Abstract
Findings on the role of plasma miR-21 expression in colorectal cancer are contradictory. Before reaching a peripheral vein (PV), microRNAs released by the tumor are dispersed throughout the body. We hypothesized that blood drawn from the mesenteric vein (MV) near the site of the primary tumor could provide more homogeneous information than blood drawn from the PV.
We have analyzed miR-21 expression in matched samples of tumor tissue, normal tissue, MV plasma, and PV plasma in 57 surgically resected patients with colon cancer and correlated our findings with clinical characteristics and disease-free survival (DFS).
miR-21 expression was higher in MV than PV plasma (P = 0.014) and in tumor than in normal tissue (P < 0.001). Patients with high levels of miR-21 in MV plasma had shorter DFS (P = 0.05) than those with low levels, and those with high levels in both MV and PV plasma had shorter DFS than all other patients (P = 0.01).
Our findings suggest that the primary tumor in colon cancer releases high concentrations of miR-21 in the MV but that these concentrations are later diluted in the circulatory system. MV expression of miR-21 may be a stronger prognostic marker than PV expression.
INTRODUCTION
Colorectal cancer (CRC) is the third most common type of cancer and the second cause of cancer death worldwide.1 The main prognostic factor for relapse and survival in CRC is disease stage, and patients with stage III disease have a higher risk of relapse than those with stage II. Surgery is the standard treatment for stage I to III, and adjuvant treatment has been shown to be effective in stage III but less so in stage II.2 Prognostic and predictive biomarkers can provide a useful tool for selecting treatment and improving outcome in these patients. The analysis of biomarkers in the plasma or serum of CRC patients is a noninvasive yet effective way to determine prognosis, detect occult tumors, and monitor treatment.
MicroRNAs (miRNAs), noncoding RNAs that play a key role in the regulation of mRNA expression, are promising diagnostic and prognostic biomarkers in several cancers.3 Numerous studies have shown that miRNAs are aberrantly expressed during tumor development and can act either as oncogenes or tumor suppressors.4,5 The specific mechanism whereby tumor cells release miRNAs into the blood is not completely understood. Recent studies have shown that exosomes and microvesicles can act as miRNA transporters,6–8 whereas other studies have found that miRNAs circulate freely in blood by binding to the AGO-2 protein complex, which prevents the digestion of RNase in plasma.9
miR-21 was the first tumor-related miRNA to be identified, detected in the serum of a patient with B-cell lymphoma.10 Since then, miR-21 has been widely studied in tumor, plasma, and serum samples, both in CRC and in other tumors, where it controls carcinogenesis by targeting different genes, including TPM1,11 PDCD4,12,13 PTEN,14 and BTG2.15 In hepatocellular carcinoma16 and non–small-cell lung cancer,17 plasma and serum levels of miR-21 have been identified as reliable biomarkers for both diagnosis and prognosis. In addition, postoperative levels of miR-21 were lower than baseline levels in both gastric cancer18 and squamous cell carcinoma of the esophagus.19
In CRC, some studies have identified miR-21 in serum20,21 or plasma22,23 as a useful diagnostic and prognostic biomarker. However, in other studies, miR-21 expression was not detected in the peripheral blood of CRC patients, although other miRNAs, including miR-17–3- miR-92,24 miR-29a,25 miR92a,26 and miR-221,27 were identified as circulating tumor biomarkers. These contradictory findings may be due to various causes, including differences in patient characteristics, internal controls, and cutoff values. Importantly, all previous studies of circulating miR-21 in CRC have consistently obtained circulating miRNAs from an area far from the primary tumor, generally from a peripheral vein (PV) located in the forearm. However, before reaching the PV of the forearm, the miRNAs released by the tumor are diluted and dispersed in other parts of the body, which may explain the inconsistency between miRNA expression levels in the tumor itself and those detected in peripheral blood.
In CRC, venous return occurs through the superior mesenteric vein (MV) if the tumor is located in the right colon, through the inferior MV if the tumor is located in the left colon, and through the iliac veins if the tumor is located in the middle or lower third rectum. Therefore, we can hypothesize that in colon cancer, blood samples drawn from the MV near the site of the primary tumor can provide more homogeneous and effective information than blood drawn from the PV of the forearm. To test this hypothesis, we have analyzed miR-21 expression in paired samples of tumor tissue, normal tissue, plasma obtained by blood drawn from the MV, and plasma obtained by blood drawn from the PV in 57 surgically resected patients with colon cancer and correlated our findings with the clinical characteristics and disease-free survival (DFS) of these patients.
METHODS
Eligibility and Patient Evaluation
From August 2009 to August 2013, samples were obtained from 57 patients with stage I to IV colon cancer who underwent surgical resection at the Municipal Hospital of Badalona. Approval for the study was obtained from the institutional review board of the hospital, and signed informed consent was obtained from all patients and controls in accordance with the Declaration of Helsinki.
All 57 patients underwent a complete history and physical examination including routine hematological and biochemical analyses, chest radiographs, and computed tomography (CT) of the thorax and abdomen. Target lesions detected by abdominal ultrasound were also assessed by CT or magnetic resonance imaging.
Samples
For all 57 patients, we obtained tumor tissue, paired normal tissue, MV blood, and PV blood. Normal tissue was obtained from the area of the colon farthest from the tumor. Both tumor and normal tissue samples were analyzed and confirmed by a pathologist and frozen at −80°C for further use.
On the day of surgery, 5 mL of blood was drawn from the PV and stored in heparinized tubes. During surgery, with vascular ligation before tumor resection, an additional 5 mL of blood was drawn from either the superior or the inferior MV, according to the anatomic location of the tumor. Blood samples from 18 healthy individuals (young male athletes) were obtained from the blood bank of the Hospital Clinic for use as controls. All blood samples were centrifuged at 5000g during 5 min, and plasma was centrifuged at 10,000g during 10 minutes at 4°C to eliminate remaining cells. Plasma samples were frozen at −80°C for further use.
RNA Extraction and miRNA Quantification
Total RNA was extracted from fresh tumor and paired normal tissue and from PV plasma and paired MV plasma using miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. miRNA detection was performed using commercial assays (TaqMan MicroRNA assays, Life Technologies, Grand Island, NY, USA) for miR-21, in the 7500 Sequence Detection System (Life Technologies). The appropriate negative controls (non-template control) were also run in each reaction. All reactions were performed in duplicate. Relative quantification was calculated using the formula 2−ΔΔCt. Normalization was performed with miR-191.
Statistical Analyses
Differences between ≥2 groups were calculated using the Mann–Whitney U test or the Kruskal–Wallis test as appropriate. miR-21 expression levels were dichotomized according to the fixed threshold method using the maxstat package of R to determine the optimal cutoff that best discriminated between different groups of patients for DFS. Fifty-two patients were evaluable for DFS; the 5 stage IV patients in whom only the primary tumor—but not the metastasis—was removed were not included in the analysis of DFS. DFS was calculated from the date of surgery to the date of death, relapse, or last follow-up. The univariate analysis of DFS was performed with the Kaplan–Meier method and compared using the log-rank test. All statistical analyses were performed with SPSS 14.0 (SPSS Inc, Chicago, IL) and R 2.6.0 Software (Vienna, AU). Statistical significance was set at P ≤ 0.05.
RESULTS
miR-21 Expression in Plasma and Tissue
Median miR-21 expression (fold change) was 0.2874 in MV plasma, 0.1805 in PV plasma, and 0.0201 in plasma from healthy controls. Median miR-21 expression in tumor and normal tissue was 0.3907 and 0.1733, respectively. miR-21 expression was significantly higher in MV plasma compared with PV plasma (P = 0.005) (Figure 1A). miR-21 expression was also significantly higher in tumor than in normal tissue (P < 0.001) (Figure 1A).
FIGURE 1.
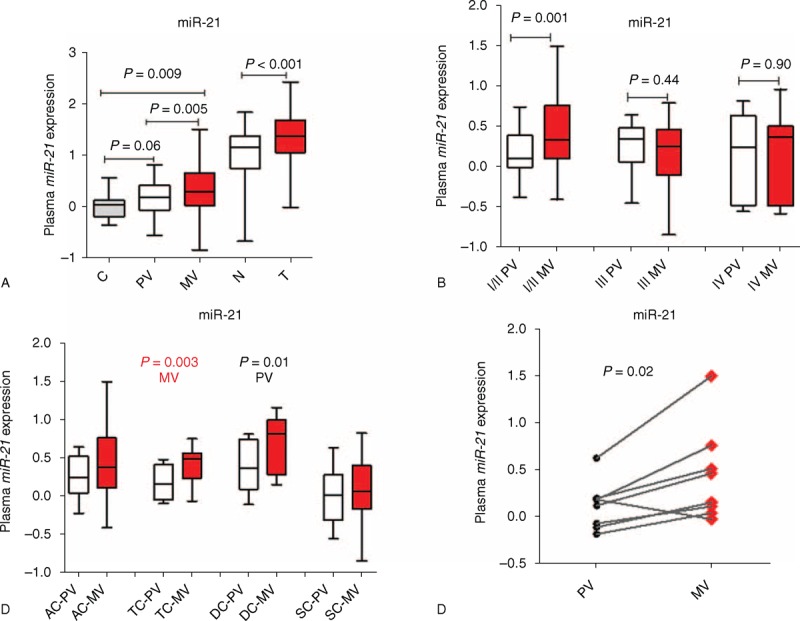
miR-21 expression levels (fold change) in plasma and tumor. (A) In plasma from healthy C and from the PV and MV of colon cancer patients and in matched T and N tissue samples from the same patients. (B) In plasma from the PV and MV of colon cancer patients classified by disease stage. (C) In plasma from the PV and MV of colon cancer patients classified by the anatomic location of the tumor. (D) In plasma from the PV and MV in 8 patients who developed locoregional metastases. AC = ascending colon, C = controls, DC = descending colon, MV = mesenteric vein, N = normal, PV = peripheral vein, SC = sigmoid colon, T = tumor; TC = transverse colon.
Patient Characteristics and miR-21 Expression
Table 1 shows the clinicopathologic characteristics of the 57 patients included in the study. Median age was 70 years. Thirty-four patients were males and 23 females. At diagnosis, 35 patients were having stage I to II of cancer, 15 stage III, and 7 stage IV. All patients underwent surgical resection; in 2 of the 7 stage IV patients, both the primary tumor and the metastasis were removed. Twenty-eight patients received adjuvant therapy with fluoropyrimidines.
TABLE 1.
Patient Characteristics
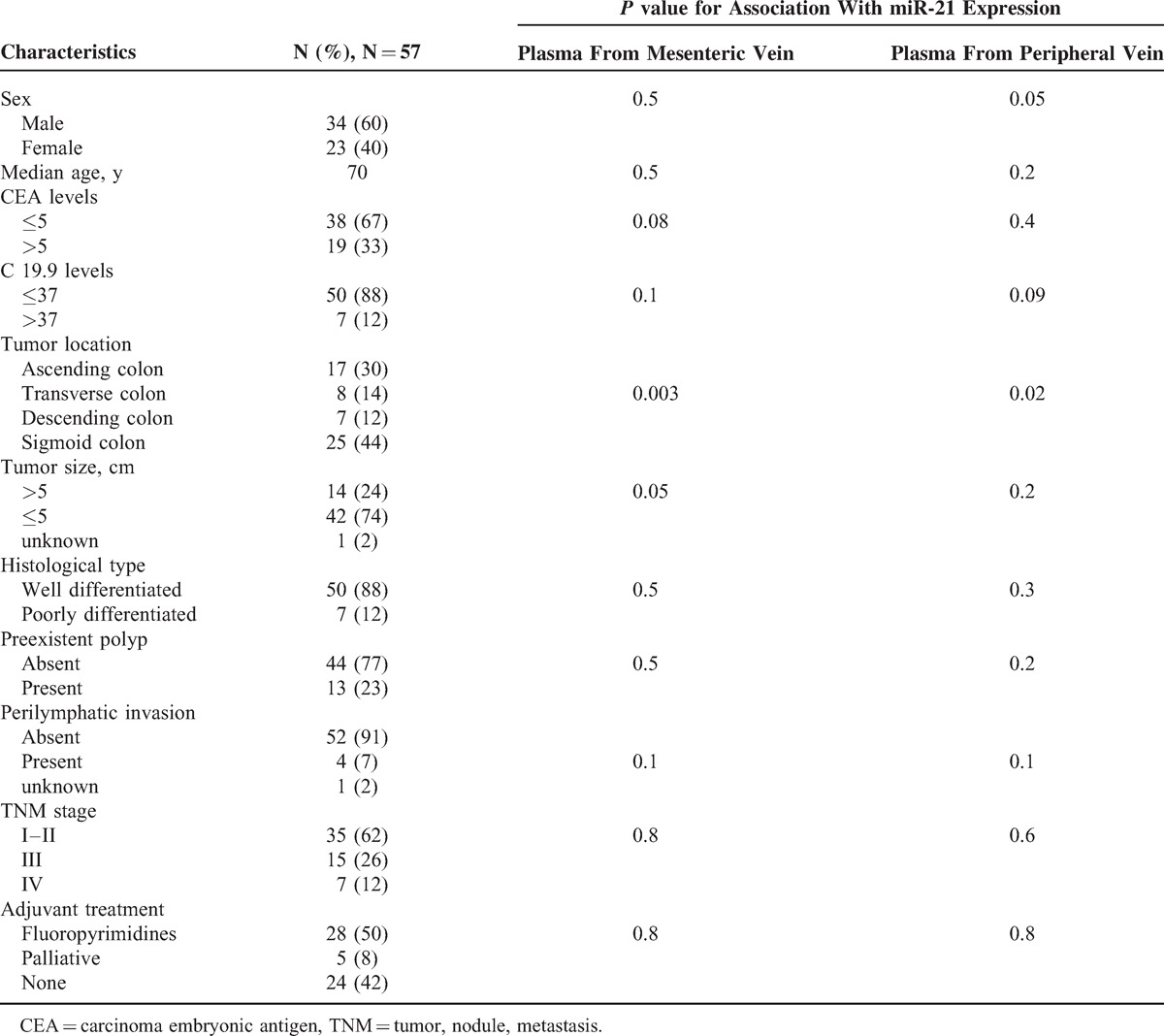
MV miR-21 levels correlated positively with tumor size (P = 0.04) and showed a trend toward correlation with carcinoma embryonic antigen levels (P = 0.08). Among patients with stage I to II disease, miR-21 levels were higher in MV plasma than in PV plasma (P = 0.001), whereas no significant differences were observed among patients with stage III to IV disease (Figure 1B). A highly significant association was observed between MV miR-21 levels and the anatomic location of the tumor (P = 0.003), whereas the association with PV miR-21 levels was less significant (P = 0.01) (Table 1, Figure 1C).
miR-21 Expression and Metastases
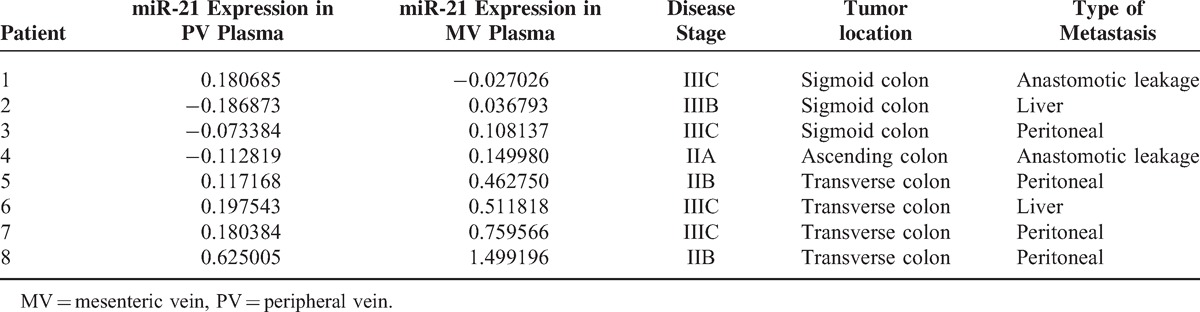
Of the 13 patients who developed metastases during the course of the disease, 8 had metastases in areas drained by the MV of the colon: 4 peritoneal metastases, 2 liver metastases, and 2 anastomotic. Of these 8 patients, 7 had higher miR-21 expression levels in MV plasma than in PV plasma (P = 0.02) (Table 2, Figure 1D).
TABLE 2.
Characteristics of 8 Patients Who Developed Locoregional Metastases
miR-21 Expression and DFS
Fifty-two patients were evaluable for DFS. Median DFS was not reached among the 8 patients with low levels of miR-21 in MV plasma, compared with 32.2 months (95% confidence interval [CI] 27.9–37.5) for the 44 patients with high levels of miR-21 (P = 0.05; Figure 2A). Median DFS was 38.1 months (95% CI 32.1–44.2) for the 24 patients with low levels of PV miR-21 and 30.1 months (95% CI 23.5–36.7) for the 28 patients with high levels (P = 0.07; Figure 2B).
FIGURE 2.
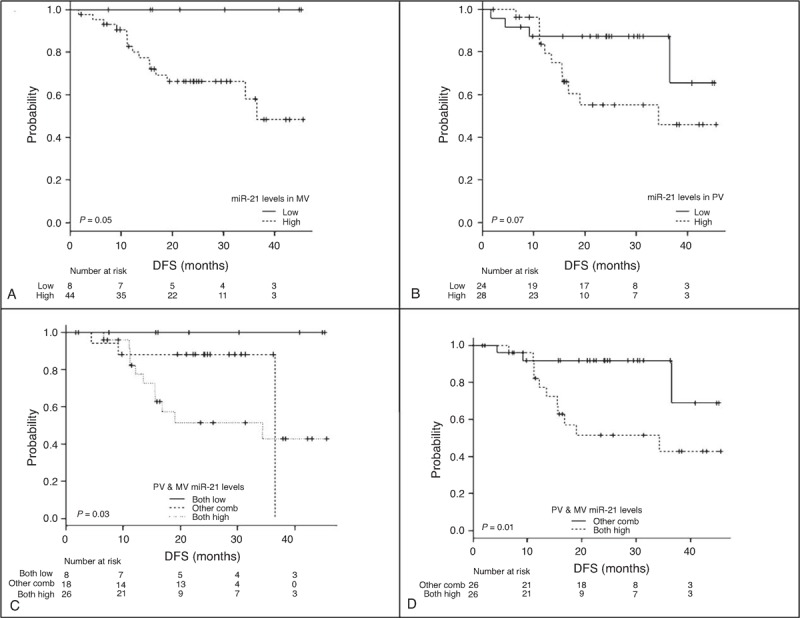
DFS according to miR-21 levels (fold change). (A) DFS according to miR-21 expression in plasma from theMV. (B) DFS according to miR-21 expression in plasma from thePV. (C) DFS in patients with high miR-21 expression in both MV and PV plasma compared with those with low expression in both the MV and the PV and those with other combinations. (D) DFS in patients with high miR-21 expression in both MV and PV plasma compared with all other patients. DFS = disease-free survival, MV = mesenteric vein, PV = peripheral vein.
To further evaluate whether the levels of miR-21 in both MV and PV plasma could have a combinatory effect on DFS, we classified patients in 3 groups: those with high miR-21 levels in both MV and PV plasma, those with low levels in both MV and PV plasma, and those with other combinations of miR-21 levels. Median DFS for the 8 patients with low MV and PV miR-21 was not reached, compared with 29.1 months (95% CI 22.2–35.9) for the 26 patients with high MV and PV miR-21 and 31.2 months (95% CI 24.9–37.5) for the remaining 18 patients (P = 0.03; Figure 2C). Based on these findings, we then compared DFS in the 26 patients with high miR-21 expression in both MV and PV plasma with all other patients. Median DFS was 29.1 months (95% CI 22.2–35.9) for these 26 patients versus 40 months (95% CI 34.8–45.2) for the remaining patients (P = 0.01; Figure 2D).
DISCUSSION
The main cause of death in patients with solid tumors is the development of metastases. The analysis of plasma and serum from cancer patients can help identify reliable biomarkers to predict relapse and metastasis in these patients. Recent findings suggest that the primary tumor can release proteins and miRNAs into the blood, which will organize a microenvironment known as a premetastatic niche in an area far from the primary tumor. This premetastatic niche will then provide support for the nesting and growth of metastatic tumor cells.28,29 Logically, the veins that are nearest the primary tumor would be most likely to contain the greatest concentration of these proteins and miRNAs.
The present study shows that miR-21 expression levels are significantly higher in MV plasma than in PV plasma. This finding suggests that the primary tumor in colon cancer releases high concentrations of miR-21 in the MV, but that these concentrations are later diluted in the circulatory system. This would explain why in other studies, only approximately 30% of miRNAs detected in PV plasma or serum mirrored those found in the primary tumor.30
Furthermore, miR-21 levels in MV plasma showed a trend toward correlation with CEA5 levels. In fact, previous studies have shown that in patients with CRC, CEA5 levels are higher in the MV than in the PV.31,32 A previous study with a large cohort of patients20 found a correlation between tumor size and miR-21 expression in PV plasma; our findings are similar, but we observed this correlation only in MV plasma. We also found an association between the anatomic location of the tumor and miR-21 expression levels in both MV and PV plasma, although miR-21 expression was higher in MV than in PV plasma. In addition, recent studies in CRC patients have observed circulating tumor cells in blood obtained from MVs and from hepatic veins.33 Taken together with these previous results, our findings indicate that veins near the tumor are the best source of biomarkers.
Similar to our findings, previous studies found no association between miR-21 expression and tumor stage, which may have been due to the use of different internal controls in these studies.21 However, among patients with stage I to II disease, we did observe a significant overexpression of miR-21 in MV plasma compared with PV plasma, which would confirm previous reports20,21 that miR-21 is overexpressed in the early stages of tumor development.
At 4 years of follow-up, patients with high miR-21 levels in MV plasma had a significantly worse prognosis than those with low levels; in contrast, no differences were observed according to miR-21 levels in PV plasma, which suggests that miR-21 is more easily detected in MV than in PV plasma. Interestingly, however, patients with high miR-21 expression in both MV and PV plasma had a significantly shorter DFS than those with low levels in either MV or PV plasma. We can speculate that whether initial MV and PV levels are high; PV levels that remain high throughout follow-up may well be used to identify patients with a high risk of relapse.
Interestingly, the 8 patients who relapsed and developed metastases in areas drained by the MV of the colon—the liver and intestines—had higher levels of MV miR-21 than PV miR-21. In contrast, metastases in areas not drained by the MV of the colon—such as the lung—were not associated with miR-21 expression levels. In fact, miR-21 has been associated with hepatocellular tumors in several studies, which have shown that miR-21 targets several genes—such as MAP2K3,34 PDCD4,35 and PTEN36—leading to the development of hepatocellular carcinomas. In a large cohort of patients with hepatocellular carcinomas, array analysis identified a panel of 7 miRNAs (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801), wherein miR-21 expression levels were able to distinguish patients from healthy controls.37
It has recently been reported that miRNAs released by the primary tumor can play a key role in preparing the premetastatic niche.38,39 miR-21 and miR-29a are released by tumor cells through exosomes, which bind to the Toll-like receptors of immune cells. The activation of the Toll-like receptors leads to the release of tumor necrosis factor-α and interleukin-6, which in turn prepare the extracellular environment for tumor growth and dissemination.40 Our findings suggest that the miRNAs released in the MV may be retained in areas near the primary tumor (liver and intestines), where they could work to build the premetastatic niche. To validate these findings, however, further research on other miRNAs associated with colon cancer should compare expression levels in MV and PV plasma and examine the potential association between MV plasma expression and locoregional metastases. The findings of the present study will hopefully act as a springboard to strengthen collaboration among surgeons, medical oncologists, and molecular biologists with the aim of improving outcome in colon cancer patients.
Footnotes
Abbreviations: CRC = colorectal cancer, CT = computed tomography, DFS = disease-free survival, miRNA = microRNA, MV = mesenteric vein, PV = peripheral vein.
This study was partially supported by a grant from SDCSC (Servei de Donació del Cos a la Ciència). RT is recipient of an APIF (Ajuts de Personal Investigador Predoctoral en Formació) grant from the Universitat de Barcelona. Neither of these funding bodies had a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11–30. [DOI] [PubMed] [Google Scholar]
- 2.Gray R, Barnwell J, McConkey C, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet 2007; 370:2020–2029. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Lee Y, Fan H, et al. Identification of common microRNA-mRNA regulatory biomodules in human epithelial cancers. Chin Sci Bull 2010; 55:3576–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014; 11:145–156. [DOI] [PubMed] [Google Scholar]
- 5.Allegra A, Alonci A, Campo S, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int J Oncol 2012; 41:1897–1912. [DOI] [PubMed] [Google Scholar]
- 6.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18:883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbings DJ, Ciaudo C, Erhardt M, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 2009; 11:1143–1149. [DOI] [PubMed] [Google Scholar]
- 8.Roberts CT, Jr, Kurre P. Vesicle trafficking and RNA transfer add complexity and connectivity to cell-cell communication. Cancer Res 2013; 73:3200–3205. [DOI] [PubMed] [Google Scholar]
- 9.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011; 108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008; 141:672–675. [DOI] [PubMed] [Google Scholar]
- 11.Zhu S, Si ML, Wu H, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 2007; 282:14328–14336. [DOI] [PubMed] [Google Scholar]
- 12.Chang KH, Miller N, Kheirelseid EA, et al. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol 2011; 37:597–603. [DOI] [PubMed] [Google Scholar]
- 13.Qi L, Bart J, Tan LP, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 2009; 9:163.doi:10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coppola V, Musumeci M, Patrizii M, et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene 2013; 32:1843–1853. [DOI] [PubMed] [Google Scholar]
- 16.Bihrer V, Waidmann O, Friedrich-Rust M, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One 2011; 6:e26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei J, Gao W, Zhu CJ, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer 2011; 30:407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma GJ, Gu RM, Zhu M, et al. Plasma post-operative miR-21 expression in the prognosis of gastric cancers. Asian Pac J Cancer Prev 2013; 14:7551–7554. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu S, Ichikawa D, Takeshita H, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 2011; 105:104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 2013; 105:849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol 2012; 138:1659–1666. [DOI] [PubMed] [Google Scholar]
- 22.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg 2012; 256:544–551. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Gao X, Wei F, et al. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene 2014; 533:389–397. [DOI] [PubMed] [Google Scholar]
- 24.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58:1375–1381. [DOI] [PubMed] [Google Scholar]
- 25.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010; 127:118–126. [DOI] [PubMed] [Google Scholar]
- 26.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol 2012; 36:e61–e67. [DOI] [PubMed] [Google Scholar]
- 27.Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 2010; 25:1674–1680. [DOI] [PubMed] [Google Scholar]
- 28.Sceneay J, Smyth MJ, Moller A. The pre-metastatic niche: finding common ground. Cancer Metastasis Rev 2013; 32:449–464. [DOI] [PubMed] [Google Scholar]
- 29.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer 2009; 9:239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pigati L, Yaddanapudi SC, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 2010; 5:e13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivankovics IG, Fernandes LC, Saad SS, et al. Peripheral and mesenteric serum levels of CEA and cytokeratins, staging and histopathological variables in colorectal adenocarcinoma. World J Gastroenterol 2008; 14:6699–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rezende Junior HC, Palma RT, Toloi GC, et al. Carcinoembryonic antigen levels in the peripheral and mesenteric venous blood of patients with rectal carcinoma. Arq Gastroenterol 2013; 50:264–269. [DOI] [PubMed] [Google Scholar]
- 33.Steinert G, Scholch S, Niemietz T, et al. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res 2014; 74:1694–1704. [DOI] [PubMed] [Google Scholar]
- 34.Xu G, Zhang Y, Wei J, et al. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer 2013; 13:469.doi: 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Q, Wang Z, Hu Y, et al. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep 2012; 27:1660–1668. [DOI] [PubMed] [Google Scholar]
- 36.Damania P, Sen B, Dar SB, et al. Hepatitis B Virus Induces Cell Proliferation via HBx-Induced microRNA-21 in Hepatocellular Carcinoma by Targeting Programmed Cell Death Protein4 (PDCD4) and Phosphatase and Tensin Homologue (PTEN). PLoS One 2014; 9:e91745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 2011; 29:4781–4788. [DOI] [PubMed] [Google Scholar]
- 38.Ghasemi R, Grassadonia A, Tinari N, et al. Tumor-derived microvesicles: the metastasomes. Med Hypotheses 2013; 80:75–82. [DOI] [PubMed] [Google Scholar]
- 39.Squadrito ML, Etzrodt M, De Palma M, et al. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol 2013; 34:350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A 2012; 109:E2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]


