Abstract
We have developed a compartmentalised culture model for the purification of axonal mRNA from embryonic, neonatal and adult rat dorsal root ganglia. This mRNA was used un-amplified for RT-qPCR. We assayed for the presence of axonal mRNAs encoding molecules known to be involved in axon growth and guidance. mRNAs for β-actin, β-tubulin, and several molecules involved in the control of actin dynamics and signalling during axon growth were found, but mRNAs for microtubule-associated proteins, integrins and cell surface adhesion molecules were absent. Quantification of β-actin mRNA by means of qPCR showed that the transcript is present at the same level in embryonic, newborn and adult axons. Using the photoconvertible reporter Kaede we showed that there is local translation of β-actin in axons, the rate being increased by axotomy. Knock down of β-actin mRNA by RNAi inhibited the regeneration of new axon growth cones after in vitro axotomy, indicating that local translation of actin-related molecules is important for successful axon regeneration.
Keywords: Axon growth, Axon regeneration, Axonal transport, Axon guidance, Growth cone, mRNA, Translation, Actin, Integrins, Tubulin
Introduction
Over the last two decades it has become clear that dendrites and also certain types of axons contain mRNA (Piper and Holt, 2004; Twiss and van Minnen, 2006, Giuditta et al., 2002; Kiebler and Bassell, 2006; Vogelaar and Fawcett, 2008a,b). This was first shown in invertebrate axons from squid and snails which contain numerous mRNAs, mainly encoding cytoskeletal and metabolic proteins, and proteins involved in local translation, as well as neuropeptides (VanMinnen et al., 1997; Gioio et al., 2001; Giuditta et al., 2002; Gioio et al., 2004; van Kesteren et al., 2006). There have been various studies on vertebrate neurites from developing neurons, axons from adult dorsal root ganglia (DRGs) that received a conditioning lesion, and also from cytoplasm squeezed out from adult nerves (axoplasm). These have identified localised cytoskeletal mRNAs, such as β-actin, β-tubulin, neurofilament, vimentin, mRNAs encoding small GTPases and CREB (Bassell et al., 1998; Eng et al., 1999; Lee and Hollenbeck, 2003; Sotelo-Silveira et al., 2006; Yao et al., 2006; Leung et al., 2006, Giuditta et al., 2002; Perlson et al., 2005; Wu et al., 2005; Cox et al., 2008). A study of pre-conditioned adult DRG neurons identified 27 mRNAs in these axons (Willis et al., 2005). While several mRNAs have been identified in immature mammalian axons, and in axons from developing Xenopus and goldfish, mRNAs appear to be present in only a few types of axons in the adult mammalian CNS. Studies on adult rat olfactory and hypothalamic axons demonstrated the presence of mRNA (Vassar et al., 1994; Wensley et al., 1995; Mohr and Richter, 2000; Nedelec et al., 2005). However, adult retinal axons do not contain detectable amounts of ribosomal protein, and in the spinal cord the only axons in which ribosomal protein is detectable are the central branches of DRG axons, suggesting that most adult CNS axons are not capable of local translation (Verma et al., 2005, Verma and Fawcett, unpublished results).
Several functions of local translation of axonal mRNA have been established. In Xenopus retinal axons it was shown that asymmetrical translation of β-actin mRNA is essential for growth cone turning (Yao et al., 2006; Leung et al., 2006). Knock-out mice for the RNA binding protein SMN1 show decreased axonal β-actin mRNA and protein in motor neuron axons in vitro, causing decreased axon growth and a reduction in growth cone size (Rossoll et al., 2003). siRNA directed against axonal rhoA abolished Sema3A-induced growth cone collapse of embryonic DRG axons in vitro (Wu et al., 2005), and local translation of CREB is involved in NGF signalling (Cox et al., 2008). Many types of axon are able to regenerate after axotomy, but their regenerative ability varies greatly, with PNS axons showing a strong regenerative response and many CNS axons showing little regeneration, even when presented with a permissive environment. The different regenerative ability of CNS and PNS axons can be modelled in vitro where DRG axons of all developmental stages are usually capable of regenerating a new growth cone after transection but adult retinal axons often fail to regenerate (Chierzi et al., 2005). The regenerative ability of DRG axons is much reduced by protein synthesis inhibitors and these axons contain ribosomal proteins and translation elongation factor at all developmental stages (Verma et al., 2005). However, the poorly regenerating adult retinal axons do not contain ribosomal protein, and their limited regeneration is not reduced by protein synthesis inhibitors (Verma et al., 2005). Local axonal translation of vimentin and importins play an important part in retrograde signalling from damaged sensory axons to the cell body (Hanz et al., 2003; Perlson et al., 2006).
PNS axotomy affects axonal transport, including probably that of mRNA (Willis et al., 2007). Currently, no localised mRNA identification data exist on axons from adult mammalian non-conditioned DRGs. In the present study we describe a new compartmented system for obtaining pure axonal material from DRG explants. We have used it to look for the presence of candidate mRNAs encoding proteins involved in the cytoskeleton, cytoskeletal control, signalling pathways and cell surface molecules. We have investigated axonal translation of β-actin mRNA and demonstrated its importance for successful axonal regeneration.
Results
A new culture method for isolation of axon-only RNA
Obtaining sufficient axonal mRNA for quantitative studies, free of glial and neuronal cell body contamination, is challenging. We have developed a new compartmented culture system for extracting axonal material from adult, neonatal and embryonic rat DRG explants. In Fig. 1, we compare our method with the currently used compartmented culture systems. Because the compartment divider is not placed in the culture dish until robust axon growth has begun, the method overcomes the inability of some axons to grow through silicone grease barriers and the need of DRGs to be held down by surface tension until adherent. It also allows for the culture of 20 or more DRG explants, and therefore the collection of significant quantities of axonal material. In the method DRG explants are placed in a line next to parallel scratches to direct axon growth and allowed to adhere and begin axon growth for one or two days. After this, triangular barriers cut from silicon elastomer with walls 1 mm thick are placed next to the DRGs without the application of silicon grease. Axons from embryonic, newborn and adult DRGs grow under the barrier for around 1 cm, but almost all fibroblasts and Schwann cells are excluded (Fig. 2). Fresh silicon elastomer is slightly sticky, and forms an effective seal between the compartments as long as fluid levels in the barrier-enclosed compartment and the rest of the dish are kept equal. To demonstrate the separation of compartments, ink was placed in the central compartment and showed almost no diffusion underneath the barrier over 12 h (Fig. 2b). After mitomycin-C treatment to the outer compartment to kill any Schwann cells or fibroblasts, mRNA could be extracted by tilting the dishes and scraping off the axons into extraction solutions. This provided sufficient RNA to detect β-actin mRNA in axons of all developmental stages without pooling or amplification. From dishes containing around 20 DRGs, between 100 ng and 150 ng of total RNA were isolated from the each culture dish, measured by nanodrop.
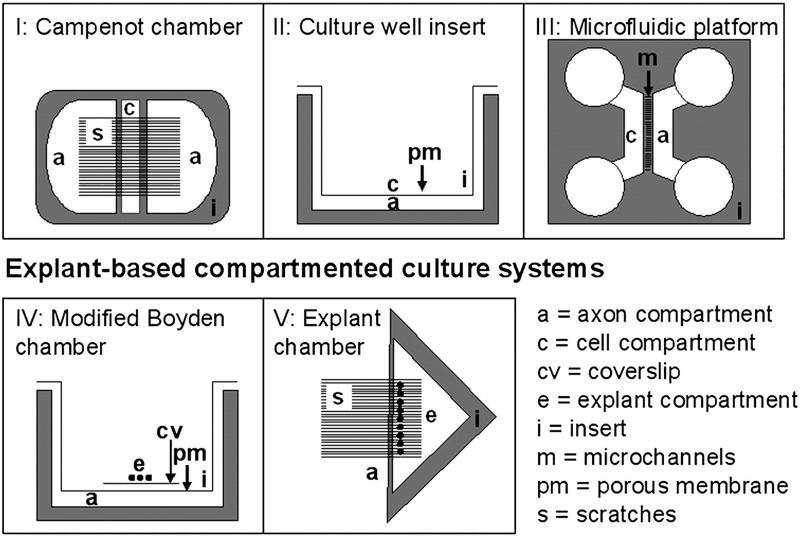
Fig. 1.
Schematic representation of current compartment culture models I–IV (Eng et al., 1999; Zheng et al., 2001; Wu et al., 2005; Taylor et al., 2006). Our newly developed “explant chamber” is shown in (V).
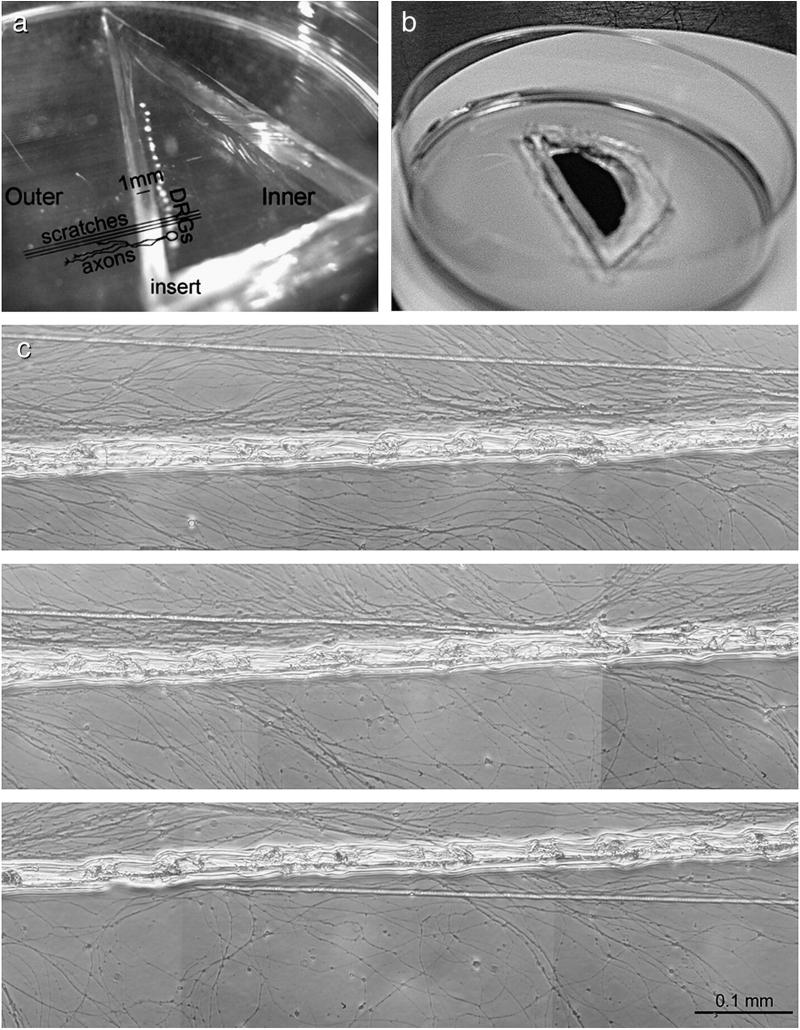
Fig. 2.
P1 DRGs cultured in the compartmentalised explant chamber. (a) Explants were plated in a row on top of the scratches and the silicon insert placed perpendicular to the scratches. (b) Solutions placed in one compartment remain in that compartment; india ink was placed in the central compartment, the experiment photographed 4 h later. (c) Axons extended from the explants, crossed the insert and grew into the outer compartment, using the scratches for guidance. Axons were harvested from the outer compartment after 7–9 days. (c) P1 DRG axons in culture at 5–7.5 mm from the insert. Numerous axons can be seen lining up with the scratches. No cells were observed in most cultures. Scale bar=0.1 mm.
Axon-only preparations were devoid of cell contamination
The silicon inserts prevented almost all Schwann cell and fibroblast migration into the axon compartment. Mitomycin C was used to eliminate any cells that had got under the barrier. After RNA extraction we tested all preparations for the presence of P0 and DNA polymerase mRNA by qPCR to ensure there was no cell contamination. The sensitivity of the P0 qPCR for detecting contaminating Schwann cells was examined using a dilution series of 100 ng to 1 pg of Schwann cell RNA, corresponding to 104 to 10−2 cells (106 cells yielded 10 μg of total RNA). We were able to detect P0 mRNA in cDNAs corresponding to as little as 10−1 cells per preparation (data not shown). This indicates that our method was sensitive enough to detect contamination of less than a single cell. Around 80% of all axon-only preparations were negative for both P0 and DNA polymerase, the remaining 20% being rejected.
Quantification of β-actin mRNA in axons at three developmental stages
The clean axon-only preparations were first subjected to cytochrome c oxidase subunit I (coxI) qPCR in order to judge whether the amount of RNA isolated was sufficient to detect target mRNAs. Samples with coxI cycle threshold (Ct) values higher than 22 (i.e. with lower levels of coxI) were not used for target gene identification. Controls in which reverse transcriptase was omitted were performed in order to exclude the possibility of DNA contamination. We used the mitochondrial enzyme coxI as a marker for the quantity of axons within our cultures, because using an axonally transported housekeeping mRNA for this purpose would clearly be meaningless in the context of an investigation of differential mRNA transport. CoxI mRNA is synthesised in mitochondria and therefore assumed to be independent of axonal mRNA transport. We estimated the number of mitochondria per length of axon by immunocytochemistry for coxI protein and by staining with mitotracker, then measuring the intensity of the staining. These estimates showed no significant differences in the density of mitochondria per unit length in the axons of the three developmental stages (data not shown).
In order to demonstrate that axonal mRNA could be detected and quantified using our technique, we first examined the presence of β-actin mRNA, which has been demonstrated in axons in previous studies (Giuditta et al., 2002, Bassell et al., 1998; Eng et al., 1999; Willis et al., 2005). In samples with coxI Ct values between 18 and 22, β-actin was readily detected between 28 and 32. β-actin levels in axons of E16.5, neonatal and adult (2- to 4-months-old) DRGs were normalised to coxI. First, the detection and quantification limits were determined by performing RT-qPCR on tenfold dilutions from 100 ng to 1 pg of total DRG RNA. Both β-actin and coxI were reproducibly detectable even in 1 pg of total RNA (Fig. 3). The ratio between β-actin and coxI was plotted over the log of the RNA concentration. The ratios measured in each RNA concentration were not significantly different from a trendline with slope zero positioned at the average ratio 0.044, indicating that even in the lowest concentrations we were able to quantify the levels with the same efficiency as in the higher concentrations (Fig. 3).
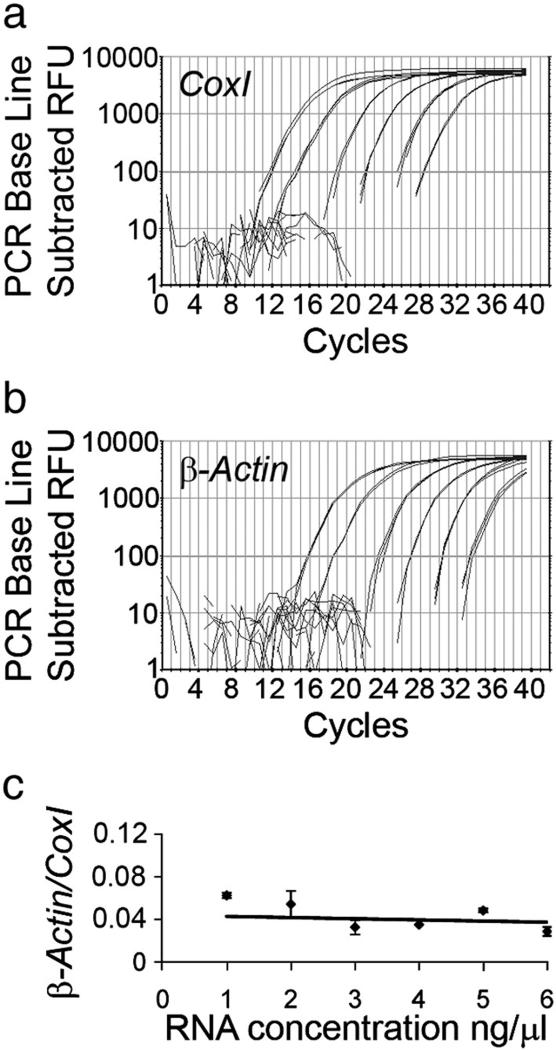
Fig. 3.
Sensitivity of the RT-qPCR for the detection of β-actin and coxI. RT-qPCR curves on DRG total RNA dilution series from 100 ng to 1 pg (in the graphs from left to right). Both coxI (a) and β-actin (b) were reliably and reproducibly amplified even from 1 pg of total RNA. Efficiencies of the coxI and β-actin PCRs were 95 and 90%, respectively. (c) Quantification of β-actin levels relative to coxI resulted in similar values for all RNA concentrations. The data points did not significantly differ from the average 0.044 (trendline) (ANOVA, p>0.05).
The β-actin Ct values used for quantification were within this linear detection range (Ct usually around 30). We quantified β-actin levels relative to coxI in clean axon preparations of E16.5 (n=5), neonatal (n=20) and 2–4-month-old rat DRGs (n=5) (Fig. 4). No significant differences in β-actin mRNA levels were found (ANOVA p=0.357) (Fig. 4).
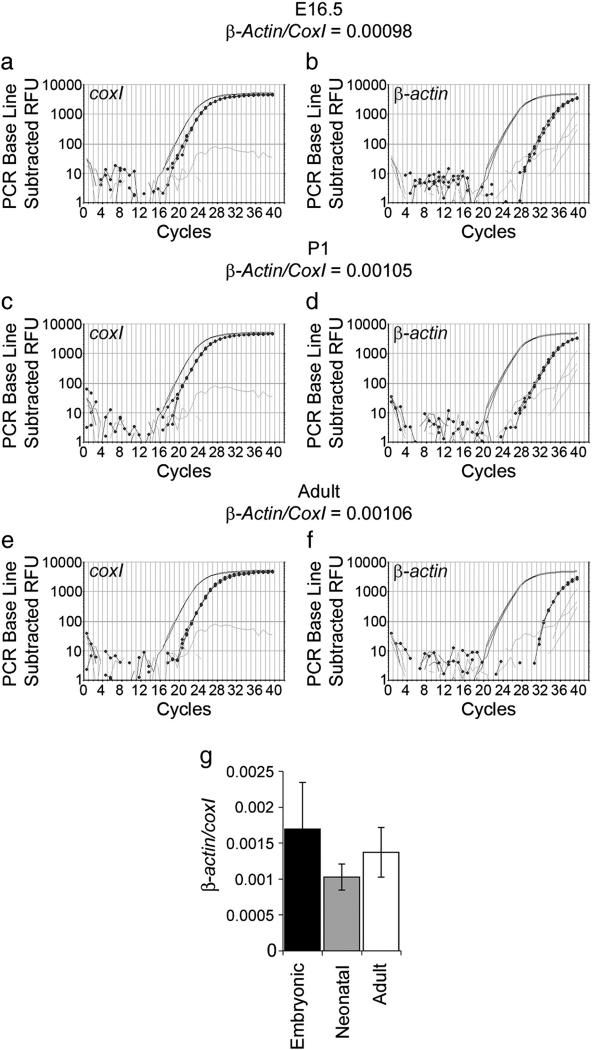
Fig. 4.
Quantification of β-actin mRNA in embryonic, neonatal and adult rat axon-only preparations. Examples of qPCR graphs for E16.5 (a, b), P1 (c, d), and adult (e, f) axon preparations. The positive control is shown in plain black, the axon-only preparation in dotted black, and the water control in grey. (g) Quantification of β-actin (b, d, f) levels relative to coxI (a, c, e) showed no significant difference between the developmental stages (ANOVA p=0.35).
We conclude, therefore, that adult DRG axons do not systematically downregulate their axonal mRNA levels relative to those seen during development.
Presence of axonal mRNA of candidate molecules involved in axon growth
The axon growth cone may be some distance from the neuronal cell body, imposing a significant delay in responses to the environment that involve synthesis of new proteins unless the synthesis is local within the axon tip. One might therefore expect that molecules involved in growth cone outgrowth and guidance might have their mRNAs present within growing or regenerating axons. We therefore assayed for the presence of some key molecules within four groups of molecules, cytoskeleton, cytoskeletal remodelling, signalling pathways and cell surface receptors. The results of this study are summarised in Table 1 and typical qPCR results are shown in Fig. 5.
Table 1.
List of target mRNA species investigated for presence in neonatal axonal preparations.
| Axonal #/ n | Ct axons | Ct DRG 10−3 | |
|---|---|---|---|
| Cytoskeletal | |||
| β-Actin | 35/35 | 29 | 23 |
| γ-Actin | 3/9 NM/ND | >34 | 28 |
| βIII-Tubulin | 10/11 | 30 | 24 |
| MyosinII | 4/4 | 32 | 26 |
| NF-L | 3/3 NM | >34 | 23 |
| NF-M | 3/3 NM | >34 | 24 |
| NF-H | 0/4 ND | x | 23 |
| Peripherin | 5/5 | 32 | 26 |
| Vimentin | 13/13 | 32 | 25 |
| Cytoskeleton-remodelling | |||
| Calmodulin | 3/3 | 32 | 28 |
| CamKIIa | 0/3 ND | x | 29 |
| CAP-23 | 3/4 | 33 | 29 |
| Cofilin1 | 5/5 | 32 | 23 |
| GAP-43 | 9/9 | 33 | 28 |
| Profilin1 | 5/5 | 31 | 26 |
| SCG10 | 5/5 | 30 | 25 |
| Microtubule-associated | |||
| MAP1A | 0/4 ND | x | 26 |
| MAP1B | 1/4 NM/ND | >35 | 24 |
| MAP2 | 0/3 ND | x | 27 |
| Tau | 0/6 ND | x | 28 |
| Signalling molecules | |||
| Cdc42 | 5/9 | 34 | 26 |
| Rac | 5/5 NM | >34 | 28 |
| Rho | 5/5 | 33 | 26 |
| Adhesion molecules | |||
| Integrin α6 | 0/3 ND | x | 29 |
| Integrin α7 | 2/4 NM/ND | >34 | 28 |
| Integrin β1 | 3/9 NM | 33/>35 | 27 |
| Integrin β4 | 0/3 | x | 27 |
| L1 | 0/3 | x | 27 |
| N-cadherina | 0/3 | x | 29 |
| Receptors | |||
| TrkA | 0/3 | x | 30 |
An mRNA was considered positive in axons when (1) it was found in at least 3 consecutive axon preparations, (2) at least 2 of the 3 replicates were positive and reproducible, and (3) the average Ct value was lower than 34. The column “Axonal #/ n” specifies the number of times an mRNA was positive out of n axonal preparations tested. mRNAs that were positive but did not match the inclusion criteria are designated NM (non-matching). Target mRNAs that were negative are designated ND (non-detectable). The approximate Ct for each mRNA in the axonal preparations and in the positive control DRG 10−3 are shown in order to indicate the abundance of the mRNA in the axons as compared to the source tissue.
ND = Not detectable.
NM = Not matching selection criteria.
Performed on mouse axons.
Fig. 5.
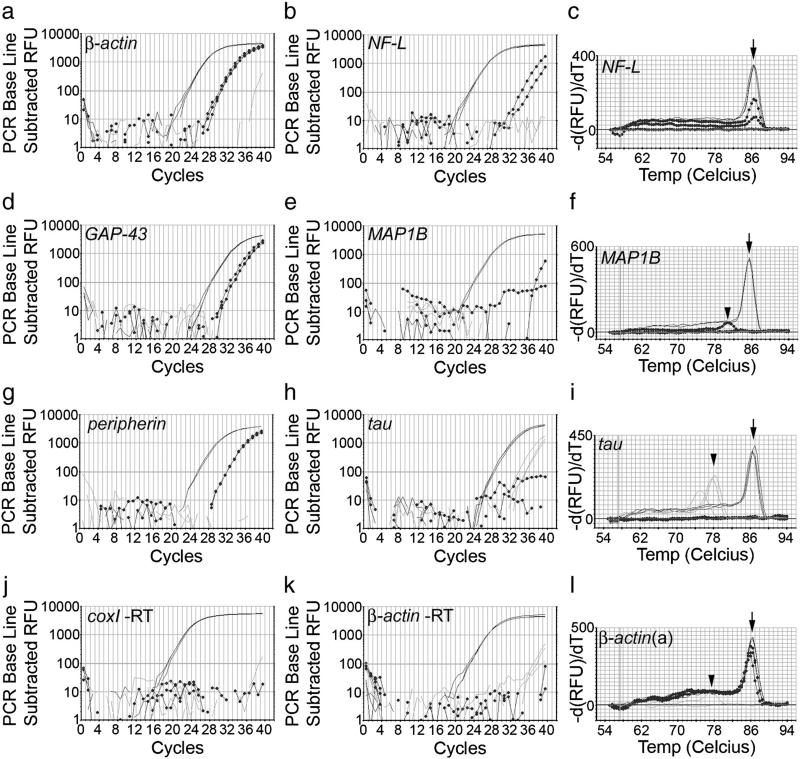
Examples of RT-qPCR curves in axon-only preparations with similar coxI levels for 3 positive target mRNAs and 3 target mRNAs that were not detectable or did not match the selection criteria. The positive control is shown in plain black, the axon-only preparation in dotted black, and the water control in grey. β-Actin (a), GAP-43 (d) and peripherin (g) mRNAs were reliably detected in axon-only preparations. NF-L (b) did not match inclusion criteria, as the curves were not reproducible and the Ct values were too high. MAP1B (e) was not detected, although the positive control shows that it is highly abundant in the DRG itself. Tau (h) was not detectable. No signal was detected in minus RT controls for CoxI (j) and β-actin (k). Melt curves (c, f, i, l) were performed in order to confirm that the axonal product peak matches with the positive control product peak (arrows). The water controls in the β-actin (l) and tau (i) PCRs contain primer dimers (arrowheads) only.
Cytoskeleton
The cytoskeletal genes β-actin, β-III-tubulin, peripherin, vimentin and myosinII were consistently present in axons, however, neurofilament mRNAs were not detected. Neurofilament H was consistently absent, whereas neurofilament M and L (Fig. 5b) both did not match selection criteria. They were occasionally positive, but the replicates never matched each other and the Ct values were around 35, more than 11 Ct values higher (i.e. >2000 times lower) than the 10−3 diluted DRG cDNA. In general, reproducibly detectable axonal target mRNAs had Ct values between 4 and 8 away from the positive control (Fig. 5). γ-actin did not match the selection criteria and was therefore regarded as absent as well. mRNAs for microtubule-associated proteins MAP1A, -1B (Fig. 5e, -2, and tau (Fig. 5h) were not detectable in axon-only preparations. MAP2 mRNA is present in dendrites, suggesting transport mechanisms that differentiate between axonal and dendritic mRNA transport. MAP2 and γ-actin mRNA has been used in several studies as a negative control (Zheng et al., 2001; Willis et al., 2005). The mRNAs for MAP2 and tau are of fairly low abundance in the DRG itself (see Table 1), but both were sufficiently highly expressed in DRGs to be theoretically detectable in axons had they been present.
Cytoskeletal remodelling
Next on our list of targets were genes associated with cytoskeletal remodelling and axon regeneration. SCG10, GAP-43 (Fig. 5d), CAP-23, calmodulin, cofilin and profilin mRNAs were present in axon-only preparations. Cofilin mRNA has been detected in axons in previous studies (Lee and Hollenbeck, 2003; Willis et al., 2005, Piper et al., 2006). mRNAs of molecules that are important for actin remodelling are therefore localised in axons. GAP-43 and CAP-23, both of whose mRNAs are present in axons, are axon growth associated proteins that link signalling pathways to axon growth, and are stimulators of axon growth when overexpressed.
Signalling molecules
As shown before in embryonic DRG axons by Wu et al. (2007), mRNA for the signalling molecule rhoA was present, whereas rac was not reliably detectable (not matching selection criteria) and cdc42 was detectable in 5 out of 9 cultures. It is probable that rac and cdc42 are present at a low concentration. Calmodulin kinase IIa (CamKIIa) mRNA was absent. This further demonstrates the difference between the complement of dendritic and axonal mRNAs, because CamKIIa is one of the key dendritic mRNA molecules (Paradies and Steward, 1997).
Cell surface molecules
As representatives of cell surface adhesion molecules and growth factor receptors we searched for mRNAs encoding several integrins, L1, N-cadherin and TrkA. All were absent except for beta1 integrin which was seen in some preparations but did not meet our criteria.
The data described above are based on qPCRs from neonatal axon-only preparations. In order to determine whether there were significant differences in the mRNAs in axons of different developmental stages, we assayed for the presence of β-actin, β-III-tubulin, peripherin and GAP-43, MAP1A, -1B, -2, and Tau in axons from embryonic day 16, neonatal and adult DRG axons. mRNAs coding for all the MAPs and tau were absent at all ages, while the other mRNAs were consistently present in axons of all developmental stages.
Some mRNAs appear to be enriched in axons
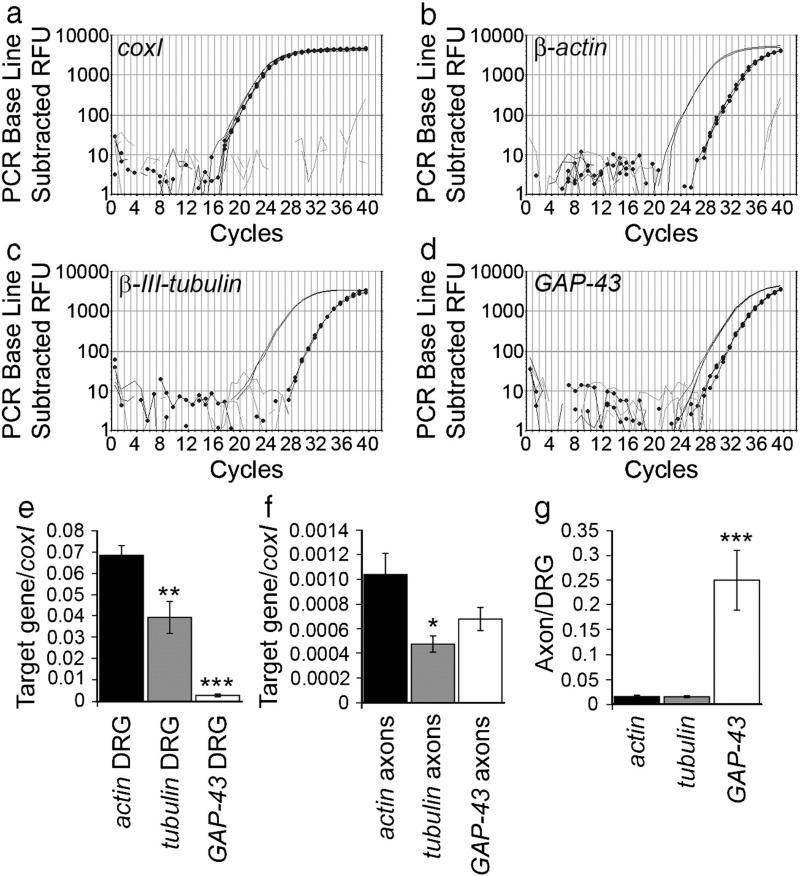
For all genes we compared axonal mRNA with 10−3 diluted DRG cDNA, to give a positive control. This also allowed us to compare mRNA levels in axons and DRGs. We quantified the levels of β-actin (n=9), β-III-tubulin (n=6) and GAP-43 (n=5) mRNA relative to coxI in DRGs and axons (Fig. 6). In both DRGs and axons actin mRNA is about twice as abundant as tubulin mRNA, axonal levels being 1.5% of their DRG levels (Figs. 6e–g). This correlates with earlier observations in squid axons (Chun et al., 1996). GAP-43 mRNA, however, was about 25 times lower than actin in DRGs, but not significantly different from actin in axons (ANOVA p=0.169) (Figs. 6e, f). This would correspond to axonal GAP-43 mRNA levels being 25% of DRG levels (Fig. 6g). The average Ct values of the target genes in the axons and positive controls are shown in Table 1. Because DRG samples contain both glial and neuronal mRNA we cannot say definitively that these differences demonstrate preferential transport of GAP43 mRNA into axons compared with actin and tubulin, but the data are suggestive.
Fig. 6.
Quantification of β-actin, β-III-tubulin and GAP-43 mRNA in neonatal rat axon-only preparations. Examples of qPCR graphs for β-actin (b), β-III-tubulin (c) and GAP-43 (d). The positive control is shown in plain black, the axon-only preparation in dotted black, and the water control in grey. Quantification relative to coxI (a) showed that β-III-tubulin mRNA levels were about half the levels of β-actin, in both DRGs (**p<0.01) and axons (*p<0.05) (e, f). GAP-43 mRNA, in contrast, was about 25 times lower than actin in DRGs (***p<0.0001), but only 1.5 times lower than actin in axons (e, f). Axonal levels of actin and tubulin were about 1.5% and 1.3% respectively of the DRG levels, whereas, the levels of GAP-43 in axons was 25% of DRG levels (g).
Actin is synthesised in axons, the rate being increased after axotomy
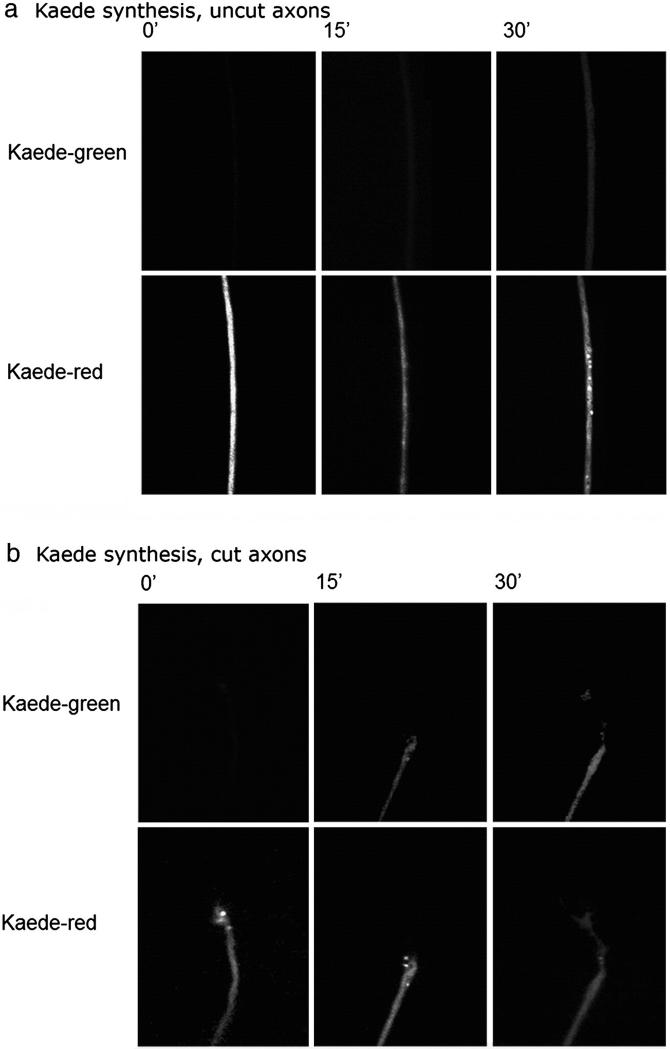
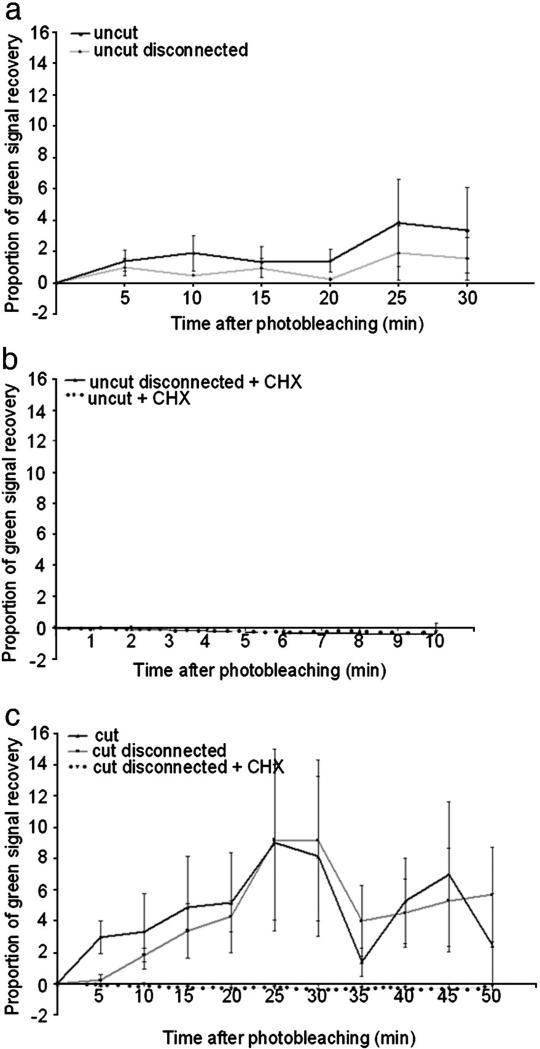
We wished to establish whether actin mRNA is translated in DRG axons, and whether the rate of synthesis of new actin is affected by axotomy. In order to do this we used the fluorescent protein CoralHue Kaede, which fluoresces green unless photoconverted with UV light after which it fluoresces red. It is therefore possible to measure the rate of appearance of newly synthesised Kaede by observing the reappearance of green fluorescence after photoconversion (Leung et al., 2006; Leung and Holt, 2008). In order to measure actin translation, we expressed Kaede under the 3′UTR of rat β-actin. In previous experiments, synthesis of reporter proteins expressed under the actin 3′UTR has been shown to parallel that of actin (Zhang et al., 2001). Dissociated adult DRG neurons were transfected with the Kaede construct using a microporator device, then plated on PLL-laminin-coated tissue culture plastic coverslips. We examined the return of newly synthesised green Kaede following photobleaching by taking pictures every 5 min (every 2 min in the case of uncut CHX treated axons) on a spinning disc confocal microscope. We photo-converted the entire neuron in order to be certain that all newly-appearing Kaede were newly translated. First, we investigated the synthesis of Kaede in uncut axons. Following photoconversion of the whole neuron there was a steady accumulation of green signal, indicating newly synthesised protein in the axons (Figs. 7a, 8a). This accumulation was blocked by application of cycloheximide, proving that it was due to new translation (Fig. 8b). In order to see whether any of this new protein was synthesised in the cell body, we disconnected the axons from the cell bodies just before starting to record the appearance of green Kaede. Newly synthesised Kaede still appeared steadily in these isolated axons demonstrating that there is protein translation driven by the 3′UTR of rat β-actin in DRG axons (Fig. 8a). Next, we asked whether the rate of synthesis was affected by cutting the axons near to the growth cone. The rate of re-appearance of Kaede was significantly increased compared with uncut axons (Figs. 7b, 8c). Prior disconnection of the axons from the cell body did not significantly alter the rate of re-appearance of Kaede in the axons, which was elevated compared with uncut axons. These differences were statistically significant by regression analysis.
Fig. 7.
Synthesis of Kaede driven by 3′UTR of rat β-actin in axons. At time 0 all the Keade is photobleached to red, then newly synthesised Kaeda appears in the green channel. In cut axons the newly synthesised Kaede is detected more rapidly and at a higher level.
Fig. 8.
Local synthesis of β-actin measured by recovery of photoconverted Kaede driven by 3′UTR of rat β-actin. (a) Kaede green signal recovery in uncut axons. Recovery of green signal was observed in uncut axons attached to, but also disconnected from the cell body, demonstrating that β-actin protein is produced by local axonal synthesis. No significant difference between attached or disconnected axons was observed. (b) Recovery of green signal in uncut axons was abolished by treatment with CHX (c) Axotomy increases the rate of local β-actin synthesis in regenerating axons attached to, but also disconnected from the cell body. Regression analysis showed that the rate of synthesis is greater in cut axons compared to the uncut axons shown in (a) (97.5% confidence), and in disconnected cut axons compared to disconnected uncut axons (99% confidence). Recovery of green signal in regenerating axons expressing Kaede-β-actin 3′ UTR was abolished by treatment with CHX.
Actin synthesis in axons is required for efficient regeneration
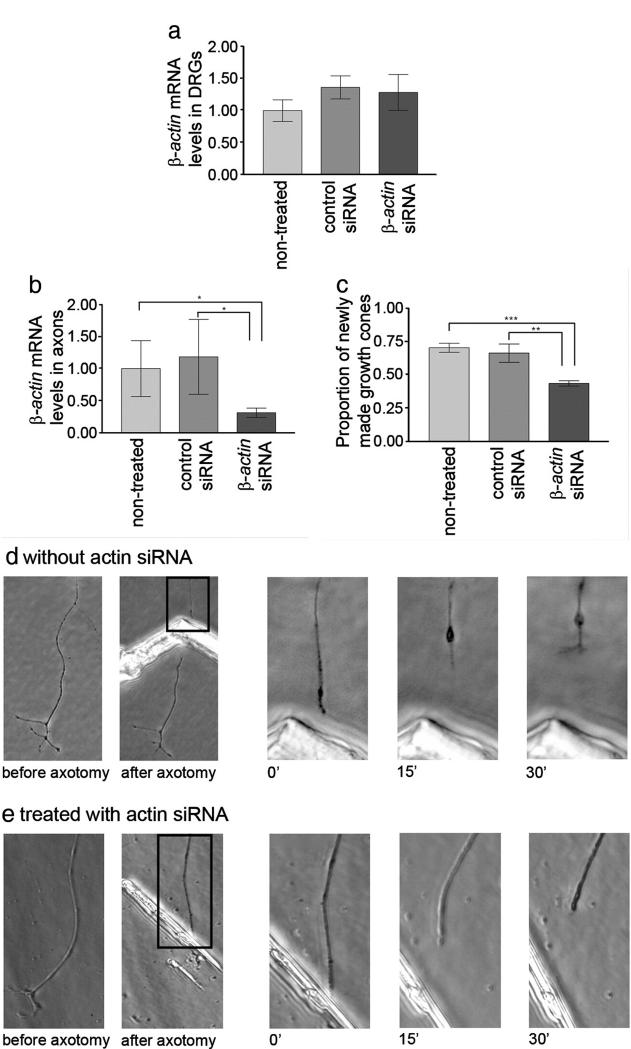
While previous work has established that mRNA translation overall is required for efficient axon regeneration in vitro, there is no information about which particular proteins need to be synthesised. We tested the hypothesis that new β-actin synthesis is needed for efficient growth cone regeneration. We first established that siRNA treatment applied to the axonal compartment reduces β-actin mRNA levels in axons within 24 h, but does not affect levels in the DRGs (Fig. 9a). Fluorescein-labeled double-stranded RNA oligomers were used to assess the efficiency of transfection. About 75% of axons in the axonal compartment were labeled while no fluorescent signal was detected in the cell body compartment, indicating that siRNAs were not retrogradely transported to the cell body during the time-course of the experiment. This observation is in accordance with similar findings by Hengst et al., 2006. We then performed axotomy of DRG axons within the axonal compartment, and assessed the ability of axons to regenerate their growth cones within 1 h of axotomy, as in previous experiments (Chierzi et al., 2005; Verma et al., 2005) (Figs. 9c, e). We observed an approximately 50% decrease in the proportion of axons that were able to form a new growth cone, while a control siRNA did not affect growth cone regeneration (Figs. 9b, d). We first examined axons at 30, 90 and 240 min to determine the best time to score regenerative success. If an axon had not regenerated by 30 min it was extremely unusual for it to begin to regenerate later at 90 or 240 min, so we scored growth cone regeneration at 30 min. Growth cone formation was therefore blocked rather than just delayed in β-actin siRNA-treated axons.
Fig. 9.
Knock down of β-actin mRNA in axons reduces the ability of axons to regenerate after axotomy in vitro. β-actin siRNA treatment delivered to the axonal compartment did not reduce the β-actin mRNA levels (levels shown relative to non-treated samples) in the DRGs in the cell body compartment (a), but did reduce β-actin mRNA levels in the axons T-test *p<0.05 (b). (c) The number of regenerating axons is shown as a proportion of axomotised axons. Treatment of the axonal compartment with β-actin siRNA reduced the ability of axons to form a new growth cone after axotomy in vitro. Treatment with a control siRNA does not lead to significant decrease of regeneration ability. ANOVA, Bonferroni post-test **p<0.01 ***p<0.001, error bars are SEM. (d and e) show representative experiments in which axons that either had or had not been treated with β-actin siRNA were cut. In (d) the axon retracts with a retraction bulb, then generates a new growth cone within 30 min, while the treated axon in (e) retracts but does not form a new growth cone.
Discussion
Although the presence of mRNA molecules in adult mammalian sensory axons is well established, only limited information is available as to their identity, and only in axons conditioned by a previous lesion (Vogelaar and Fawcett, 2008a,b). There is also a lack of data on comparisons between different developmental stages of mammalian axons. We have established a compartmentalised culture method which allows us to isolate rodent DRG axons of all developmental stages without the need for a conditioning lesion or the need to pool axons from different cultures. The conditioning lesion, used in the experiments of Willis et al. (2005) for culturing adult DRGs, may lead to changes in axonal RNA transport as compared to non-conditioned DRGs. We aimed to compare axonal RNA levels of embryonic, neonatal and adult DRGs and, therefore, developed these new chambers, allowing us to culture axons of all developmental stages at the same conditions. We used the axon-only preparations to examine expression of several candidate genes, and quantify some of these. We have also undertaken an in depth analysis of the distribution of β-actin mRNA and demonstrated that axonal β-actin translation is important for efficient axon regeneration.
Neurons are unique in the length of their processes, particularly in adulthood. This means that any communication between the axon tip and the cell soma through transport of molecules will take a long time with transport speeds of up to 250 μm/min for fast axonal transport, up to 15 μm/min for slow transport. Where axons are growing through the complex terrain of the developing embryo some degree of growth cone autonomy might therefore be desirable. In the adult peripheral nervous system, mild trauma that does not cut the axons but disrupts the cytoskeleton in the mid-part of axons within the limbs may occur frequently, requiring axonal repair a very long way from the cell body. The damage would be most efficiently repaired through local processes as opposed to protein transport from the cell body. Damage that cuts axons will also generally occur far from the cell body. In order to find out which processes requiring new protein synthesis in PNS axons might be locally autonomous, we examined candidate mRNAs representing the key mechanisms in axon growth and cytoskeletal repair. We found that axonal mRNAs are highly selected, and different from those present in and dendrites. Actin, tubulin and molecules involved in controlling actin dynamics such as cofilin and profilin all had mRNAs present in the axons. Also present were the mRNAs for Rho and cdc42, while rac was not present at a sufficiently high level to meet our inclusion criteria. Also present were mRNAs for molecules involved in growth cone control signalling and regeneration, including GAP-43, CAP23, and SCG10 and calmodulin. However, there were some interesting absences; mRNAs for microtubule-associated proteins, some of which are present in dendrites, were absent in axons, as was mRNA for neurofilament although two other intermediate filament mRNAs, vimentin and peripherin were present. The absence of neurofilament mRNAs is in contrast to studies in Goldfish Mauthner axons, chicken sympathetic axons and conditioning-lesioned DRGs (Weiner et al., 1996; Lee and Hollenbeck, 2003; Willis et al., 2005) where either NF-M or NF-L mRNA was found. Overall, the picture is that molecules involved in dynamic regulation of the cytoskeleton, particularly the actin cytoskleton, can be locally translated in axons. Of these, GAP-43 (also called B-50) is well-known for its role in growth cone motility and axon regeneration, interacts with F-actin and plays a role in membrane extension (Oestreicher et al., 1997). GAP-43 mRNA has been detected in vitro in growth cones of PC12 cells (Smith et al., 2004). Calmodulin, when released from GAP-43 by phosphorylation, stimulates polymerisation of actin and tubulin. CAP-23 has been shown to have similar functions as GAP-43 (Wiederkehr et al., 1997). SCG10 is a tubulin-destabilising molecule and is upregulated after axon injury (Mason et al., 2002; Grenningloh et al., 2004). Cofilin and Profilin are involved in actin polymerisation and de-polymerisation, and therefore in actin dynamics. The absence of mRNAs for microtubule-associated proteins is interesting. mRNAs for some of these molecules are present in dendrites, where they play a part in dynamic change (Szebenyi et al., 2005). Individual microtubule-associated proteins are not necessary for normal axon growth, but deletion of two of them blocks it, suggesting that at least some of the molecules need to be present for axon regeneration and repair (Takei et al., 2000). Perhaps the most surprising finding in our mRNA screen is the absence of mRNAs for several integrins, L1, N-cadherin and trkA. In principle, growth cones as they progress will encounter changes in the extracellular environment to which they will have to react by changing the molecules on their surface. Changes such as this have been seen in axonal surface integrin levels when axons are placed on different concentrations of extracellular matrix molecules (Condic and Letourneau, 1997). These changes take some time to occur, because growing axons pause for many minutes when they encounter a change in substrate (Burden-Gulley et al., 1995). The implication of our result is that these changes in cell surface molecules must either involve new synthesis in the cell body followed by axonal transport, or there must be a pool of the molecules at the growth cone whose insertion into the membrane is modulated by the outside environment. A possible reason for the absence in axons of mRNAs encoding cell surface molecules was the need for a Golgi apparatus to enable them to be inserted into the membrane. It now appears that there is a Golgi-like structure in PNS axons that can perform this task (Merianda et al., 2009). Exactly how this compartmentalisation of mRNAs is achieved is not clear. A single mechanism that allows sorting of RNAs has not been identified and it is likely that regulatory mechanisms are multiple (Kiebler and Bassell, 2006). The presence of a code on the sequence of the RNA (cis-acting element) recognised by a trans-acting factor is a mechanism to achieve partition. A cis-acting element has been identified for β-actin, Eph, Tau and RhoA (Kislauskis et al., 1994; Eom et al., 2003; Tiruchinapalli et al., 2003; Huttelmaier et al., 2005; Pan et al., 2007; Aronov et al., 2001), but they seem to be specific to the RNA transported, rather than being shared by all axonal RNAs. Moreover, mRNAs fold to form secondary and tertiary structures and localisation signals may operate at these levels (Hamilton and Davis, 2007) so that an analysis of the primary structure of mRNAs will identify localisation sequences only in a minority of cases. These secondary and tertiary structures may be difficult to predict making it hard to identify localisation motifs. It is also possible that there is differential degradation of mRNAs, and components necessary for RNA interference have been shown to be present in axons (Hengst et al., 2006).
It is not surprising, therefore, that we find that the abundance of axonal mRNAs relative to the levels in DRGs varies between mRNA species, with the mRNA for GAP-43 being particularly abundant. It is also probable that influences such as growth factors, activity and substrate will influence the transport of different mRNA species into axons (Zhang et al., 2001; Tiruchinapalli et al., 2003).
Our previous work has shown that sensory axons, unlike adult retinal axons from the CNS, contain translational machinery, and that inhibiting translation inhibits axon regeneration (Verma et al., 2005). In the present paper we have examined the role of one specific mRNA, that encoding β-actin, in growth cone regeneration. Previous studies have shown that local translation of β-actin is important for growth cone turning (Leung et al., 2006; Yao et al., 2006; Leung and Holt, 2008). The response of the growth cone to guidance cues is preceded by and dependent on the local transport and translation of mRNAs to the turning side of the growth cone. In the case of a chemo-attraction by netrin-1 or BDNF β-actin mRNA is transported to the stimulated side of the growth cone and translated there (Leung et al., 2006; Yao et al., 2006). In the case of repulsion β-actin mRNA was transported and translated at the far side of the growth cone (Yao et al., 2006). However, local translation of β-actin is not necessary for continuing axonal elongation. Similarly, we showed previously that local translation is not needed for continuing axon growth, but is needed for regeneration of cut axons (Eng et al., 1999; Verma et al., 2005). These data indicate that local protein synthesis from axonal mRNA plays a role in very specific local processes involving cytoskeletal remodelling. We examined local synthesis of β-actin in axons using the fluorescent reporter molecule Kaede, which photoconverts to a different fluorescent wavelength when exposed to UV light. By expressing this molecule under the control of the β-actin 3′ UTR, we were able to monitor β-actin translation in axons (Leung et al., 2006; Leung and Holt, 2008). We found that there is a low baseline rate of translation in normally growing adult DRG axons, which is increased following axotomy. As with growth cone turning, this local translation is important for efficient growth cone regeneration, because knock down of axonal mRNA levels in compartmentalised cultures partially blocked axon regeneration. Local β-actin translation is therefore important for changes in growth cone behaviour, which are largely controlled by actin dynamics, but growth can be maintained in its absence. The lack of translational machinery in adult CNS axons means that they are not able to respond to axotomy with local translation of β-actin and other molecules involved in cytoskeletal dynamics, and if the axotomy is some distance from the cell body it will take a considerable time for newly synthesised molecules to arrive from the cell body. This may partly explain why the vigour of CNS axon regeneration decreases as axotomy occurs further from the cell body, while this relation does not exist for PNS axons, which are capable of local translation (Fernandes et al., 1999). Local translation of proteins also plays another key part in PNS regeneration in signalling back to neuronal cell bodies. Thus at sites of injury several mRNAs, including vimentin and importins, are translated enabling retrograde signalling by Erk, MAPKinases and other molecules to mediate the conditioning effect, which increases the vigour of axon regeneration from sensory neurons from about 1 day after a first axotomy (Hanz et al., 2003; Perlson et al., 2006; Yudin et al., 2008).
Overall, our results show that mRNA transport into axons is restricted to particular mRNA species that are different from those found in dendrites. The mRNAs that we detected are particularly focused on cytoskeletal dynamics, particularly relating to the actin cytoskeleton. Our demonstration that β-actin translation is necessary for efficient growth cone regeneration suggests that the initiation of regeneration is locally autonomous. The presence of mRNAs involved in modulation of the actin cytoskeleton suggests that these are important for axon regeneration. The observation that adult DRG axons still contain significant levels of mRNA, suggests that this local control of regeneration remains active in sensory axons and persists throughout life. The lack of translational machinery in most CNS axons means that they are probably unable to remodel the cytoskeleton effectively after axotomy, leading to a poor regenerative ability.
Experimental methods
Explant chambers
Dorsal root ganglia (DRGs) were dissected from embryonic (E16), neonatal (P0–P2), and adult (2 to 4-months-old) rats and nerve stumps were carefully cut off. Adult DRGs were cut into 2–3 smaller pieces. DRG explants (20–25 per dish) were plated in a row on top of scratches made with a Campenot pin rake (Tyler Research Corporation) in Nunclon dishes (NUNC) coated with PDL (20 μg/ml, Sigma) and laminin (1 μg/ml, Sigma) in a minimal amount of medium containing ITS+ (1:100, BD Biosciences), penicillin-streptomycinfungicide (1:100, Sigma), supplemented with 10 μg/ml human recombinant insulin (Roche) and 10–100 ng/ml NGF. For the first days the volume of medium was kept low so that surface tension holds the explants to the dish. At 1 day (E16/P1) or 2 days (adult) in culture a triangular insert, cut from silicon elastomer (Dow Corning) was placed into the dish (Fig. 2) and 0.5 μg/ml mitomycin-C (MMC, Sigma) was added to the culture medium in order to abolish cell proliferation. The axons were maintained for 7–9 days, with MMC treatment every other day. A picture of the culture system is shown in Fig. 2.
Axon-only RNA isolation and reverse transcription
When the axons had grown more than 1 cm into the outer compartment, they were washed with cold PBS and scraped into RNA lysis buffer, the dish being tilted so that the buffer avoided the insert. Total RNA was isolated using the RNeasy Micro kit, according to the manufacturer’s protocol (QIAGEN). An on-column DNase step was performed. The complete RNA was then immediately processed into cDNA using random hexamer primers (50 ng) and Superscript III reverse transcriptase, according to the manufacturer’s protocol (Invitrogen), increasing the reaction volume to 25 μl to enable complete use of the 12 μl RNA eluate. Finally, 100 μl UltraPure™ DNase/RNase-Free Distilled Water (Invitrogen) was added to increase the pipetting accuracy for the real-time qPCR analysis.
DRG and Schwann cell RNA isolation and reverse transcription
Total RNA was isolated as previously described (Vogelaar et al., 2004) with minor modifications from adult (3-months-old) rat DRGs from the inner compartment. Briefly, DRGs were detached from the plate and homogenized in TRIzol®, using the Ultra-Turrax T8 homogeniser (IKA). Total RNA was isolated according to the manufacturer’s protocol (Invitrogen) with minor changes. An additional chloroform step was performed to increase purity and RNA was precipitated by centrifugation for 1 h at 4 °C. In-solution treatment with RNase-free DNaseI (Roche) was performed and purification was achieved using the RNeasy MinElute RNA cleanup kit (QIAGEN). 2 μg of RNA was used for reverse transcription with a final volume of 200 μl as previously described (Vogelaar et al., 2004). A 10−3 dilution of this cDNA was used as a positive control throughout the study. A tenfold dilution series of the total RNA, ranging from 100 ng to 1 pg, was used as a template for cDNA synthesis to study the combined sensitivity of the reverse transcription reaction and the real-time PCR. Schwann cells derived from an immortalised line were grown in the presence of 2 μM forskolin and 10 μg/ml bovine pituitary extract (BPE) and 106 cells were lysed in TRIzol. Total RNA was isolated as described above. A tenfold dilution series from 100 ng to 1 pg was used as a template for cDNA synthesis and real-time PCR in order to study the minimum number of contaminating cells that we could detect.
Real-time quantitative PCR
Primers were selected using Premier Biosoft Beacon Designer software. Selection criteria included a Ta between 52 and 60 °C, minimal secondary structures of primers and products (ΔG values between 0.0 and −2.0) and an overall score greater than 70. Optimisations were performed on DRG cDNA using iQ SYBR-Green Supermix and the iCycler iQ Thermal Cycler (Biorad). Reactions were run in triplicate, using 5 μl of template cDNA. First, a temperature gradient was performed on 1:1000 diluted DRG cDNA in order to find the optimal temperature and primer concentration. Then, standard curves were produced using tenfold DRG cDNA dilution series. The qPCR reactions were regarded as optimised when they reached an efficiency between 90 and 105% (occasional exceptions were 88–89% and 109%) with a correlation coefficient greater than 0.994 and worked well even at the lowest cDNA concentrations (10−4 or 10−5, depending on the abundance of the genes). Melt curves and sequencing were performed to ensure the absence of primer dimers and the product specificity. Table 2 shows the optimisation results for all the primers used.
Table 2.
Real-time PCR primer sequences and optimisation data.
| Gene | Sequence | Conc (nM) |
Temp (°C) |
Efficiency (%) |
|---|---|---|---|---|
| Contamination controls | ||||
|
DNA
polymerase β |
CCAAGGACAGGAGTGAATGAC AAGCACAGAGAAGAGGCAATC |
200 | 56 | 98.6 |
| P0 | GACGCCAGTGCTGTATGC GCCCGCTAACCGCTATTTC |
200 | 54 | 92.2 |
| Positive control | ||||
| CoxI | CCAGTATTAGCAGCAGGTATC CGAAGAATCAGAATAGGTGTTG |
200 | 60 | 92.7 |
| Target genes | ||||
| β-Actin | GTCCACCTTCCAGCAGATG CTCAGTAACAGTCCGCCTAG |
200 | 60 | 96.9 |
| γ-Actin | GTATTAACCAACAGCAGACTTC GAACAGAAACTGGGTCCTAAG |
100 | 60 | 99.0 |
| βIII-Tubulin | CCGCCTGCCTCTTCGTCTC TAGTTGCCGCTGGGGTCTATG |
200 | 60 | 90.7 |
| Cdc42 | TGTCAAGTATGTGGAGTGTTC CTGGAGATGCGTTCATAGC |
100 | 56 | 97.5 |
| Cofilin1 | GCGGCTCTGTTCTTCTGTAG CCTTCTTGCGTTTCTTCACTTC |
200 | 53 | 94.5 |
| GAP43 | AGTGCCCGACAGGATGAG CAGGACAGGAGAGGAAACTTC |
100 | 56 | 92.4 |
| Integrin α6 | CAGGGACTTACAACTGGAAAGG CAGGAACGGGCACGAGAC |
200 | 60 | 98.2 |
| Integrin α7 | CGGAGACTTGACCTTGAATAGC GAATGACCACAGCCCCTTTG |
200 | 60 | 94.3 |
| Integrin β1 | TGCCTTGCTGCTGATTTGG ACTTCGGATTGACCACAGTTG |
400 | 56 | 102 |
| Integrin β4 | CTCCTTCTTGGTGGCTCATTC AGTCCCAGGCAGGCTTTG |
200 | 60 | 101 |
| L1 | CTGGCTGTGAAGACTAATGG CCCTTGCTGCGTTTGATG |
200 | 56 | 94.3 |
| MAP1A | GAGTCGGAGAAGAAAGGAGAG CACCAAACACAGCAGCATAC |
200 | 52 | 90.0 |
| MAP1B | CGGACAGTGCTTTGAGAAC GTGAGGCTGGCTTTGTTG |
200 | 52 | 88.7 |
| MAP2 | ATCACCGACCTGCTTGTTC TCCCAACTCTCCCTCATCAG |
200 | 56 | 98.0 |
| MyosinI | ACAGATGAGGAAGTGGATGAG TTTCGCTCCGTGCTTGAG |
200 | 56 | 90.6 |
|
N-cadherin (mouse primers) |
CGCCATCATCGCTATCCTTCTG CGCTCTTTATCCCGCCGTTTC |
100 | 58 | 109 |
| NF-L | CAGCGTGGGTAGCATAAC CGAGCAGACATCAAGTAGG |
200 | 54 | 90.0 |
| NF-M | GAGGAGGAGGAGCAGGAAAC ACCACCACTTTGTCATCACTTC |
200 | 60 | 95.6 |
| NF-H | TGACAGAAGAGGAGGACAAAG TTTCACAGGAGACTTGGTTTC |
200 | 53 | 93.9 |
| Peripherin | TGCCCGTTCATTCCTTTG ACTCTGTCACCACCTTCTC |
200 | 54 | 89.2 |
| Profilin1 | GGTTGGTGTCCTGGTAGG CATTGTAAATTCCCCGTCTTG |
100 | 56 | 95.4 |
| Rac1 | TCATCAGTTACACGACCAATG ACGCAGTCTGTCATAATCTTC |
200 | 53 | 98.7 |
| RhoA | CACACAAGGCGGGAGTTAG CGTCTTTGGTCTTTGCTGAAC |
200 | 55 | 101 |
| SCG10 | AGACTCAGTGCCTTACTCAG TTTAGCCATTGTAGGGATGTG |
200 | 53 | 93.6 |
| Tau (HMW) | GGCTACACACCAACCTTTG CCACCTCCTCCTCACTTC |
100 | 56 | 97.0 |
| TrkA | GATGCTGGTGGCTGTCAAG ACGATGTGTTGGTGCTGTAG |
100 | 54 | 105 |
| Vimentin | TTTCTCTGCCTCTTCCAAAC ATCACCTGTCCGTCTCTG |
100 | 60 | 89.3 |
Target gene qPCRs
In order to identify and quantify target genes in axon preparations, 5 μl of the axon-only cDNA preparations were subjected to real-time quantitative PCR. Each run also included the positive control cDNA (DRG 10−3 dilution) and a water control, and reactions were performed in triplicate. Minus RT controls were performed to ensure that there was no DNA contamination. Melt curves were performed in order to confirm that the product from the axon-only cDNA was the same as the product from the diluted DRG cDNA. First, P0 and DNA polymerase qPCRs were performed to show that the axons were not contaminated with any cellular mRNA. Second, the positive control cytochrome oxidase c subunit I (coxI) was run to check whether there were sufficient amounts of axons in the preparation. CoxI is a mitochondrially encoded gene, which will be used later to normalise axonal RNA levels with. β-Actin qPCRs were performed on all axon cDNAs of all developmental stages in order to quantify its levels. The other target gene qPCRs were performed on selected samples with high enough levels of β-actin (cycle threshold (Ct) <32). A target gene was regarded present in axons if (1) it was found in at least 3 consecutive axon preparations, (2) at least 2 of the 3 replicates were positive and reproducible, and (3) the average Ct value was lower than 34.
Quantifications
In axon preparations where the β-actin Ct values were reproducible and within the sensitivity range (as determined by the RNA dilution series described above) the levels of target mRNAs relative to coxI were calculated using the following equation: X0,target/X0,CoxI = E−Ct,target/E−Ct,CoxI in which X0=mRNA levels at cycle 0, E=efficiency, Ct=cycle threshold (Vogelaar et al., 2004). β-Actin was quantified in axon preparations from all developmental stages. In neonatal axons, the levels of β-actin, β-tubulin and GAP-43 mRNA were calculated and compared.
β-actin mRNA knock down
P0–P2 DRGs were dissected and cultured as described above. Axons growing into the outer compartment were incubated with 3 different sequences of rat β-actin siRNA (Stealth RNAi, Invitrogen) or a control siRNA not specific for any known mammalian sequence (Stealth RNAi Negative Control, Invitrogen). siRNAs were mixed with lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency in axons was assessed by transfecting axons with fluorescein-labeled double-stranded RNA oligomers (Block-it Fluorescent Oligos, Invitrogen). Gene knock down was assessed 24 h after transfection by means of qPCR. Actin levels relative to Cox1 were calculated using the equation stated above.
Immunofluorescence staining was used to assess β-actin protein knock down. Axons from dissociated or whole DRGs explants were fixed in PFA 4%, permeabilised in 3% Triton in PBS, and blocked in 5% NGS. Phalloidin was used to visualise β-actin. Anti-Neurofilament H was used as positive control and normaliser. Images were taken using a Leica microscope and a high resolution CCD camera (Hamamatsu). All images were taken at the same settings for light and exposure. Images were analysed by Image J. Regions along the axons were drawn using the software, and the average pixel intensity per unit area was calculated. The intensity of background was similarly measured in areas adjacent the axons and subtracted from the axon value. To control for axonal dimensions, intensity of β-actin was normalised to the intensity of neurofilament-H staining, calculated as for β-actin.
Regeneration assay
The regeneration assay was performed 24 h after transfection with siRNA. Axons grown beyond the silicone insert were axotomised with a fine needle under a phase-contrast microscope (Nikon). Cut axons were photographed at the time of axotomy, and then every 5 min during the first hour. Some cultures were observed for 1.5 h and again at 4 h after axotomy. Images were acquired using a CCD camera (Nikon) and the Nikon Act-1 software. During the recordings cultures were kept at 37 °C in L-15 medium supplemented with ITS+, NGF, insulin and PSF. The percentage of axons regenerating a new growth cone within 1 h was calculated.
Live visualisation of actin synthesis by using Kaede fluorescence recovery
CoralHue Kaede was amplified from pKaede-S1 (MBL, Japan; Accession no. AB085641) using the following primers: 5′tttttggatccgccaccatggtga3′ and 5′ttttttttgcggccgcttacttgacgttgtccggcaaca3′. The amplified fragment was digested by BamHI/NotI restriction enzymes and cloned into the BamHI/NotI sites of pcDNA 3.1+ Hygro expression vector (Invitrogen). The 3′UTR of rat β-actin (Accession no. V01217) was amplified from rat brain cDNA using the following primers: 5′ttttttctagatgcgcaagttaggttttgtc3′ and 5′ttttttctagaagccatgccaaatgtctcat3′. The amplified fragment was digested with NotI and XbaI restriction enzymes and cloned in the NotI/XbaI sites of pcDNA 3.1+ Hygro.
Dissociated adult DRG neurons were transfected with Kaede-β-actin 3′UTR using a microporator (Digital Bio Technology) and grown on μ-Dishes (Ibidi) coated with PDL and laminin. A regeneration assay was performed 24 h to 48 h after transfection on axons expressing the plasmid. The whole axon together with the neuronal soma was photoconverted by 5 second exposure to 405 nm light and then axotomised as described above. Occasionally, axons were separated from their cell body. Axotomised axons were observed at 100× with a Nikon inverted microscope. Images were captured with a Hamamatsu cooled CCD camera by exposure to 491 nm and 561 nm light. In order to measure fluorescence intensity regions were drawn along the axons using Metamorph software (Molecular Devices), and the average pixel intensity per unit area was calculated. The intensity of background was similarly measured in areas adjacent the axons and subtracted from the axon value. Recovery of fluorescence was expressed as a proportion of the first frame.
Acknowledgments
This work was supported by grants from Action Medical Research, the Medical Research Council, the Wellcome Trust, the Henry Smith Charity and The Christopher and Dana Reeve Foundation.
References
- Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. J. Neurosci. 2001;21:6577–6587. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Zhang HL, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley SM, Payne HR, Lemmon V. Growth cones are actively influenced by substrate-bound adhesion molecules. J. Neurosci. 1995;15:4370–4381. doi: 10.1523/JNEUROSCI.15-06-04370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chierzi S, Ratto GM, Verma P, Fawcett JW. The ability of axons to regenerate their growth cones depends on axonal type and age, and is regulated by calcium, cAMP and ERK. Eur. J. Neurosci. 2005;21:2051–2062. doi: 10.1111/j.1460-9568.2005.04066.x. [DOI] [PubMed] [Google Scholar]
- Chun JT, Gioio AE, Crispino M, Giuditta A, Kaplan BB. Differential compartmentalization of mRNAs in squid giant axon. J. Neurochem. 1996;67:1806–1812. doi: 10.1046/j.1471-4159.1996.67051806.x. [DOI] [PubMed] [Google Scholar]
- Condic ML, Letourneau PC. Ligand-induced changes in integrin expression regulate neuronal adhesion and neurite outgrowth. Nature. 1997;389:852–856. doi: 10.1038/39878. [DOI] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of beta-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J. Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KJ, Fan DP, Tsui BJ, Cassar SL, Tetzlaff W. Influence of the axotomy to cell body distance in rat rubrospinal and spinal motoneurons: differential regulation of GAP-43, tubulins, and neurofilament-M. J. Comp. Neurol. 1999;414:495–510. doi: 10.1002/(sici)1096-9861(19991129)414:4<495::aid-cne6>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Eyman M, Zhang HS, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J. Neurosci. Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Lavina ZS, Jurkovicova D, Zhang H, Eyman M, Giuditta A, Kaplan BB. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur. J. Neurosci. 2004;20:865–872. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Kaplan BB, van Minnen J, Alvarez J, Koenig E. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 2002;25:400–404. doi: 10.1016/s0166-2236(02)02188-4. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Soehrman S, Bondallaz P, Ruchti E, Cadas H. Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J. Neurobiol. 2004;58:60–69. doi: 10.1002/neu.10279. [DOI] [PubMed] [Google Scholar]
- Hamilton RS, Davis I. RNA localization signals: deciphering the message with bioinformatics. Semin. Cell. Dev. Biol. 2007;18:178–185. doi: 10.1016/j.semcdb.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J. Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hollenbeck PJ. Organization and translation of mRNA in sympathetic axons. J. Cell Sci. 2003;116:4467–4478. doi: 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- Leung KM, Holt CE. Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nat. Protoc. 2008;3:1318–1327. doi: 10.1038/nprot.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MR, Lieberman AR, Grenningloh G, Anderson PN. Transcriptional upregulation of SCG10 and CAP-23 is correlated with regeneration of the axons of peripheral and central neurons in vivo. Mol. Cell. Neurosci. 2002;20:595–615. doi: 10.1006/mcne.2002.1140. [DOI] [PubMed] [Google Scholar]
- Merianda TT, Lin A, Lam J, Vuppalanchi D, Willis DE, Karin N, Holt CE, Twiss JL. A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol. Cell. Neurosci. 2009;40:128–142. doi: 10.1016/j.mcn.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr E, Richter D. Axonal mRNAs: functional significance in vertebrates and invertebrates. J. Neurocytol. 2000;29:783–791. doi: 10.1023/a:1010987206526. [DOI] [PubMed] [Google Scholar]
- Nedelec S, Dubacq C, Trembleau A. Morphological and molecular features of the mammalian olfactory sensory neuron axons: what makes these axons so special? J. Neurocytol. 2005;34:49–64. doi: 10.1007/s11068-005-5047-7. [DOI] [PubMed] [Google Scholar]
- Oestreicher AB, de Graan PN, Gispen WH, Verhaagen J, Schrama LH. B-50, the growth associated protein-43: modulation of cell morphology and communication in the nervous system. Prog. Neurobiol. 1997;53:627–686. doi: 10.1016/s0301-0082(97)00043-9. [DOI] [PubMed] [Google Scholar]
- Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell. Biol. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies MA, Steward O. Multiple subcellular mRNA distribution patterns in neurons: a nonisotopic in situ hybridization analysis. J. Neurobiol. 1997;33:473–493. doi: 10.1002/(sici)1097-4695(199710)33:4<473::aid-neu10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Perlson E, Michaelevski I, Kowalsman N, Ben-Yaakov K, Shaked M, Seger R, Eisenstein M, Fainzilber M. Vimentin binding to phosphorylated Erk sterically hinders enzymatic dephosphorylation of the kinase. J. Mol. Biol. 2006;364:938–944. doi: 10.1016/j.jmb.2006.09.056. [DOI] [PubMed] [Google Scholar]
- Piper M, Holt C. RNA translation in axons. Annu. Rev. Cell Dev. Biol. 2004;20:505–523. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van HF, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J. Neurobiol. 2004;61:222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Kun A, Koenig E, Sotelo JR. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Bollati F, Bisbal M, Sheridan S, Faas L, Wray R, Haferkamp S, Nguyen S, Caceres A, Brady ST. Activity-driven dendritic remodeling requires microtubule-associated protein 1A. Curr. Biol. 2005;15:1820–1826. doi: 10.1016/j.cub.2005.08.069. [DOI] [PubMed] [Google Scholar]
- Takei Y, Teng JL, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J. Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Rhee SW, Jeon NL. Microfluidic chambers for cell migration and neuroscience research. Methods Mol. Biol. 2006;321:167–177. doi: 10.1385/1-59259-997-4:167. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss JL, van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J. Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- van Kesteren RE, Carter C, Dissel HMG, van Minnen J, Gouwenberg Y, Syed NI, Spencer GE, Smit AB. Local synthesis of actin-binding protein beta-thymosin regulates neurite outgrowth. J. Neurosci. 2006;26:152–157. doi: 10.1523/JNEUROSCI.4164-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanMinnen J, Bergman JJ, VanKesteren ER, Smit AB, Geraerts WPM, Lukowiak K, Hasan SU, Syed NI. De novo protein synthesis in isolated axons of identified neurons. Neuroscience. 1997;80:1–7. doi: 10.1016/s0306-4522(97)00137-1. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory-bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J. Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar CF, Fawcett JW. Axonal mRNA in regeneration. In: Müller HW, editor. Neural Degeneration and Repair. Gene Expression Profiling, Proteomics, and Systems Biology. WILEY-VCH Verlag GmbH and co.; Weinheim: 2008a. pp. 135–151. [Google Scholar]
- Vogelaar CM, Fawcett JW. Axonal mRNA in regeneration. In: Muller HW, editor. Neural Degeneration and Repair. Wiley; 2008b. [Google Scholar]
- Vogelaar CF, Hoekman MF, Brakkee JH, Bogerd J, Burbach JP. Developmental regulation of homeobox gene expression in dorsal root ganglion neurons is not recapitulated during regeneration of the crushed sciatic nerve. Neuroscience. 2004;125:645–650. doi: 10.1016/j.neuroscience.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Zorn AM, Krieg PA, Bittner GD. Medium weight neurofilament mRNA in goldfish Mauthner axoplasm. Neurosci. Lett. 1996;213:83–86. doi: 10.1016/0304-3940(96)12860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensley CH, Stone DM, Baker H, Kauer JS, Margolis FL, Chikaraishi DM. Olfactory marker protein messenger-RNA is found in axons of olfactory receptor neurons. J. Neurosci. 1995;15:4827–4837. doi: 10.1523/JNEUROSCI.15-07-04827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, Staple J, Caroni P. The motility-associated proteins GAP-43, MARCKS, and CAP-23 share unique targeting and surface activity-inducing properties. Exp. Cell Res. 1997;236:103–116. doi: 10.1006/excr.1997.3709. [DOI] [PubMed] [Google Scholar]
- Willis DE, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J. Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CWK, Zeng FY, Eberwine J. mRNA transport to and translation in neuronal dendrites. Anal. Bioanal. Chem. 2007;387:59–62. doi: 10.1007/s00216-006-0916-1. [DOI] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss JL, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang BS, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J. Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]