR mutations. The aforementioned results are supported by another study reporting that erlotinib can pass through the blood–brain barrier.21,22
Thus, knowledge of the EGFR mutation status of brain metastases is valuable in the treatment planning for NSCLC patients with brain metastases. However, numerous studies have shown that there is discordance in the EGFR mutation status between the primary tumors and metastases.12,23–29 Whereas a metastasis develops from a single cell of the original tumor, EGFR-activating mutations arise during tumor formation.27,28 Because it is impossible in most cases to obtain a tissue sample of brain metastases, and blood or cerebrospinal fluid cannot be used to determine the EGFR mutation status of brain metastases, methods to predict the EGFR mutation status of metastases would aid in determining the proper treatment for NSCLC patients with brain metastases.
Support vector machines (SVMs) have been widely used to support the construction of prediction models.30,31 Linear discriminant analysis (LDA) is also a well known technique in statistical pattern classification for improving discrimination and compressing information content.32–34 Thus, the purpose of this study was to use LDA combined with SVM to develop a model to predict the EGFR mutation status of brain metastasis in NSCLC patients based on their clinical features and the EGFR mutation status of the primary lung tumor.
METHODS
Patient Characteristics
The study included 31 patients with NSCLC and brain metastases from 6 medical centers in Southwest China, and was conducted between January 2003 and December 2011. Eligibility requirements were histological proof of NSCLC, as well as sufficient quantity of paraffin-embedded lung and brain metastases specimens for EGFR mutation detection using direct DNA sequencing and amplification refractory mutation system analysis. The demographic and clinical characteristics of the patients and their tumors were used to develop and test the LDA-SVM-based EGFR mutation models for NSCLC brain metastases. Written informed consent was obtained from each patient or their family member, and the study was approved by the institutional review board of the Research Institute of Surgery, Daping Hospital, Third Military Medical University (Chongqing, PR China).
EGFR Mutation Analysis
Mutation analysis of the EGFR–TK domain was based on Zhao et al.35 Briefly, genomic DNA was extracted using a QIAamp DNA FFPE Tissue Kit (No. 56404; Qiagen, Valencia, CA), and exons 18 to 21 were amplified with 4 pairs of primers.35 Polymerase chain reaction (PCR) products were purified using a PyroMark Q96 Vacuum Workstation (No. 9001529, Qiagen), and sequenced in a PyroMark Q96 ID (Qiagen). All sequence variations were confirmed by multiple independent PCR amplifications and repeated sequencing reactions.
For lung cancer and brain metastases with inconsistent EGFR mutations confirmed by sequencing, the same DNA samples used for sequencing were also tested for EGFR mutations using an amplification refractory mutation system in the ADx-ARMS EGFR Mutation Test Kit (AmoyDx; Amoy Diagnostics Co., LTD., Xiamen, FuJian, 361026, China) and following the manufacturer's instructions. A total of 29 mutations in the EGFR gene can be detected using this kit. All quantitative PCR reactions were performed using a Light Cycler 480 II instrument (Roche Applied Science, Indianapolis, IN).
Feature Transformation and Model Construction
SVM is a type of supervised learning method belonging to the family of generalized linear classifiers; it is a popular classifier based on the structural risk minimization principle, whose object is to minimize the generalization error of the classifier. LDA and the related Fisher's linear discriminant are well studied classification algorithms used in statistics, pattern-recognition, and machine-learning to find a linear combination of features, which characterizes or separates 2 or more classes of objects or events.
In this study, for the purpose of better classification and prediction, LDA and SVM were integrated to generate the model. A detailed description of the LDA and SVM methods are described in Appendix A, http://links.lww.com/MD/A130.
Validation Model and Best Fit Method
The data were divided into 5 equal and distinct subsets. Four subsets were combined and used for training, and the remaining set was used for testing. This validation process was repeated 5 times, allowing each subset to serve once as the test dataset. The validation procedure was based on prior study,36–38 and is briefly summarized as follows. The data were divided into 5 groups, that is, the 31 patients were divided into group 1 (n = 6), group 2 (n = 6), group 3 (n = 6), group 4 (n = 6), and the group of 5 (n = 7). Due to the small sample size, we further modified the validation to a “leave-one-out cross-validation.” We selected 1 set of the 5 groups as a “validation” group; 3 out of the 4 remaining groups were designated as the “training group,” and the last group served as the “test group” (TV: training–validation). Training–testing–validation (3 : 1 : 1) processes were applied to determine the best fit. Using this method, every validation group will generate data of 4 SVMs (TV groups).
The capability of the created models for the input datasets was evaluated by validation, according to accuracy, sensitivity (true positive rate), and specificity (true negative rate). This tool provides an estimate of how well a particular model might perform on a new dataset drawn from the same statistical distribution. The best models were selected according to the capabilities.
Coding of Clinical Features
The dimensions and coding of 13 original clinical features included the following: mutation status of lung tumor (0: mutation-negative; 1: mutation positive); node stage (eg, N2 stage = 2); sex (0: male; 1: female); age 0 to 100 years (eg, 25 years = 25); cancer cell type (0: adenocarcinoma; 1: others); tumor stage (eg, T2 stage, = 2); Karnofsky performance status (eg, a Karnofsky performance status of 50 = , 50); smoking history (0: nonsmoker; 1: ever smoker; 2: current smoker); brain metastasis time in months (eg, metastasis at 2 years = 24); number of brain metastases (eg, 1 brain metastasis = 1); tumor diameter in cm (eg, a tumor diameter of 2.3 cm = 2.3); extracranial metastases (0: others; 1: no); and lung tumor (0: others; 1: controlled).
Pilot Study and Assessments
After failure of systemic chemotherapy, a TKI was used as second- or third-line treatment for patients with stage IIIB/IV NSCLC who were EGFR mutation-negative, and as first-line therapy for those with stage IIIB/IV NSCLC who were EGFR mutation positive. In this study, patients who developed brain metastases during the course of TKI treatment received a TKI plus WBRT at the Cancer Center of Third Military Medical University. WBRT was administered at a dosage of 40 Gy/20f/4w. Gefitinib was administered orally (250 mg) once daily until disease progression, according to Response Evaluation Criteria in Solid Tumors criteria, intolerable toxicity, or patient refusal. Before radiotherapy, all patients underwent enhanced brain magnetic resonance imaging to identify brain metastasis number and size. Each patient also underwent physical examination, laboratory tests, electrocardiogram, and chest and upper abdomen computed tomography scans before treatment initiation. The medical history and smoking status of each patient were documented. The objective tumor response was assessed at 4 weeks after WBRT, and then every 8 weeks thereafter. Additional assessments could be performed at any time when symptoms or signs suggested that the disease might be progressing.
Statistical Analysis
Logistic regression was performed to determine the factors associated with EGFR mutation status in the primary tumors and metastases of NSCLC patients with brain metastases. For the purpose of better classification and best fit, LDA and SVM were integrated to generate the model (Figure 1). A receiver operating characteristic curve was employed to obtain the area under the curve (AUC), sensitivity, and specificity. SVM analysis was done with the LibSVM (a library for support vector machines. 2001, Software available at http://www.csie.ntu.edu.tw/∼cjlin/libsvm) software package39 in a validation design, as previously described, and predictive accuracy was measured. In order to assess the replicability of the main results, the outcomes after input of random combinations of training objects was examined 100 times with 20 individuals each time. All statistic assessments were 2 sided and evaluated at the 0.05 level of significance. Statistical analyses were performed using SPSS 15.0 statistics software (SPSS Inc., Chicago, IL).
FIGURE 1.
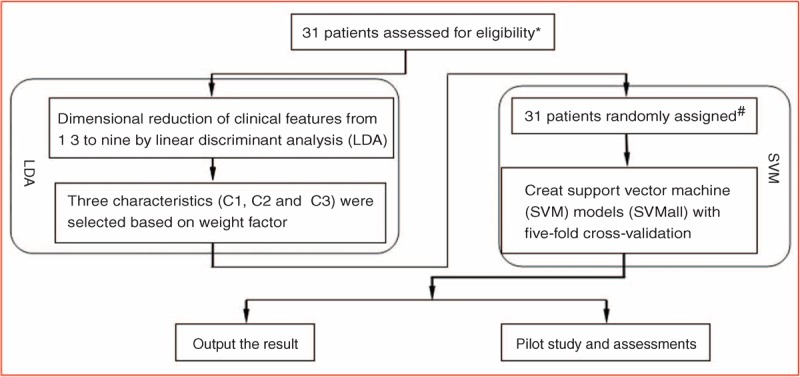
Flowchart describing the construction of the non-small cell lung cancer brain metastasis epidermal growth factor receptor mutation prediction model. ∗All patients in the study had paraffin-embedded specimens of their pulmonary/brain metastases. #All patients were randomly assigned to 5 groups (6 patients in 4 groups, and 7 patients in the fifth group).
RESULTS
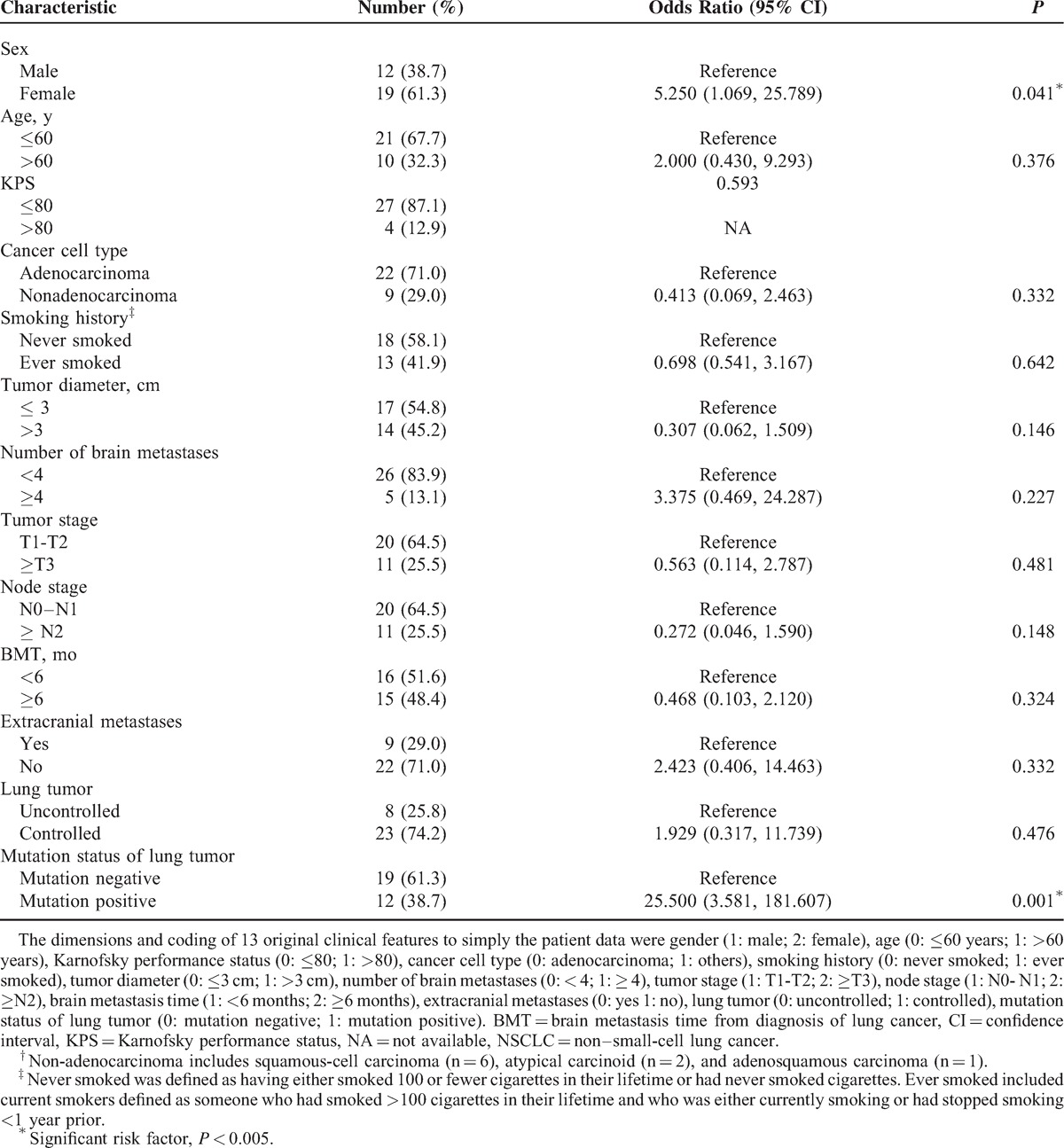
A total of 31 patients with NSCLC and brain metastases were screened and met the inclusion criteria. Of the 31 lung cancer samples examined, direct DNA sequencing detected 8 deletion mutations in exon 19 and 4 L858R point mutations in exon 21. In the 31 brain metastasis samples, there were also 8 deletion mutations in exon 19, but only 3 L858R point mutations in exon 21. Thus, an 83.87% concordance between lung cancer and brain metastases was observed. As shown in Table 1, logistic regression analysis indicated that sex (odds ratio = 5.250; 95% confidence interval 1.069–25.789; P = 0.041) and mutation status of the lung tumor (odds ratio = 25.500; 95% confidence interval, 3.581 to 181.607; P = 0.001) were associated with EGFR mutation status of brain metastases.
TABLE 1.
Logistic Regression Analysis for Determining Factors Predictive of the EGFR Mutation Status of Brain Metastases in Patients With NSCLC (N = 31)
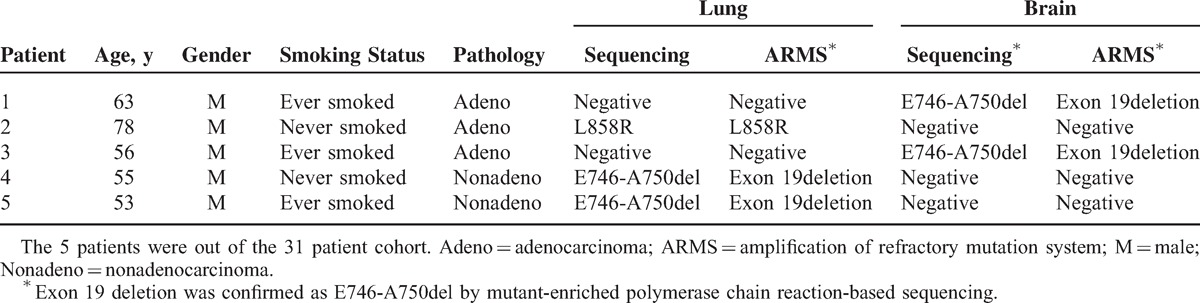
In 5 male patients, a discordant mutations status between the lung cancer and brain metastases was detected. Of the 5 patients, 3 had adenocarcinomas. Two of these patients had brain metastases with a deletion mutation of exon 19 and no mutations of the lung cancer, and 1 patient had no mutations noted in the brain metastases and a point mutation of exon 21 in the lung cancer. One of the 5 patients had squamous cell carcinoma and 1 atypical carcinoid, and in both cases no mutations of the brain metastases were found, and both had a deletion mutation of exon 19 of the lung cancer. All discordant mutations were all confirmed by ARMS (Table 2). No significant difference was observed in the distribution of mutation type between lung cancer and brain metastases (P = 0.192).
TABLE 2.
Analysis of Discordant EGFR Mutations Between Primary Lung Carcinoma and Brain Metastases
The LDA method was used to reduce the dimensions of the 13 original clinical features to simplify the patient data. There is redundancy in 9 characteristics, and a feature selection method was used to select an optimal subset (C1, C2, C3) for the classification of patient input data. The data of the 31 patients were projected to 3-dimensional space as shown in Figure 2. The results indicated that after the LDA, the between-class distances were greater, whereas the within-class differences were closer, and the data can be divided much easier. Even though, there were still 2 points that were wrongly classified.
FIGURE 2.
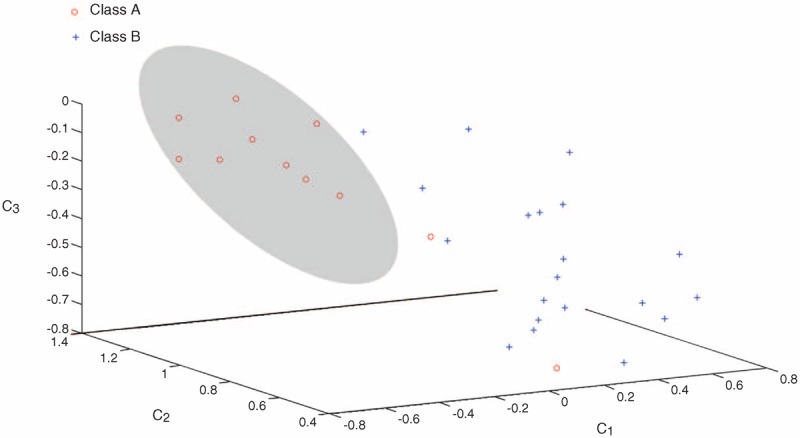
Spatial projection of patient data. Nine feature values obtained from dimensionality reduction of the data of the 31 patients were projected to 3-dimensional space. In 3-dimensional, the corresponding axis value was the feature values of C1, C2, and C3. The hyperplane on each of the projections relates to the dimensionality to which the projection was made against. Class A: individuals with epidermal growth factor receptor mutations; Class B: individuals without epidermal growth factor receptor mutations.
Figure 3 presents the results of the SVM algorithm of factors predictive of brain metastases being EGFR mutation positive. The results indicated that the mutation status of the lung tumor (AUC = 0.790; P < 0.001 in positive vs negative) was associated with EGFR mutations of brain metastases.
FIGURE 3.
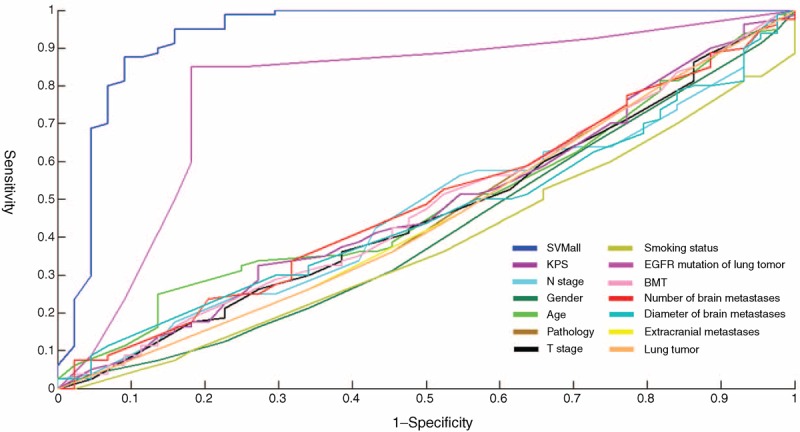
Receiver operating characteristics curve of SVM models.
In our experiment, a validation procedure was used for training and testing the SVM classifier under various model and parameter settings, and the values of the parameters that produced the best result in the validation were used for SVM testing. A total of 20 collaborate submodels were obtained, and collectively called SVMall in the study, that is, the ultimate prediction model. Through validation, SVMall was more reliable for predicting the results (AUC = 0.9186). When the probability was >0.6288, EGFR mutations of brain metastases were predicted with a sensitivity of 88.6% and specificity of 87.5%.
Twelve patients with brain metastases diagnosed by magnetic resonance imaging received EGFR–TKI and WBRT treatment (Table 3). There were 10 patients with EGFR mutation-positive lung tumors and 2 patients (#4 and #11) for whom their lung tumors negative for EGFR mutations. After dimensionality reduction of the 13 clinical features of the 12 patients in the validation test set by LDA, the corresponding feature values of C1, C2, and C3 were input into SVMall.
TABLE 3.
Clinical Outcomes of the 12 Patients Who Received Whole-Brain Radiotherapy and Tyrosine Kinase Inhibitor Treatment
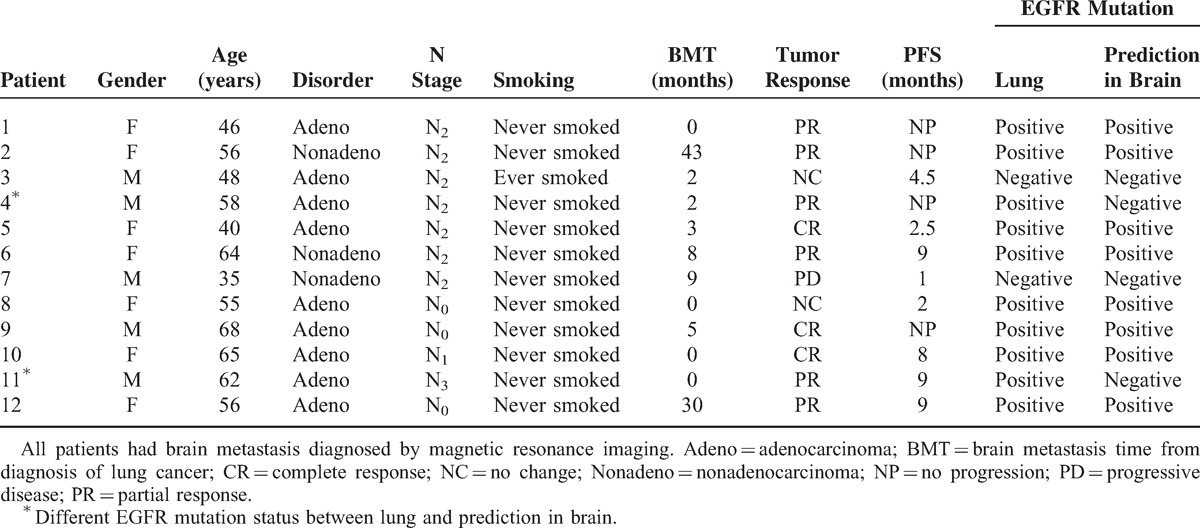
Using the model created, the EGFR mutation status of the brain metastases was determined to be different from that of the primary lung cancer in 2 patients. Eight patients (8/12) were found to have EGFR mutation-positive brain metastases, of whom 7 achieved a complete response or partial response with EGFR–TKI and WBRT. Four patients (4/12) were found to have EGFR mutation-negative mutation metastases, of whom only 2 patients achieved a partial response after EGFR–TKI and WBRT. No statistical difference was found in the treatment efficacy between EGFR mutation-positive and -negative brain metastases (P = 0.236).
To examine the replicability of the LDA-SVM-based EGFR mutation model, we analyzed the outcomes with different input values consisting of random combinations of training objects. After analyzing 100 different combinations using a random selection subgroup with 20 individuals each time, the majority of the results were distributed in the same area, confirming both the replicability and the stability of our model and procedure (Figure 4).
FIGURE 4.
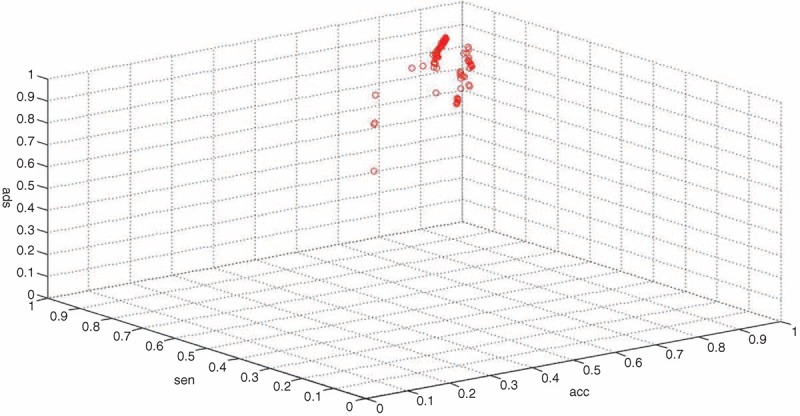
Replicability of the linear discriminant analysis–support vector machine-based epidermal growth factor receptor Mutation Model. spe = specificity, sen = sensitivity, acc = accuracy.
DISCUSSION
The present study represents an attempt to develop a model using patient clinical features and the EGFR mutation status of a primary lung tumor to predict the EGFR mutation status of brain metastases in patients with NSCLC. An LDA-SVM-based approach was able to predict the EGFR mutation status of brain metastases in a small cohort of patients with NSCLC.
The present study has shown that tumors with EGFR-activating mutations demonstrate a better response to EGFR-TKIs than those without mutations.10,11,40 Mutation status has also been confirmed to be a better predictor of TKI treatment efficacy than EGFR protein expression or EGFR gene copy number.10,11,42 In addition, an in vitro study has shown a 500- to 1000-fold reduction in the clonogenic survival of mutant EGFR-expressing lung cancer cell lines in response to ionizing radiation compared with those expressing wild-type EGFR.15 Both EGFR mutations and the administration TKIs have also been noted to be independent predictors of response to WBRT in the treatment of brain metastases from NSCLC.13,15 Thus, knowledge of EGFR mutation status is important for guiding treatment decisions; however, it has been clearly shown that there is considerable discordance between the EGFR mutation status of primary tumors and that of metastases.12,23–29
The gold standard for determining EGFR mutation status is analysis of a tissue specimen; however, obtaining tissue from brain metastases is associated with obvious difficulties. In addition, there is a lack of evidence supporting the analysis of blood or cerebrospinal fluid specimens to determine the EGFR mutation status of NSCLC brain metastases. Furthermore, the clinical features of NSCLC patients, such as gender, histological type, smoking status, and clinical stage, have no clear correlation with the EGFR mutation status of brain metastases.12,23–25
Predictive models have been used in the diagnosis and prognosis of a number of cancers, including breast, colon, and prostate cancers,31–34 as well as other disease states, such as attention-deficit hyperactivity disorder.41 In this study, we defined 13 candidate clinical features of NSCLC patients with brain metastases, and then applied a LDA-SVM-based method to integrate these clinical features to predict the EGFR mutation status of NSCLC brain metastases. A model for determining the EGFR mutation status of NSCLC brain metastases was devised, and the model was tested in a cohort of 12 NSCLC patients with brain metastases. Compared with obtaining brain metastases specimens for analysis to guide treatment decisions, it is much simpler and obviously less invasive to use a model based on clinical features. In comparison with previously published gene-based predictive models,30,42 our clinical feature-based model is also simpler to perform. In addition, although different gene sets are used, they each track a common set of biologic characteristics that are present in different groups of patients with NSCLC brain metastases, resulting in similar outcome predictions.
Compared with other machine learning algorithms, such as decision trees and artificial neural networks, SVM is well-suited to managing predictive problems, including high-dimensional data and a limited number of training samples. LDA is a powerful tool for dimensionality reduction before later classification, which has been utilized for statistical prediction. With LDA-SVM integration, more clinical features can be combined to predict the EGFR mutation status of brain metastases. Moreover, the method can also exclude the interaction of clinical features to some extent.
The primary limitation of this study is the small dataset. A training-testing-validation (3 : 1 : 1) process was used to find the best fit. However, this is not a real cross-validation procedure as for a cross-validation mechanism, the final result is an average of the 5 subgroups for measuring the classifiers not for parameter setting. We treated 1 of the 5 subgroups as a validation set, which is easily misunderstood for mixing the 5-fold one and the leave-one-out one. In real parameter setting, 3 subgroups (18 samples) out of 5 are regarded as a training set for constructing a model. It is arguable that this is not sufficient data for building such a complex SVM model. The finding of no statistical difference between the treatment effects of EGFR mutation-positive and -negative brain tumors is likely due to the small sample size. In addition, the strategy is difficult to carry out, and the issue of overfitting the model is difficult to address. Lastly, the set only included deletions and point mutations; insertion and substitution mutations were not determined. However, deletions within exon 19 and the L858R mutation in exon 21 together account for approximately 90% of EGFR mutations, and insertion and substitution mutations are relatively rare.43
In conclusion, the present study showed that a LDA-SVM-based approach was able to determine the EGFR mutation status of brain metastases in a small cohort of patients with NSCLC. Further studies with larger cohorts should be carried out to validate our findings in the clinical setting.
Footnotes
Abbreviations: AUC = area under the curve, CI = confidence interval, EGFR = epidermal growth factor receptor, LDA = linear discriminant analysis, NSCLC = non-small cell lung cancer, PCR = polymerase chain reaction, PR = partial response, SVMs = support vector machines, TKIs = tyrosine kinase inhibitors, TV = training–validation, WBRT = whole-brain radiation therapy.
NH and GW contributed equally to this work.
Author summary: Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors are used for treating non-small lung cancer (NSCLC), and tumors with EGFR-activating mutations have a better response to the drugs than those without mutations. However, in many cases the mutation status of the metastases is different than that of the primary tumor. Although tyrosine kinase inhibitors are used for treating brain metastases in patients with NSCLC, their efficacy varies, and this variation may in part be due to the mutation status of the metastases. Whereas it is relatively simple to obtain a tissue sample of the primary lesion in patients with NSCLC, obtaining a biopsy of a brain metastasis is difficult, if not impossible. For this reason a method for predicting the mutation status of brain metastases would be of value. It is known that EGFR-activating mutations occur more frequently in nonsmokers, females, and people of Asian ethnicity, as well as in those with adenocarcinoma. In this study, we developed a model to predict the EGFR mutation status of brain metastasis in patients with NSCLC based on patient clinical features and the EGFR mutation status of the primary lung cancer.
Z-ZY designed the study and wrote the protocol, performed the research/study, managed the literature searches and analyses, and wrote the first draft of the manuscript; NH performed the research/study, managed the literature searches and analyses, and wrote the first draft of the manuscript; GW performed the research/study and managed the literature searches and analyses; Y-HW performed the research/study and managed the literature searches and analyses; S-FC performed the research/study, managed the literature searches and analyses, and undertook the statistical analysis; G-DL wrote the first draft of the manuscript; CC performed the research/study; DW wrote the first draft of the manuscript; Z -SH undertook the statistical analysis; X-QY wrote the first draft of the manuscript; YH managed the literature searches and analyses; H-LX performed the research/study; D-DH performed the research/study; K-LX managed the literature searches and analyses; YW managed the literature searches and analyses; MH wrote the first draft of the manuscript.
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin 2007; 57:43–66. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer 1995; 75 suppl 1:191–202. [DOI] [PubMed] [Google Scholar]
- 3.Sorensen JB, Hansen HH, Hansen M, Dombernowsky P. Brain metastases in adenocarcinoma of the lung: frequency, risk groups, and prognosis. J Clin Oncol 1988; 6:1474–1480. [DOI] [PubMed] [Google Scholar]
- 4.Wen PY, McLaren Black P, Loeffler JS. Hellman S, Rosenberg SA, De Vita VT., Jr Metastatic brain cancer. Cancer: Principles and Practice of Oncology 6th ed.Philadelphia, PA: Lippincott Williams & Wilkins; 1999. 2655–2670. [Google Scholar]
- 5.Weissman DE. Glucocorticoid treatment for brain metastases and epidural spinal cord compression: a review. J Clin Oncol 1988; 6:543–551. [DOI] [PubMed] [Google Scholar]
- 6.Diener-West M, Dobbins TW, Phillips TL, Nelson DF. Identification of an optimal subgroup for treatment evaluation of patients with brain metastases using RTOG study 7916. Int J Radiat Oncol Biol Phys 1989; 16:669–673. [DOI] [PubMed] [Google Scholar]
- 7.Borgelt B, Gelber R, Kramer S, et al. The palliation of brain metastases: final results of the first two studies by the Radiation Therapy Oncology Group. Int J Radiat Oncol Biol Phys 1980; 6:1–9. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361:958–967. [DOI] [PubMed] [Google Scholar]
- 9.Wu YL, Zhong WZ, Li LY, et al. Epidermal growth factor receptor mutations and their correlation with gefitinib therapy in patients with non-small cell lung cancer: A meta-analysis based on updated individual patient data from six medical centers in Mainland China. J Thorac Oncol 2007; 2:430–439. [DOI] [PubMed] [Google Scholar]
- 10.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304:1497–1500. [DOI] [PubMed] [Google Scholar]
- 11.Sholl LM, Xiao Y, Joshi V, et al. EGFR mutation is a better predictor of response to tyrosine kinase inhibitors in non-small cell lung carcinoma than FISH, CISH, and immunohistochemistry. Am J Clin Pathol 2010; 133:922–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Roca C, Raynaud CM, Penault-Llorca F, et al. Diferential expression of biomarkers in primary non-small cell lung cancer and metastatic sites. J Thorac Oncol 2009; 4:1212–1220. [DOI] [PubMed] [Google Scholar]
- 13.Zeng YD, Zhang L, Liao H, et al. Gefitinib alone or with concomitant whole brain radiotherapy for patients with brain metastasis from non-small-cell lung cancer: a retrospective study. Asian Pac J Cancer Prev 2012; 13:909–914. [DOI] [PubMed] [Google Scholar]
- 14.Porta R, B Sánchez-Torres JM, Paz-Ares L, et al. Brain metastases from lung cancer responding to erlotinib: the importance of EGFR mutation. Eur Respir J 2011; 37:624–631. [DOI] [PubMed] [Google Scholar]
- 15.Gow CH, Chien CR, Chang YL, et al. Radiotherapy in lung adenocarcinoma with brain metastases: effects of activating epidermal growth factor receptor mutations on clinical response. Clin Cancer Res 2008; 14:162–168. [DOI] [PubMed] [Google Scholar]
- 16.Shimato S, Mitsudomi T, Kosaka T, et al. EGFR mutations in patients with brain metastases from lung cancer: association with the efficacy of gefitinib. Neurooncology 2006; 8:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das AK, Sato M, Story MD, et al. Non-small cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res 2006; 66:9601–9608. [DOI] [PubMed] [Google Scholar]
- 18.Wu YL, Zhou C, Cheng Y, et al. Erlotinib as second-line treatment in patients with advanced non-small-cell lung cancer and asymptomatic brain metastases: a phase II study (CTONG-0803). Ann Oncol 2013; 24:993–999. [DOI] [PubMed] [Google Scholar]
- 19.Welsh JW, Komaki R, Amini A, et al. Phase II trial of erlotinib plus concurrent whole-brain radiation therapy for patients with brain metastases from non-small-cell lung cancer. J Clin Oncol 2013; 31:895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin DY, Na II, Kim CH, et al. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J Thorac Oncol 2014; 9:195–199. [DOI] [PubMed] [Google Scholar]
- 21.Togashi Y, Masago K, Fukudo M, et al. Cerebrospinal fluid concentration of erlotinib and its active metabolite OSI-420 in patients with central nervous system metastases of non-small cell lung cancer. J Thorac Oncol 2010; 5:950–955. [DOI] [PubMed] [Google Scholar]
- 22.Weber B, Winterdahl M, Memon A, et al. Erlotinib accumulation in brain metastases from non-small cell lung cancer: visualization by positron emission tomography in a patient harboring a mutation in the epidermal growth factor receptor. J Thorac Oncol 2011; 6:1287–1289. [DOI] [PubMed] [Google Scholar]
- 23.Park S, Holmes-Tisch AJ, Cho EY, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol 2009; 4:809–815. [DOI] [PubMed] [Google Scholar]
- 24.Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol 2009; 20:696–702. [DOI] [PubMed] [Google Scholar]
- 25.Kalikaki A, Koutsopoulos A, Trypaki M, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer 2008; 99:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid K, Oehl N, Wrba F, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res 2009; 15:4554–4560. [DOI] [PubMed] [Google Scholar]
- 27.Monaco SE, Nikiforova MN, Cieply K, et al. A comparison of EGFR and KRAS status in primary lung carcinoma and matched metastases. Hum Pathol 2010; 41:94–102. [DOI] [PubMed] [Google Scholar]
- 28.Han HS, Eom DW, Kim JH, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer 2011; 12:380–386. [DOI] [PubMed] [Google Scholar]
- 29.Sun L, Zhang Q, Luan H, et al. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Cancer Res 2011; 30:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabatier R, Finetti P, Cervera N, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat 2011; 126:407–420. [DOI] [PubMed] [Google Scholar]
- 31.Zeng T, Liu J. Mixture classification model based on clinical markers for breast cancer prognosis. Artif Intell Med 2010; 48:129–137. [DOI] [PubMed] [Google Scholar]
- 32.Liang S, Singh M, Dharmaraj S, Gam LH. The PCA and LDA analysis on the differential expression of proteins in breast cancer. Dis Markers 2010; 29:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanmohammadi M, Bagheri Garmarudi A, Samani S, et al. Application of linear discriminant analysis and Attenuated Total Reflectance Fourier Transform Infrared microspectroscopy for diagnosis of colon cancer. Pathol Oncol Res 2011; 17:435–441. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Do KA, Wen S, et al. Merging microarray data, robust feature selection, and predicting prognosis in prostate cancer. Cancer Inform 2006; 2:87–97. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Zhao J, Zhao X, et al. Detection of EGFR gene mutations in 100 non-small cell lung cancer clinical samples by a real-time polymerase chain reaction method using amplification refractory mutation system specific primers and Taqman fluorescence probes]. Zhongguo Fei Ai Za Zhi 2013; 16 1:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Zhou S, Yin FF, et al. Investigation of the support vector machine algorithm to predict lung radiation-induced pneumonitis. Med Phys 2007; 34:3808–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu ZH, Sun BY, Ma Y, et al. Three immunomarker support vector machines-based prognostic classifiers for stage IB non-small-cell lung cancer. J Clin Oncol 2009; 27:1091–1099. [DOI] [PubMed] [Google Scholar]
- 38.Das SK, Chen S, Deasy JO, et al. Combining multiple models to generate consensus: application to radiation-induced pneumonitis prediction. Med Phys 2008; 35:5098–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang CC, Lin CJ. LIBSVM: a library for support vector machines; 2001. http://www.csie.ntu.edu.tw/cjlin/libsvm 2001. [Accessed June 19, 2013]. [Google Scholar]
- 40.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361:947–957. [DOI] [PubMed] [Google Scholar]
- 41.Mueller A, Candrian G, Grane VA, et al. Discriminating between ADHD adults and controls using independent ERP components and a support vector machine: a validation study. Nonlinear Biomed Phys 2011; 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes DN, Monti S, Parmigiani G, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol 2006; 24:5079–5090. [DOI] [PubMed] [Google Scholar]
- 43.Li AR, Chitale D, Riely GJ, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn 2008; 10:242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]


