Abstract
Neurosarcoidosis is a clinical subtype of sarcoidosis characterized by the presence of granulomas in the nervous system. Here, we report a highly significant association with a variant (rs75652600, P = 3.12 × 10−8, odds ratios = 4.34) within a zinc finger gene, ZNF592, from an imputation-based fine-mapping study of the chromosomal region 15q25 in African-Americans with neurosarcoidosis. We validate the association with ZNF592, a gene previously shown to cause cerebellar ataxia, in a cohort of European-Americans with neurosarcoidosis by uncovering low-frequency variants with a similar risk effect size (chr15:85309284, P = 0.0021, odds ratios = 5.36).
Introduction
Sarcoidosis is a multisystem disorder characterized by granulomatous inflammation in one or more affected organs.1 Though the etiology of sarcoidosis remains elusive, studies have implicated both environmental and genetic components, including a recent common-variant study that identified a suggestive association on chromosome 15q25.2 While up to 90% of patients diagnosed with sarcoidosis have lung involvement, prevalence of nervous system involvement, known as neurosarcoidosis, has been reported as high as 10%.3 Specific studies that examine the phenotypic variance leading to a diagnosis of neurosarcoidosis could elucidate the genetic and environmental architecture leading to sarcoidosis in general.3
Though sarcoidosis impacts individuals of all races, ages, and genders, African-Americans (AA) are more commonly and severely affected in the United States than European Americans (EA).1,3 To date, no study has identified the genetic or environmental etiology of neurosarcoidosis. Using a subset of AA neurosarcoidosis cases from a recent genome-wide association (GWA) study,2 we aimed to identify genetic variation associated with risk of neurosarcoidosis and validate these associations in an EA cohort. Rather than performing a genome-wide analysis that could lead to inflated results due to small samples, we focused our analysis to a particular region of chromosome 15 to determine possible association with neurosarcoidosis. We hypothesized that this region could be in association with neurosarcoidosis from two previous results. First, a linkage mapping association was uncovered near this same region using the neurosarcoidosis and lymph subphenotype.4 Additionally, a suggestive association signal on this region of chromosome 15 from a recent sarcoidosis GWA common variant study.2
Subjects and Methods
Our AA sample originally comprised 1487 cases and 1684 controls taken from four different studies. Sample quality control measures removed duplicate patients, individuals that were extreme population outliers, and samples with a low genotyping call rate (<90%) among other criteria previously described.2 The final AA sample was comprised of 1273 sarcoidosis cases and 1645 health controls. Of the 1273 cases, 83 were sub classified with neurosarcoidosis. This combined dataset comprised A Case Control Etiologic Study of Sarcoidosis (ACCESS), Sarcoidosis Genetic Analysis (SAGA), Henry Ford Health System in Detroit Michigan, healthy controls taken from the Oklahoma Medical Research Foundation (OMRF) Lupus Family Registry and Repository, and HapMap controls from Yoruba in Ibadan, Nigeria (YRI), and of African ancestry in Southwest USA (ASW) obtained from the Illumina HumanOmni1-Quad iControlDB (http://www.illumina.com/science/icontroldb.ilmn). Table1 summarizes the composition of our dataset from these four sources, specifying the neurosarcoidosis cases used in later analyses. For the EA validation sample, 32 of the 442 cases that passed identical quality control measures were classified as neurosarcoidosis cases, which were compared against 2284 EA controls. Full details of our sample collection have previously been reported.2
Table 1.
Summary of African-American samples after quality control
| Study | Sarcoidosis cases | Neurosarcoidosis cases | Healthy controls |
|---|---|---|---|
| ACCESS | 222 | 25 | 251 |
| SAGA | 566 | 37 | 482 |
| Henry Ford | 485 | 21 | 482 |
| OMRF | 0 | 0 | 557 |
| HapMap YRI-ASW | 0 | 0 | 180 |
| Total | 1273 | 83 | 1645 |
A summary of the AA sample composition for the sarcoidosis cases, neurosarcoidosis cases, and healthy controls. ACCESS, A Case Control Etiologic Study of Sarcoidosis; SAGA, Sarcoidosis Genetic Analysis; OMRF, Oklahoma Medical Research Foundation; YRI, Yoruba in Ibadan, Nigeria; ASW, African ancestry in Southwest USA.
Genotyping was performed at OMRF using the Illumina HumanOmni1-Quad array for over 1.1 million variants across the genome. The standard quality control methods, including a 1% variant minor allele frequency (MAF) and 95% call rate, were applied to the common-variant dataset as described in Adrianto et al.2 To determine the effect of low-frequency variants driving the signal in 15q25, we performed targeted resequencing of this region using the Illumina HiSeq 2000 platform with Illumina Pipeline software version 1.7 using standard procedures.5 Genomic DNA (3–5 μg) from 187 Sarcoidosis cases (12 neurosarcoidosis) and 293 healthy controls of AA decent was prepared for sequencing using an Illumina Paired-End Genomic DNA Sample Prep Kit. No additional EA samples were resequenced. We compared sequence-based variant calls with single-nucleotide polymorphisms (SNPs) previously genotyped on the Illumina HumanOmni1-Quad array platform and found >99% concordance between platforms. Imputation was performed spanning the hg19 chromosomal coordinates chr15:85,275,210–85,494,027 with the results of targeted sequencing experiment as a reference panel along with the 1000 Genomes Project Phase I integrated variant set. The result of the imputation was 1783 variants that passed quality control (information measure >0.5; average maximum posterior genotype call probability >0.9) measures, totaling 4703 variants evaluated in the analyses of 15q25.
Single-marker association analyses were performed using the EMMAX6 and PLINK7 logistic regression. As EMMAX implements a variance component approach to the linear mixed model, the software simultaneously adjusts for both pairwise genetic relatedness between individuals and corrects for population stratification. As EMMAX does not compute odds ratios (OR) or confidence intervals, PLINK logistic regression was employed to further analyze single-marker associations. The Local ancestry in AdMixed Populations (LAMP) program8 was then utilized to estimate local ancestry defined as the probability of carrying zero, one, or two copies of West African (or European) ancestral allele at each SNP for each individual.9 Ancestral allele frequency estimates for Illumina Omni-Quad SNPs were obtained from the HapMap Yoruba and CEPH European Utah catalog database. Variants strongly associated with the neurosarcoidosis phenotype (P < 1 × 10−6) were examined for functional activity using a standard query into the RegulomeDB webserver.10
Results
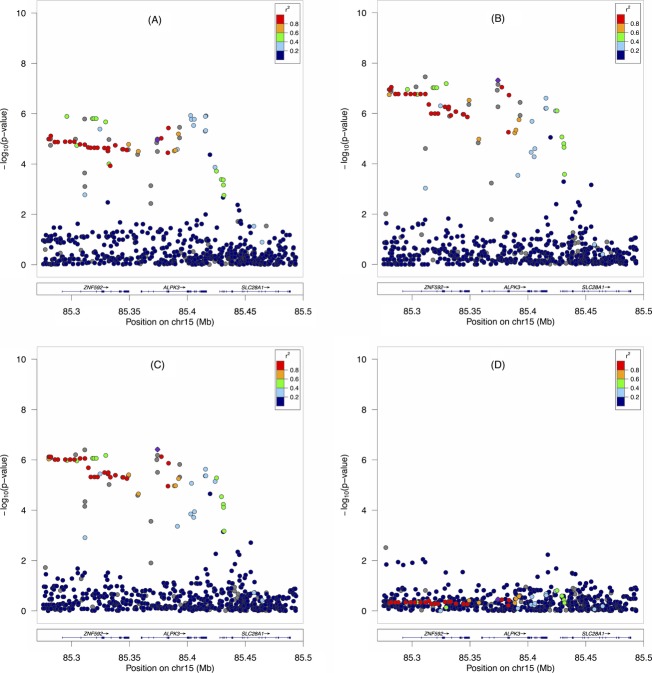
In our previous genome-wide common variant analysis for AA sarcoidosis cases and controls,2 we observed suggestive association in the 15q25 region spanning two genes, ZNF592 and ALPK3, for all AA sarcoidosis cases and controls (Fig.1A). Figure1B shows the results of association between only neurosarcoidosis cases and healthy controls. Although the sample size dropped considerably from our first association analysis (1273 cases/1645 controls) to our second association analysis (83 neurosarcoidosis cases/1645 controls), we observed a substantial increase in statistical significance. While no variant in the first analysis had a P-value less than 1 × 10−6, several of the variants in the second analysis surpassed genome-wide significance (P < 5 × 10−8 from EMMAX) with the most significant SNP at rs75652600 (P = 3.12 × 10−8; OR = 4.34, 95% CI = 2.30–8.20; MAF = 4.6%). We note that rs75652600 was imputed with strong linkage disequilibrium (LD) with rs62019469 (r2 = 0.98; P = 9.05 × 10−8; OR = 3.43, 95% CI = 2.12–5.54; MAF = 4.9%), which was present in the original genotyping. When assessing potential functionality from our highly significant variants using RegulomeDB,10 we determined that a genotyped variant with a strong association signal in ZNF592, rs3748376 (P = 5.5 × 10−7; OR = 3.64, 95% CI = 2.12–6.25; r2 = 0.75 with rs75652600), was likely to affect binding between USF1 and ZNF592 in the K562 cell line evidenced by a transcription factor-binding site and a DNase hypersensitivity peak.10 RegulomeDB revealed no other functionality for highly significant variants.
Figure 1.
A regional plot of variant-sarcoidosis association in the AA sample. Plots show the −log10(P-value) of case/control associations using EMMAX for (A) 1273 sarcoidosis cases and 1645 controls, (B) 83 neurosarcoidosis cases and 1645 controls, (C) 83 neurosarcoidosis cases and 1645 controls adjusted for West African and European descent local ancestry estimates, and (D) 83 neurosarcoidosis cases and 1645 controls conditioned on the most significant SNP, rs75652600, in ZNF592. AA, African-Americans; SNP, single-nucleotide polymorphism.
As the AA genome is admixed for African and European descent, genetic association signals could result from an association with local ancestry of common alleles instead of disease. Thus, we adjusted for not only global but also local ancestry to protect against this source of false positives. As shown in Figure1C, our result indicates that this signal in 15q25 is not due to ancestry, but the result of a statistical association between the variants in this region and neurosarcoidosis as rs75652600 (P = 3.99 × 10−7; OR = 4.27, 95% CI = 2.44–7.49) was still strongly significant, particularly noting the minimal change in OR.
While we were able to establish a strong association between neurological involvement and 15q25, two genes, ALPK3 and ZNF592, each contained variants strongly associated with sarcoidosis. Many SNPs in the ALPK3-ZNF592 150 kb region, including rs75652600, are in strong LD with each other. This implies that while the association signal persists across both genes, there is likely only one effect in this region. To verify the presence of a single association signal in this region, we used multivariate logistic regression adjusting for the most significant SNP, rs75652600, within ZNF592 and found the significance of the remaining association signals within this region significantly diminished (minimum P = 0.0036, chr15:85277267:D, Fig.1D). This is not true when conditioning on SNPs within ALPK3. Thus, we suggest that the broad association effect is a product of a single association in ZNF592 and strong LD in the region. Though we had specifically hypothesized that the association with this region could be better characterized with the neurosarcoidosis subphenotype, the same association models were performed for 11 other subphenotypes (lung, eye, thoracic, cardiac, liver, spleen, renal, skin, parotid/salivary gland, muscle, and bone marrow), and none of these secondary analyses surpassed genome-wide significance as in the neurosarcoidosis sample.
After characterizing the association in the AA cohort, we examined variants in ZNF592 within the EA neurosarcoidosis samples. Performing an identical case/control analysis with a smaller subset of cases (32 EA neurosarcoidosis cases/2284 EA controls), we identified two variants with a similar risk effect size, chr15:85301273 (P = 0.018; OR = 3.80; 95% CI = 1.158–12.45; MAF = 1.4%) and chr15:85309284 (P = 0.0021; OR = 5.36; 95% CI = 1.616–12.45; MAF = 1.0%). Notably, these two implicated variants represented independent effects (r2 = 0.0057) of one another. Common variants implicated in the AA analysis were not significant in the EA analysis (rs62019469 P = 0.304; rs75652600 not imputed in EA sample), highlighting the importance of the fine-mapping analysis performed in this study. Our results suggest a multi-ethnic association between the ZNF592 gene and risk of neurosarcoidosis. Table2 summarizes the results of the variant associations in both ethnicities.
Table 2.
Summary of variants in ZNF592 associated with neurosarcoidosis
| Variant | AA | EA | ||||
|---|---|---|---|---|---|---|
| P-value | OR | MAF (%) | P-value | OR | MAF (%) | |
| rs62019469 | 9.05 × 10−8 | 3.43 | 4.9 | 0.30 | 1.42 | 28.4 |
| rs75652600 | 3.12 × 10−8 | 4.34 | 4.6 | – | – | – |
| chr15:85301273 | – | – | – | 0.018 | 3.8 | 1.4 |
| chr15:85302984 | – | – | – | 0.0021 | 5.36 | 1.0 |
A summary of the P-values, OR, and MAF for the key variants identified in this study. The genotyped SNP, rs62019469, was called in both the EA and AA samples while rs75652600 was imputed only in the AA sample, chr15:85301273 and chr15:85302984 were called in the EA sample after targeted resequencing. AA, African-Americans; EA, European-Americans; OR, odds ratios; MAF, minor allele frequency.
Discussion
Though our AA discovery sample represented a subset of a larger genetic association, the association analysis using the 83 neurosarcoidosis cases and 1645 controls had 96.7% power to explain 10% of the heritability for a marker at the genome-wide significance level.11 From our neurosarcoidosis case/control analysis, we observed variants in ZNF592, notably rs75652600, that achieved genome-wide significance. Zinc finger protein 592 plays a role in cerebral development and is expressed throughout the central nervous system at relatively high levels.12 Notably, ZNF592 mutations have been shown to cause a cerebral ataxia.12
To further substantiate the role of ZNF592, we examined genetic factors that interact with this gene. Malovannaya et al. showed that ZNF592 functions in a multiple-protein complex with ZMYND8.13 Notably, ZMYND8 showed significant chromatin modification patterns in CD4+ T cells, suggesting a role for ZMYND8 in T-cell immune response.14 Additionally, as the results of our RegulomeDB query uncovered a potential interaction between USF1 and ZNF592, we note that the selective interaction between USF1 and STAT1 mediates the activation of CIITA (a MHC Class II transactivator) by IFNγ, suggesting that USF1 has a defined role in MHC Class II activity.15 Both CD4+ T Cells and MHC Class II activity have been implicated in sarcoidosis, and these interactions support a role for ZNF592 in the existing immune-based genetic understanding of the systemic disease. Finally, a strong interaction between ZNF592 and SMAD9 was originally uncovered in a study to determine additional proteins involved in SMAD signaling.16 SMAD9, like many proteins in this pathway, plays an important role in TGF-β signaling.17 As TGF-β has been associated with the severity of pulmonary sarcoidosis,18 we hypothesize that the interaction between ZNF592 and SMAD9 could be significant in granuloma formation.
Though sarcoidosis is thought to aggregate in families, the risk to relatives of neurosarcoidosis patients is largely unknown. From our 83 neurosarcoidosis cases in the AA sample, only one sib pair and one parent pair was observed. Noting the familial risk ratio (λ) of sarcoidosis has been reported as 2.49 for parents and sibs combined, an accurate measure for the degree of familial risk for neurosarcoidosis must be examined in larger samples.19
Our identification of a genetic effect driven by common variant association in one ethnicity but low-frequency variants in another is unusual but has been observed in other disease states. This effect could be explained due in part to the difference in allele frequencies of the common variant association. The genotyped SNP, rs62019469, has a MAF in the EA sample at 28.4% while only 4.9% in the AA sample. Though the conditional analysis verified the AA signal was not driven by particular ethnic differences in the samples, the imbalance of the proportion of risk alleles between these two ethnicities likely differentiates the disparity in association. We note that a similar risk variant architecture has been observed in interferon induced with helicase C domain 1 (IFIH1) in systemic lupus erythematosus (SLE). Similar to our association of ZNF592 in neurosarcoidosis, common and low frequency variants from IFIH1 were differently associated with the EA and AA samples in SLE, suggesting a differential role in protection through an evolutionary mechanism.20
While our approach compared neurosarcoidosis cases against healthy controls, we note an association analysis was performed comparing neurosarcoidosis cases against sarcoidosis cases that were not diagnosed with nervous system involvement. This analysis yield no strongly associated variants in 15q25 as the prevalence of undiagnosed neurosarcoidosis patients likely confounded the signal. Though the prevalence of neurological symptoms was 6.5% in our sample, previous studies have shown that the true prevalence may be as high as 14% after assessing sarcoidosis patients postmortem.3 Uncovering the genetic basis of neurosarcoidosis will aid in the understanding of this subtype of sarcoidosis. As our novel association of ZNF592 with neurosarcoidosis explains only a fraction of the phenotypic variance of the systemic disease, additional studies to augment this association are needed to sufficiently characterize the genetic etiology of neurosarcoidosis.
Acknowledgments
We recognize the NHLBI-funded ACCESS and SAGA research groups for the original data collection efforts that contributed the majority of the patient samples to this study. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS), Heart, Lung and Blood Institute (NHLBI), and National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under award numbers: P20GM103456 and P30GM110766 to I. A., R01-HL54306 and U01-HL060263 to M. C. I., R56-AI072727 and R01-HL092576 to B. A. R., and RC2HL101499 and R01HL113326 to C. G. M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
All authors contributed substantively to this work. C. A. L., I. A., A. M. L., M. C. I., B. A. R., and C. G. M. were involved in study conception and design. C. A. L., I. A., and A. M. L. were involved in the data organization and statistical analysis. M. C. I., B. A. R., and C. G. M. coordinated the data collection and processing. C. A. L., I. A., and C. G. M drafted the manuscript and prepared the figures. All authors were involved in the reviewing and editing of the manuscript.
Conflict of Interest
None declared.
References
- Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- Adrianto I, Lin CP, Hale JJ, et al. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One. 2012;7:e43907. doi: 10.1371/journal.pone.0043907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki K, Judson MA. Neurosarcoidosis: clinical manifestations, diagnosis and treatment. Presse Med. 2012;41(6 Pt 2):e331–e348. doi: 10.1016/j.lpm.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Sinha R, Iyengar S, et al. Genetic linkage analysis of sarcoidosis phenotypes: the sarcoidosis genetic analysis (SAGA) study. Genes Immun. 2007;8:379–386. doi: 10.1038/sj.gene.6364396. [DOI] [PubMed] [Google Scholar]
- Levin AM, Adrianto I, Datta I, et al. Performance of HLA allele prediction methods in African Americans for class II genes HLA-DRB1, -DQB1, and -DPB1. BMC Genet. 2014;15:72. doi: 10.1186/1471-2156-15-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Sankararaman S, Kimmel G, Halperin E. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25:i213–i221. doi: 10.1093/bioinformatics/btp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AM, Iannuzzi MC, Montgomery CG, et al. Admixture fine-mapping in African Americans implicates XAF1 as a possible sarcoidosis risk gene. PLoS One. 2014;9:e92646. doi: 10.1371/journal.pone.0092646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR. Novel genetic analysis for case-control genome-wide association studies: quantification of power and genomic prediction accuracy. PLoS One. 2013;8:e71494. doi: 10.1371/journal.pone.0071494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Poitelon Y, Chouery E, et al. CAMOS, a nonprogressive, autosomal recessive, congenital cerebellar ataxia, is caused by a mutant zinc-finger protein, ZNF592. Eur J Hum Genet. 2010;18:1107–1113. doi: 10.1038/ejhg.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Li Y, Bulynko Y, et al. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci USA. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- Colland F, Jacq X, Trouplin V, et al. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaous J, Hata A. TGF-beta signalling through the SMAD pathway. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- Limper AH, Colby TV, Sanders MS, et al. Immunohistochemical localization of transforming growth factor-beta 1 in the nonnecrotizing granulomas of pulmonary sarcoidosis. Am J Respir Crit Care Med. 1994;149:197–204. doi: 10.1164/ajrccm.149.1.8111583. [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Kirkey KL, Major M, et al. Familial risk ratio of sarcoidosis in African-American sibs and parents. Am J Epidemiol. 2001;153:188–193. doi: 10.1093/aje/153.2.188. [DOI] [PubMed] [Google Scholar]
- Molineros JE, Maiti AK, Sun C, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet. 2012;9:e1003222. doi: 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]