Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) are a frequent cause of bacteremia and sepsis, but the role of ExPEC genetic virulence factors (VFs) in sepsis development and outcome is ill-defined. Prospective study including 120 adult patients with E. coli bacteremia to investigate the impact of bacterial and host factors on sepsis severity and mortality. Patients' clinical and demographic data were registered. Phylogenetic background of E. coli isolates was analyzed by SNP pyrosequencing and VFs by PCR. The E. coli isolates presented an epidemic population structure with 6 dominant clones making up to half of the isolates. VF gene profiles were highly diverse. Multivariate analysis for sepsis severity showed that the presence of cnf and blaTEM genes increased the risk of severe illness by 6.75 (95% confidence interval [CI] 1.79–24.71) and 2.59 (95% CI 1.04–6.43) times respectively, while each point in the Pitt score increased the risk by 1.34 (95% CI 1.02–1.76) times. Multivariate analysis for mortality showed that active chemotherapy (OR 17.87, 95% CI 3.35–95.45), McCabe-Jackson Index (OR for rapidly fatal category 120.15, 95% CI 4.19–3446.23), Pitt index (OR 1.78, 95% CI 1.25–2.56) and presence of fyuA gene (OR 8.05, 95% CI 1.37–47.12) were associated to increased mortality while the presence of P fimbriae genes had a protective role (OR 0.094, 95%IC 0.018–0.494). Bacteremic E. coli had a high diversity of genetic backgrounds and VF gene profiles. Bacterial VFs and host determinants had an impact on disease evolution and mortality.
Keywords: bloodstream infection, E coli, mortality, outcome, systemic inflammatory response syndrome, virulence factor
Background
Escherichia coli is the gram-negative organism most frequently isolated in adult patients with bacteremia.1,2 Though it is a commensal of the normal intestinal microbiota of mammals and birds,3 some strains of E. coli are pathogenic, either intestinal or extraintestinal.4,5
Extraintestinal pathogenic E. coli (ExPEC) strains, cause a variety of infections, including urinary tract infections (UTI), sepsis and neonatal meningitis.4 ExPEC have several virulence factors (VFs) that may play a role in infection by enabling the bacterial cells to move into the host and disseminate. ExPEC VFs include adhesion molecules, iron acquisition systems, host defense-subverting mechanisms and toxins. Several VFs have been associated with the source of infection6 or bloodstream invasion,7 but their role in determining a patient´s prognosis is ambiguous.8-12
In order to investigate the relevance of ExPEC VFs in sepsis, we have prospectively studied the characteristics of E.coli causing bacteremia in our hospital and their relationship with sepsis severity and mortality.
Patients and Methods
Setting and design
One-year prospective observational study of all the episodes of bacteremia due to E. coli in our hospital from May 2009 to May 2010. Cases were detected through the daily review of blood culture results from adult patients (aged >18 years). The study was approved by the Hospital La Paz Ethics Committee. Written informed consent was obtained before including patients in the study.
Variables and data collection
We collected, from medical records, demographic characteristics, underlying diseases, reason for admission, diagnosis, source of bacteremia, antimicrobial therapy and outcome. Data were recorded in a standardized form and anonymised according to the Hospital regulations.
Definitions
Bacteremia episodes were primarily classified as nosocomially-acquired (NA) or community-acquired (CA), in accordance with CDC criteria.13 CA episodes were further classified as healthcare-associated (HCA) if any of the following criteria were present14: a 48-hours hospital admission during the previous 90 days, hemodialysis, intravenous medication or home wound care in the previous 30 days or residence in a nursing home or long-term care facility.
Charlson comorbidity index (range 0–37)15 and McCabe-Jackson classification (non-fatal, ultimately fatal or rapidly fatal)16 were used to evaluate comorbidity and prognosis. Acute severity of illness at presentation was evaluated with the Pitt Bacteremia Score.17 Sepsis level was graded as sepsis, severe sepsis and septic shock according to the criteria of systemic inflammatory response syndrome (SIRS).18
Immunosuppression was defined as use of immunosuppressive therapy (steroid treatment, other immunosupresor drugs, immunomodulating agents or chemotherapy) and/or presence of any immunosuppressive illness (cancer, hematological malignancy, HIV infection).
The source of bacteremia was determined according to the clinical presentation and/or the evidence of an identical strain cultured near to or on the same date as the onset of bacteremia. If the source of bacteremia could not be identified, it was classified as primary bacteremia.
Antimicrobial therapy was considered appropriate if the patient received at least one agent with in vitro activity against the isolate according to the results of antimicrobial susceptibility testing (AST), and if the dosage and route of administration were correct according to current clinical guidelines. Time from the onset of the bacteremia episode to the initiation of the treatment was obtained from medical and nurse records.
Outcome parameters
Crude mortality during hospitalization and within 30 days after onset of bacteremia was registered. Relationship between death and bacteremia—directly related, indirectly related and non-related—was assessed by two different investigators by reviewing hospital medical records. In those patients who had been discharged, mortality was evaluated through local electronic health records. Readmission within one month after discharge and its relatedness to the previous episode of bacteremia were also registered as indicator of patient outcome.
Bacterial determinants
Blood cultures were incubated in Bactec (BACTEC TM, Becton Dickinson, Franklin Lakes, NJ, USA) or BacT/ALERT® (bioMérieux, Marcy l’Etoile, France) systems. Identification and antibiotic susceptibility was determined using Wider® (Fco. Soria Melguizo, Madrid, Spain) or Vitek2® (bioMérieux) systems. Susceptibility assays were interpreted according to CLSI guidelines (CLSI, 2011). ESBL production was confirmed by E-test ESBL strips (bioMérieux).
The phylogenetic groups were determined by multiplex PCR20 and by single nucleotide polymorphism (SNP) pyrosequencing.21 The pyrosequencing method generates SNP allelic profiles or SAPs. Each SAP may include one or more MLST types, and is numbered with the lowest ST number included in the profile, e.g., SAP131 is numbered after ST131 and includes ST131, as well as other STs with higher numbers (see ref. 21). Presence of 25 VF genes and 4 betalactamase families (TEM, CTX-M, VIM and OXA-48) was determined by PCR as described elsewhere.22,23 Genetic analysis of betalactamase families (TEM, CTX-M, VIM and OXA-48) was performed regardless of their expression in the Etest. Virulence score for each isolate was defined as the number of VF genes present.
Statistical analysis
Univariate regression analyses were performed for clinical and bacteriological factors. Percentages were compared using the χ2 test or the Fisher exact test, as appropriate and continuous variables using the Mann-Whitney U test. All variables achieving a P value <0.10 were then entered into the multivariate logistic regression models for sepsis severity and mortality respectively. A stepwise forward process method was used to obtain the models in which all clinical risk factors had a P value <0.05. As our study was exploratory, all VFs and betalactamases were included in the models irrespective of their p values in univariate analysis. Significant VFs were afterward removed to know their weight on multivariate models. All the tests were performed using SPSS version 17.0 (SPSS, Chicago, IL).
Results
The study included 120 adult patients with E. coli bacteremia. Data regarding SIRS were available for 119 patients.
Clinical and epidemiological characteristics
Patients were predominantly elderly (68.9% > 65 years old), with frequent comorbidities (median Charlson score 3). The major comorbidities were solid or hematological malignancies (50%), immunosuppression (28.3%, including 20 patients on chemotherapy) and diabetes mellitus (20% of the patients). There was an equal gender distribution, with 49.6 % of male patients. Overall, 44.2% of infection episodes were NA, 11.7% were HCA and 44.2% were CA (Table 1; Table S4).
Table 1.
Patient and pathogen characteristics according to sepsis severity and outcome
| All patients n = 120 | Mild illness n = 74 | Severe illness n = 45 | P value + | Survivors n = 87 | Non-survivors n = 33 | P value ++ | |
|---|---|---|---|---|---|---|---|
| Host Characteristics | |||||||
| Sex, male | 59 (49.6) | 34 (45.9) | 25 (55.6) | — | 37 (62.7) | 22 (37.3) | 0.025 |
| Sex, female | 49 (81.7) | 11 (18.3) | — | ||||
| Age, mean (range) | 70.77 (20–98) | 72.65 (30–96) | 67.69 (20–98) | — | 71.78 (20–98) | 68.03 (33–96) | — |
| Age >65 years | 82 (68.9) | 54 (73) | 28 (62.2) | — | 64 (73.6) | 18 (56.3) | — |
| Charlson Index, median (IQR) | 3 (1–4) | 3 (1–4) | 3 (1–4) | — | 2 (1–4) | 4 (2.5–4.5) | 0.001 |
| Charlson Index >2 | 65 (54.2) | 39 (52) | 26 (57.8) | — | 40 (46) | 25 (75.8) | 0.004 |
| Diabetes mellitus | 24 (20) | 12 (16) | 12 (26.7) | — | 16 (18.4) | 8 (24.2) | — |
| Cancer | 48 (40) | 31 (41.3) | 17 (37.8) | — | 30 (34.5) | 18 (54,5) | 0.060 |
| Hematological malignancy | 12 (10) | 10 (13.3) | 2 (4.4) | — | 7 (8) | 5 (15.2) | — |
| Inmunosupression | 34 (28.3) | 24 (32) | 10 (22.2) | — | 19 (21.8) | 15 (45.5) | 0.013 |
| HIV infection | 2 (1.7) | 2 (2.7) | 0 | — | 1 (1.1) | 1 (3) | — |
| Steroid treatment | 11 (9.2) | 10 (13.3) | 1 (2.2) | 0.051 | 8 (9.25) | 3 (9.1) | — |
| Other inmunosupresor drugs | 7 (5.8) | 7 (9.3) | 0 | 0.044 | 3 (3.4) | 4 (12.1) | 0.090 |
| Immunomodulating agents | 1 (0.8) | 1 (1.3) | 0 | — | 1 (1.1) | 0 | — |
| Chemotherapy | 20 (16.7) | 11 (14.7) | 9 (20) | — | 9 (10.3) | 11 (33.3) | 0.005 |
| Admission Service | — | — | |||||
| Medical | 77 (65.8) | 52 (72.2) | 25 (55.6) | 54 (64.3) | 23 (69.7) | ||
| Surgical | 26 (22.2) | 13 (18.1) | 13 (28.9) | 22 (26.2) | 4 (12.1) | ||
| Critical Care Units | 14 (12) | 7 (9.7) | 7 (15.5) | 8 (9.5) | 6 (18.2) | ||
| Mc Cabe Jackson Index: | — | 0.001* | |||||
| Non-fatal | 47 (39.5) | 29 (39,2) | 18 (40) | 42 (48.8) | 5 (15.2) | ||
| Ultimately fatal | 53 (44.5) | 31 (41.9) | 22 (48.9) | 35 (40.7) | 18 (54.5) | ||
| Rapidly fatal | 19 (16) | 14 (18.9) | 5 (11.1) | 9 (10.5) | 10 (30.3) | ||
| Global death | 33 (27.5) | 19 (25.3) | 14 (31.3) | — | NA | NA | |
| Length of stay, mean (SD) | 25.61 (27) | 22.87 (22.69) | 30.11 (34.25) | — | 22.74 (26.56) | 33.09 (29.64) | — |
| Days of stay prior to BSI, mean (SD)* | 9.26 (15.11) | 9.06 (14.28) | 9.59 (16.57) | — | 8.09 (15.06) | 12.5 (15.03) | — |
| BSI-Episode Characteristics | |||||||
| Pitt Index, median (IQR) | 1 (0–1.25) | 0 (0–1) | 1(0–2) | 0.005 | 0.5 (0–1) | 1 (0–3) | 0.02 |
| Pitt >1 | 27 (24,5) | 9 (13.2) | 18 (42.9) | 0.001 | 16 (19.5) | 11 (39.3) | 0.044 |
| BSI origin | — | — | |||||
| Nosocomial | 53 (44,2) | 32 (42.7) | 21 (46.7) | 35 (40.2) | 18 (54.4) | ||
| Community-acquired | 53 (44,2) | 35 (46.7) | 18 (40) | 44 (50.6) | 9 (27.3) | ||
| Healthcare associated | 14 (11,7) | 8 (10.7) | 6 (13.3) | 8 (9.25) | 6 (18.2) | ||
| BSI Source | — | 0.033 | |||||
| Urinary | 40 (33,3) | 26 (34.7) | 14 (31.1) | 33 (37.9) | 7 (21.2) | ||
| Intra-abdominal | 57 (47,5) | 36 (48) | 21 (46.7) | 42 (48.3) | 15 (45.5) | ||
| Other or unknown | 23 (19.2) | 13 (17.3) | 10 (22.2) | 12 (13.8) | 11 (33.3) | ||
| Hours from BSI to treatment | 7.31 (12.26) | 7.31 (12.26) | 6.7 (13.23) | — | 7.05 (11.89) | 6.7 (13.71) | — |
| Adequate treatment** | 111 (100) | 69 (100) | 42 (100) | — | 81 (100) | 30 (100) | — |
Note: Data are displayed as number (%) unless otherwise noted. BSI blood stream infection. IQR: interquartile range. SD: standard deviation. + Comparisons for mild and severe illness groups. ++ Comparisons for survivors and non-survivors—Not significant. *Only for patients with nosocomial BSI episodes. **From treated patients.
The source of infection was intra-abdominal in 47.5% of the cases, urinary in 33.3%, primary in 14.2%, catheter-related in 3.3%, and skin and soft tissue in 2 (Table 1). There were more women, more patients with cancer and higher mortality in the intra-abdominal-source group (57.1% vs. 32.5%, P = 0.023; 56.1% vs. 25%, P = 0.003; 26.3% vs. 17.5% P = 0.033, respectively) (Table S3).
One hundred and 11 patients received antibiotic treatment and it was considered appropriate in all cases. Nine patients did not receive any antibiotic treatment. Fifty-one per cent of the patients received the first dose of antibiotic during the first hour since the clinical presentation of the bacteremia episode and 79.6% during the first 8 hours. Patients were treated empirically with a betalactam/betalactamase inhibitor in 44.14% cases, with a carbapenem in 33.3%, with a 3rd- or 4th-generation cephalosporin in 12.6%, with a quinolone in 7.2% and one patient was treated with colistin.
Bacterial determinants
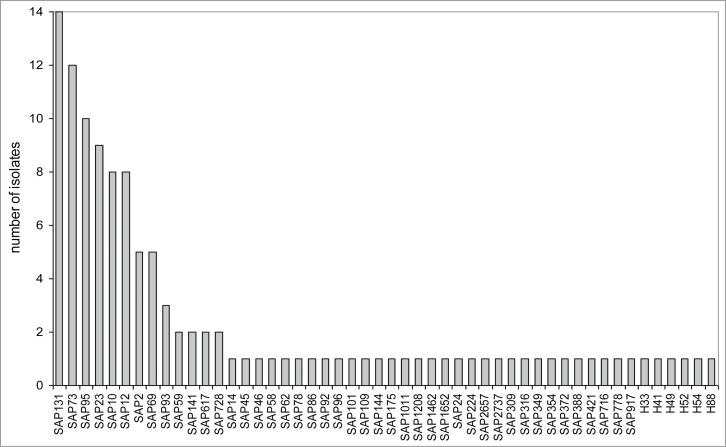
Multiplex PCR typing was done to determine the distribution of the isolates among the major E. coli phylogenetic groups.20 Phylogroup B2 was the most common with 41.6% of the isolates, followed by group A with 30%, group D with 17.5%, and group B1 with 10.8%. MLST-based SNP typing showed a diverse population with 50 different SNP allelic profiles (SAPs). Each SAP is equivalent to one or a few closely related MLST sequence types (ST).21 Among the 50 profiles found, 37 appeared once and only 6 appeared in more than 5 isolates. These six allelic profiles accounted for 50.8% of the isolates: SAP131 (n = 14), SAP73 (n = 12), SAP95 (n = 10), SAP23 (n = 9), SAP12 (n = 8) and SAP10 (n = 8) (Fig. 1).
Figure 1.
Distribution of allelic profiles in the bacteremic population. The histogram shows the SNP allelic profiles (SAP) detected in our study population, and the number of isolates belonging to each one. The six profiles labeled as Hx were not found in the MLST database. Profile numbering taken from ref. 21
PCR analyses of VF genes showed that the 120 isolates presented 108 different VF gene profiles, of which 99 were unique, 7 appeared twice, 1 appeared three times, and another 1 appeared four times. The frequencies of the individual VF ranged from no presence of papG allele I or 2.5% of afaB/C to 95.8% for fimH (Table 2). Virulence scores were calculated as the sum of the VF genes present in each isolate. The median virulence score was 9 (range 0 to 18, IQR 6 to 13). The median virulence scores for each phylogroup were 13, 8, 9 and 6.5, for B2, B1, D and A, respectively (P < 0.001) (Table 2).
Table 2.
Virulence factors distribution according to phylogenetic group
| Phylogenetic group |
||||||
|---|---|---|---|---|---|---|
| All isolates | A | B1 | B2 | D | ||
| Virulence-related gene | n = 120 | n = 36 | n = 13 | n = 50 | n = 21 | P |
| Adhesins | ||||||
| papC | 58 (48.3) | 10 (27.8) | 4 (30.8) | 32 (64) | 12 (57.1) | 0.004 |
| papG allele I | 0 | 0 | 0 | 0 | 0 | — |
| papG allele II | 36(30) | 4 (11.1) | 2 (15.4) | 23 (46) | 7 (33.3) | 0.003 |
| papG allele III | 12 (10) | 2 (5.6) | 0 | 10 (20) | 0 | 0.018 |
| afaB/C | 3 (2.5) | 0 | 0 | 2 (4) | 1 (4.8) | — |
| fimH | 115(95.8) | 32 (88.9) | 13 (100) | 50 (100) | 21 (95.2) | — |
| iha | 42 (35) | 7 (19.4) | 4 (30.8 | 19 (38) | 12 (57.1) | — |
| sfa/focDE | 24(20) | 2 (5,6) | 2 (15,4) | 20 (40) | 0 | <0.0001 |
| Toxins | ||||||
| cdtB | 7 (5.8) | 0 | 0 | 5 (10) | 2 (9.5) | — |
| cnf1 | 17 (14.2) | 0 | 0 | 17 (34) | 0 | <0.0001 |
| hlyA | 28 (23.3) | 4 (11.1) | 2 (15.4) | 20 (40) | 2 (9.5) | 0.004 |
| Sat | 32 (26.7) | 2 (5.6) | 4 (30.8) | 16 (32) | 10 (47.6) | 0.003 |
| Usp | 69 (57.5) | 5 (13.9) | 6 (46.2) | 50 (100) | 8 (38.1) | <0.0001 |
| Protectins | ||||||
| kpsMTII | 66 (55) | 8 (22.2) | 6 (4.2) | 38 (76) | 14 (66.7) | <0.0001 |
| ompT | 90 (75) | 17 (47.2) | 8 (61.5) | 50 (100) | 15 (71.4) | <0.0001 |
| Iron-uptake | ||||||
| iroN | 61 (50.8) | 18 (80) | 10 (76.9) | 31 (62) | 2 (9.5) | <0.0001 |
| fyuA | 91 (75.8) | 19 (52.8) | 8(61.5) | 50 (100) | 14 (66.7) | <0.0001 |
| iutA | 83 (69.2) | 23 (63.9) | 10 (76.9) | 36 (72) | 14 (66.7) | — |
| iucD | 77 (64.2) | 21 (58.3) | 10 (76.9) | 33 (66) | 13 (61.9) | — |
| ireA | 23 (19.2) | 3 (8.3) | 4 (30.8) | 13 (26) | 3 (14.3) | — |
| Betalactamases | ||||||
| TEM | 67 (55.8) | 25 (69.4) | 7 (53.4) | 26 (52) | 9 (42.9) | — |
| CTX-M | 5 (4.2) | 1 (2.7) | 1 (7.7) | 3 (6) | 0 | — |
| VIM | 1 (0.8) | 0 | 0 | 1 (2) | 0 | — |
| Miscellaneous | ||||||
| malX | 68 (56.7) | 6 (16.7) | 6 (46.2) | 49 (98) | 7 (33.3) | <0.0001 |
| svg | 12 (10) | 1 (2.8) | 1 (7.7) | 9 (18) | 1 (4.8) | — |
| ibeA | 16 (13.3) | 1 (2.8) | 0 | 11 (22) | 4 (19) | — |
| tratT | 76 (63.3) | 20 (55.6) | 10 (76.9) | 36 (72) | 10 (47.6) | — |
| cvaC | 25 (20.8) | 13 (36.1) | 2 (15.4) | 10 (20) | 0 | — |
| Median Virulence score (IQR) | 9 (6–13) | 6.50 (4–8.75) | 8 (4–12) | 13 (10–15) | 9 (6–10) | |
Note: Data are displayed as number (%) of isolates. papC, P fimbrial; papG allele I, II y III, P fimbrial adhesin molecule, with variants I, II, and III; afaB/C, afimbrial adhesin; fimH, type 1 fimbrial adhesion, iha, bifunctional enterobactin receptor/adhesin; sfa/foc, S and F1C fimbriae; cdtB, cytolethal distending toxin; cnf, cytotoxic necrotizing factor; hlyA, hemolysin; sat, secreted autotransporter toxin; usp, uropathogenic specific protein; kpsMTII, group II capsule synthesis; iroN, putative catecholate siderophore receptor; fyuA, yersiniabactin receptor; iutA, aerobactin receptor; iucD, aerobactin biosynthesis; TEM and CTX-M betalactamases; VIM metallobetalactamase; malX, marker for pathogenicity-associated island from strain CFT073; svg, specific for virulence subgroup; ibeA, invasion of brain endothelium A; traT, serum-resistance associated outer membrane protein; cvaC, colicin V receptor. -, not significant.
The rates of antibiotic resistance were low (Table S1) and no statistically significant differences between NA and CA infections were observed. Six (5.1%) isolates showed ESBL phenotype (4 cases NA, 1 HCA, 1 CA), 1 associated a VIM metallo-betalactamase, and another one an OXA-48 carbapenemase (NA cases).
Phylogroup distribution did not have significant associations with patient characteristics (Tables S2 and S3), or antibiotic resistance, but were strongly correlated with VF genes, mostly through significant, and often strong, associations between VF genes and group B2 (Table 2). The strongest association was found for the cnf gene, which was detected exclusively among B2 isolates. In addition, all B2 isolates had the usp, ompT and fyuA genes.
According to the origin of infection, the only significant association was a higher frequency of the usp, kpsMTII and malX genes in CA infection. According to the source, only the iutA gene was found to be significantly less frequent in intra-abdominal infections. And in relation to the immunological status, the iucD gene was more frequent in immunosuppressed patients (79.4% vs. 58.1%, P = 0.035) (Table S4).
SIRS response and risk factors for sepsis severity
The SIRS response was graded in three categories: sepsis was found in 62.2% of the patients, severe sepsis in 28.6%, and septic shock in 9.2%. Because the latter group was too small for statistical analysis, the two more severe conditions were merged, and the response categorized as mild illness (sepsis, 61.6% of the patients) or severe illness (severe sepsis or septic shock, 37.55% of the patients) (Table 1). No differences between these two groups were found with respect to age, sex, comorbidity or immunosuppression. Similar distributions were also found in relation to the origin and the source of bacteremia, antibiotic resistance profiles and phylogroups. Pitt scores were higher in severe disease than in mild disease (mean 1.66 vs. 0.77, P = 0.005). Patients were empirically treated with a betalactam/betalactamase inhibitor in 39.1% and 52.4% of mild and severe illness respectively, in 36.2% and 28.6% with a carbapenem, in 14.5% and 9.5% with a 3rd- or 4th-generation cephalosporin, and in 6.7% and 4.8% with a quinolone. One patient in each group received an aminoglycoside and one patient in the severe illness group was treated with colistin.
Median virulence scores did not differ between the two groups (9 for mild disease and 10 for severe disease, P = 0.296) (Table S1), and only the cnf gene was significantly more frequent in the severe sepsis group (24.4% vs. 8%, P = 0.016) (Table S2).
Multivariate analysis revealed that the presence of cnf and blaTEM genes was associated to a 6.65 (95% CI 1.79–24.71) and a 2.59 (95% CI 1.04–6.43) increased risk of severe illness respectively (Table 3). Each point in the Pitt score increased risk by 1.34 (95% CI 1.02–1.76). The area under the curve to predict sepsis severity for this model was 0.763 (95% CI 0.673–0.853). No other independent factors were found to be related with the severity of the disease. The predictive ability of the model when removing all the VFs was reduced to a ROC area under the curve of 0.700 (95% CI 0.597–0.802).
Table 3.
Multivariate logistic regression analysis for sepsis severity
| Variable | SE () | OR | 95% CI | P | ||
|---|---|---|---|---|---|---|
| Pitt index | 0.292 | 0.140 | 1.34 | 1.02 | 1.76 | 0.037 |
| cnf presence | 1.894 | 0.670 | 6.65 | 1.79 | 24.71 | 0.005 |
| blaTEM presence | 0.950 | 0.464 | 2.59 | 1.04 | 6.43 | 0.04 |
| Constant | −1.528 | 0.397 | 0.22 | 0.000 | ||
Mortality and risk factors for mortality
Total 30-day mortality was 27.7% (33 patients). In 63.6% of these patients the bacteremia episode was not considered to be directly related to death, in 21.2% it was considered the main cause for death, and in 15.2% it was considered possible/probable cause.
The main features of survivors and non-survivors are shown in Table 1. Comorbidity was remarkably higher in the non-survivors group as reflected in their higher Charlson index (2.62 vs. 4.06, P = 0.001), more malignancies (34.5 vs. 54.5%, P = 0.06) and higher frequency of chemotherapy (10.3% vs. 33.3%, P = 0.001). McCabe-Jackson Index was different between both groups with a significant trend, with more rapidly-fatal prognosis in the non-survivors group (30.3% vs. 10.5%, P = 0.001),
The origin of infection in deceased patients was NA in 54.8% (18 patients), HCA in 27.3% (6) and CA in 27.3% (6). There were less infections of urinary source in the non-survivors group (21.2% vs. 37.9%, P = 0.033).
Although Pitt index values were higher in non-survivor patients (mean 0.84 vs. 1.9, P = 0.02), the fraction of patients with septic shock or severe sepsis in both groups did not differ statistically: 24 patients had severe sepsis and 7 septic shock among the survivors, while 10 patients had severe sepsis and 4 septic shock among the non-survivors.
ESBL-producing isolates were found only among the non-survivors (6 strains, P < 0.001). All treated patients in both groups received adequate antibiotics, although it was started later in the non-survivors group (mean time to treatment starting: 8.09 hours in survivors and 12.51 hours in non-survivors).
VF gene frequencies did not differ between survivors and non-survivors in the univariate analysis. The set of VF genes included 3 genes enconding components of P fimbriae, papC, papGII and papGIII. These were analyzed separately and grouped as P fimbriae.
Multivariate analysis revealed that active chemotherapy (OR 17.87, 95% CI 3.35–95.45), McCabe-Jackson Index (OR for rapidly-fatal category 120.15, 95% CI 4.19–3446.23), Pitt index (OR 1.78, 95% CI 1.25–2.56) and presence of the fyuA (OR 8.05, 95% CI 1.37–47.12) were positively associated with mortality while the presence of any of the genes that encode for P fimbriae components had a protective role (OR 0.094, 95% CI 0.018–0.494). The ROC area under the curve to predict mortality for this model was 0.853 (95% CI 0.769–0.936). No other independent factors were found to be associated with mortality (Table 4). The predictive capacity of the model when removing all the VFs was reduced to a ROC area under the curve of 0.820 (95% CI 0.732–0.908).
Table 4.
Multivariate logistic regression analysis for mortality
| Variable | SE () | OR | 95% CI | P | ||
|---|---|---|---|---|---|---|
| Active chemotheraphy | 2.883 | 0.855 | 17.871 | 3.346 | 95.458 | 0.001 |
| McCabe-Jackson Index | ||||||
| No fatal | — | — | — | — | — | 0.013 |
| Ultimately fatal | 2.990 | 1.497 | 19.894 | 1.058 | 374.124 | 0.046 |
| Rapidly fatal | 4.789 | 1.712 | 120.153 | 4.189 | 3446.225 | 0.005 |
| Pitt Index | 0.577 | 0.178 | 1.781 | 1.256 | 2.526 | 0.001 |
| P fimbria genes | −2.362 | 0.845 | 0.094 | 0.018 | 0.494 | 0.005 |
| fyuA gene | 2.085 | 0.902 | 8.047 | 1.374 | 47.125 | 0.021 |
| Constant | −5.971 | 1.779 | 0.003 | 0.001 | ||
Discussion
In our study of 120 E. coli bloodstream infections we have found that host and pathogen factors were associated with severity and outcome of infection. Our analysis shows an association between certain E. coli VFs with disease severity (cnf and blaTEM), increased mortality (fyuA) and decreased mortality (papC, papGII and/or papGIII in any combination, all them encoding components of P fimbriae). One of the strengths of our study is that we carefully characterized all E. coli bloodstream infections epidemiologically, clinically and genotypically. E. coli isolates were analyzed for a broad range of VFs, phylogenetic background and antibiotic susceptibility.
In our series, the bacterial isolates had a high genetic diversity and a mixed population structure, with a few epidemic clonal complexes making up to half of the isolates, and several sporadic isolates, most of them unique, making up the other half. Remarkably, the major allelic profile was SAP131, which is equivalent to the pandemic ST131. This is similar to the population structure found previously in our hospital isolates.21 The genetic diversity was much higher for the VF genes, where just a few profiles appeared more than once.
The association of cnf with E. coli bloodstream infection severity has not been reported before, although Jauréguy et al.9 reported a higher, but non-significant, frequency of cnf in patients with septic shock. A plausible biological explanation for this association is that cnf codes for a toxin, the cytotoxic necrotizing factor 1. This is a 113 kDa protein that activates small GTP-binding proteins in host cells, conferring E. coli the ability to affect cellular functions including inflammatory response and inhibition of phagocytic and chemotactic activities of neutrophils.28,29 The frequency of isolates containing cnf in our cohort—14.2%—was slightly lower than in prior series which have reported frequencies of around 20–30%.8,9 In most studies, the higher frequencies are found in phylogroup B224 and isolates from urinary sources,6,25 while they have been reported in less than 5% of commensal strains.26
The association of blaTEM with sepsis severity is difficult to explain because it appears to be independent of the betalactamase activity (since all patients received appropriate antibiotics) and it is possible that the presence of blaTEM might be a marker of the presence of some other VFs.
The mortality rate in our cohort was 27.7%, which is within expectations, considering the frequent nosocomial nature and non-urinary sources of our episodes. Prior studies have reported mortality rates ranging from 5% to 30%.27 In spite of the association of cnf and blaTEM genes with sepsis severity neither of these 2 VF were associated with mortality. Multivariate analysis showed 2 different E. coli factors associated with mortality: fyuA increased the risk, while any combination of genes encoding for P fimbria components had a protective role.
The presence of the yersiniabactin siderophore receptor encoded by fyuA in previously reported series ranges from 51% in an ESBL E. coli collection 22 to 96% in children with UTI due to E. coli.7 The fyuA gene product is involved in the efficient uptake of iron from bloodstream 28 and in invasion of the bloodstream from urinary tract.29 Previous studies have reported contradictory results regarding fyuA and mortality. Lefort et al.8 found fyuA more frequently in survivors (78.09% vs. 66.18%, p = 0.0025), but this relationship was lost in multivariate analysis. On the contrary, Jauréguy et al.9 found fyuA slightly more frequently in non-survivors (87.5% vs. 77.7%), but this difference was not significant.
The P fimbriae, encoded by the pap genes, mediate bacterial adhesion to host cells.30 Previous data regarding P fimbriae and mortality are also conflicting, i.e Jauréguy et al.9 found that the papGIII gene was related to fatal outcome, but in the COLIBAFI study8 the same genes played a protective role. A protective role was found also for papGII in a multicentric study of ESBL E. coli bacteremia.31 The mechanism for the protective effect is unknown, but is likely related to the immunogenicity of the P fimbriae, that might help to develop an earlier or stronger immune response.
As expected, multivariate analysis found host risk factors associated with the outcome of the bacteraemia: the McCabe-Jackson Index, active chemotherapy treatment and Pitt Index, all well documented in the literature.8,11,16 Pitt index is a well-established sepsis predictor17 that gives internal validity to the model.
We acknowledge some limitations in our study, mainly related to the relatively small sample size. Our cohort included a limited number of bacteremia episodes which had different sources and few cases of severe sepsis and fatal cases. The high genetic heterogeneity of the E. coli population, together with the low bacteremia-related-mortality limits our ability to establish associations with VFs. We have to acknowledge that the predictive value of the models to predict sepsis severity and mortality changed modestly, and with an overlap on CIs, when we removed the contribution of VF, suggesting limited prognostic importance.
Our study also shares with prior studies 8,9,12 the difficulties derived from the high diversity of genetic backgrounds, VF gene profiles and the heterogeneity in patient populations. The wide diversity in host and pathogen factors might be the major reason for the disparity of conclusions drawn among different studies. In our opinion these methodological difficulties call for different approaches, that might include multicentric studies with larger sample sizes, analyses of VF gene profiles instead of single VF genes,22 and whole-genome sequencing with genome-wide association studies 32 in order to identify E. coli determinants of sepsis severity or mortality that could be targets for specific intervention in the future. Probably, the most promising approach is whole-genome sequencing. These technologies are now becoming broadly accessible to microbiology laboratories, and it is very likely that soon this approach will be faster, cheaper and much more informative than PCR of selected genes.32
In summary, in our series of E. coli bacteraemia we found a mixed bacterial population structure, with a few epidemic clones accounting for half of the isolates, and several sporadic isolates making up the other half. The heterogeneity of the patient population and the extremely high genetic diversity in VF gene profiles complicate the analysis, but in spite of these, both, host and bacterial factors showed an impact on sepsis severity and mortality. The presence of the cnf and blaTEM genes was associated with sepsis severity while the presence of the fyuA gene was associated with mortality and the presence of P fimbria genes with improved survival. A multivariate logistic regression model found that the major parameters were related to the host status, while the VF genes had a modest, though significant, contribution to the model predictive capacity.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Our thanks are due to Martin J. Smyth, B. A., for his help in correcting the English.
Funding
This research was supported by a public grant to J. M. from Fondo de Investigaciones Sanitarias, Spain [FIS PI10/00795].
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. De Kraker MEA, Jarlier V, Monen JCM, Heuer OE, van de Sande N, Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect 2013; 19(9): 860-8. [DOI] [PubMed] [Google Scholar]
- 2. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004; 39:309-17; PMID:15306996; http://dx.doi.org/ 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 3. Yan F, Polk D. Commensal bacteria in the gut: learning who our friends are. Curr Opin Gastroenterol. 2004; 565-71; PMID:15703684; http://dx.doi.org/ 10.1097/00001574-200411000-00011 [DOI] [PubMed] [Google Scholar]
- 4. Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis 2000; 181:1753-4; PMID:10823778; http://dx.doi.org/ 10.1086/315418 [DOI] [PubMed] [Google Scholar]
- 5. Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol 2004; 2:123-40; PMID:15040260; http://dx.doi.org/ 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 6. Wang M-C, Tseng C-C, Chen C-Y, Wu J-J, Huang J-J. The role of bacterial virulence and host factors in patients with Escherichia coli bacteremia who have acute cholangitis or upper urinary tract infection. Clin Infect Dis 2002; 35:1161-6; PMID:12410475; http://dx.doi.org/ 10.1086/343828 [DOI] [PubMed] [Google Scholar]
- 7. Bonacorsi S, Houdouin V, Mariani-Kurkdjian P, Mahjoub-Messai F, Bingen E. Comparative prevalence of virulence factors in Escherichia coli causing urinary tract infection in male infants with and without bacteremia. J Clin Microbiol 2006; 44:1156-8; PMID:16517919; http://dx.doi.org/ 10.1128/JCM.44.3.1156-1158.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lefort A, Panhard X, Clermont O, Woerther PL, Branger C, Mentré F, Fantin B, Wolff M, Denamur E; COLIBAFI Group . Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J Clin Microbiol 2011; 49:777-83; PMID:21177892; http://dx.doi.org/ 10.1128/JCM.01902-10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jauréguy F, Carbonnelle E, Bonacorsi S, Clec'h C, Casassus P, Bingen E, Picard B, Nassif X, Lortholary O. Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis. Clin Microbiol Infect 2007; 13:854-62; PMID:17617183; http://dx.doi.org/ 10.1111/j.1469-0691.2007.01775.x. [DOI] [PubMed] [Google Scholar]
- 10. Landraud L, Jauréguy F, Frapy E, Guigon G, Gouriou S, Carbonnelle E, Clermont O, Denamur E, Picard B, Lemichez E, et al. Severity of Escherichia coli bacteraemia is independent of the intrinsic virulence of the strains assessed in a mouse model. Clin Microbiol Infect 2013; 19(1):85-90; http://dx.doi.org/ 10.1111/j.1469-0691.2011.03750.x [DOI] [PubMed] [Google Scholar]
- 11. Hekker TA, Groeneveld B, Simoons-Smit AM, de Man P, Connell H, MacLaren DM. Role of bacterial virulence factors and host factors in the outcome of Escherichia coli bacteraemia. Eur J Clin Microbiol Infect Dis 2000; 19:312-6; PMID:10834824; http://dx.doi.org/ 10.1007/s100960050483 [DOI] [PubMed] [Google Scholar]
- 12. Skjøt-Rasmussen L, Ejrnæs K, Lundgren B, Hammerum AM, Frimodt-Møller N. Virulence factors and phylogenetic grouping of Escherichia coli isolates from patients with bacteraemia of urinary tract origin relate to sex and hospital- vs. community-acquired origin. Int J Med Microbiol 2012; 302:129-34; PMID:22571989; http://dx.doi.org/ 10.1016/j.ijmm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 13. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control 1988; 16:128-40; PMID:2841893 [DOI] [PubMed] [Google Scholar]
- 14. Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL. Health care-Associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 2002; 137:791-7; PMID:12435215; http://dx.doi.org/ 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 15. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373-83; PMID:3558716; http://dx.doi.org/ 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 16. McCabe WR, Jackson GG. Gram-negative bacteremia I. Etiology and Ecology. Arch Intern Med 1962; 110:847-55. [Google Scholar]
- 17. Chow JW, Yu VL. Combination antibiotic therapy versus monotherapy for gram-negative bacteraemia: a commentary. Int J Antimicrob Agents 1999; 11:7-12; PMID:10075272 [DOI] [PubMed] [Google Scholar]
- 18. Bone RC, Sprung CL, Sibbald WJ. Definitions for sepsis and organ failure. Crit Care Med 1992; 20:724-6; PMID:1597021; http://dx.doi.org/ 10.1097/00003246-199206000-00002 [DOI] [PubMed] [Google Scholar]
- 19. Institute C and LS Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. CLSI M100-S22. Wayne, PA: CLSI; 2012. [Google Scholar]
- 20. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the escherichia coli phylogenetic group. Appl Environ Microbiol 2000; 66; 4555-8; PMID:11010916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernández-Romero N, Romero-Gómez MP, Gómez-Gil MR, Mingorance J. Epidemic population structure of extraintestinal pathogenic Escherichia coli determined by single nucleotide polymorphism pyrosequencing. Infect. Genet Evol 2011; 11:1655-63; PMID:21723423; http://dx.doi.org/ 10.1016/j.meegid.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 22. Rodríguez-Baño J, Mingorance J, Fernández-Romero N, Serrano L, López-Cerero L, Pascual A. Virulence profiles of bacteremic extended-spectrum β-lactamase-producing Escherichia coli: association with epidemiological and clinical features. PLoS One 2012; 7:e44238; PMID:22970186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aubert D, Naas T, Héritier C, Poirel L, Nordmann P. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J Bacteriol 2006; 188:6506-14; PMID:16952941; http://dx.doi.org/ 10.1128/JB.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson JR, Delavari P, Kuskowski M, Stell AL. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 2001; 183:78-88; PMID:11106538; http://dx.doi.org/ 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- 25. Ruiz J, Simon K, Horcajada J. Differences in virulence factors among clinical isolates of Escherichia coli causing cystitis and pyelonephritis in women and prostatitis in men. J Clin Microbiol 2002; 40:4445-9; PMID:12454134; http://dx.doi.org/ 10.1128/JCM.40.12.4445-4449.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duriez P, Clermont O, Bonacorsi S. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 2001; 147: 1671-6; PMID:11390698 [DOI] [PubMed] [Google Scholar]
- 27. Laupland KB, Gregson DB, Church DL, Ross T, Pitout JDD. Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. Clin Microbiol Infect 2008; 14:1041-7; PMID:19040476; http://dx.doi.org/ 10.1111/j.1469-0691.2008.02089.x [DOI] [PubMed] [Google Scholar]
- 28. Ananias M, Yano T. Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Brazilian J Med Biol Res 2008; 41:877-83; PMID:19030710; http://dx.doi.org/ 10.1590/S0100-879X2008001000008 [DOI] [PubMed] [Google Scholar]
- 29. Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000; 181:261-72; PMID:10608775; http://dx.doi.org/ 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 30. Wiles T, Kulesus R, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol 2008; 85:11-9; PMID:18482721; http://dx.doi.org/ 10.1016/j.yexmp.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodríguez-Baño J, Mingorance J, Fernández-Romero N, Serrano L, López-Cerero L, Pascual A; ESBL-REIPI Group . Outcome of bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli: impact of microbiological determinants. J Infect 2013; 67:27-34; PMID:23588104; http://dx.doi.org/ 10.1016/j.jinf.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 32. Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 2015; 25(1):119-28; http://dx.doi.org/10.1101/gr.180190.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.