Supplemental Digital Content is available in the text.
Keywords: critically ill, culture-independent, early diagnosis, infection, microbiology, molecular detection
Objective:
Early identification of causative microorganism(s) in patients with severe infection is crucial to optimize antimicrobial use and patient survival. However, current culture-based pathogen identification is slow and unreliable such that broad-spectrum antibiotics are often used to insure coverage of all potential organisms, carrying risks of overtreatment, toxicity, and selection of multidrug-resistant bacteria. We compared the results obtained using a novel, culture-independent polymerase chain reaction/electrospray ionization-mass spectrometry technology with those obtained by standard microbiological testing and evaluated the potential clinical implications of this technique.
Design:
Observational study.
Setting:
Nine ICUs in six European countries.
Patients:
Patients admitted between October 2013 and June 2014 with suspected or proven bloodstream infection, pneumonia, or sterile fluid and tissue infection were considered for inclusion.
Interventions:
None.
Measurements and Main Results:
We tested 616 bloodstream infection, 185 pneumonia, and 110 sterile fluid and tissue specimens from 529 patients. From the 616 bloodstream infection samples, polymerase chain reaction/electrospray ionization-mass spectrometry identified a pathogen in 228 cases (37%) and culture in just 68 (11%). Culture was positive and polymerase chain reaction/electrospray ionization-mass spectrometry negative in 13 cases, and both were negative in 384 cases, giving polymerase chain reaction/electrospray ionization-mass spectrometry a sensitivity of 81%, specificity of 69%, and negative predictive value of 97% at 6 hours from sample acquisition. The distribution of organisms was similar with both techniques. Similar observations were made for pneumonia and sterile fluid and tissue specimens. Independent clinical analysis of results suggested that polymerase chain reaction/electrospray ionization-mass spectrometry technology could potentially have resulted in altered treatment in up to 57% of patients.
Conclusions:
Polymerase chain reaction/electrospray ionization-mass spectrometry provides rapid pathogen identification in critically ill patients. The ability to rule out infection within 6 hours has potential clinical and economic benefits.
The availability of rapid and reliable infectious disease diagnostics that can provide results directly from patient specimens represents a major unmet need in managing critically ill patients. Current sepsis guidelines recommend initiation of IV antibiotic therapy as early as possible, ideally within the first hour (1), as any delay in effective antimicrobial therapy may result in decreased survival (2). Effective therapy requires that the identity of causative pathogens and their resistance patterns are known. However, the current standard-of-care, which depends on blood culture-based initial diagnosis, often takes at least 48–72 hours to provide a result. Furthermore, cultures often remain negative even when bacterial or fungal infections are strongly suspected (3), in part, related to concurrent antibiotic treatment (4).
Molecular diagnostic techniques that do not depend on growth of organisms in culture may offer a distinct advantage over current methods. Most of the recently described molecular methods, however, rely on culture amplification as a precursor to diagnosis (5–8). Although these techniques may accelerate diagnosis for positive cultures, they do not address the significant proportion of false-negative cultures observed in patients with sepsis. In addition, many of these methods also use targeted pathogen detection with limited pathogen coverage such that negative results are often not highly predictive.
Polymerase chain reaction followed by electrospray ionization-mass spectrometry (PCR/ESI-MS) can detect more than 800 bloodstream infection (BSI)-relevant pathogens in a single assay and in approximately 6 hours (9–13). It can also identify three classes of antibiotic resistance markers associated with resistance to methicillin (mecA), vancomycin (vanA/vanB), and carbapenems (KPC). Using this technique, we recently demonstrated 83% sensitivity and 94% specificity compared with culture for direct detection of pathogens in whole-blood specimens from patients with suspected BSIs (13).
Here, we describe findings from the multicenter observational Rapid Diagnosis of Infections in the Critically Ill (RADICAL) study. The primary objective was to compare results obtained using the novel culture-independent PCR/ESI-MS technology with those obtained from standard microbiological testing as a measure of clinical performance. Secondarily, to broadly address the clinical value of PCR/ESI-MS detections, a panel of independent clinical adjudicators was used to identify changes in patient management that may have occurred had the results from the PCR/ESI-MS technology been available for clinical use and assumed to be correct.
PATIENTS AND METHODS
All adult patients (≥ 18 yr) admitted between October 2013 and June 2014 to one of nine ICUs in six European countries for the management of suspected or proven sepsis or severe infection were considered for inclusion in this prospective study. Ethical approval was obtained in each participating center. Informed consent was obtained from the patient or patient’s legal representative.
Patient Inclusion and Exclusion Criteria
Patients were considered for inclusion if they had 1) suspected or proven severe infection or sepsis and or 2) suspected or proven healthcare-associated pneumonia (HAP/HCAP), ventilator-associated pneumonia (VAP), or severe community-acquired pneumonia (sCAP). Because pneumonia is the most common precipitating cause of sepsis, there may be an overlap between these two populations, but patients were included in one of the two groups, not both. Pneumonia (HAP/HCAP, VAP, and sCAP) was diagnosed in patients with an endotracheal tube in situ and a new infiltrate on chest radiograph plus temperature more than 38°C or less than 35°C, increased sputum production, increased or decreased WBC count (> 12 or < 4 cells/mL3), or a clinical suspicion of pneumonia, and the treating clinician expected the patient to still be intubated the next day.
The following exclusion criteria were used: the treating clinician expected the patient to be discharged from the ICU on the day of evaluation or the following day, the treatment intent was palliative, the clinician was not committed to aggressive treatment, or death was deemed imminent and inevitable. Patients who had previously been included, but were readmitted to the ICU during the same hospitalization, were not included a second time.
Specimen Collection and Processing
Blood samples were collected whenever physicians ordered blood cultures because of clinical suspicion of a BSI, pneumonia, or secondary site infection at a normally sterile site. Standard-of-care microbiology testing was performed according to usual practice in each institution. For testing using PCR/ESI-MS, a minimum of 5-mL EDTA whole blood was drawn from the same venipuncture as for standard-of-care microbiology testing. An extra 5 mL was collected from an additional site whenever applicable. Lower respiratory tract (LRT) specimens from bronchoalveolar lavage (BAL) or endotracheal aspirate (ETA) were collected per standard clinical protocol, and aliquots used for PCR/ESI-MS testing. For sterile fluids and tissues (SF&T), specimens tested included cerebrospinal (CSF), pleural, peritoneal, ascitic, or synovial fluid and surgical tissue/biopsies. Urine samples were not considered. A minimum of 0.5-mL fluid specimen or a minimum of 35-mg solid specimen was taken.
Specimens were cooled to 4°C within 30 minutes of collection and maintained at 4°C or frozen at –20°C until analysis. Genomic DNA was extracted from 5 mL of the EDTA-treated whole-blood clinical specimens as described previously (13). For other specimens (BAL, ETA, and other normally SF&T), total volume was brought up to 5 mL with buffered saline and processed as above. Eluates from the extraction were transferred into 16 wells (30 μL/well) of a custom polymerase chain reaction (PCR) assay strip prefilled (25 μL/well) with 18 unique primer pairs and concentrated PCR master mix. The gene targets, primer sequences, and configuration have been described in detail previously (14). General PCR formulations and thermocycling conditions have also been described elsewhere (10).
Clinical Data Collection
Orion Clinical Services (Berkshire, United Kingdom), a clinical research organization, provided electronic case report forms for collecting clinical data from enrolled patients. All pertinent patient data were de-identified and securely stored to maintain anonymity and confidentiality.
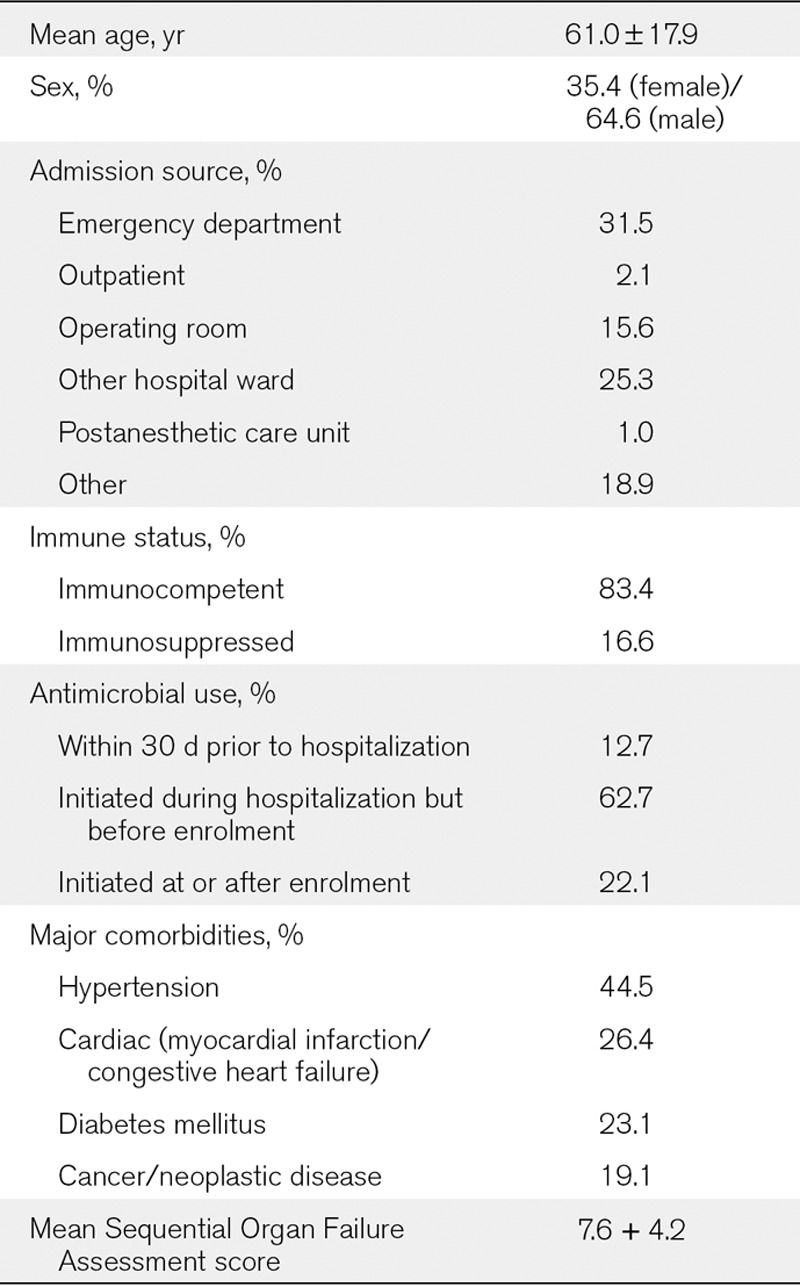
The following baseline data were collected upon enrolment: patient demographics (e.g., age and sex), date and time of hospital and ICU admission, admission source (e.g., emergency department, outpatient clinic/referral, operating room, postanesthesia care unit, and other hospital unit), major comorbid conditions, immune status (immunosuppressed/immunocompetent), site of suspected or confirmed infection, antimicrobial course prior to study enrolment, surgery/procedures for suspected site of infection prior to enrolment, and Sequential Organ Failure Assessment Score (15).
Clinical data collected during admission included pertinent laboratory data, use of mechanical ventilation, and antimicrobial/antibiotic therapy including duration of therapy, and date therapy was initiated and discontinued. Vasoactive therapy, renal replacement therapy, surgical and other procedures for diagnosis/treatment of infection, radiological testing for diagnosis/evaluation of potential infection, indwelling vascular access devices, and vital status at 28 days were also recorded. Discharge data included date of discharge (ICU and hospital), discharge destination (general hospital floor, skilled nursing facility, and home), and vital status at discharge (survival/death).
Specimen Data Collection
The following specimen-related data were recorded: specimen type (whole blood, body fluid, tissue, BAL, and ETA); date and time of collection for each specimen taken for use with PCR/ESI-MS; total amount of fluid collected; sample handling and storage; standard-of-care microbiological test results for specimens taken concurrently; time to obtain positive results (and or determination of negative results) for standard-of-care microbiological testing; data for secondary objectives and to adjudicate potential discrepant results; recent microbiological testing within prior 30 days (if available); and additional microbiological testing following enrolment in study until hospital discharge.
Sample Size
This exploratory study was not statistically powered. We estimated that approximately 500 patients would have to be enrolled to enable a reasonable assessment of the performance of the PCR/ESI-MS suite of assays when compared with standard-of-care microbiology.
Data Analysis
Results obtained with the PCR/ESI-MS technology for each specimen were compared with those obtained using conventional microbiology methods for the same sample. If multiple specimens were taken from a patient per standard-of-care protocols, each was independently analyzed in this study. Agreement and concordance were assessed using a McNemar test (16) and Cohen κ (17). All percentages and CIs for proportions were calculated using the exact method and are rounded to the nearest percentage. Direct comparison of positive and negative results was conducted with organism identification for each method (conventional microbiology vs PCR/ESI-MS) for each specimen type. Coagulase-negative staphylococcus and other common skin contaminants were annotated as “potential contaminants” for both methods and excluded from the overall analysis, as previously described (13).
Discrepant results between the PCR/ESI-MS and culture cannot be directly confirmed by an independent method, as previously described (13). Two approaches were used to resolve discrepancies. In a subset of patients, multiple samples were collected per standard-of-care. This included two independent fresh venipunctures (left arm vs right arm) or one venipuncture plus one sample collected from an indwelling line. Paired analysis of PCR/ESI-MS testing results between these independently collected samples was conducted to indicate the likelihood of true infection. In addition, independent clinical adjudication (described below) was performed using all the clinical data collected as part of the study, including standard-of-care microbiology results and PCR/ESI-MS results.
All statistical tests were performed using SAS version 9.3 (SAS Institute, Cary, NC) or Minitab16 (Minitab, State College, PA). A p value of less than 0.05 was considered statistically significant.
RESULTS
Of 543 patients enrolled in the study, 14 did not have matching PCR/ESI-MS or standard-of-care microbiology results and were excluded from the final analysis. Table 1 shows the patient demographics, reflecting a typically heterogeneous ICU population: one third of the patients were admitted from the emergency department; 75% were exposed to one or more antibiotics prior to study enrolment. Overall mortality was 29%, with cardiac arrest, septic shock, multiple organ failure, and acute respiratory distress syndrome accounting for ~62% of deaths.
TABLE 1.
Patient Demographics and Baseline Clinical Characteristics
BSI Analysis
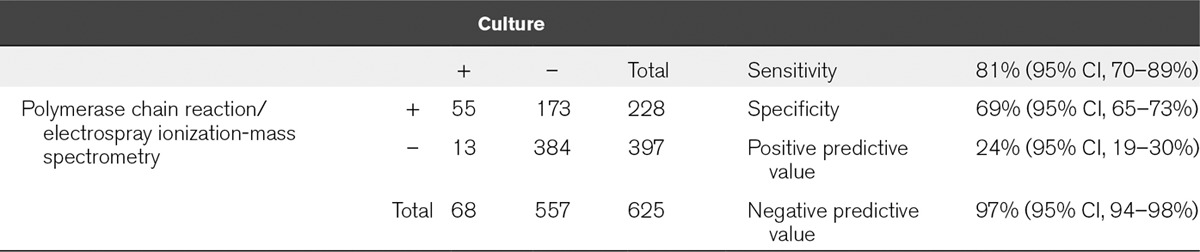
A total of 616 direct whole-blood specimens from the 529 patients were tested to assess the accuracy of organism identification. PCR/ESI-MS results from analysis of blood using the bacteria, antibiotic resistance, and Candida BSI assay were compared with results from standard clinical microbiology cultures. As shown in Table 2, there were 228 PCR/ESI-MS positive specimens (36.5%) for at least one pathogen compared with 68 positive specimens by culture (10.9%). The total number of positive tests for each method was statistically different (McNemar test statistic = 137.6; df = 1; p < 0.0001). There were 55 samples that were positive for the same organism with both techniques (Table 2), yielding an overall concordance of identification (calculated sensitivity) of 81% (95% CI, 70–89%) and a κ value of 0.25 (95% CI, 0.18–0.31). In 13 instances, culture identified an organism that was either negative by PCR/ESI-MS (6/13) or the identity of the organism reported by PCR/ESI-MS did not match the organism identified by microbiology testing (7/13) (Table S1, Supplemental Digital Content 1, http://links.lww.com/CCM/B418). In contrast, PCR/ESI-MS reported a BSI-relevant organism in 173 additional specimens that were culture negative, resulting in a calculated assay specificity of 69% (Table S2, Supplemental Digital Content 1, http://links.lww.com/CCM/B418). Finally, there were 384 concordant negative specimens, yielding a negative predictive value (NPV) of ~97% (95% CI, 94–98%).
TABLE 2.
Bloodstream Infection Assay Performance
The frequencies of organisms detected from BSI specimens are shown in Figure 1. The distributions of the top 10 species detected by microbiology and those detected by PCR/ESI-MS were similar. The largest single discrepancy between the two methods by sheer volume of detections was in the identification of Escherichia coli. Although culture and PCR/ESI-MS techniques both reported E. coli as the most abundant species, PCR/ESI-MS detection was 4-fold higher (89 vs 21). Other organisms in which blood culture performed less well included the enterococcus species, Enterococcus faecalis (1 vs 10) and Enterococcus faecium (2 vs 25), Candida albicans (2 vs 13), and Staphylococcus aureus (14 vs 31). In contrast, Pseudomonas aeruginosa detection was comparable between the two methods (6 vs 8). Additional analysis of the PCR/ESI-MS results showed that the levels (genome equivalent/mL) of organisms reported in the majority of these PCR/ESI-MS positive, but culture-negative, cases were similar to cases in which culture matched PCR/ESI-MS detections (data not shown).
Figure 1.
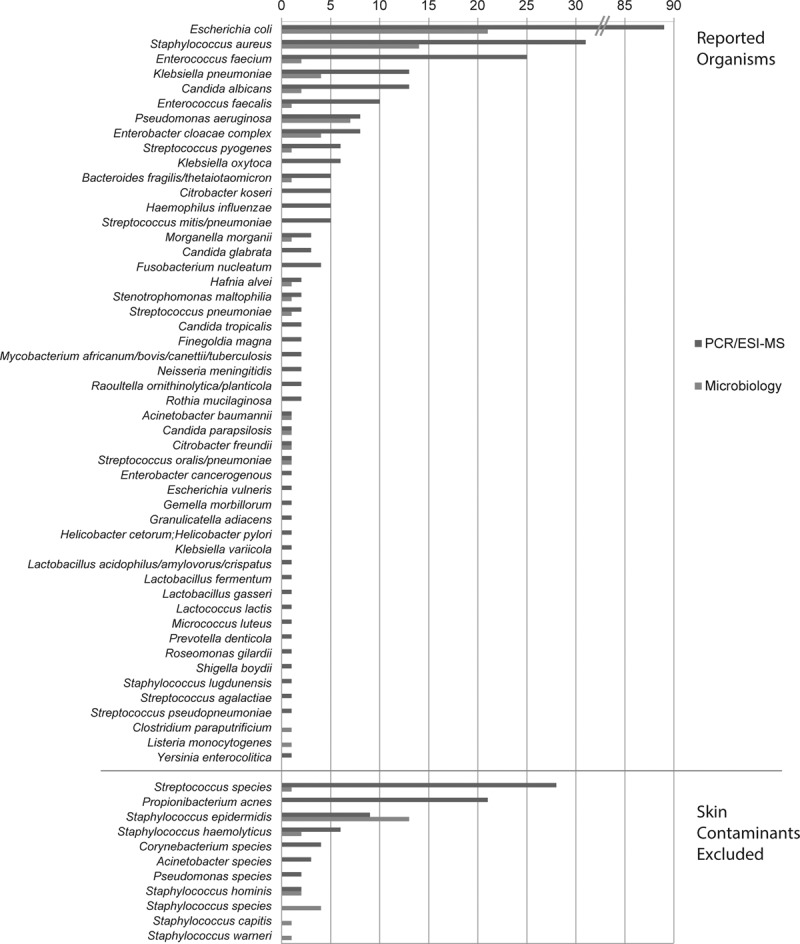
Bacteria and Candida detected in the Rapid Diagnosis of Infections in the Critically Ill (RADICAL) study. Distribution of organisms reported by polymerase chain reaction/electrospray ionization-mass spectrometry (PCR/ESI-MS) (blue bar) and culture (red bar) observed in the RADICAL study are shown, sorted by decreasing order of PCR/ESI-MS reported organisms. Both methods showed similar distribution for the top eight reportable organisms that were seen >5 times by PCR/ESI-MS, with some minor reshuffling of the order. PCR/ESI-MS showed a longer tail of reportable organisms that were infrequent (≤5 times). Normal skin flora are shown below the line were not included in further analysis by either method.
When subsets of specimens were tested in replicate using independently collected samples, 52 of 168 replicate samples were positive in both PCR/ESI-MS tests (Table S3, Supplemental Digital Content 1, http://links.lww.com/CCM/B418). In contrast, culture was positive in the two replicates in only 12 of 158 samples, all of which were also PCR/ESI-MS positive in both samples. In three instances, culture was positive in one of two replicates and was negative by PCR in both samples tested (false negative by PCR), whereas in three and four cases, respectively, when culture was positive in one replicate, PCR was positive in one or both replicates (Table S4, Supplemental Digital Content 1, http://links.lww.com/CCM/B418).
Nonbloodstream Infections
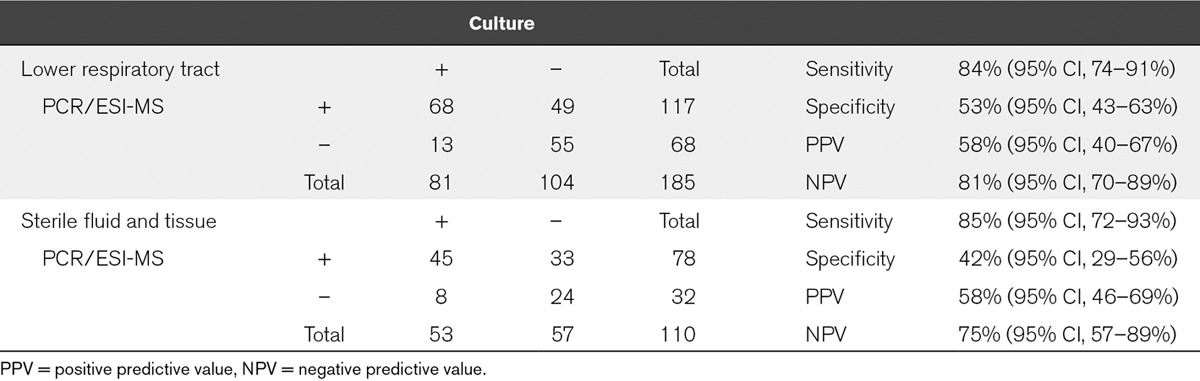
Heterogeneous samples from patients with suspected pneumonia or sterile site infections were also obtained in several cases. Overall, there were 185 LRT samples (88 BAL, 96 ETA, and 1 other) and 110 SF&T samples (36 intraperitoneal fluid, 14 pleural fluid, 11 CSF, 13 tissue, and 36 other fluid types). Results from the analysis of these specimens are shown in Table 3. LRT and SF&T specimens often had multiple detections reported by both methods in several samples. Only the primary detections by either method were included in the analysis. The overall sensitivities for concordance between standard-of-care and PCR/ESI-MS were 84% (95% CI, 74–91%) and 85% (95% CI, 72–93%), respectively. As for the bloodstream infection data, the McNemar test for both the LRT and the SF&T sample data showed that the total number of samples considered positive was significantly different for culture versus PCR/ESI-MS (McNemar test statistic = 20.9 for LRT and 15.2 for SF&T; p < 0.0001 in both cases). Also similar to the bloodstream infection data, there was more agreement in the contingency table comparing culture to PCR/ESI-MS than would be expected by chance (LRT κ = 0.35; 95% confidence limits, 0.23, 0.47 and SF&T κ = 0.27; 95% confidence limits, 0.11, 0.43). For LRT specimens, there was no statistically significant difference in sensitivity (p = 0.677) or specificity (p = 0.444) when testing the hypothesis that the BAL proportion – the ETA proportion was equal to zero.
TABLE 3.
Assay Performance in Samples From Lower Respiratory Tract and Sterile Fluid and Tissue Infections
In 151 patients, two or more specimen types (BSI plus LRT and/or SF&T) were obtained and analyzed. In 86 of these 151 cases (57%), the same organisms were reported by PCR/ESI-MS in all samples tested from an individual patient (data not shown). In comparison, culture concordance between the sample types was seen in only 19 cases (12%), driven largely by no detection reported in the BSI culture results.
Resistance Markers
There were no identified cases of Klebsiella-associated carbapenemase. There was a single report of vancomycin-resistant Enterococci, which was matched across the two methods. There were 23 reports of mecA+ staphylococcus organisms (seven in BSI samples, 13 in LRT samples, and three in SF&T samples), with the following agreement between PCR/ESI-MS and culture: for BSI samples, results were concordant in four cases, and PCR/ESI-MS was positive and culture negative in three; for LRT samples, results were concordant in three cases, PCR/ESI-MS was positive with culture negative in nine, and PCR/ESI-MS was negative with culture positive in one; and the three cases in the SF&T samples were concordant across PCR/ESI-MS and culture.
Clinical Value Assessment
To assess the clinical significance of the PCR/ESI-MS detections, each case was adjudicated by a panel of three independent clinical expert reviewers randomly selected from a pool of seven infectious disease, microbiology, and intensive care specialists not associated with the study sites. The panel was provided with clinical case summaries, and PCR/ESI-MS results and standard-of-care results from all samples tested. To identify potential changes in antimicrobial management that may have occurred if the results from the PCR/ESI-MS technology had been available for clinical use, the panel was provided with a questionnaire (Table 4). Analysis of the reviewers’ independent responses was performed using a majority rule such that two of three responses for a given patient determined the outcome for that patient. Table 4 provides a breakdown of the responses by each question. Combining all responses per patient and eliminating overlapping answers so that only a single response was recorded per patient gave an overall recommendation for change based on PCR/ESI-MS compared with standard-of-care in 178 patients (41%). This percentage increased to 57% for cases in which PCR/ESI-MS results were positive and microbiology results were negative.
TABLE 4.
Clinical Value Adjudication by Independent Reviewers

DISCUSSION
The important findings of the RADICAL study are that PCR/ESI-MS detected BSI pathogens with high overall sensitivity and NPV; PCR/ESI-MS was three times more likely to identify an organism than standard culture; and, if available, PCR/ESI-MS results may have altered the treatment regimen in as many as 57% of patients.
Sepsis affects a large proportion of the critically ill population. Despite improvements in recent years, morbidity and mortality rates remain high (18). The importance of initiating treatment as soon as possible has been highlighted and shown to be associated with improved outcomes (2), yet this finding needs to be balanced against the direct risks and stewardship issues arising from overzealous or inappropriate antibiotic use.
Rapid diagnosis of severe infection or sepsis is thus crucial not only to optimize a patient’s chances of survival but also to encourage responsible antibiotic use. However, diagnosing infection accurately in critically ill patients is challenging. Characteristic clinical and laboratory signs of severe infection, such as tachycardia, fever, and altered WBC count, are nonspecific and are often present in other acute conditions. Biomarkers, such as C-reactive protein and procalcitonin, are also nonspecific and are of more value in ruling out infection than in making a definite diagnosis (19). Microbiological culture results are negative in many patients with sepsis, largely because prior antimicrobial therapy affects ex vivo growth in culture medium (1). Certain microorganisms are also particularly difficult to culture, requiring specific growth media or a particular environment. As culture results often require several days to become available, patients with suspected severe infection are, therefore, often started on empiric broad-spectrum antibiotics to increase the likelihood that a pathogenic organism will be adequately covered. This approach, although valid in terms of preventing delays in starting treatment with currently available diagnostic techniques, has several negative aspects, including the potential for toxicity with multiple antibiotics, the high-associated costs, and the effects of antibiotic pressure on the development of antimicrobial resistance (20).
Availability of a technique that could provide more rapid pathogen identification directly from patient samples could, therefore, represent a marked improvement in terms of enabling more rapid diagnosis and earlier initiation of appropriate antimicrobial therapy, with associated beneficial effects on outcomes, antimicrobial resistance, costs, and toxicity. Various methods have been suggested for this purpose, including single pathogen assays, which are of limited use in patients with suspected sepsis in whom multiple organisms may be involved; selected-pathogen assays, which use specific molecular targets to identify some 20–35 species (21–23); and broad-range pathogen assays, which use universal or conserved targets to identify many hundreds of species, but for which earlier versions lacked sensitivity due to the small volumes of blood extracted for analysis (24, 25).
The PCR/ESI-MS test used in RADICAL can detect more than 800 bacterial and candida species from a single 5-mL EDTA blood sample, with results available within 6 hours from when the sample enters the testing laboratory. This is much faster than standard culture techniques that frequently take 48–96 hours for full identification with susceptibility profile. As expected, the PCR/ESI-MS assay identified significantly more positive samples than standard culture, possibly because it does not rely on bacterial growth and thus can identify organisms even in the presence of ongoing antimicrobial therapy. In the present study, over 75% of the patients were exposed to antibiotic treatment prior to enrolment (Table 1). This may explain in part the significant lack of microbiological growth in a large number of the PCR/ESI-MS positive cases. The simple κ statistic was used to estimate the agreement between culture and PCR/ESI-MS, but it underrepresents this agreement because of the requirement that the PCR/ESI-MS assay and blood culture results matched in terms of organism identity in order to be considered a true positive in the contingency table. The data were simplified and samples categorized as either an organism match or no match; hence, all organism agreement can be considered as being unlikely to have happened by chance. Furthermore, the paucity (< 1–10) of microbial colony-forming units/mL of blood, as reflected by higher culture yields through taking large volumes of blood (26–28), could be expected to significantly increase the risk of a false-negative result with a 20-mL blood sample. Despite this, the PCR/ESI-MS and culture methods both showed similar agreement on replicate testing using the κ statistic.
Importantly, in 41% of cases, the panel of independent experts would have recommended a change in management, including initiation of therapy, altered antimicrobial spectrum, and/or change in duration of therapy, based on the PCR/ESI-MS results. This percentage increased to 57 when PCR/ESI-MS tests were positive.
This study has several limitations. First, PCR/ESI-MS cannot provide detailed antimicrobial susceptibility information, unlike culture techniques. Hence, although it can provide sensitive and rapid identification of causative microorganisms, PCR/ESI-MS cannot currently replace culture methods. The results reported here need to be confirmed in studies that can directly determine the impact of this approach on clinical and economic outcomes, including length of stay and survival, but also on resistance patterns.
Second, the greater detection rate of E. coli, S. aureus, E. faecium, C. albicans, and Klebsiella pneumoniae by PCR/ESI-MS compared with routine culture was unanticipated, and the explanation is unclear. Prior to study inclusion, most patients were exposed to combinations of two or more antibiotics active against Gram-positive and Gram-negative organisms and were often receiving one or more antifungals in addition. As stated above, the bacterium/fungus may have been largely cleared with preexisting antibiotics, hence the negative culture results, but remaining DNA remnants in the circulation may have been sufficient to give a positive PCR/ESI-MS. The sensitivity of the technique increases the risk of identifying contaminants and commensals; however, the pathogens most frequently detected in the study are those associated with infection. Accepting the validity of these data, the PCR/ESI-MS test could be of importance to help target antimicrobial therapy in patients who have already started antimicrobials and have negative cultures (salvage microbiology) (29). Further, ideally interventional, studies are warranted to confirm and further explore these findings.
ACKNOWLEDGMENTS
We acknowledge the help and assistance of the following members of the RADICAL team:
Clinical Co-Investigators:
Agata Adamczyk, Child Christ Hospital, Warsaw, Poland.
Karolina Daniel, Child Christ Hospital, Warsaw, Poland.
Niall Samy MacCallum, University College London Hospitals, London, and supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Centre, United Kingdom.
Dominque Durand, Hospital Erasme, Brussels, Belgium.
Christophe Lelubre, Hospital Erasme, Brussels, Belgium.
Stephane Merat, Military Hospital Begin, Paris, France.
Thierry Vieilleville, Military Hospital Begin, Paris, France.
Vincent Peigne, Military Hospital Percy, Paris, France.
Jérôme Pugin, University Hospital Geneva, Geneva, Switzerland.
Valerie Noquet Boyer, University Hospital Geneva, Geneva, Switzerland.
Jessica Raic, University Hospital Frankfurt, Frankfurt, Germany.
Patrick Meybohm, University Hospital Frankfurt, Frankfurt, Germany.
Polymerase chain reaction/electrospray ionization-mass spectrometry and Microbiology Laboratory Testing:
Ronan Murphy, The Royal London Hospital, London, United Kingdom.
Michael Ronald Millar, The Royal London Hospital, London, United Kingdom.
Severine Mercier Delarue, Hospital Saint-Louis, Paris, France.
Marine Minier, Hospital Saint-Louis, Paris, France.
Gesuele Renzi, University Hospital Geneva, Geneva, Switzerland.
Independent Adjudicators:
Peter E. Spronck, Gelre Hospital, Apeldoorn, The Netherlands.
Mario Luchetti, A. Manzoni General Hospital, Lecco, Italy.
Guiseppe Nattino, A. Manzoni General Hospital, Lecco, Italy.
Manu Shankar Hari, Guy’s and St. Thomas Hospital, London, United Kingdom.
Veerappan Chithambaran, Royal Oldham Hospital, Manchester, United Kingdom.
Federicia Franco, Barnet Hospital, Barnet, United Kingdom.
Vincenc Jaume Mestre Saura, Hospital de Sabadell, Sabadell, Spain.
Clinical Research Organization:
Steve Hietschold, Orion Clinical Services, Slough, United Kingdom (Program Manager).
Valerie Kite, Orion Clinical Services, Slough, United Kingdom (electronic case report forms [eCRF]/database/data analysis).
Miranda Goodchild, Orion Clinical Services, Slough, United Kingdom (eCRF/database/data analysis).
Nacer Arbaoui, Orion Clinical Services, Slough, United Kingdom (clinical monitor).
Joanna Pellegrini, Orion Clinical Services, Slough, United Kingdom (clinical monitor).
Thijs Schreuder, Orion Clinical Services, Slough, United Kingdom (clinical monitor).
Abbott Europe
Doris Krams (study operations).
Martina Rost (application support).
Steffen Koch (service engineer).
Guillaume Vivies (service engineer).
Dr. D. Brealey and Prof. M. Singer are supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Supplementary Material
Footnotes
See also p. 2504.
Rapid Diagnosis of Infections in the Critically Ill Team members are listed in the acknowledgment.
Drs. Picard-Maureau, Chalfin, Ecker, and Sampath designed the study. Drs. Simon, Schrenzel, and Wilks tested all samples using polymerase chain reaction/electrospray ionization-mass spectrometry. Drs. Sampath and Brealey performed the data analysis. Dr. Vincent wrote the draft article. Drs. Brealey, Libert, Abidi, O’Dwyer, Zacharowski, Mikaszewska-Sokolewicz, Schrenzel, Simon, Wilks, Picard-Maureau, Chalfin, Ecker, Sampath, and Singer provided critical revision.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccmjournal).
Supported, in part, by Ibis Biosciences, Abbott. The funder of the study designed the protocol in collaboration with participating investigators, provided the polymerase chain reaction/electrospray ionization-mass spectrometry devices, and had overall responsibility for the conduct of the study.
Dr. Vincent’s institution received financial support from Abbott. Dr. Brealey received financial support from Abbott to conduct and present this study at international meetings (travel expenses and lecture fees) and disclosed off-label product use (now Conformité Européene [CE] marked for use in Europe alone). His institution received financial support. Dr. Libert’s institution received financial support from Abbott (received support for travel to meetings for the study). Dr. Abidi disclosed work for hire. Dr. O’Dwyer’s institution received grant support from Ibis Biosciences. Dr. Zacharowski received lecture honoraria from Abbott in the past 3 years and disclosed off-label product use (machine from Abbott). His institution received grant support from Abbott for a clinical trial (Frankfurt was study center and received case money for enrollment of patients. No other payment was received). Dr. Mikaszewska-Sokolewicz disclosed off-label product use (three research assays and three classes of matrices). Her institution received financial support (plane ticked and hotel costs covered for study initiation meeting; hospital was to be paid for study assignment). Dr. Schrenzel received grants from Abbott during the course of the study. Dr. Schrenzel’s institution received grant support. Dr. Simon served as a board member for Beckman Coulter and received grants and personal fees from Ibis Abbott during the conduct of the study. He and his institution consulted for Abbott, bioMérieux, bioRad, and GlaxoSmithKline. He received grant support, support for travel, and support for participation in review activities. Dr. Wilks received support (Abbott paid for installation and running of PlexID platform), disclosed off-label product use (Abbott PlexID now Iridica is CE marked but not the Food and Drug Administration), and received support for article research from Abbott. His institution received grant and funding support from Abbott. His institution received refund of traveling expenses (never claimed, dating back for a couple of years). Dr. Picard-Maureau disclosed off-label product use (product now for diagnostic use in the European Union [CE-In-vitro Diagnostics]) and received funding from Abbott (full-time employee of Abbott and worked on that project as part of job). Dr. Chalfin received funding (employee of Abbott and also received stock grants as part of employment). Dr. Ecker received funding (employee of Abbott that manufactures the technology). Dr. Sampath disclosed relationships (full-time employee of Abbott). Dr. Singer received grant support and funding from Abbott.
REFERENCES
- 1.Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign Guidelines Committee including The Pediatric Subgroup. Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 3.Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents. 2007;30(Suppl 1):S7–15. doi: 10.1016/j.ijantimicag.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Lamy B, Roy P, Carret G, et al. What is the relevance of obtaining multiple blood samples for culture? A comprehensive model to optimize the strategy for diagnosing bacteremia. Clin Infect Dis. 2002;35:842–850. doi: 10.1086/342383. [DOI] [PubMed] [Google Scholar]
- 5.Dodémont M, De Mendonça R, Nonhoff C, et al. Performance of the Verigene Gram-negative blood culture assay for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2014;52:3085–3087. doi: 10.1128/JCM.01099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beal SG, Ciurca J, Smith G, et al. Evaluation of the nanosphere Verigene Gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J Clin Microbiol. 2013;51:3988–3992. doi: 10.1128/JCM.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altun O, Almuhayawi M, Ullberg M, et al. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan KE, Ellis BC, Lee R, et al. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: A bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol. 2012;50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ecker DJ, Sampath R, Li H, et al. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn. 2010;10:399–415. doi: 10.1586/erm.10.24. [DOI] [PubMed] [Google Scholar]
- 10.Eshoo MW, Crowder CD, Li H, et al. Detection and identification of Ehrlichia species in blood by use of PCR and electrospray ionization mass spectrometry. J Clin Microbiol. 2010;48:472–478. doi: 10.1128/JCM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaleta EJ, Clark AE, Johnson DR, et al. Use of PCR coupled with electrospray ionization mass spectrometry for rapid identification of bacterial and yeast bloodstream pathogens from blood culture bottles. J Clin Microbiol. 2011;49:345–353. doi: 10.1128/JCM.00936-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaleta EJ, Clark AE, Cherkaoui A, et al. Comparative analysis of PCR-electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles. Clin Chem. 2011;57:1057–1067. doi: 10.1373/clinchem.2011.161968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacconi A, Richmond GS, Baroldi MA, et al. Improved sensitivity for molecular detection of bacterial and Candida infections in blood. J Clin Microbiol. 2014;52:3164–3174. doi: 10.1128/JCM.00801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laffler TG, Cummins LL, McClain CM, et al. Enhanced diagnostic yields of bacteremia and candidemia in blood specimens by PCR-electrospray ionization mass spectrometry. J Clin Microbiol. 2013;51:3535–3541. doi: 10.1128/JCM.00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 16.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
- 17.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 18.Kaukonen KM, Bailey M, Suzuki S, et al. Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000-2012. JAMA. 2014;311:1308–1316. doi: 10.1001/jama.2014.2637. [DOI] [PubMed] [Google Scholar]
- 19.Vincent JL, Teixeira L. Sepsis biomarkers. Value and limitations. Am J Respir Crit Care Med. 2014;190:1081–1082. doi: 10.1164/rccm.201410-1895ED. [DOI] [PubMed] [Google Scholar]
- 20.Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: Causes, consequences, and management. Front Public Health. 2014;2:145. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber J, Nierhaus A, Braune SA, et al. Comparison of three different commercial PCR assays for the detection of pathogens in critically ill sepsis patients. Med Klin Intensivmed Notfmed. 2013;108:311–318. doi: 10.1007/s00063-013-0227-1. [DOI] [PubMed] [Google Scholar]
- 22.Bloos F, Sachse S, Kortgen A, et al. Evaluation of a polymerase chain reaction assay for pathogen detection in septic patients under routine condition: An observational study. PLoS One. 2012;7:e46003. doi: 10.1371/journal.pone.0046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang SS, Hsieh WH, Liu TS, et al. Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis—A systemic review and meta-analysis. PLoS One. 2013;8:e62323. doi: 10.1371/journal.pone.0062323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordana-Lluch E, Giménez M, Quesada MD, et al. Improving the diagnosis of bloodstream infections: PCR coupled with mass spectrometry. Biomed Res Int. 2014;2014:501214. doi: 10.1155/2014/501214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordana-Lluch E, Carolan HE, Giménez M, et al. Rapid diagnosis of bloodstream infections with PCR followed by mass spectrometry. PLoS One. 2013;8:e62108. doi: 10.1371/journal.pone.0062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall MM, Ilstrup DM, Washington JA., 2nd Effect of volume of blood cultured on detection of bacteremia. J Clin Microbiol. 1976;3:643–645. doi: 10.1128/jcm.3.6.643-645.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenney JH, Reller LB, Mirrett S, et al. Controlled evaluation of the volume of blood cultured in detection of bacteremia and fungemia. J Clin Microbiol. 1982;15:558–561. doi: 10.1128/jcm.15.4.558-561.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cockerill FR, 3rd, Wilson JW, Vetter EA, et al. Optimal testing parameters for blood cultures. Clin Infect Dis. 2004;38:1724–1730. doi: 10.1086/421087. [DOI] [PubMed] [Google Scholar]
- 29.Farrell JJ, Sampath R, Ecker DJ, et al. “Salvage microbiology”: Detection of bacteria directly from clinical specimens following initiation of antimicrobial treatment. PLoS One. 2013;8:e66349. doi: 10.1371/journal.pone.0066349. [DOI] [PMC free article] [PubMed] [Google Scholar]


