Abstract
Recently, several thousand people have been killed by the Ebolavirus disease (EVD) in West Africa, yet no current antiviral medications and treatments are available. Systematic investigation of ebolavirus whole genomes during the 2014 outbreak may shed light on the underlying mechanisms of EVD development. Here, using the genome-wide screening in ebolavirus genome sequences, we predicted four putative viral microRNA precursors (pre-miRNAs) and seven putative mature microRNAs (miRNAs). Combing bioinformatics analysis and prediction of the potential ebolavirus miRNA target genes, we suggest that two ebolavirus coding possible miRNAs may be silence and down-regulate the target genes NFKBIE and RIPK1, which are the central mediator of the pathways related with host cell defense mechanism. Additionally, the ebolavirus exploits the miRNAs to inhibit the NF-kB and TNF factors to evade the host defense mechanisms that limit replication by killing infected cells, or to conversely trigger apoptosis as a mechanism to increase virus spreading. This is the first study to use the genome-wide scanning to predict microRNAs in the 2014 outbreak EVD and then to apply systematic bioinformatics to analyze their target genes. We revealed a potential mechanism of miRNAs in ebolavirus infection and possible therapeutic targets for Ebola viral infection treatment.
The genus Ebolavirus belongs to the family Filoviridae and order Mononegavirales, where the members of this genus are called ebolaviruses1. The Ebolavirus (EBOV, formerly designated “Zaire ebolavirus”) is one of the five known Ebolaviruses. EBOV cause Ebolavirus disease (EVD) in humans and other mammals. EVD is a type of hemorrhagic fever having a high case fatality rate2. From March 2014 to October 2014, the Ebola outbreak in West Africa has sickened 8,399 people, killing 4,033 of them3. This is the largest EBOV outbreak in the history4. Each month, several thousand people from affected areas travel to the North America, and even more travellers enter and leave Europe, other parts of Africa, and Asia5. Such air travel situations increase the possibility of EVD transmission. The United States and France have reported that several patients tested positive for Ebola3,4,5,6. Moreover, no current antiviral medications are available for EBOV7, and no effective EVD treatment exists at this time8.
MicroRNAs (miRNAs) are small regulatory non-coding RNAs, ranging from 19 to 24 nucleotides, that post-transcriptionally regulate target gene expression by inhibiting the translation of mRNA transcripts or cleaving them9,10,11,12. While encoded not only by cellular genomes but also by viral genomes13,14,15, miRNAs play a vital role in numerous cellular processes, including cell metabolism, viral infection, and antiviral immune response16,17. Viral miRNAs are mostly identified by traditional cloning from virus-infected cells18,19,20. Computational prediction and hybridization analysis are also applied to viral miRNA identification21,22. For known viral miRNAs, the majority of them are encoded by DNA viruses while only a few derived from RNA viruses23,24. To date, the RNA virus-encoded miRNAs have been identified in a few retroviruses including bovine leukemia vius (BLV)25,26, human immunodeficiency virus (HIV)25, West Nile virus (WNV)27, Dengue virus28 and hepatitis A virus (HAV)23. Recently, Liang et al.24 have identified two EBOV miRNAs, which means that EBOV can encode functional miRNAs.
Unlike the method used by Liang et al.24, based on bioinformatics whole genome-wide scanning and screening, we predicted the potential mature 2014 EBOV miRNAs, their target genes, and related signaling pathways. To the best of our knowledge, this is the first paper to systemic genome-wide analyze and predict the potential non-coding RNAs in the 2014 outbreak EBOVs along with their target genes. The results show several target genes regulated by the possible miRNAs may have important functions in human immune and antiviral response systems. Our research helps to further assess the roles of the 2014 outbreak EBOV miRNAs and their potential targets during viral infection and virus-host interactions, and thus to speed up the process of effective EVD treatment development.
Results
Alignment of EBOV whole genomes and prediction of mature EBOV miRNAs
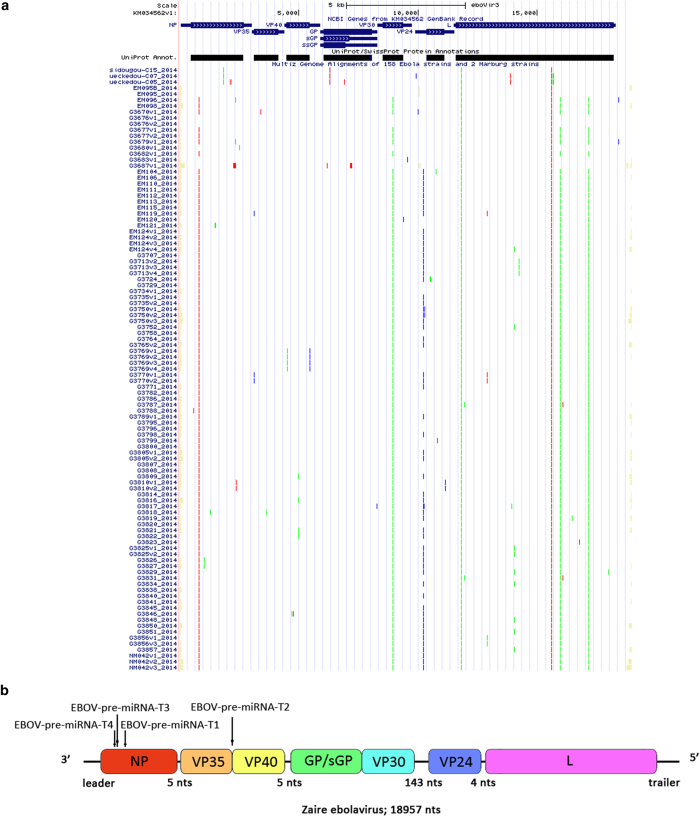
As shown in Fig. 1A, we first aligned the complete genome sequences of the 102 human EBOV strains acquired from the 2014 outbreak29. The mutation sites were all highlighted in different colors30. Four real microRNA precursor candidates, with the names of EBOV-pre-miRNA-T1, EBOV-pre-miRNA-T2, EBOV-pre-miRNA-T3 and EBOV-pre-miRNA-T4, were picked out. Their sequences are listed in Table 1. EBOV-pre-miRNA-T1 is 136-base long, EBOV-pre-miRNA-T2 is 80-base long, EBOV-pre-miRNA-T3 is 99-base long, and EBOV-pre-miRNA-T4 is 99-base long. Their locations were marked in Fig. 1B. The start position of EBOV-pre-miRNA-T1 is 653, while its end position is 788. EBOV-pre-miRNA-T2 is located between the position of 4373 and of 4452. EBOV-pre-miRNA-T3 is at the range between 488 and 586. EBOV-pre-miRNA-T4 is located from the position of 479 to 568. Seven different mature EBOV miRNA candidates were predicted using the four EBOV miRNA precursor candidates. The sequences of the eight mature EBOV miRNAs are listed in Table 2, where EBV-miR-T3-5p has the same sequence with EBV-miR-T4-5p. Their lengths are all 22-base long. By aligning and blasting the seven miRNAs, including EBV-miR-T1-5p, EBV-miR-T1-3p, EBV-miR-T2-5p, EBV-miR-T2-3p, EBV-miR-T3-5p, EBV-miR-T3-3p, EBV-miR-T4-5p and EBV-miR-T4-3p, to other species of viral or host miRNAs, only EBV-miR-T3-5p has some similar miRNAs. We analyzed the conservation between EBV-miR-T3-5p and its similar miRNAs, which is described in the Table 3. A flowchart describing the computational prediction of the putative miRNAs is shown in Fig. 2.
Figure 1.
The 102 complete genome sequences alignment of the 2014 outbreak EBOV. (A) Full alignment map of all 102 EBOV complete genomes. (B) Locations and putative target sites of the four real miRNA precursor candidates.
Table 1. The predicted pre-miRNAs in EBOV.
| Pre-miRNA | Strand | Start Position | End Position | Length | Sequence (5′-3′) |
|---|---|---|---|---|---|
| EBOV-pre-miRNA-T1 | Plus | 653 | 788 | 136 | UUGAUUUUCAAGAGAGUGCGGACAGUUUCCUUCUCAUGCUUUGUCUUCAUCAUGCGUACCAAGGAGAUUACAAACUUUUCUUGGAAAGUGGCGCAGUCAAGUAUUUGGAAGGGCACGGGUUCCGUUUUGAAGUCAA |
| EBOV-pre-miRNA-T2 | Plus | 4373 | 4452 | 80 | AACCAAAAAUGAUGAAGAUUAAGAAAAACCUACCUCGACUGAGAGAGUGUUUUUUCAUUAACCUUCAUCUUGUAAACGUU |
| EBOV-pre-miRNA-T3 | Plus | 488 | 586 | 99 | GGAUGACGCCGAGUCUCACUGAAUCUGACAUGGAUUACCACAAGAUCUUGACAGCAGGUCUGUCCGUUCAACAGGGGAUUGUUCGGCAAAGAGUCAUCC |
| EBOV-pre-miRNA-T4 | Plus | 479 | 568 | 99 | GGAUGACGCCGAGUCUCACUGAAUCUGACAUGGAUUACCACAAGAUCUUGACAGCAGGUCUGUCCGUUCAACAGGGGGUUGUUCGGCAAAGAGUCAUCC |
Table 2. The predicted mature miRNAs in EBOV. The EBV-miR-T3-5p and EBV-miR-T4-6p have the same sequences, which are highlighted in grey color.
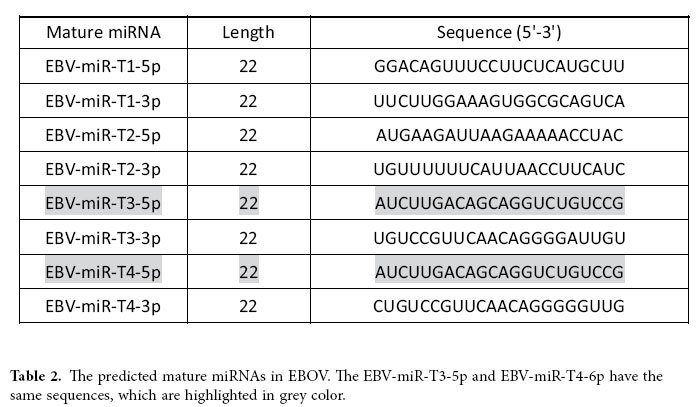
Table 3. Alignment of Query (EBV-miR-T3-5p) to mature miRNAs.
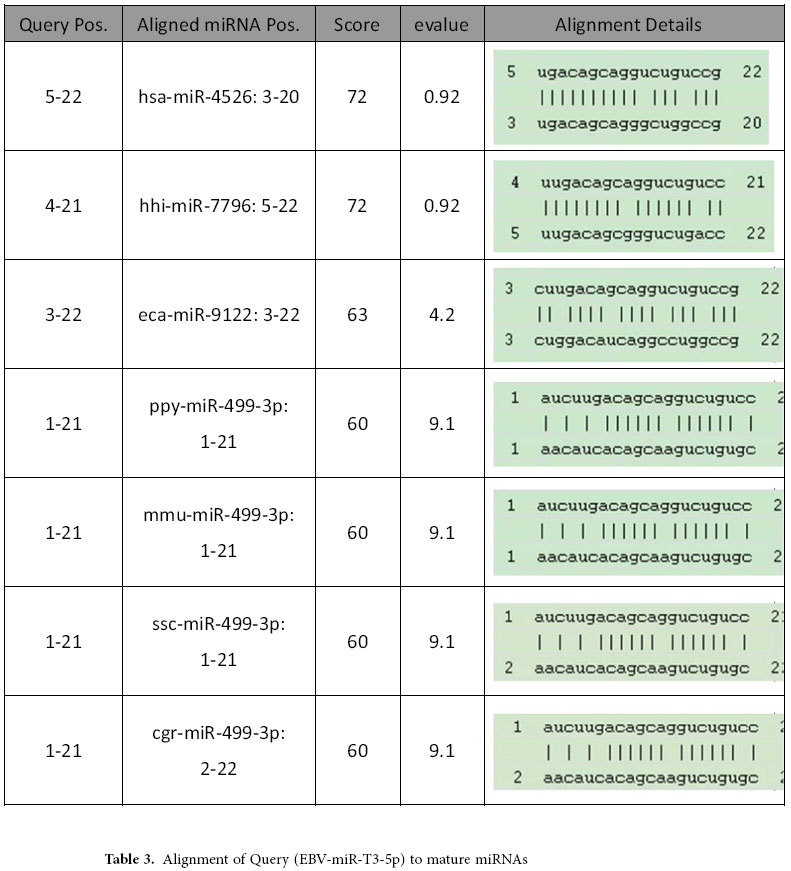
Figure 2.

Workflow of the 2014 outbreak EBOV-encoded miRNA prediction. The MiPred algorithm was used to identify genuine pre-miRNAs, and the MatureBayes tool was used to predict the mature miRNA sequences.
Structures of potential EBOV pre-miRNAs and mature miRNAs
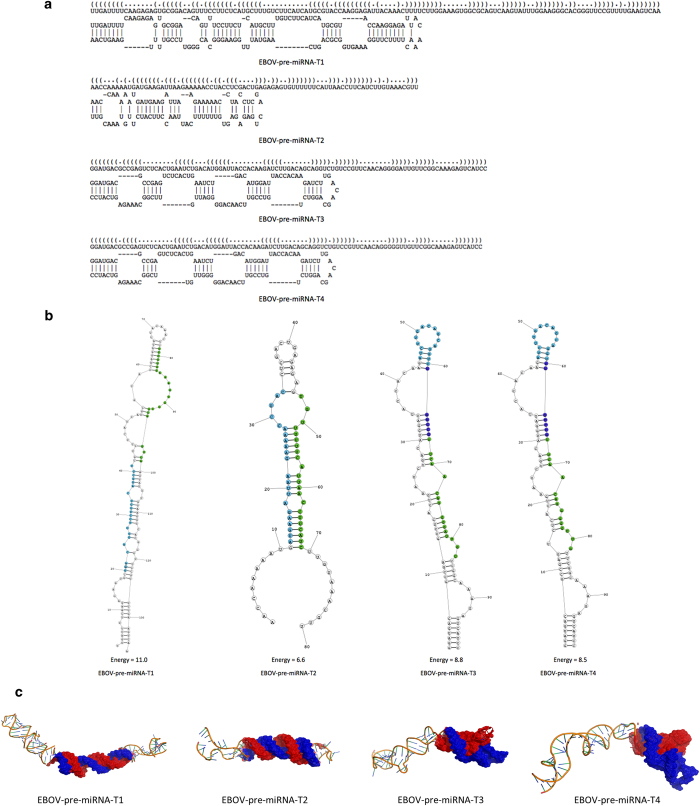
In order to achieve more accurate prediction, we predicted the primary, secondary, and tertiary structure of the four potential EBOV pre-miRNAs31,32. RNAfold was used to predict consensus secondary structures of the pre-miRNAs. Attempts to fold these pre-miRNAs, along with their flanking sequences, into the expected hairpin structures were successful in candidates EBOV-pre-miRNAs-T1, EBOV-pre-miRNAs-T2, EBOV-pre-miRNAs-T3 and EBOV-pre-miRNAs-T4, while other pre-miRNAs failed to fold into any form of stable RNA structures because that they could only give rise to stem-loops that were too short to be authentic pre-miRNA stem-loops. These findings led us to believe that EBOV-pre-miRNAs-T1, EBOV-pre-miRNAs-T2, EBOV-pre-miRNAs-T3 and EBOV-pre-miRNAs-T4 are indeed authentic EBOV pre-miRNAs. The optimal secondary structures in dot-bracket notation with a minimum free energy of −39.4 kcal/mol of EBOV-pre-miRNA-T1, −18.8 kcal/mol of EBOV-pre-miRNA-T2, −33.51 kcal/mol of EBOV-pre-miRNA-T3 and −31.91 kcal/mol of EBOV-pre-miRNA-T4 are given in Fig. 3A,B shows the predicted hairpin structures of confirmed EBOV pre-miRNAs. Hairpins longer than 100 nt were truncated; these hairpins are indicated by a double slash preceding the stem. Furthermore, the 3D structures of the potential EBOV pre-miRNAs were computationally predicted. Even though EBOV-pre-miRNA-T3 and EBOV-pre-miRNA-T4 have similar 2D structures31, they own different 3D structures (Fig. 3C). The positions of possible mature miRNAs were colored in second and tertiary structures (Figs. 3B,C)32. For EBOV-pre-miRNAs-T1, EBOV-pre-miRNAs-T2, EBOV-pre-miRNAs-T3 and EBOV-pre-miRNAs-T4 alternative predictions, which are likely to represent the authentic stem-loop structures recognized and cleaved by Drosha are shown underneath the minimal energy structures.
Figure 3.
Predicted hairpin structures of potential EBOV pre-miRNAs. (A) The primary structures of the four EBOV pre-miRNAs. (B) The secondary structures of the four EBOV pre-miRNAs. (C) The tertiary structures of the four EBOV pre-miRNAs.
Bioinformatics analysis of mature EBOV miRNAs and prediction of their target genes
Based on the sequences of the seven EBOV miRNAs, EBV-miR-T1-5p, EBV-miR-T1-3p, EBV-miR-T2-5p, EBV-miR-T2-3p, EBV-miR-T3-5p, EBV-miR-T3-3p, EBV-miR-T4-5p and EBV-miR-T4-3p, their target genes were searched by TargetScan33 and the other miRNA regulation database (the integration database of miRecords, TarBase, starbase, and miR2Disease)34,35,36,37. All these potential mature miRNAs have total 138 possible target genes in human genome (Table 4). Interestingly, the genes P11, EFNA3, FKRP, AMBRA1, HEPACAM, HCCA2, LPHN1 and PHF21B were all targeted by at least two potential mature EBOV miRNAs.
Table 4. The list of possible target genes.
| Gene Name | Gene ID | Human Symbol |
|---|---|---|
| ABCC13 | 150000 | ABCC13 (ATP-binding cassette, sub-family C (CFTR/MRP), member 13, pseudogene) |
| ADNP2 | 22850 | ADNP2 (ADNP homeobox 2) |
| ADRA2B | 151 | ADRA2B (adrenoceptor alpha 2B) |
| ADTRP | 84830 | ADTRP (androgen-dependent TFPI-regulating protein) |
| AHR | 196 | AHR (aryl hydrocarbon receptor) |
| ALDH3B1 | 221 | ALDH3B1 (aldehyde dehydrogenase 3 family, member B1) |
| AMBRA1 | 55626 | AMBRA1 (autophagy/beclin-1 regulator 1) |
| AMICA1 | 120425 | AMICA1 (adhesion molecule, interacts with CXADR antigen 1) |
| ARFIP1 | 27236 | ARFIP1 (ADP-ribosylation factor interacting protein 1) |
| ARHGAP27 | 201176 | ARHGAP27 (Rho GTPase activating protein 27) |
| ATXN10 | 25814 | ATXN10 (ataxin 10) |
| BEST4 | 266675 | BEST4 (bestrophin 4) |
| C2orf27A | 29798 | C2orf27A (chromosome 2 open reading frame 27A) |
| CA7 | 766 | CA7 (carbonic anhydrase VII) |
| CA9 | 768 | CA9 (carbonic anhydrase IX) |
| CABP7 | 164633 | CABP7 (calcium binding protein 7) |
| CBX4 | 8535 | CBX4 (chromobox homolog 4) |
| CCL21 | 6366 | CCL21 (chemokine (C-C motif) ligand 21) |
| CCND2 | 894 | CCND2 (cyclin D2) |
| CEP55 | 55165 | CEP55 (centrosomal protein 55kDa) |
| CERS3 | 204219 | CERS3 (ceramide synthase 3) |
| CNN2 | 1265 | CNN2 (calponin 2) |
| CPSF7 | 79869 | CPSF7 (cleavage and polyadenylation specific factor 7, 59kDa) |
| CRAMP1L | 57585 | CRAMP1L (Crm, cramped-like (Drosophila)) |
| CRYAA | 1409 | CRYAA (crystallin, alpha A) |
| CTDSPL2 | 51496 | CTDSPL2 (CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2) |
| CYB561 | 1534 | CYB561 (cytochrome b561) |
| DCLK2 | 166614 | DCLK2 (doublecortin-like kinase 2) |
| DEAF1 | 10522 | DEAF1 (DEAF1 transcription factor) |
| DMWD | 1762 | DMWD (dystrophia myotonica, WD repeat containing) |
| EFNA3 | 1944 | EFNA3 (ephrin-A3) |
| EGFLAM | 133584 | EGFLAM (EGF-like, fibronectin type III and laminin G domains) |
| EIF2AK4 | 440275 | EIF2AK4 (eukaryotic translation initiation factor 2 alpha kinase 4) |
| ELF3 | 1999 | ELF3 (E74-like factor 3 (ets domain transcription factor, epithelial-specific )) |
| ELFN2 | 114794 | ELFN2 (extracellular leucine-rich repeat and fibronectin type III domain containing 2) |
| ELOVL2 | 54898 | ELOVL2 (ELOVL fatty acid elongase 2) |
| ENDOU | 8909 | ENDOU (endonuclease, polyU-specific) |
| EVI5 | 7813 | EVI5 (ecotropic viral integration site 5) |
| EXOSC6 | 118460 | EXOSC6 (exosome component 6) |
| FAM20C | 56975 | FAM20C (family with sequence similarity 20, member C) |
| FAM213B | 127281 | FAM213B (family with sequence similarity 213, member B) |
| FASN | 2194 | FASN (fatty acid synthase) |
| FBXL7 | 23194 | FBXL7 (F-box and leucine-rich repeat protein 7) |
| FKRP | 79147 | FKRP (fukutin related protein) |
| FLJ46026 | 400627 | FLJ46026 (FLJ46026 protein) |
| FOXK1 | 221937 | FOXK1 (forkhead box K1) |
| G2E3 | 55632 | G2E3 (G2/M-phase specific E3 ubiquitin protein ligase) |
| GCOM1 | 145781 | GCOM1 (GRINL1A complex locus 1) |
| GMEB2 | 26205 | GMEB2 (glucocorticoid modulatory element binding protein 2) |
| GNAT1 | 2779 | GNAT1 (guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1) |
| GPR64 | 10149 | GPR64 (G protein-coupled receptor 64) |
| GRB10 | 2887 | GRB10 (growth factor receptor-bound protein 10) |
| GTPBP1 | 9567 | GTPBP1 (GTP binding protein 1) |
| HDAC5 | 10014 | HDAC5 (histone deacetylase 5) |
| HENMT1 | 113802 | HENMT1 (HEN1 methyltransferase homolog 1 (Arabidopsis)) |
| HEPACAM | 220296 | HEPACAM (hepatic and glial cell adhesion molecule) |
| HES3 | 390992 | HES3 (hes family bHLH transcription factor 3) |
| JARID2 | 3720 | JARID2 (jumonji, AT rich interactive domain 2) |
| JRK | 8629 | JRK (Jrk homolog (mouse)) |
| KIAA1244 | 57221 | KIAA1244 (KIAA1244) |
| KIAA1598 | 57698 | KIAA1598 (KIAA1598) |
| LINGO1 | 84894 | LINGO1 (leucine rich repeat and Ig domain containing 1) |
| LOC284395 | 284395 | LOC284395 (uncharacterized LOC284395) |
| LOC388022 | 388022 | LOC388022 (uncharacterized LOC388022) |
| LOC388942 | 388942 | LOC388942 (uncharacterized LOC388942) |
| LOC401134 | 401134 | LOC401134 (uncharacterized LOC401134) |
| LOC440600 | 440600 | LOC440600 (uncharacterized LOC440600) |
| LOC441455 | 441455 | LOC441455 (makorin ring finger protein 1 pseudogene) |
| LPGAT1 | 9926 | LPGAT1 (lysophosphatidylglycerol acyltransferase 1) |
| LPHN1 | 22859 | LPHN1 (latrophilin 1) |
| LRRC27 | 80313 | LRRC27 (leucine rich repeat containing 27) |
| MAPKAPK3 | 7867 | MAPKAPK3 (mitogen-activated protein kinase-activated protein kinase 3) |
| MBIP | 51562 | MBIP (MAP3K12 binding inhibitory protein 1) |
| MMP25 | 64386 | MMP25 (matrix metallopeptidase 25) |
| MOB2 | 81532 | MOB2 (MOB kinase activator 2) |
| MOB3A | 126308 | MOB3A (MOB kinase activator 3A) |
| MOB3B | 79817 | MOB3B (MOB kinase activator 3B) |
| MRE11A | 4361 | MRE11A (MRE11 meiotic recombination 11 homolog A (S. cerevisiae)) |
| MTMR11 | 10903 | MTMR11 (myotubularin related protein 11) |
| MTMR6 | 9107 | MTMR6 (myotubularin related protein 6) |
| MUM1 | 84939 | MUM1 (melanoma associated antigen (mutated) 1) |
| MVB12B | 89853 | MVB12B (multivesicular body subunit 12B) |
| NAP1L4 | 4676 | NAP1L4 (nucleosome assembly protein 1-like 4) |
| NFKBIE | 4794 | NFKBIE (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon) |
| NLGN4X | 57502 | NLGN4X (neuroligin 4, X-linked) |
| NPLOC4 | 55666 | NPLOC4 (nuclear protein localization 4 homolog (S. cerevisiae)) |
| NT5E | 4907 | NT5E (5′-nucleotidase, ecto (CD73)) |
| NUBP2 | 10101 | NUBP2 (nucleotide binding protein 2) |
| PAQR4 | 124222 | PAQR4 (progestin and adipoQ receptor family member IV) |
| PBXIP1 | 57326 | PBXIP1 (pre-B-cell leukemia homeobox interacting protein 1) |
| PDPK1 | 5170 | PDPK1 (3-phosphoinositide dependent protein kinase 1) |
| PHF20 | 51230 | PHF20 (PHD finger protein 20) |
| PHF21B | 112885 | PHF21B (PHD finger protein 21B) |
| POLR2M | 81488 | POLR2M (polymerase (RNA) II (DNA directed) polypeptide M) |
| PPP4C | 5531 | PPP4C (protein phosphatase 4, catalytic subunit) |
| PRKAB2 | 5565 | PRKAB2 (protein kinase, AMP-activated, beta 2 non-catalytic subunit) |
| PRKY | 5616 | PRKY (protein kinase, Y-linked, pseudogene) |
| PTPRT | 11122 | PTPRT (protein tyrosine phosphatase, receptor type, T) |
| RABEP1 | 9135 | RABEP1 (rabaptin, RAB GTPase binding effector protein 1) |
| RAPH1 | 65059 | RAPH1 (Ras association (RalGDS/AF-6) and pleckstrin homology domains 1) |
| RBM15B | 29890 | RBM15B (RNA binding motif protein 15B) |
| RBMS3 | 27303 | RBMS3 (RNA binding motif, single stranded interacting protein 3) |
| RECQL5 | 9400 | RECQL5 (RecQ protein-like 5) |
| RHBDL1 | 9028 | RHBDL1 (rhomboid, veinlet-like 1 (Drosophila)) |
| RIPK1 | 8737 | RIPK1 (receptor (TNFRSF)-interacting serine-threonine kinase 1) |
| RNF185 | 91445 | RNF185 (ring finger protein 185) |
| RNF31 | 55072 | RNF31 (ring finger protein 31) |
| S100A10 | 6281 | S100A10 (S100 calcium binding protein A10) |
| SAMD4B | 55095 | SAMD4B (sterile alpha motif domain containing 4B) |
| SCRT1 | 83482 | SCRT1 (scratch family zinc finger 1) |
| SELPLG | 6404 | SELPLG (selectin P ligand) |
| SETD8 | 387893 | SETD8 (SET domain containing (lysine methyltransferase) 8) |
| SGTA | 6449 | SGTA (small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha) |
| SLITRK1 | 114798 | SLITRK1 (SLIT and NTRK-like family, member 1) |
| SOCS4 | 122809 | SOCS4 (suppressor of cytokine signaling 4) |
| STC2 | 8614 | STC2 (stanniocalcin 2) |
| SYNGR1 | 9145 | SYNGR1 (synaptogyrin 1) |
| TBL2 | 26608 | TBL2 (transducin (beta)-like 2) |
| THOC3 | 84321 | THOC3 (THO complex 3) |
| TM9SF4 | 9777 | TM9SF4 (transmembrane 9 superfamily protein member 4) |
| TMEM63A | 9725 | TMEM63A (transmembrane protein 63A) |
| TRIM35 | 23087 | TRIM35 (tripartite motif containing 35) |
| UPP1 | 7378 | UPP1 (uridine phosphorylase 1) |
| USP45 | 85015 | USP45 (ubiquitin specific peptidase 45) |
| UTS2B | 257313 | UTS2B (urotensin 2B) |
| VAX2 | 25806 | VAX2 (ventral anterior homeobox 2) |
| VPS37B | 79720 | VPS37B (vacuolar protein sorting 37 homolog B (S. cerevisiae)) |
| WASF2 | 10163 | WASF2 (WAS protein family, member 2) |
| WFIKKN2 | 124857 | WFIKKN2 (WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2) |
| XDH | 7498 | XDH (xanthine dehydrogenase) |
| YY1AP1 | 55249 | YY1AP1 (YY1 associated protein 1) |
| ZCCHC24 | 219654 | ZCCHC24 (zinc finger, CCHC domain containing 24) |
| ZFP91 | 80829 | ZFP91 (ZFP91 zinc finger protein) |
| ZMIZ2 | 83637 | ZMIZ2 (zinc finger, MIZ-type containing 2) |
| ZNF444 | 55311 | ZNF444 (zinc finger protein 444) |
| ZNF672 | 79894 | ZNF672 (zinc finger protein 672) |
| ZNF767P | 79970 | ZNF767P (zinc finger family member 767, pseudogene) |
| ZNF804A | 91752 | ZNF804A (zinc finger protein 804A) |
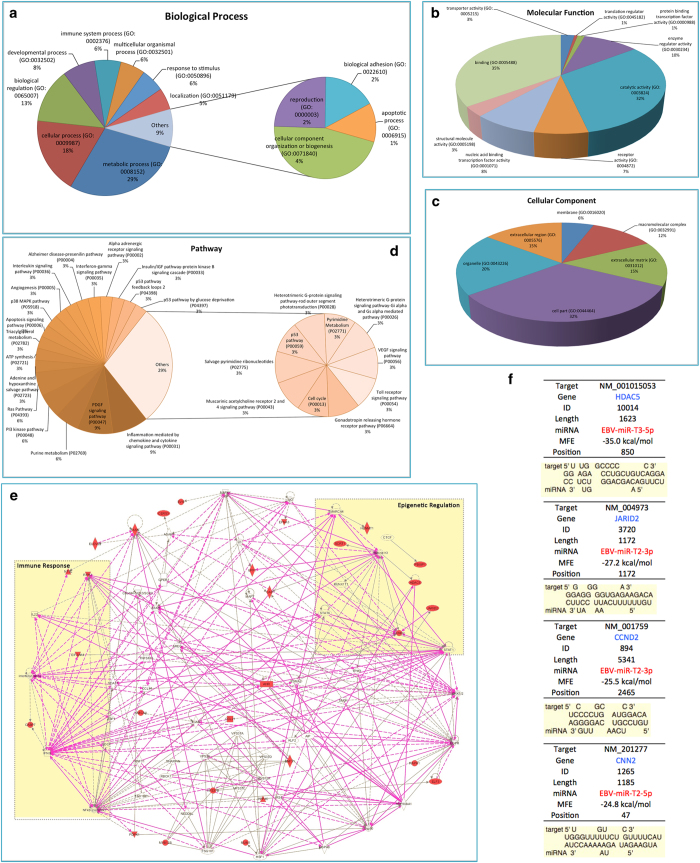
Using the GO and DAVID databases38,39, identified proteins were clustered into groups based on their biological processes, molecular function, and cellular compartment. Such clustering allowed us to determine how the differentially expressed proteins were distributed according to biological process (Fig. 4A), molecular function (Fig. 4B) and cellular compartment (Fig. 4C). Clusters with the biological process included proteins that are involved in immune system process (GO:0002376), multicellular organismal process (GO:0032501) and response to stimulus (GO:0050896), which are important for human antiviral response (Fig. 4A). This analysis grouped proteins into nine molecular functional classes (Fig. 4B), which were associated with transcription regulation, such as transporter activity (GO:0005215), translation regulator activity (GO:0045182), protein binding transcription factor activity (GO:0000988) and enzyme regulator activity (GO:0030234). Protein clusters were then further categorized into six subcellular distributions according to the cellular compartment (Fig. 4C). Fig. 4D shows the pathway enrichment analysis of the target genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using the DAVID bioinformatics tool40,41. The top enriched pathways are listed here (p-value < 0.01). Fig. 4D clearly shows that these proteins are functionally closely related, and are involved in multiple pathways. Inflammation are mediated by chemokine and cytokine signaling pathway (P00031), PDGF signaling pathway (P00047), Purine metabolism (P02769), PI3 kinase pathway (P00048) and Ras Pathway (P04393), which are important in human immune response to virus infection.
Figure 4.
Bioinformatic analysis of target gene prediction and related signaling pathways of the potential mature miRNAs in EBOV. The predicted target gene of potential mature EBOV miRNAs were classified by the GO and DAVID databases based on biological process (A), molecular function (B) and cellular compartment (C). (D) The pathway enrichment analysis of candidate genes. Top enriched pathways are listed (p value < 0.01). (E) The gene regulation network analysis of the potential target genes. (F) The detail information of target genes of miRNA EBV-miR-T3-5p, EBV-miR-T2-3p, EBV-miR-T2-3p and EBV-miR-T2-5p.
The gene regulation network (GRN) of the target genes42,43 is shown in the Fig. 4E. We find that the possible target genes, HDAC5 and JARID244, are important epigenetic factors in transcription regulation, which have strong interaction with the transcription regulator in human cells, such as Histone h3, STAT1, CTCF, CCND2 and CTCF. It is illustrated that the EBOV down-regulated the epigenetic factor to large-scale epigenetic alterations in the host gene expressions45, which will damage the normal molecular regulation in immune signaling and thus disorder human immune system to block the anti-viral response. The target proteins, NFKBIE and RIPK, are the key co-regulator with IL22 and IFNG or the part of complex of NFκB in human immune system46,47. It also suggests that the EBOV affects the signaling pathway of immune system by the non-coding RNA to inhibit the infection response. Further network analysis was conducted to see the potential biological pathways and processes in which these differentially expressed proteins were involved. In addition, Fig. 4F illustrates the detail information of target genes which play key roles in important immune and epigenetic responses.
Signaling pathway analysis of the target genes
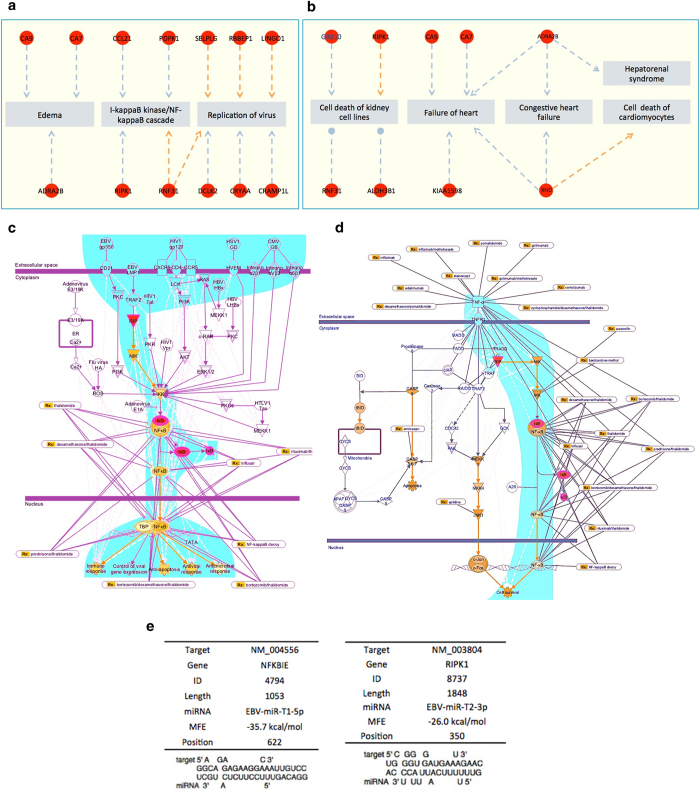
According to our gene regulation network analysis, NFKBIE and RIPK1 are two target genes of possible mature miRNAs, EBOV-miR-T1-5p and EBOV-miR-T2-3p, which plays important roles in immune system. Fig. 5A,B show the cytotoxin-associated target genes. NFKBIE48 and RIPK49 genes are both involved in the NF-κB and TNF signaling pathways (Fig. 5C,D). NFKBIE encodes IκB epsilon (IκBε), a member of the IκB family. Its binding to NF-κB inhibits the nuclear translocation of NF-κB50. RIPK1, Receptor-interacting protein 1, is a key effector molecule in the TNFα-induced activation of the transcription factor NF-κB51. The signaling pathway analysis shows that the TNF-a–mediated NF-kB signaling pathway plays a key role in inflammatory response (Fig. 5C,D). Through analyzing the molecular mechanism in the NF-kB and TNF signaling pathways, those two pathways were discovered to participate in the host immune response, thus they are also attractive targets of viral pathogens52. The EBOV coding possible miRNAs, EBOV-miR-T1-5p and EBOV-miR-T2-3p, may be silence and down-regulate the NFKBIE and RIPK1, which are the central mediator of those pathways. In addition, the EBOV exploits the non-coding RNA to inhibit the NF-kB and TNF factor to evade the host defense mechanisms that limit replication by killing infected cells, or to conversely trigger apoptosis as a mechanism to increase virus spreading. NF-kB is activated by multiple families of viruses, including HIV-1, HTLV-1, hepatitis B virus (HBV), hepatitis C virus (HCV), EBV, and influenza virus53. This activation may serve several functions of promoting viral replication, preventing virus-induced apoptosis, and suprressing the immune response to the invading pathogen54. The genes NF-kB and TNF are affected by synthetic dsRNAs. It suggests that viruses that generate dsRNA replicative intermediates employ a common mechanism to enhance viral replication. The EBOV glycoprotein (GP) and the other viral proteins also induce the NF-kB and TNF signaling pathways55. The miRNA and target gene details of EBV-miR-T1-5p and EBV-miR-T2-3p are listed in Fig. 5E. The target gene of EBV-miR-T1-5p is named NFKBIE with ID No. 4794, and 1053-bp long. The target gene of EBV-miR-T2-3p is named RIPK1 with ID No. 8737, and 1848-bp long. Normally when a virus enters a human body, the target genes mentioned above should increase their expression levels. Based on our analysis, once EBOV enters the human body, the miRNAs EBV-miR-T1-5p and EBV-miR-T2-3p down-regulate the target genes of NFKBIE and RIPK1 to evade host innate immune responses and finally leads to multiple organ dysfunction syndrome (MODS).
Figure 5.
Signaling pathway analysis of the potential mature EBOV miRNA target genes. (A) The associated genes of Edema, □-kappaB kinase/ NF-kappaB cascade and replication of virus. (B) The associated genes of cell death of kidney cell lines, failure of heart, congestive heart failure, hepatorenal syndrome and cell death of cardiomyocytes. (B) TNF signaling pathway. (C) NF-κB signaling pathway. (D) Detail information of target gene NFKBIE. (E) Detail information of target gene RIPK1.
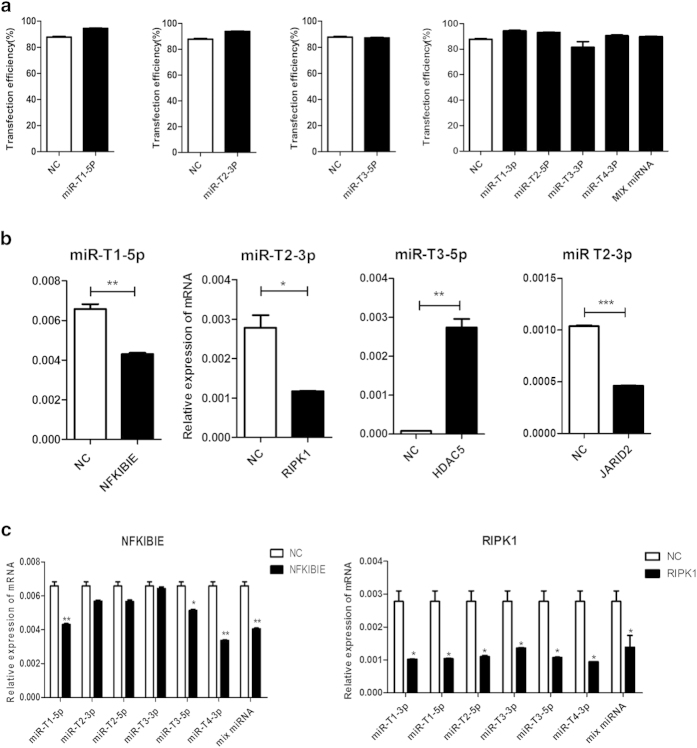
EBOV miRNA regulated the expression of target genes NFKIBIE, RIPIK1, HDAC5 and JARID2
The miRNA mimics was synthesized and transfected into Hela cells for 24 h/37 °C. The results showed that EBOV-miRNA mimics directly regulate the expression of target genes, NFKIBIE, RIPIK1, JARID2 and HDAC5, in Hela cells. Real time (RT)-PCR was used to quantify and compare the mRNA expression of target genes in the transfected Hela cells with the EBOV-miRNA mimics and the scramble mimics as negative control. The transfection efficiency of miRNA mimics, including miR-T1-3p, miR-T2-5p, miR-T3-3p, miR-T4-3p and their mixed miRNA (MIX miRNA), and the scrambled miRNA mimics were detected by fluorescence activated cell sorter (FACS), which were more than 80% (Fig. 6A). The RT-PCR analysis indicated that expression of NFKIBIE was significantly down-regulated by 1.526 times in miR-T1-5p mimics-transfected Hela cells and the expression of RIPIK1 and JARID2 were down-regulated by miR-T2-3p mimics with 1.947 times and 2.25 times separately (Fig. 6B). Interestingly, in the transfected Hela cells with miR-T3-5p mimics, the expression of HDAC5 is increasing compared with the transfected Hela cells with scrambled miRNA mimics (Fig. 6B). We also found that the mRNA expression of NFKIBIE were inhibited by miR-T1-3p, miR-T3-5p, miR-T4-3p and MIX miRNA mimics, while all EBOV-miRNA mimics significantly can block the expression of RIPK1 (Fig. 6C). Such results illustrated that these miRNA may have important functions in human immune and antiviral response systems.
Figure 6.
Transfection efficiency of EBOV-miRNA mimics and effect of the EBOV-miRNA mimics on the expression of target genes in Hela cells. (A) Transfection efficiency 24 h post-transfection in Hela cells as percentage of fluorescent dye labeled miRNA positive cells. (B) Relative expression levels of NFKIBIE, RIPK1, HDAC5 and JARID2, which were regulated by EBOV-miRNA mimics miR-T1-5p, miR-T2-3p and miR-T3-5p, respectively. (C) Relative expression levels of NFKIBIE and RIPK1, which were regulated by six EBOV-miRNA mimics and their mixed miRNA (MIX miRNA). *: P < 0.05, **: P < 0.01, ***: P < 0.001. Label “NC” means negative control.
Conclusion
From March 2014 to October 2014, the largest EBOV outbreak in the history has killed several thousand people in West Africa, yet no current specific treatment is validated for EBOV. miRNAs has been demonstrated to circulate in a highly stable, cell-free form in body fluids thus circulating miRNAs can be used as non-invasive biomarkers for molecular diagnostics and prognostics24. Since the majority of known viral miRNAs are encoded by DNA viruses, RNA-virus-encoded miRNAs remains controversial. Recently, despite that several novel miRNAs have been experimentally identified in the West Nile virus (WNV)28, Dengue virus (DENV)56, hepatitis A virus (HAV)23, the viral miRNAs in numerous viruses have been identified using computational prediction followed by experimental validation23,57,58,59. This year H. Liang et al.24 have successfully identified two EBOV miRNAs, which provide new evidence that EBOV can encode functional miRNAs.
In this paper, based on the whole genome alignment of the 2014 outbreak EBOVs29, which is different from the analysis method of Liang et al.24, we predicted and experimentally verified seven mature EBOV miRNAs and their regulated target genes. By analyzing the signaling pathways of the target genes, we hypothesized that EBOV miRNAs, especially EBV-miR-T1-5p and EBV-miR-T2-3p, down-regulate the target genes of NF-KB and TNF expression which are involved in virus-cell interaction, immune escape, and cell apoptosis48,49. As viruses evolve under the highly selective pressures of the immune system, they acquire the capacity to target critical steps in the host cell life, hijacking vital cellular functions to promote viral pathogenesis. Many viruses have evolved mechanisms to target the NF-kB pathway to facilitate their replication, cell survival, and evasion of immune responses. In addition, some viruses use the NF-kB pathway either for its anti-apoptotic properties to evade the host defense mechanisms or to trigger apoptosis as a mechanism of virus spread. Not surprisingly, recent studies focusing on the interrelations between NF-kB and virus pathogenesis reveal that many viral products bypass signal-induced stimulation and/or receptor-proximal steps to directly interface with the IKK complex. Continuous activation of NF-kB as a consequence of viral persistence in some instances leads to oncogenic transformation, which is an area of increasing scientific study and fascination.
It is noteworthy that this is the first paper to describe a strategy that the EBOV non-coding RNA may be evolved to modulate the NF-kB and TNF signaling pathways, to enhance viral replication, host cell survival, and evasion of immune responses. According to our prediction results, the EBOV-miRNA mimics of miR-T1-5p can significantly inhibit the expression of NFKIBIE, while the miR-T2-3p mimics can block the expression of RIPK1 significantly.
Such results illustrated that these miRNA may have important functions in human immune and antiviral response systems. Ongoing research will undoubtedly continue to reveal other novel interactions between viruses and the NF-kB pathway that will permit a more precise molecular dissection of this parasitic fatal attraction. Considering that the target genes regulated by our predicted miRNAs play key roles in human immune response, we highlight our prediction work raises the hope of quickly finding an effective EBOV disease treatment.
Materials and Methods
EBOV whole genome sequencing
To show the genome-wide nucleotide and amino acid signatures, we retrieved full genome sequences (as of October 22, 2014) from the genome browser at NCBI Database. One-hundread-and-two human EBOV genomes were derived from the 2014 outbreak29,30.
Multiple genome-wide alignments
The multiple sequence alignment tools ClustalW and MUSCLE were applied for the alignment of the EBOV genomes. The alignments were then analyzed to identify the characteristic sites as potential signatures to distinguish different virus genomes and proteins30.
Bioinformatics prediction of the miRNAs
A flowchart describing the computational prediction of the putative miRNAs is shown in Fig. 2. Briefly, the viral genome was scanned for stem-loop structures of miRNA precursor (pre-miRNA) using VMir60 (http://www.hpi-hamburg.de/research/ departments-and-research-groups/antiviral-defense-mechanism/software-down- load.html), a computational analyzer program for the prediction of putative pre-miRNAs. The complete genome sequences of 138 different human EBOV strains were acquired from the PubMed, where 102 human EBOV strains belong to 2014 EBOV outbreak. VMir predictions were carried out using the default parameters. The putative pre-miRNAs that satisfied the filter parameters of a VMir score ≥150 and a window count ≥35 were selected for further assessment. Then, the source code of MiPred61 was used to distinguish real and pseudo miRNA precursors from the obtained sequences with a prediction confidence equal to or greater than 70%. Subsequently, mature miRNA sequences were predicted from the pre-miRNA stem-loops. Then we used the MatureBayes tool62 to extend the prediction coverage of the mature miRNAs. The default conditions were used for the MatureBayes tool.
Prediction of Novel Viral miRNA Targets
Human target genes of novel EBOV miRNAs were predicted using TargetScan33 custom miRNA prediction methods. Putative targets within the viral genome were predicted using TargetScan Perl script34,36,37,60.
Gene ontology (GO) analysis
GO analysis of the significant probe list was performed using PANTHER (http://www.pantherdb.org/)38,39, using text files containing the Gene ID list and accession numbers of the Illumina probe ID. All data analysis and visualization of differentially expressed genes were conducted using R 2.4.1 (www.r-project.org). In addition, the DAVID Functional Annotation Bioinformatics Microarray Analysis tools (http://david.abcc.ncifcrf.gov/) were used to study the biological function of the regulated genes.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG)40,41 is a collection of online databases dealing with genomes, enzymatic pathways, and biological chemicals. The PATHWAY database records networks of molecular interactions in the cell that includes organism-specific network maps (http://www.genome.jp/kegg/). A total of 9 pathways, involving 49 genes, were collected from KEGG.
Constructing Gene Regulation Network
The integrated regulatory network was constructed based on systematic integration of various high-throughput datasets42,43. The target genes associated with candidate miRNAs were selected from miRNA regulation database (the integration database of miRecords, TarBase, starbase, and miR2Disease) and TFs associated with those target genes were selected from Transcriptional Regulatory Element Database (TRED). The integrated target genes-TF regulatory network was constructed by using Cytoscape software (http://cytoscape.org/), which is an open source software for visualizing complex networks and integrating these networks with any type of attribute data.
Cell culture and transfection
Hela cells at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone, USA) . Synthetic duplex EBV-miRNA mimics, scramble oligonucleotides used as negative control (NC) (GenePharma, Shanghai, China) at a final concentration of 50 nM were introduced into Hela cells by siPORT™ NeoFX™ transfection agent (AM4511, Applied Biosystems Inc., USA). according to the manufacturer’s instructions. Cells were harvested at 24 hours after transfection.
RNA extraction and quantitative real-time PCR analysis
The harvested cells were placed in TRIzolH Reagent (Invitrogen, Carlsbad,CA), and total RNAs were extracted according to the TRIzol manufacturer’sinstructions. Total RNA (3 μg) was primed by oilgodT and converted intocDNA using SuperScript III (Invitrogen). The SYBR green-based real-time PCR was performed in Light Cycle 2.0 System(Roche) and Relative quantification of mRNA was measured using 2-ΔΔCt method which normalized to GAPDH. Each PCR assay was performed in a final volume of 20 ul, containing 2 ul of DNA template, 25 pmol of each type-specific primer set, 2X Maxima SYBR Green (Thermo Scientific), Reactions were incubated at 50 °C for2 minutes, followed by PCR amplification under the following conditions: 95 °C for10 minutes, 95 °C for 30 seconds, 57 °C for 30 seconds, 72 °C for 30 minute for a total of 40 cycles, Following the PCR step, a melt curve analysis was performed by increasing the temperature from 65 °C to 95 °C at increments of 0.1 °C/s for each fluorescence reading. The primer sequences for qPCR analysis were as follows: GAPDH, forward 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse 5′- AGGGGCCATCCACAGTCTTC-5′; NFKBIE, forward 5′- TGCCAACAGATGGCCCATAC-3′ and reverse 5′- TGTTCTTTTCACTAGAGGCACCA-3′; RIPK1, forward 5′-TGGGCGTCATCATGAGGAAG-3′ and reverse 5′-CGCCTTTTCCATGTAAGTAGCA-3′ ; MAPKAP3, forward 5′- ATGAGAACATGCACCATGGCAAGC-3′ and reverse 5′- GGGCAATGTTATGGCTGTGCAGAA -3′; HDAC5, forward 5′- TTCTTTGGACCAGAGTTCCC-3′ and reverse 5′- GTTGGGTTCAGAGGCTGTTT-3′; JARID2, forward 5′- GAGCATGTGTTTCAGCAAGG-3′ and reverse 5′- CTTCTCTTCCACTAGCCTCCAG-3′
Flow cytometry
Green fluorescence after fluorescent dye labeled miRNA delivery into Hela cells were assessed in an Accuri C6 BD Biosciences flow cytometer, excited by an argon 480 nm laser and detected by use of a 520 nm optical filter. The mean fluorescence intensity (MFI) data were collected from these cell populations. All data was analyzed with a minimum of 15000 events setting the respective negative control to less than 1% in BD CFlow software. Post-transfection in Hela cells as percentage of fluorescent dye labeled miRNA positive cells.
Additional Information
How to cite this article: Teng, Y. et al. Systematic Genome-wide Screening and Prediction of microRNAs in EBOV During the 2014 Ebolavirus Outbreak. Sci. Rep. 5, 9912; doi: 10.1038/srep09912 (2015).
Acknowledgments
This work was supported by a grant from the National Hi-Tech Research and Development (863) Program of China (No. 2012AA022003 and No.2014AA021402), the China Mega-Project on Infectious Disease Prevention (No. 2013ZX10004605, No. 2011ZX10004001, No. 2013ZX10004607-004 and No. 2013ZX10004217-002-003) and the State Key Laboratory of Pathogen and BioSecurity Program (No. SKLPBS1113).
Footnotes
Author Contributions Y.T., Y.Z.W. and B.H.L. characterized the materials and designed the experiments, under the supervision of Y.G.T. and W.C.C. Y.Z.W., W.L.L. and P.Z. implemented the experiments under the supervision of W.J.Y. X.L.L.Z. wrote the manuscript with further contributions from Y.T. and Y.G.T. Y.T., H.F. and H.W.Y. analyzed the data. All authors reviewed the manuscript.
References
- Kuhn J. H. et al. Proposal for a revised taxonomy of the family Filoviridae: Classification, names of taxa and viruses, and virus abbreviations. Archies of Virology , 155, 2083–2103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T. G. Filoviruses: Marburg and Ebola. Viral Infections of Humans . 14, 337–350 (2014). [Google Scholar]
- Lupkin S., Timeline of the Ebolavirus in America. WABC-TV New York. (2014). Available at: http://7online.com/news/timeline-of-the-ebola-virus-in-america-/348789/. (Accessed 18th October 2014)
- World Heath Organization. Ebola response roadmap. (28th Angust 2014)
- Frieden T. R., Damon I., Bell B. P., Kenyon T. & Nichol S. Ebola 2014--New Challenges, New Global Response and Responsibility. N. Eng. J. Med. 371, 1117–1180 (2014). [DOI] [PubMed] [Google Scholar]
- Dixon M. G. & Schafer I. J. Ebola Viral Disease outbreak--West Africa, 2014 Morbidity and Mortality Weekly Report (MMWR). 63, 548–551 (2014). [PMC free article] [PubMed] [Google Scholar]
- World health Organization. EbolaVirus Disease. (September 2014) Available at: http://www.who.int/mediacentre/factsheets/fs103/en/ (Accessed: 12th Febrary 2015).
- Turner C. EbolaVirus Disease: An emerging Threat. Nursing . 44, 68–69 (2014). [DOI] [PubMed] [Google Scholar]
- Kim V. N., Han J. & Siomi M. C. Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cell Biol. 10, 128–139 (2009). [DOI] [PubMed] [Google Scholar]
- Carthew R. W. & Sontheimer E. J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 136, 642–655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P. & Voinnet O. Revisiting the Principles of microRNA Target Recognition and Mode of Action. Nat. Rev. Mol. Cell Biol. 10, 141–148 (2009). [DOI] [PubMed] [Google Scholar]
- Almeida M. I., Reis R. M. & Calin G. A. MicroRNAhistory: discovery, recentapplications, and nextfrontiers. Mutat Res. 717, 1–8 (2011). [DOI] [PubMed] [Google Scholar]
- Seo G. J., Chen C. J. & Sullivan C. S. Merkel Cell Polyomavirus Encodes a microRNA with the Ability to Autoregulate Viral Gene Expression. Virology . 383, 183–187 (2009). [DOI] [PubMed] [Google Scholar]
- Grundhoff A. & Sullivan C. S. Virus-encoded microRNAs. Virology . 411, 325–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. Viruses and microRNAs. Nat. Genet. 38, S25–S30 (2006). [DOI] [PubMed] [Google Scholar]
- Kincaid R. P. & Sullivan R. P. Virus-encoded microRNAls: an overview and a look to the future. Plos Pathogy . 8, e10033018; Doi: 10.1371/journal.ppat.1003018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz N., Christalla T., Tessmer U. & Grundhoff A. A Global Analysis of Evolutionary Conservation Among Known and Predicted Gammaherpesvirus microRNAs. J. Virol. 84, 716–728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. et al. Identification of Virus-encoded microRNAs. Science. 304, 734–736 (2004). [DOI] [PubMed] [Google Scholar]
- Pfeffer S. et al. Identification of microRNAs of the Herpesvirus Family. Nat. Methods. 2, 269–276 (2005). [DOI] [PubMed] [Google Scholar]
- Samols M. A., Hu J., Skalsky R. L. & Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 79, 9301–9305 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C. Prediction and Identification of Herpes Simplex Virus 1- Encoded microRNAs. J. Virol. 80, 5499–5508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C. S., Grundhoff A. T., Tevethia S., Pipas J. M. & Ganem D. SV40-encoded microRNAs Regulate Viral Gene Expression and Reduce Susceptibility to Cytotoxic T Cell. Nature. 435, 682–686 (2005). [DOI] [PubMed] [Google Scholar]
- Shi J. Identification and Validation of a Novel microRNA-like Molecule Derived from a Cytoplasmic RNA Virus Antigenome by Bioinformatics and Experimental Approaches. Virol. J. 11, 121:1–121:14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H. Identification of EbolaVirus microRNAs and Their Putative Pathological Function. Sci. China Life Sci. 57, 973–81 (2014). [DOI] [PubMed] [Google Scholar]
- Kincaid R. P., Burke J. M. & Sullivan C. S. RNA Virus microRNA that Mimics a B-Cell oncomiR. Proc. Natl. Acad. Sci. USA, . 109, 3077–3082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewick N. Deep Sequencing Reveals Abundant NonCanonical Retroviral microRNAs in B-cell Leukemia/Lymphoma. Proc. Natl. Acad. Sci. USA . 110, 2306–2311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul D., Ahlawat A. & Gupta S. D. HIV-1 Genome-Encoded hiv1-mir-H1 Impairs Cellular Responses to Infection. Mol. Cell Biochem. 323, 143–148 (2009). [DOI] [PubMed] [Google Scholar]
- Hussain M. West Nile Virus Encodes a microRNA-like Small RNA in the 3′ Untranslated Region which up-Regulates GATA4 mRNA and Facilitates Virus Replication in Mosquito Cells. Nucleic. Acids Res. 40, 2210–2223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebolavirus Resource. NCBI. Available at: http://www.ncbi.nlm.nih.gov/genome/viruses/variation/ebola/. (Accessed 24th October 2014).
- UCSC Genome Browser on Ebolavirus Sierra Leone 2014 (G3683/KM034562.1 /eboVir3) Assembly. UCSC Ebola genome browser. Available at: http://genome.ucsc.edu/cgi-bin/hgTracks?db=eboVir3. (Accessed 24 October 2014).
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M. & Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature . 452, 51–55 (2008). [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed paring, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell . 120, 15–20 (2005). [DOI] [PubMed] [Google Scholar]
- Li S. C., Pan C. Y. & Lin W. C. Bioinformatic discovery of microRNA precursors from human ESTs and introns. BMC Genomics . 7, 164:1–164:11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C., Shiau C. K. & Lin W. C. Vir-Mir db: prediction of viral microRNA candidate hairpins. Nucleic Acids Res. 36, 184–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37, 105–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Corda B. & Hatziqeorqiou A. G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 12, 192–197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang d. W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Huang d. W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A. & Thomas P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, 377–386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casaqrande J. T. & Thomas P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, 808–815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarsaihan D. Epigenetic mechanisms in inflammation. J. Dent. Res. 90, 9–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. Ebolavirus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 16, 187–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M. S., West A. P. & Ghosh S. NF-kappaB and the immune response. Oncogene . 25, 6758–6780 (2006). [DOI] [PubMed] [Google Scholar]
- Dannappel M. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature . 513, 90–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouzen K. Functional variants in NFKBIE and RTKN2 invovled in activation of the NF-kB Pathway are associated with rheumatoid arthritis in Japanese. Plos Genetics . 8, p. e1002949; Doi: 10.1371/journal.pgen.1002949 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature . 513, 95–99 (2014). [DOI] [PubMed] [Google Scholar]
- Bouwmeester T. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 6, 97–105 (2004). [DOI] [PubMed] [Google Scholar]
- Lee R. E., Walker S. R., Savery K., Frank D. A. & Gaudet S. Fold change of nuclear NF-kB determines TNF-induced transcription in single cells. Mol. Cell . 53, 867–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoya Y. Positive feedback between NF-kB and TNF-alpha promotes leukemia-initiating cell capacity. J. Clin. Invest. 124, 528–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. M. & McFadden G. Modulation of NF-kB signalling by microbial pathogens. Nat. Rev. Microbiol. 9, 291–306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J., Kwon H. & Genin P. Hostile takeovers: viral appropriation of the NF-kB pathway. J. Clin. Invest. 107, 143–151 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R. E. & Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 (2008). [DOI] [PubMed] [Google Scholar]
- Hussain M. & Asgari S. MicroRNA-like viral small RNA from dengue virus 2 antoregulates its replication in mosquito cells. Proc. Natl. Acad. Sci. USA . 111, 2746–2751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Singh C. P., Bhavani A. & Nagaraju J. Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology . 407, 120–128 (2010). [DOI] [PubMed] [Google Scholar]
- Hussain M., Taft R. J. & Asgari S. An insect virus-encoded microRNA regulates viral replication. J. Virol. 82, 9164–9170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besecker M. I., Harden M. E., Li G., Wang X. & Griffiths A. Discovery of herpes B virus-encoded microRNAs. J. Virol. 82, 3413–3416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C., Shiau C. K. & Lin W. C. Vir-Mir db: prediction of viral microrna candidate hairpins. Nucleic Acids Res. 36, 184–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P. MiPred: classfication of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 35, 339–344 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkirtzou K., Tsamardinos I., Tsakalides P. & Poirazi P. MatureBayes: a probabilistic algorithm for identifying the mature miRNA within novel precursors. Plos One . 5, e11843; Doi: 10.1371/journal.pone.0011843. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]