Supplemental Digital Content is available in the text
Abstract
Long non-coding RNAs (lncRNAs) are recently discovered RNA transcripts that are aberrantly expressed in many tumor types. Numerous studies have suggested that lncRNAs can be utilized for cancer diagnosis and prognosis. LSINCT5 (long stress-induced non-coding transcript 5) is dramatically upregulated in breast and ovarian cancer and affects cellular proliferation. However, the expression pattern of LSINCT5 in gastrointestinal cancer and the association between aberrant expression of LSINCT5 in gastrointestinal cancer and malignancy, metastasis, or prognosis remain unknown.
LSINCT5 expression was detected in gastrointestinal cancer and paired adjacent normal tissue samples or cell lines using reverse transcription quantitative PCR (RT-qPCR). We also investigated the potential relationship between tumor LSINCT5 levels and clinicopathological features of gastrointestinal cancer. Finally, we assessed whether LSINCT5 influences in vitro cell proliferation.
The expression of LSINCT5 is significantly upregulated in gastrointestinal cancer tissues and cell lines relative to their normal counterparts. In addition, increased LSINCT5 expression was correlated with a larger tumor size, deeper tumor depth, and advanced clinical stage. Kaplan–Meier analysis indicated that gastric cancer (GC) and colorectal cancer (CRC) patients with higher LSINCT5 expression levels have worse disease-free survival (DFS) and disease-specific survival (DSS) rates. Moreover, multivariate analysis revealed that increased expression of LSINCT5 is an independent predictor of DFS and DSS rates in GC patients. The ectopic expression of LSINCT5 in gastrointestinal cancer cell lines resulted in an increase in cellular proliferation; conversely, knock down of LSINCT5 significantly inhibited proliferation.
These results suggest that LSINCT5 may represent a novel prognostic indicator and a target for gene therapy in gastrointestinal cancer.
INTRODUCTION
The human transcriptome contains large numbers of protein-coding messenger RNAs (mRNAs) and a large set of non-coding RNA (ncRNA) that have structural, regulatory, or unknown functions. In general, ncRNAs are grouped into two major classes based on their length. Those transcripts shorter than 200 nucleotides (nt) are usually recognized as small ncRNAs, which include Piwi-interacting RNAs, small-interfering RNAs, microRNAs (miRNAs) and some bacterial regulatory RNAs. The well-documented miRNAs are naturally occurring small ncRNAs that are capable of regulating target mRNAs at the post-transcriptional level.1,2 Overwhelming evidence suggests that in cancer, miRNAs are often dysregulated and may serve as oncogenes or tumor suppressors.3,4 Long non-coding RNAs (lncRNAs) are a newly discovered class of non-coding RNA transcripts that are longer than 200 nt.5 Similar to small non-coding RNAs, lncRNAs are involved in a number of biological functions and pathological processes, such as stem cell pluripotency, development, cell growth, and apoptosis. Moreover, emerging studies have revealed that some differentially expressed lncRNAs are important regulatory molecules in tumorigenesis and development, demonstrating their potential roles in both oncogenic and tumor-suppressive pathways.6
Several studies have documented a link between the aberrant expression of lncRNAs and the pathogenesis/prognosis of cancer.7 For example, the increased expression of the oncogenic lncRNA HOTAIR is associated with metastasis in breast cancer patients,8 and CRC patients with higher HOTAIR expression levels have a relatively worse prognosis.9 We previously demonstrated that the aberrant expression of LOC285194, a p53-regulated tumor suppressor, correlates with several clinicopathological factors such as tumor size and metastasis. Furthermore, decreased LOC285194 expression is an independent predictor of disease-specific survival (DSS) for CRC patients.10 Our results also suggested that ncRAN (non-coding RNA expressed in aggressive neuroblastoma) can contribute to CRC cell migration and invasion and predicts poor overall survival for CRC patients.11 These results suggest that lncRNAs could be used for cancer diagnosis, as prognostic biomarkers, and as potential therapeutic targets.12
LSINCT5 (long stress-induced non-coding transcript 5) is a 2.6-kb polyadenylated intergenic lncRNA that is located in the nucleus on the negative strand and is potentially transcribed by RNA polymerase III.13,14 LSINCT5 is overexpressed in breast and ovarian cancer cell lines and tumor tissues compared with their normal counterparts. In addition, an oncogenic function of LSINCT5 has been suggested by knockdown experiments that showed a decrease in cellular proliferation.14 Moreover, low LSINCT5 expression levels have been observed in less proliferative normal tissues such as the brain, whereas higher LSINCT5 expression levels have been observed in highly proliferative normal tissues, including the kidney, pancreas, colon, and spleen.13,14 However, little information is available on the expression pattern of LSINCT5 in gastrointestinal cancer, and whether the aberrant expression of LSINCT5 in gastrointestinal cancer is associated with malignancy, metastasis, or prognosis remains unknown.
In this study, we investigated whether LSINCT5 expression is altered in gastrointestinal cancer tissue or cell lines compared with adjacent normal tissue or normal cell lines. We also investigated the potential relationship between tumor LSINCT5 levels and clinicopathological features of gastrointestinal cancer, such as tumor size, growth phase, pathological stage, vascular invasion, nervous invasion, lymphatic metastasis, peritoneal metastasis, disease-free survival (DFS), and DSS. Finally, we assessed whether LSINCT5 influences in vitro cell proliferation.
MATERIALS AND METHODS
Tumor Tissue Samples
Samples of fresh gastric (GC) and colorectal cancer (CRC) tissues and samples of paired adjacent noncancerous tissues were randomly selected from patients who underwent resection of primary GC or CRC at Fudan University Shanghai Cancer Center from September 2007 to April 2008. The carcinoma diagnosis was histopathologically confirmed. None of the patients received preoperative therapy.
Data for all subjects were obtained from medical records, pathology reports, and personal interviews with the subjects. A total of 71 patients (57 males and 14 females) suffered GC; the median age was 60 years (range, 34–82 years). A total of 74 patients (35 males and 39 females) suffered CRC; the median age was 56 years (range, 30–86 years). The collected clinicopathological features included tumor size, location, histological stage, depth of invasion, lymphatic metastasis, vascular invasion, nervous invasion, clinical stage, and peritoneal/distance metastasis. Patient follow-up was performed every 2–3 months during the first year after surgery and every 3–6 months thereafter until June 30, 2013. The histological grade of the cancers was assessed according to the standard of the World Health Organization. Cancers were classified using the TNM staging system of the American Joint Committee on Cancer (2010) and the International Union against Cancer. This study was approved by The Clinical Research Ethics Committee of Fudan University Shanghai Cancer Center. Written informed consent was obtained from all participants.
Cell Lines and Culture Conditions
Human GC cell lines (AGS, MKN-45, MGC-803, BGC-823, and HGC-27), CRC cell lines (LoVo, Caco-2, DLD1, HCT-8, and HCT-116), one normal gastric epithelial cell line (GES-1), and one normal intestinal mucous cell line (CCC-HIE-2) were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) or in Dulbecco's modified Eagle's medium (Hyclone, Logan, UT, USA). All cell lines were supplemented with 10% fetal bovine serum and 2 mM L-glutamine (Invitrogen) at 37°C in a 10% CO2 atmosphere.
RNA Isolation, Reverse Transcription (RT), and Quantitative PCR (qPCR)
Total RNA was extracted from tissue samples and cells using TRIzol (Invitrogen) according to the manufacturer's protocol. RT and qPCR kits were utilized to evaluate LSINCT5 expression in the tissue samples and cells. The RT and qPCR reactions were performed as previously described,12 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control to normalize the data. The qPCR primers for LSINCT5 and GAPDH are detailed in Supplementary, http://links.lww.com/MD/A115.
Ectopic Expression and Gene Silencing of LSINCT5 in Cells
For the enhanced expression study, full-length LSINCT5 (GU228577.2) complementary DNA was cloned into pcDNA3.1 (Suntrans-bio, shanghai, China). The resulting vector was sequenced and named pcDNA-LSINCT5. To knock down LSINCT5 expression, 3 individual siRNAs (LSINCT5-siRNA1, LSINCT5-siRNA2, and LSINCT5-siRNA3; Bioneer, Daejeon, South Korea) were synthesized and optimized. An extra siRNA was designed as a negative control (NC-siRNA) (Supplementary, http://links.lww.com/MD/A115). MGC-803 and Caco-2 cells were transfected with vector or pcDNA-LSINCT5, HGC-27 and LoVo cells were transfected with control siRNA or LSINCT5-siRNA. The LSINCT5 expression levels were determined using RT-qPCR. After 48 hours, total RNA was extracted from the harvested cells using TRIzol, and LSINCT5 expression was measured by RT-qPCR as described above.
Cell Proliferation Assay
Cell proliferation was evaluated using a CCK-8 assay (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. This assay is based on the cleavage of the WST-8 tetrazolium salt by mitochondrial dehydrogenase in viable cells. For this assay, 2 × 103 cells/well were incubated in 100 μL culture medium in 96-well plates. The cells were transfected with vector or pcDNA-LSINCT5 and control siRNA or LSINCT5-siRNA2 or LSINCT5-siRNA3 for 1, 2, or 3 days before 10 μL CCK-8 (5 mg/mL) was added to the culture medium in each well. After 1 hour of incubation at 37°C, the absorbance at 450 nm was measured in a ThermoMax Microplate Reader (Molecular Devices). Each experiment was repeated 3 times, and the data represent the mean of all measurements.
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 (IBM, SPSS, Chicago, IL, USA). The significance of the differences between groups was estimated using Student's t test, the χ2 test, or the Wilcoxon test. The Kaplan–Meier method with the log-rank test for comparisons was used to calculate the DFS and the DSS rates. The DFS rate was calculated from the date of surgery to the date of progression (local and/or distal tumor recurrence) or to the date of death. The DSS rate was defined as the length of time between the diagnosis and death or last follow-up. Survival data were evaluated using univariate and multivariate Cox regression analyses. A value of P < 0.05 indicated a significant difference.
RESULTS
LSINCT5 Expression Is Significantly Upregulated in GC and CRC
Our first goal was to investigate whether LSINCT5 expression could be detected and whether its expression was altered in GC and CRC tissue compared with adjacent normal tissue. Using GAPDH as the normalization control, we determined that 54 (73%) cases displayed high expression of LSINCT5 in GC tissues (Figure 1A), whereas 45 (63%) cases displayed high expression of LSINCT5 in CRC tissues (Figure 1B). In addition, LSINCT5 expression was significantly higher in GC tissue (Figure 1C) or CRC tissue (Figure 1D) compared with adjacent normal tissue (both P < 0.001). We next examined the expression level of LSINCT5 in several GC and CRC cell lines and in two normal gastrointestinal cell lines. LSINCT5 expression was significantly higher in the GC or CRC cell lines than in the GES-1 (Figure 1E) or CCC-HIE-2 cell lines (Figure 1F).
FIGURE 1.
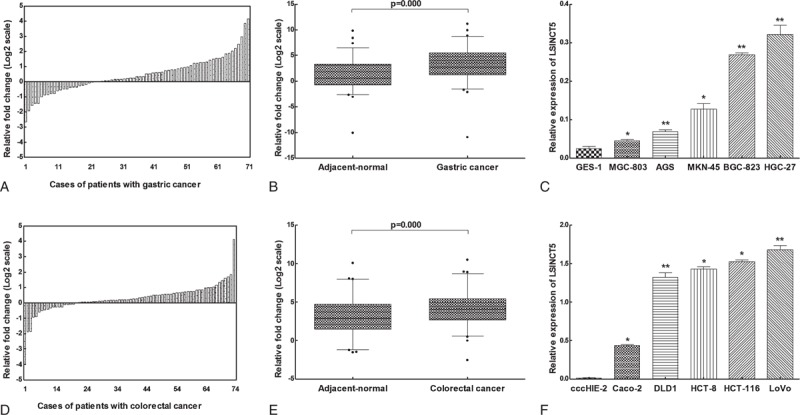
LSINCT5 expression in GC and CRC tissue samples and cell lines. High LSINCT5 expression levels were observed in GC tissues in 54 (73%) cases (A), while 45 (63%) cases showed high expression of LSINCT5 in CRC tissues when normalized to GAPDH (D). Compared with adjacent normal tissues, LSINCT5 was overexpressed in GC tissues (B) and CRC tissues (E). (C) LSINCT5 expression was quantitated in 5 GC cell lines and the GES-1 cell line. ∗Significant difference compared with GES-1 (mean ± SD; ∗P < 0.05, ∗∗P < 0.01, n = 3). (F) LSINCT5 expression was quantified in 5 CRC cell lines and the CCC-HIE-2 cell line. ∗Significant difference compared with CCC-HIE-2 (mean ± SD; ∗P < 0.05, ∗∗P < 0.01, n = 3).
LSINCT5 Expression Is Correlated With GC and CRC Clinical Pathology
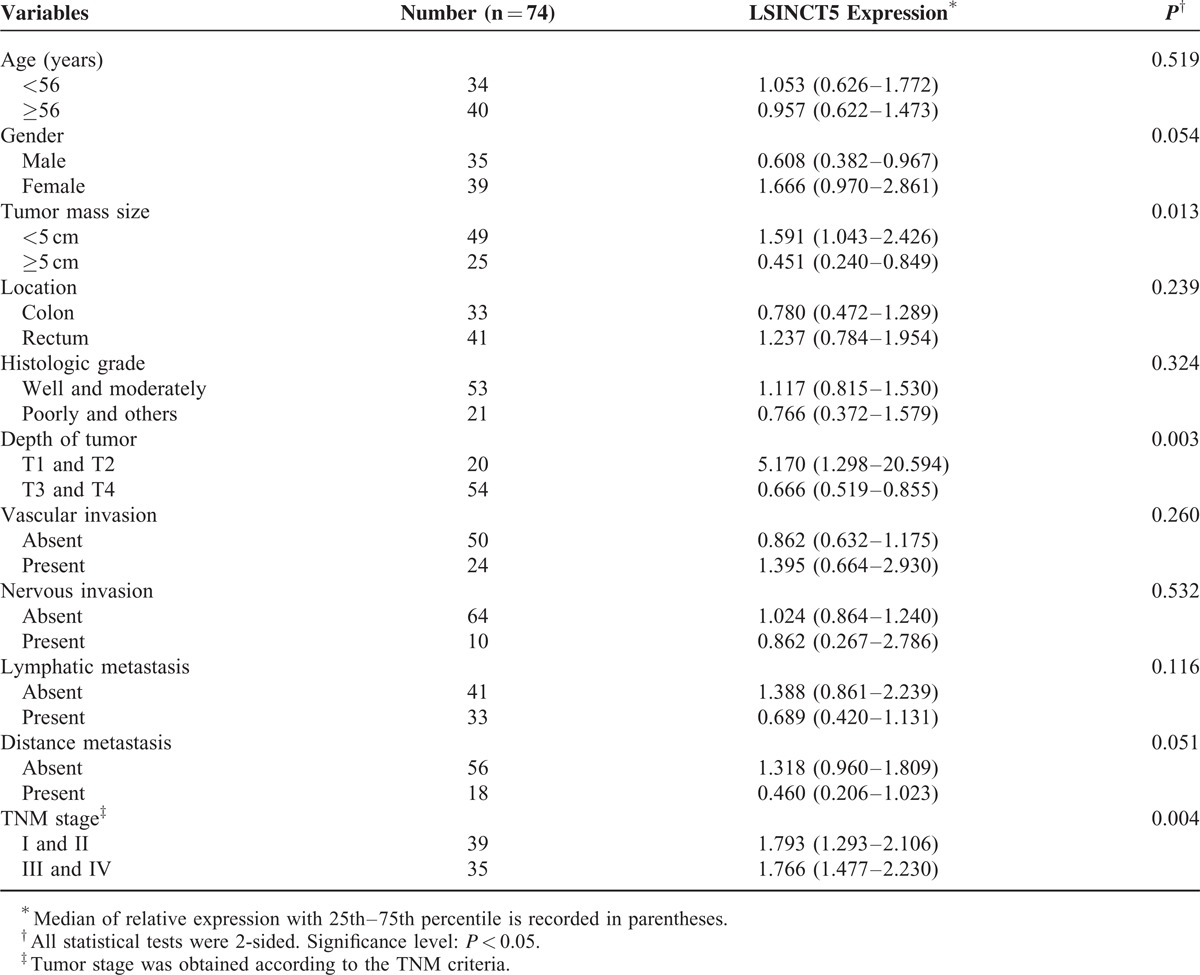
Next, we determined whether tumor LSINCT5 expression levels are associated with specific clinicopathological characteristics. GC patients with higher LSINCT5 expression levels tended to have larger tumor sizes (P = 0.025), deeper tumor invasion depth (P = 0.022), more lymphatic metastasis (P = 0.043), and advanced TNM stages (P = 0.016) (Table 1), while the high expression of LSINCT5 was associated with increased tumor size (P = 0.013), deeper tumor invasion depth (P = 0.003), and advanced TNM stage (P = 0.005) for CRC patients (Table 2). These data suggest that LSINCT5 overexpression is associated with the clinical progression and development of human gastrointestinal cancer. However, there was no significant correlation between LSINCT5 expression and other clinicopathological features such as age, gender, tumor location, histological grade, vascular invasion, nervous invasion, or distant/peritoneal metastasis (P > 0.05).
TABLE 1.
Relationship Between LSINCT5 Expression and Clinicopathologic Parameters of Gastric Cancer Patients
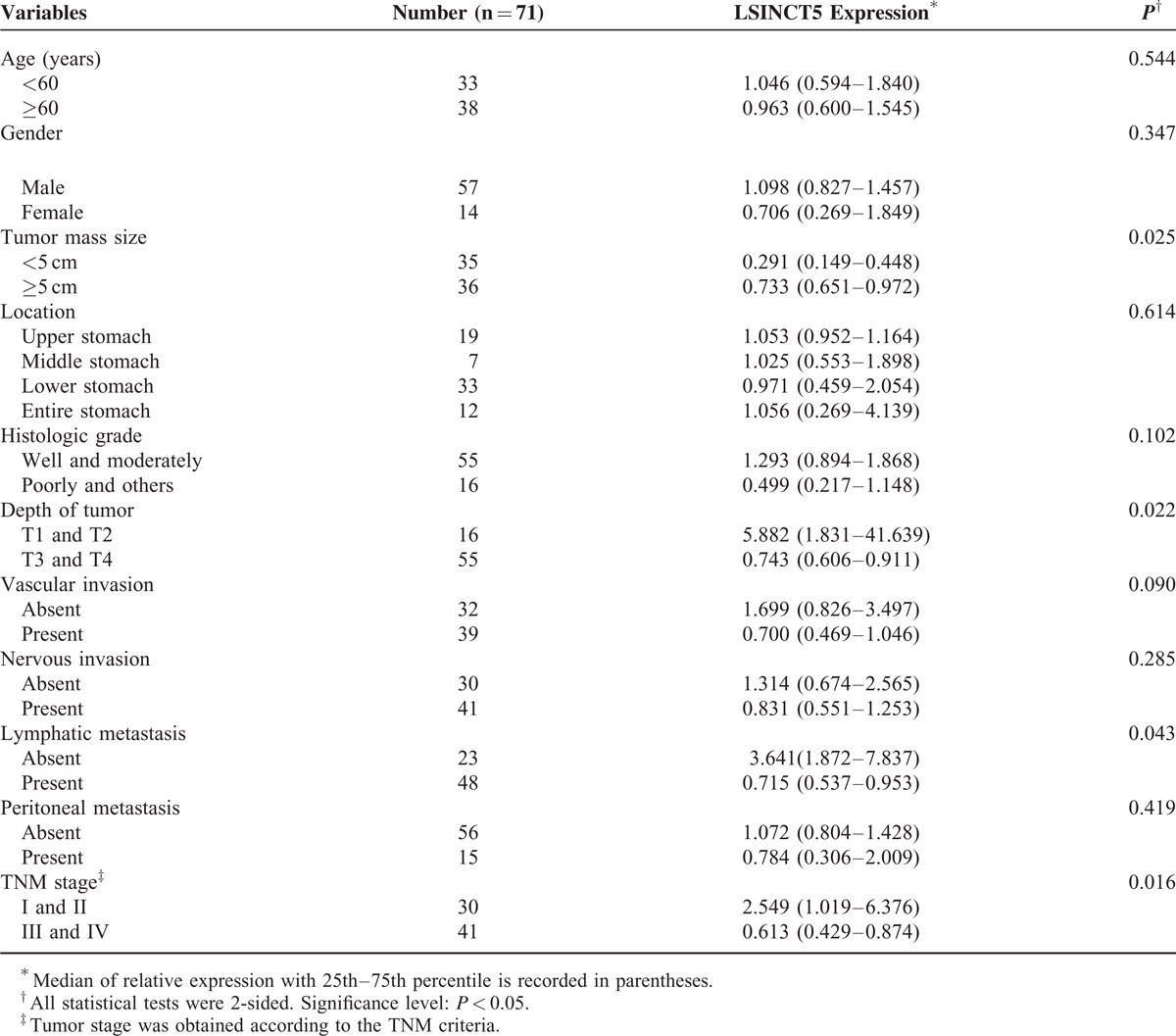
TABLE 2.
Relationship Between LSINCT5 Expression and Clinicopathologic Parameters of Colorectal Cancer Patients
LSINCT5 Expression Is Correlated With GC and CRC Patients’ Prognosis
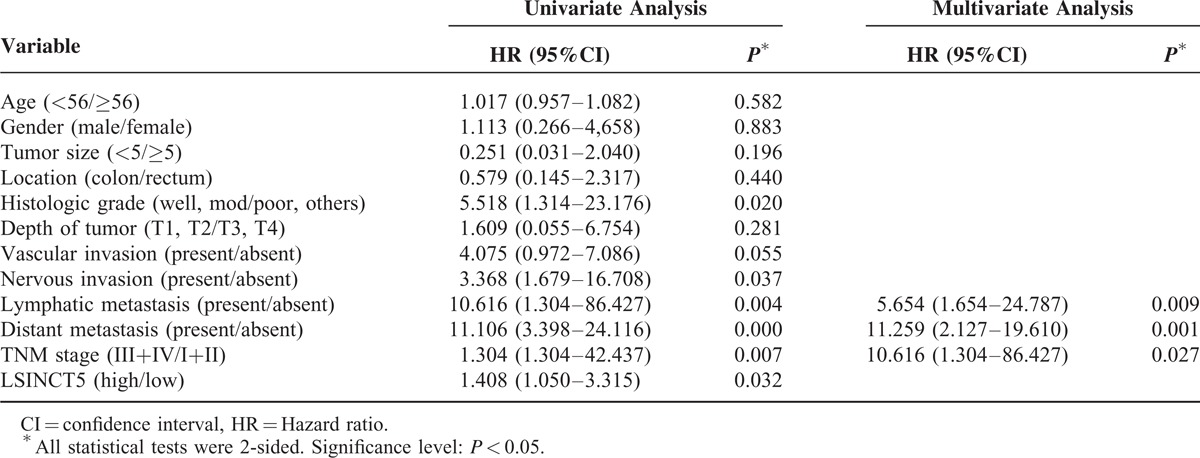
To assess the correlation between LSINCT5 expression and gastrointestinal cancer patients’ prognosis, the expression levels of LSINCT5 in tumor tissue were categorized as low or high relative to the mean. Kaplan–Meier analysis indicated that patients with higher expression levels of LSINCT5 had significantly shorter DFS and DSS than did those with lower expression levels in both the GC cohort (P = 0.003 and 0.004; log rank test, Figure 2A and B) and the CRC cohort (P = 0.012 and 0.029; log rank test, Figure 2C and D). A univariate Cox proportional hazards regression analysis of DFS also revealed that both GC (Table 3) and CRC (Table 4) patients with higher expression levels of LSINCT5 were found to have significantly worse prognoses than those with lower expression levels. However, a multivariate analysis demonstrated that the increased expression of LSINCT5 was only an independent indicator for DFS (Table 3) and DSS (Table 5) in GC patients; differences in the DFS (Table 4) or DSS (Table 6) rates in the CRC cohort did not reach statistically significant levels. These data suggest that LSINCT5 plays an important role in gastric carcinogenesis.
FIGURE 2.
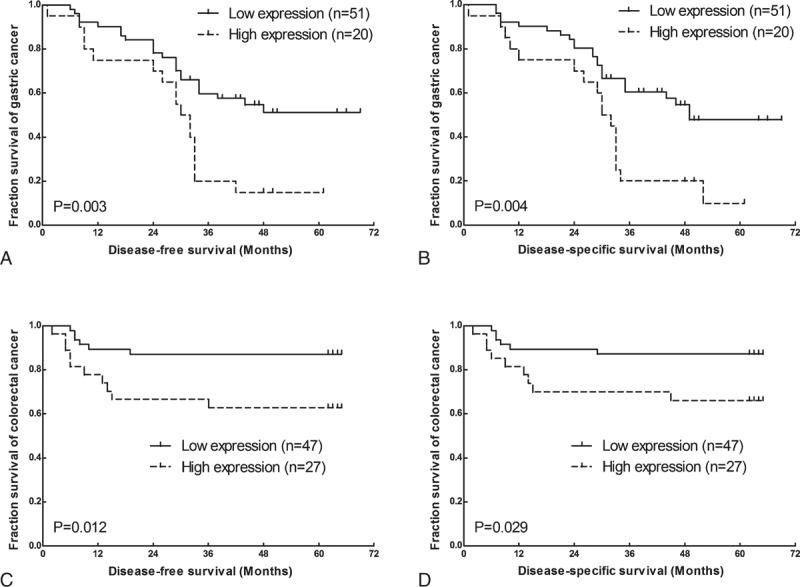
Kaplan–Meier DFS and DSS analyses in GC and CRC. The Kaplan–Meier DFS (A) and DSS (B) analyses for GC patients based on the expression of LSINCT5 (n = 71) showed that the DFS and DSS rates in the LSINCT5 high-expression group were significantly lower than those of patients with low LSINCT5 expression levels. Kaplan–Meier DFS (C) and DSS (D) analyses for CRC patients (n = 74) showed that the DFS and DSS rates in the LSINCT5 high-expression group were significantly lower than those of patients with low LSINCT5 expression levels.
TABLE 3.
Univariate and Multivariate Analysis of Clinicopathological Factors for Disease-Free Survival in Gastric Cancer
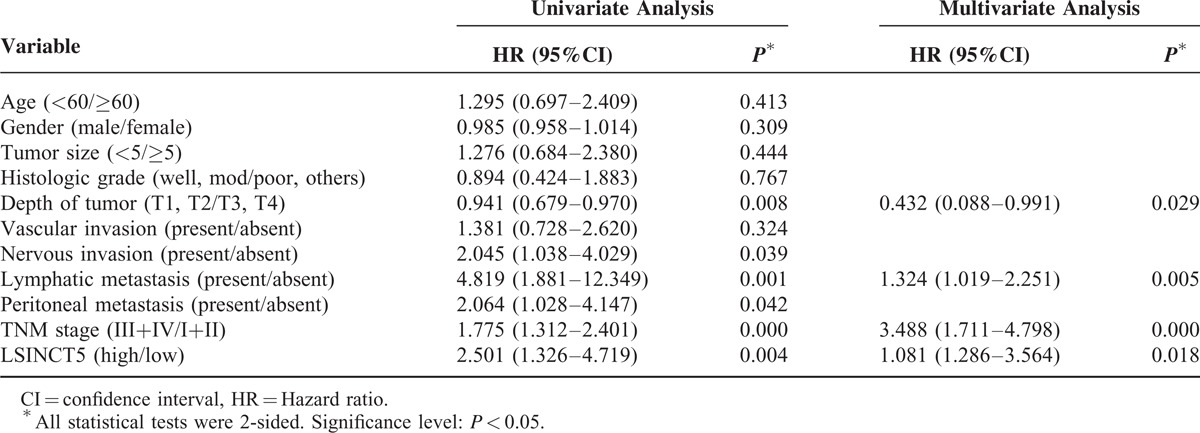
TABLE 4.
Univariate and Multivariate Analysis of Clinicopathological Factors for Disease-Free Survival in Colorectal Cancer
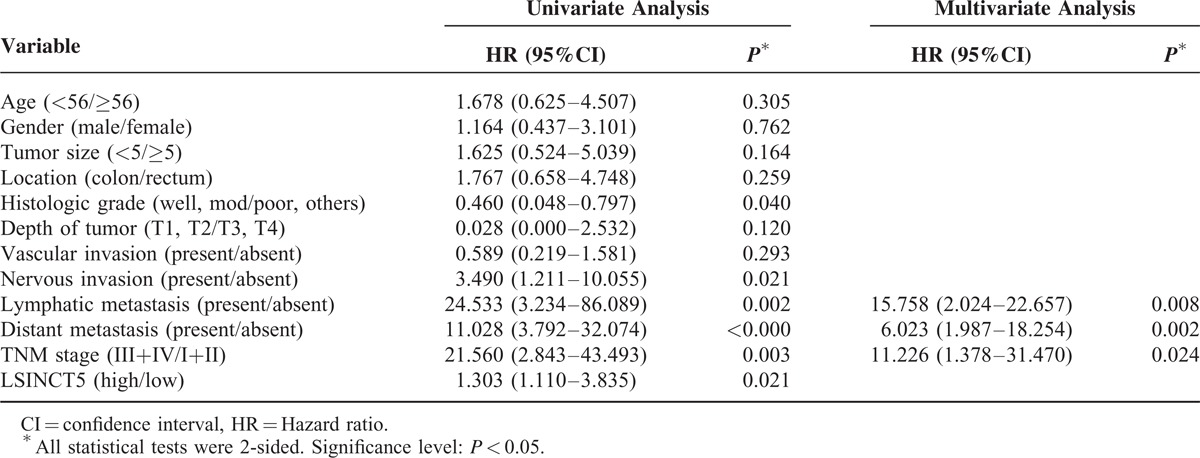
TABLE 5.
Univariate and Multivariate Analysis of Clinicopathological Factors for Disease-Specific Survival in Gastric Cancer
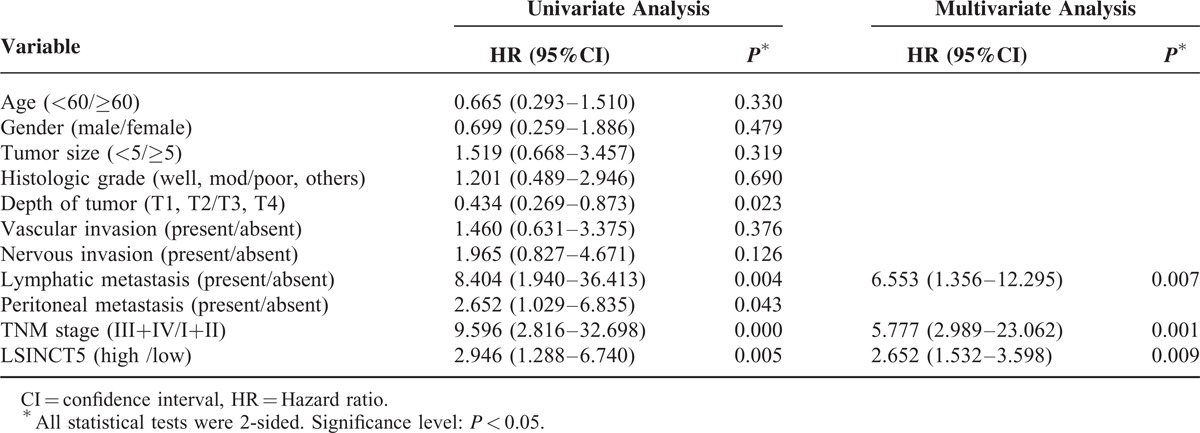
TABLE 6.
Univariate and Multivariate Analysis of Clinicopathological Factors for Disease-Specific Survival in Colorectal Cancer
LSINCT5 Promotes Tumor Cell Growth
To investigate the biological function of LSINCT5 in GC cell lines, we ectopically expressed LSINCT5 in MGC-803 and Caco-2 cells (which expressed the lowest relative levels of LSINCT5), silenced LSINCT5 in HGC-27 and LoVo cells (which expressed the highest relative levels of LSINCT5) and evaluated the resulting cell proliferation. Up-regulating LSINCT5 significantly promoted tumor cell growth (Figure 3A–C), whereas down-regulating LSINCT5 inhibited tumor cell growth, as measured using the CCK-8 assay (Figure 3D–F).
FIGURE 3.
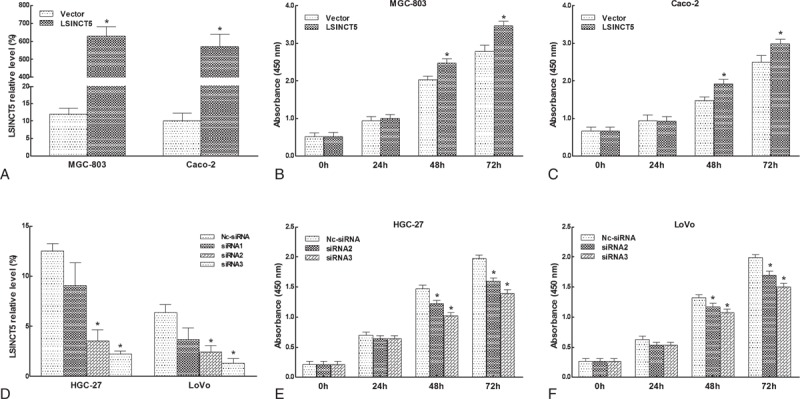
LSINCT5 promoted cell proliferation. MGC-803 and Caco-2 cells were transfected with vector or pcDNA-LSINCT5, HGC-27 and LoVo cells were transfected with control siRNA or LSINCT5-siRNA. LSINCT5 expression levels were determined using RT-qPCR. Up-regulating LSINCT5 (A) significantly promoted tumor cell growth (B, C), whereas down-regulating LSINCT5 (D) inhibited tumor cell growth, as measured using the CCK-8 assay (E, F). The error bars represent the SEM (n = 3). ∗Significant difference compared with the control (P < 0.05).
DISCUSSION
In this study, we confirmed that the expression of LSINCT5 was significantly higher in GC and CRC tissues than in adjacent normal tissues. In addition, LSINCT5 expression was markedly increased in several gastrointestinal cancer cell lines compared with normal cell lines. Clinically, the increased expression of LSINCT5 is an independent predictor of DFS and DSS rates in GC patients. Moreover, using loss-of-function and gain-of-function approaches, we determined that LSINCT5 promoted gastric cancer cell growth in vitro.
LSINCT5 has been reported to be overexpressed in breast and ovarian cancer cell lines and tumor tissues compared with their normal counterparts,13,14 and these findings are consistent with our present report of increased LSINCT5 expression in gastrointestinal cancer, indicating that LSINCT5 may be a common oncogenic lncRNA. Although a variety of lncRNAs are aberrantly expressed in several tumors, the mechanisms underlying this dysregulation are poorly understood. LSINCT5 is an intergenic lncRNA located 829,825 bp downstream of the IRX4 gene and 31,529 bp upstream of the IRX2 gene, both genes closely related to tumor development. LSINCT5's disorder in gastrointestinal tumors may be the downstream effect of these two genes. In addition, to determine how LSINCT5 expression was regulated transcriptionally, we performed a search for possible transcription factor-binding sites using online software programs MatInspector (www.genomatix.de/online_help/help_matinspector/matinspector_help.html) and TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html) in the promoter region of LSINCT5 and found that LSINCT5 may be regulated by many known tumorigenic and development related transcription factors such as C/EBPbeta, signal transducers and activators of transcription (STATx), etc. Moreover, combined with the experimental results obtained by Silva et al,13 we assumed that LSINCT5, similar to LOC285194,15 and growth arrest-specific transcript 5 (GAS5),16 ERIC,17 and JADE18, may be a product from cancer progress, for example, in response to DNA damage. The precise mechanism requires further study.
The role of LSINCT5 in cellular proliferation was also investigated in our study. Up-regulating LSINCT5 significantly promoted tumor cell growth, whereas down-regulating LSINCT5 inhibited tumor cell growth, as measured using the CCK-8 assay. Combined with the experimental results of Silva et al,14 we assumed that LSINCT5 may act as an oncogenic lncRNA by influencing many cellular processes, including proliferation, in the carcinogenesis of cancers such as breast cancer, ovarian cancer, and gastrointestinal cancer. Although LSINCT5 can facilitate the progression of some types of cancer, little is known about the underlying molecular mechanisms. The precise mechanism of how lncRNA regulates the expression of its target genes is not clear. Microarray analysis has shown that several cancer-associated genes, such as NEAT1, PSPC1, EPPK1, ACTR2, and CXCR4, display a more than 2-fold increase or decrease in expression level in response to LSINCT5 knockdown in MCF7 cells. Silva et al14 suggested that increased LSINCT5 leads to increased CXCR4, which may explain the proliferative effect of LSINCT5. The exact role of LSINCT5 in the process of GC and CRC development is not clear. We speculate that LSINCT5 may also regulate gastrointestinal cancer cell proliferation through different potential target genes. Nevertheless, as the target genes of lncRNA may differ between specific tissues and cell types and the specific target genes controlled by LSINCT5 in gastrointestinal cancer remain unknown, further studies are required to determine whether it regulates the genes identified by microarray analysis.
Gastric cancer is one of the most frequent malignancies in East Asian countries.19 Patients with gastric cancer are mostly diagnosed at advanced clinical stages and typically exhibit lymphatic tumor dissemination, with poor prognoses and a 5-year survival rate of <30%.20,21 Colorectal cancer is the third most common malignant tumor and is one of the most common cancers in China.22,23 Despite efforts in multiple fields, there has been little success in improving the DFS rate of gastrointestinal cancer patients. Identification of differential risk groups based on both clinical and molecular prognostic factors is a topic of growing interest. One promising strategy for identifying molecular markers involved in gastrointestinal cancer progression is to focus on lncRNA expression associated with patient survival. For example, CRC patients with higher PCAT-1 expression have a relatively poor overall survival rate,24 and a high expression level of HOTAIR is a predictor of poor overall survival in both GC25 and CRC9 patients. In this study, we determined that tumor LSINCT5 expression levels were associated with specific clinicopathological characteristics. For example, both GC and CRC patients with higher expression of LSINCT5 tended to have larger-sized tumors, advanced invasion depths, and advanced TNM stages. More importantly, we observed that high expression levels of LSINCT5 were significantly associated with poor DFS and DSS rates in both GC and CRC patients, and a multivariate analysis revealed that LSINCT5 is an independent prognostic indicator for the DFS and DSS rates of GC patients. Therefore, LSINCT5 may be a candidate biomarker for predicting tumor recurrence and clinical outcomes in GC patients.
Finally, it is worth noting that there were some unavoidable limitations in this study. First, the prognosis prediction reliability of LSINCT5 is limited by the small sample, which requires further confirmation in large-scale and multi-center studies. Second, the present study demonstrated that LSINCT5 is overexpressed in gastrointestinal cancer, but the mechanisms underlying this dysregulation deserve further study. Last but not least, although we confirmed that LSINCT5 promotes tumor cell growth in vitro, the precise mechanism of how LSINCT5 regulates the expression of its target genes in GC is not clear. In future work, high-throughput techniques should be applied to obtain a global view of the changes in lncRNA, miRNA and mRNA molecules and elucidate the mechanisms of LSINCT5 regulation.
In conclusion, we determined that LSINCT5 expression is significantly upregulated in gastrointestinal cancer tissues and cell lines and correlated with some clinical pathology. In addition, LSINCT5 expression may be potential for predicting the prognosis of GC patients. Moreover, we determined that LSINCT5 promotes gastric cancer cell growth in vitro. Taken together, these results suggest that LSINCT5 may represent a novel prognostic indicator and a target for gene therapy in GC.
ACKNOWLEDGMENTS
This study was supported by National Clinical Key Discipline (2013–2015), Priority of Shanghai key discipline of medicine (2013–2015), Shanghai R&D public service platform construction projects (12DZ2295100), National Natural Science Foundation of China (81472220, 81101806), Academic Award for Doctoral Candidates by Ministry of Education (2012), Mingdao Fund for Medical Graduate Student by Shanghai Medical College (2013), and Shanghai Science and Technology Development Fund (Basic Research Major Project, no. 10DJ1400500; Domestic Science and Technology Cooperation Project, no. 14495800300).
Footnotes
Abbreviations: CRC = colorectal cancer, DFS = disease-free survival, DSS = disease-specific survival, GAPDH = glyceraldehyde-3-phosphate dehydrogenase, GC = gastric cancer, lncRNAs = long non-coding RNAs, LSINCT5 = long stress-induced non-coding transcript 5, ncRAN = non-coding RNA expressed in aggressive neuroblastoma, RT-qPCR = reverse transcription quantitative PCR.
Authors’ Contributions: Conception and design: Peng Qi, Mi-Die Xu. Acquisition of data (acquired and managed patients, provided facilities, etc): Wei-Wei Weng, Xiao-Han Shen, Shu-Juan Ni, Lei Dong, Dan Huang, Cong Tan. Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis): Wei-Qi Sheng, Xiao-Yan Zhou. Writing, review, and/or revision of the manuscript: Peng Qi, Mi-Die Xu, Xiang Du.
Mi-Die Xu and Peng Qi contributed equally to this work.
The authors declare no conflict of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science 2005; 309:1519–1524. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–866. [DOI] [PubMed] [Google Scholar]
- 4.Kong YW, Ferland-McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol 2012; 13:e249–e258. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet 2010; 19:R162–R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011; 1:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez DS, Hoage TR, Pritchett JR, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet 2008; 17:642–655. [DOI] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71:6320–6326. [DOI] [PubMed] [Google Scholar]
- 10.Qi P, Xu MD, Ni SJ, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med 2013; 11:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi P, Xu MD, Ni SJ, et al. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol Carcinog 2014; Feb 12. doi: 10.1002/mc.22137. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 2013; 26:155–165. [DOI] [PubMed] [Google Scholar]
- 13.Silva JM, Perez DS, Pritchett JR, et al. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics 2010; 95:355–362. [DOI] [PubMed] [Google Scholar]
- 14.Silva JM, Boczek NJ, Berres MW, et al. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol 2011; 8:496–505. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res 2013; 41:4976–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krell J, Frampton AE, Mirnezami R, et al. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS ONE 2014; 9:e98561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldstein O, Nizri T, Doniger T, et al. The long non-coding RNA ERIC is regulated by E2F and modulates the cellular response to DNA damage. Mol Cancer 2013; 12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan G, Hu X, Liu Y, et al. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J 2013; 32:2833–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long N, Moore MA, Chen W, et al. Cancer epidemiology and control in north-East Asia—past, present and future. Asian Pac J Cancer Prev 2010; 11 Suppl 2:107–148. [PubMed] [Google Scholar]
- 20.Hundahl SA, Phillips JL, Menck HR. The National Cancer Data Base Report on poor survival of U.S. gastric carcinoma patients treated with gastrectomy: Fifth Edition American Joint Committee on Cancer staging, proximal disease, and the “different disease” hypothesis. Cancer 2000; 88:921–932. [PubMed] [Google Scholar]
- 21.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin 2001; 51:15–36. [DOI] [PubMed] [Google Scholar]
- 22.Zhao P, Dai M, Chen W, Li N. Cancer trends in China. Jpn J Clin Oncol 2010; 40:281–285. [DOI] [PubMed] [Google Scholar]
- 23.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012; 62:10–29. [DOI] [PubMed] [Google Scholar]
- 24.Ge X, Chen Y, Liao X, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol 2013; 30:588. [DOI] [PubMed] [Google Scholar]
- 25.Xu ZY, Yu QM, Du YA, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci 2013; 9:587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]


