Abstract
Prostaglandins are C20 fatty acid metabolites with diverse biological functions. In mammalian cells, prostaglandins are produced from arachidonic acid (AA) via cyclooxygenases (COX1 and COX2). Although fungi do not possess cyclooxygenase homologues, several pathogenic species are able to produce prostaglandins from host-derived arachidonic acid. In this study, we characterized the prostaglandin profile of the emerging human pathogen Candida parapsilosis with HPLC-MS and compared it to that of C. albicans. We found that both species synthesized prostaglandins (mainly PGD2 and PGE2) from exogenous AA. Furthermore, as OLE2 has been associated with prostaglandin synthesis in C. albicans, we generated homozygous OLE2 deletion mutants in C. parapsilosis and examined their PGE2 production. However, the PGE2 production of the OLE2 KO strain was similar to that of wild type (WT), indicating that OLE2 is not required for prostaglandin synthesis in C. parapsilosis. Interestingly, analyses of the fatty acid composition of WT and OLE2 KO cells by gas chromatography (GC) highlighted the accumulation of palmitoleic and oleic acid in the OLE2 deletion mutant. The OLE2 KO cells were killed more efficiently by human monocytes-derived macrophages (MDMs) as well as induced higher interleukin-10 (IL-10) secretion, indicating that OLE2 affects the virulence of C. parapsilosis. Taken together, these results contribute to the better understanding of fatty acid biosynthesis pathways in C. parapsilosis.
Keywords: Candida parapsilosis, fatty acid composition, immunomodilation, prostaglandins
Introduction
Candida parapsilosis is an emerging opportunistic human fungal pathogen, and it is currently one of the leading causes of invasive candidiasis.1-3 The populations with the highest risk for nosocomial infections with C. parapsilosis are the low-birth-weight neonates, elderly and immunocompromised patients, such as individuals with AIDS or organ transplantation.1,2,4 Unlike Candida albicans, C. parapsilosis is frequently transmitted horizontally, and it can cause invasive disease without colonizing the host before dissemination.4 The use of prosthetic devices and indwelling catheters also increases the risk for C. parapsilosis infection.4 However, despite its high clinical relevance, the pathogenesis of C. parapsilosis has remained largely obscure.
Prostaglandins (PGs) are biologically active polyunsaturated fatty acids (PUFAs) that have important signaling and immunomodulatory functions.5 In mammalian cells, they are synthesized de novo from membrane-derived arachidonic acid (AA) via cyclooxigenases (COX).5,6 Prostaglandins play an important role in the regulation of immune responses by modulating phagocytosis, cytokine and chemokine production and release, and lymphocyte proliferation.7 In particular, PGE2 inhibits T helper (Th) 1 and promotes Th2 responses by modulating the cytokine production of lymphocytes.8 Numerous pathogenic fungi, such as C. albicans and Cryptococcus neoformans, are able to produce prostaglandins along with other arachidonic acid metabolites.9 Importantly, C. albicans PGE2 is biologically active on mammalian cells, as it inhibits the proliferation of splenocytes and decreases their TNFα production while stimulating IL-10 secretion.7 Furthermore, PGE2 modulates the morphogenesis of C. albicans, further supporting the role of fungal prostaglandins as potential virulence factors.7 Although little information is available about the biosynthesis of prostaglandins in Candida spp., one study has described that the multicopper oxidase gene FET3 and the stearyl-CoA desaturase gene OLE2 are involved in PGE2 synthesis in C. albicans.10 This finding indicates a direct link between the production of fatty acids and bioactive lipid mediators, underscoring the role of de novo fatty acid biosynthesis in pathogenic yeasts.
Saturated and unsaturated fatty acids (SFA and UFA, respectively) have an essential role in eukaryotic cells as building blocks of membranes and lipid storage.11,12 De novo fatty acid biosynthesis is essential for growth and virulence of both C. albicans and C. parapsilosis.13-15 As in Saccharomyces cerevisiae, the biosynthesis of fatty acids in C. albicans and C. parapsilosis starts with the generation of malonyl-CoA from acetyl-CoA and CO2, via the biotin-bound enzyme acetyl-CoA carboxylase (AccI).,17 In the second step, the elongation of the carbon chain is catalyzed by fatty acid synthases (Fas1 and Fas2) and elongase 1 (Elo1), resulting in long-chain SFA.18,19 To produce UFA, a subsequent desaturation reaction catalyzed by stearyl-CoA desaturase (Ole1) is necessary to introduce a double bond into saturated fatty acyl-CoA substrates.17,20 In C. albicans and C. parapsilosis, there are 2 homologues with the S. cerevisiae OLE1, OLE1 and OLE2.21 While OLE1 is essential in C. albicans, deletion of OLE1 in C. parapsilosis results in UFA auxotrophic mutants.17,21 Importantly, Ole1 plays a crucial role in virulence in both species, as demonstrated by in vivo murine infection models.17,22 However, while OLE2 has been associated with prostaglandin synthesis in C. albicans, the role of OLE2 in C. parapsilosis is unknown.
In this study, our main objective was to examine whether C. parapsilosis is able to synthesize prostaglandin-like molecules from exogenous AA. Furthermore, to assess the potential role of OLE2 in prostaglandin production, we generated an OLE2 “knock-out” (KO) C. parapsilosis strain. To gain further insights into the role of OLE2 in the pathobiology of C. parapsilosis, we also analyzed the fatty acid composition and the virulence properties of the mutant strain.
Results
C. parapsilosis produces prostaglandins from exogenous arachidonic acid
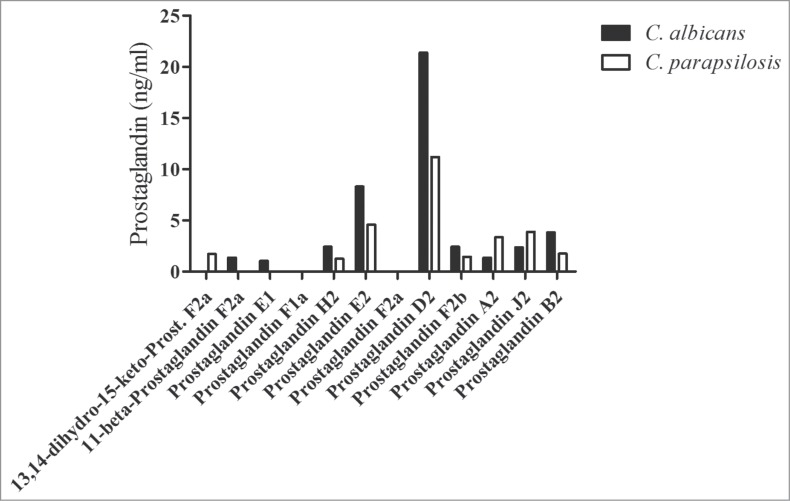
First we wanted to examine whether C. parapsilosis is able to synthesize prostaglandins from exogenous AA. To characterize the prostaglandin profiles of C. parapsilosis and C. albicans, wild type yeast cells were grown overnight in YPD medium and subsequently incubated in PBS with or without 0.5 mM AA for 24 hours. The levels of different prostaglandin derivatives (13,14-dihydro-15-keto-PGF2α, 11β-PGF2α, PGE1, PGF1α, PGH2, PGE2, PGF2α, PGD2, PGF2β, PGA2, PGJ2, PGB2) in cell-free supernatants were determined by HPLC-MS analysis (Fig. 1). Following incubation with AA, we detected a variety of prostaglandin compounds in the supernatants of C. albicans and C. parapsilosis cultures. In both species, the main secreted prostaglandins were PGD2 and PGE2. Furthermore, we also detected compounds from other prostaglandin classes, such as PGH2, PGF2β and PGA2, in the supernatants of both C. albicans and C. parapsilosis cultures. However, we did not detect any prostaglandin production in our experimental system when Candida cells were incubated solely in PBS. These results clearly demonstrate that, C. parapsilosis synthesizes prostaglandins from exogenous AA.
Figure 1.
C. albicans and C. parapsilosis produce prostaglandins from exogenous AA. C. albicans and C. parapsilosis WT strains were cultured in YPD medium overnight, washed, and subsequently incubated in PBS for 24 h with 500 µM AA. The concentration of different prostaglandin compounds in culture supernatants was determined by HPLC-MS analysis. Results are representative of 2 independent experiments.
CpOLE2 is not required for prostaglandin synthesis
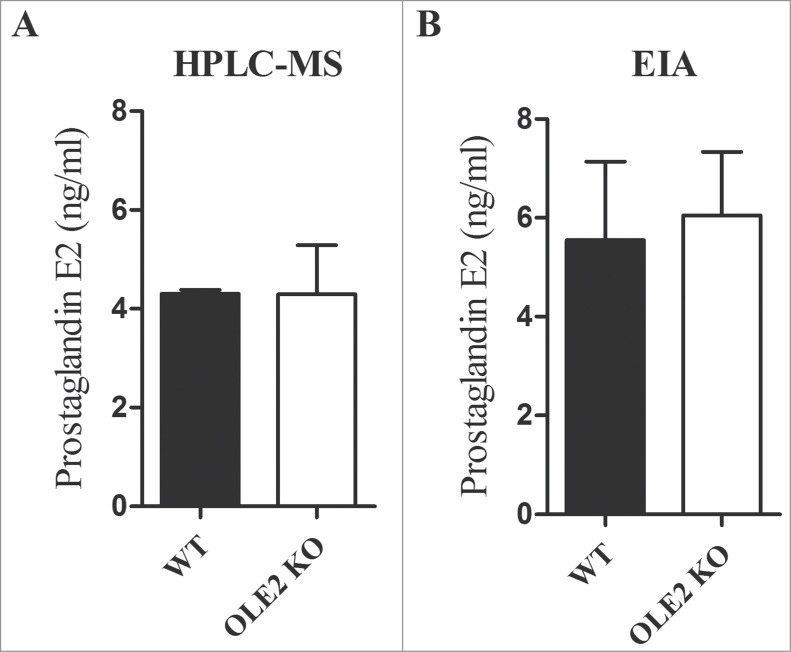
The biosynthetic pathway of fungal eicosanoid production is still not fully understood. However, several candidate enzymes, including cytochrome P450s and multicopper oxidases have been implicated in prostaglandin production.23 The disruption of the putative Δ9-desaturase gene OLE2 in C. albicans results in significantly decreased PGE2 production.10 Therefore, in order to examine whether OLE2 is involved in prostaglandin synthesis in C. parapsilosis, we constructed homozygous OLE2 deletion mutants (Fig. S1), and tested their PGE2 production using HPLC-MS as well as a monoclonal PGE2 EIA kit. However, we found that OLE2 mutants secreted PGE2 in amounts that were similar to that of the WT parental strain (Fig. 2). Therefore, OLE2 is not required for PGE2 biosynthesis in C. parapsilosis.
Figure 2.
CpOLE2 is not required for PGE2 synthesis. C. parapsilosis WT and OLE2 KO strains were cultured in YPD medium overnight, washed, and subsequently incubated in PBS for 24 h with or without 500 µM AA.The concentration of PGE2 in culture supernatants was determined by HPLC-MS (A) or EIA (B). Results are mean ± SEM and represent the average of 3 independent experiments.
Role of CpOLE2 in de novo fatty acid biosynthesis
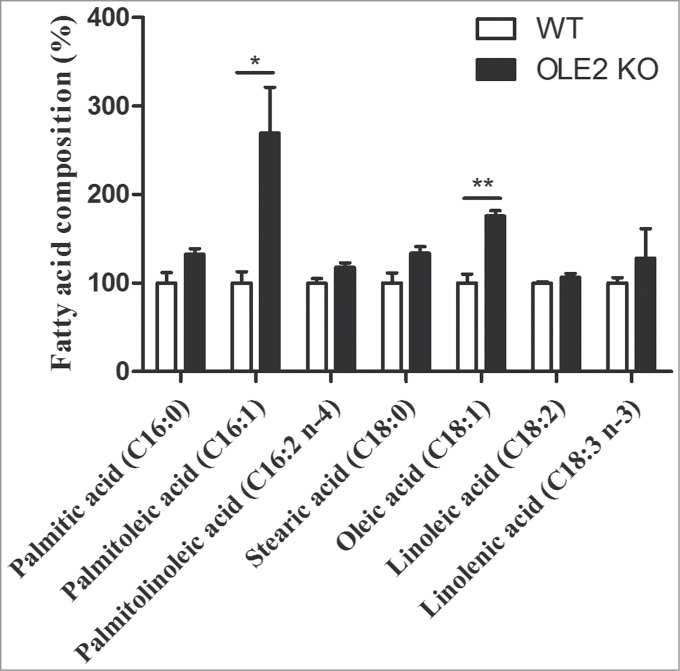
After the finding that OLE2 is not required for PGE2 production in C. parapsilosis, we asked the question whether it has a role in de novo fatty acid biosynthesis. To investigate this possibility, we compared the fatty acid profile of OLE2 mutants with that of WT C. parapsilosis. The amount of different fatty acids (palmitic acid, palmitoleic acid, palmitolinoleic acid, stearic acid, oleic acid, linoleic acid and linolenic acid) in fungal cell extracts was determined by gas chromatography. Figure 3 shows the fatty acid composition of OLE2 KO C. parapsilosis cells relative to that of WT cells. Interestingly, we found that OLE2 mutants accumulated significantly more palmitoleic and oleic acid compared to WT C. parapsilosis cells (mean ± SEM, palmitoleic acid, 615.0 ± 117.5 vs. 228.1 ± 29.1 ng/mg dry weight, P < 0.05; oleic acid, 16.3 ± 0.5 vs. 9.2 ± 0.9 µg/mg dry weight, P < 0.01). These monounsaturated fatty acids are synthesized from palmitic and stearic acid, respectively, via Δ9-desaturases, and are further converted into palmitolinoleic (16:2 n-4) and linoleic acid (18:2) by Δ12-desaturases. However, we found that the cellular levels of palmitic and stearic acids as well as that of palmitolinoleic and linoleic acids were unaffected by the absence of OLE2. Taken together, these results suggest that C. parapsilosis Ole2 is involved in fatty acid biosynthesis and the action is presumably not or not exclusively a Δ9-desaturase.
Figure 3.
OLE2 KO C. parapsilosis cells have altered fatty acid composition. WT and OLE2 KO C. parapsilosis cells were cultured in YPD medium overnight, washed, lyophilized, and subjected to analysis by gas chromatography. For better comparison, fatty acid levels were normalized to those of WT cells and expressed as %. The actual concentration of palmitoleic and oleic acid (expressed as ng/mg or µg/mg dry weight) is described in Results. Data are mean ± SEM and represent the average of 3 independent experiments. *P < 0.05, **P < 0.01 by unpaired t-test.
OLE2 C. parapsilosis mutants have decreased virulence in vitro
Next we sought to determine whether the lack of OLE2 affected the virulence of C. parapsilosis. First we examined the growth of OLE2 mutants in comparison to WT on different media (YPD, YNB, YCB/BSA; Fig. S1), at different temperatures (20°C, 30°C, 37°C; Fig. S2), as well as at different pH ranges (pH 4-8; Fig. S3). We also assessed the growth of the mutant strain under different oxidative, membrane and cell wall stress-inducing conditions (H2O2, SDS, calcofluor white, congo red, caffeine; Fig. S4). However, the growth of the mutant strain was similar to the WT for all circumstances tested. Also, OLE2 deletion did not affect the formation of pseudohyphae (Fig. S5). Next we investigated the phagocytosis and killing of OLE2 mutants by primary human monocyte-derived macrophages (MDMs) using an acridin orange staining method (Fig. 4). We found that more MDMs phagocytosed the OLE2 mutants (phagocytosis%, mean ± SEM, 69.2 ± 5.7% vs. 52.7 ± 4.9%, P < 0.05, Fig. 4C), and OLE2 deficient cells were killed with significantly higher efficiency compared to WT yeasts (killing efficiency, mean ± SEM, 50.2 ± 8.7% vs. 41.2 ± 8.4%, P < 0.05, Fig. 4D). We also examined the killing of OLE2 mutant C. parapsilosis cells by CFU determinations, which confirmed the results of the fluorescence microscopic analysis (Fig. 4E). As fungal lipids can modulate the immune response,7 we also examined the cytokine secretion of MDMs following stimulation with OLE2 mutants or WT C. parapsilosis cells. We found that while the level of TNFα, IL-1β and IL-6 were similar in MDMs stimulated with OLE2 mutant or WT C. parapsilosis (not shown), OLE2 mutants stimulated significantly higher IL-10 production than WT cells (mean ± SEM, 581.8 ± 292.2 pg/mL vs. 437.0 ± 246.9 pg/mL, P < 0.05, n = 5). Fig. 4F shows the cytokine secretion of MDMs stimulated with OLE2 KO or WT C. parapsilosis (cytokine levels were normalized for each donor to cytokine levels induced by WT cells [100%] to exclude donor-to-donor variability). Altogether, these results suggest that OLE2 affects the virulence of C. parapsilosis, although further research is warranted to determine its specific role during infection.
Figure 4.
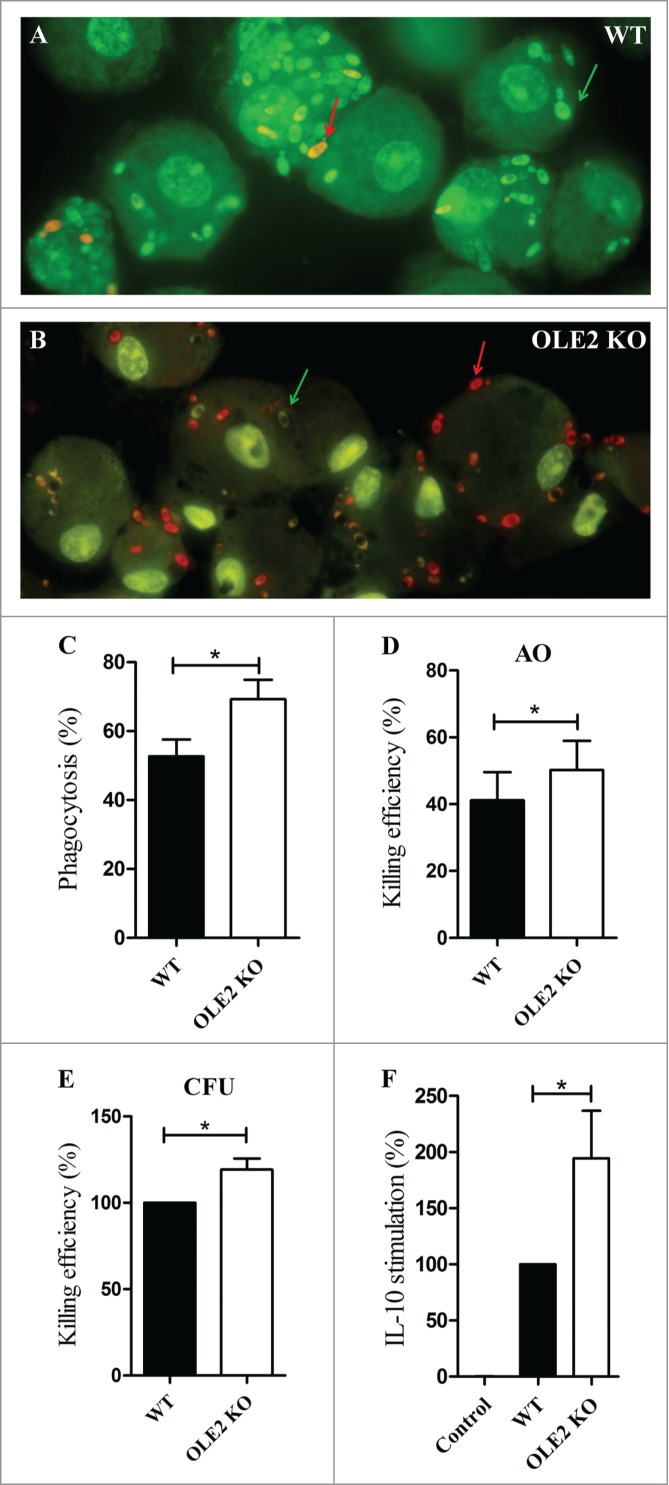
The lack of OLE2 affects the virulence of C. parapsilosis. (A–B) Acridine orange staining of MDMs co-cultured for 3 h with WT (A) or OLE2 KO (B) C. parapsilosi s. Live cells show green fluorescence (green arrow), dead cells show red fluorescence (red arrow). (C) Phagocytosis of WT and OLE2 KO C. parapsilosis by human MDMs as determined by fluorescent microscopic analysis. Phagocytosis is expressed as the percent of macrophages that have ingested at least one yeast cell (phagocytosis%). Data represent mean ± SEM for 4 donors. (D) Intracellular killing of WT and OLE2 KO C. parapsilosis by human MDMs as determined by fluorescent microscopic analysis. Results represent the percent of dead yeast cells ± SEM for 4 donors. AO, acridin orange. (E) Killing of WT and OLE2 KO C. parapsilosis by human MDMsas measured by CFU determinations. Data were normalized to WT and represent mean ± SEM for 4 donors. (F) MDMs were stimulated for 24 h with WT or OLE2 KO C. parapsilosis and the concentration of IL-10 in cell culture supernatants was determined by ELISA. Data were normalized for each donor to cytokine levels induced by the WT strain (100%) and are expressed as mean ± SEM for 5 donors. Actual cytokine levels are described in Results. *P < 0.05 as determined by paired t-test using the GraphPad Prism 5 software.
Discussion
Several pathogenic Candida spp., including C. albicans, C. tropicalis and C. dubliniensis produce prostaglandins.7,24 In this report, we demonstrate that C. parapsilosis is also capable of synthesizing prostaglandins from exogenously added AA. We show that the prostaglandin profile of C. parapsilosis is similar to that of C. albicans, with PGE2 and PGD2 being the main prostaglandins produced. In mammals, prostaglandins are important signaling molecules that are crucial for the regulation of the inflammatory response.5 They are synthesized from AA which is liberated from membrane phospholipids by phospholipase A2 upon inflammatory stimuli.8 Subsequently, AA is converted to PGH2 by cyclooxigenases, and PGH2 is further processed by tissue-specific prostaglandin synthases to give rise to a series of different classes of prostaglandins.8 Secretion of fungal prostaglandins during infection may substantially influence the host's immune response, promoting the survival of the pathogen. Notably, we found that the most abundant prostaglandin produced by both C. albicans and C. parapsilosis was PGD2, which exerts both pro-and anti-inflammatory properties, and is also involved in thermoregulation, hormone release and pain responses in mammals. Furthermore, PGD2 is converted by a non-enzymatic reaction to 15-deoxy-Δ12,14-PGJ2, which is a potent anti-inflammatory lipid mediator and has been shown to inhibit the NFκB signaling pathway.25 As a potent pro-inflammatory response is required for the successful clearance of invasive Candida, the anti-inflammatory effects of 15-deoxy-Δ12,14-PGJ2 may be beneficial for the pathogen. PGE2 was also produced in high concentrations by C. albicans and C. parapsilosis, and PGE2 mediates T-cell responses and influences other physiological functions.8 Due to its ability to block Th1-type responses and promote Th2-type immunity,8 fungal PGE2 may also help the pathogen survive in the host.
Although the pathway of eicosanoid production in mammalian cells is well studied, little is known about the biosynthesis of prostaglandins in lower eukaryotes. While C. albicans does not possess cyclooxigenases, several enzymes, such as the fatty acid desaturase Ole2 and the multicopper oxidase Fet3 have been shown to contribute to prostaglandin production.10 The presence of a conserved Δ9-desaturase domain in Ole2 protein suggested that it has Δ9-desaturase activity, while its cytochrome B domain was thought to be responsible for prostaglandin synthesis.10 However, the role of OLE2 in C. parapsilosis has never been previously investigated. To examine whether OLE2 is involved in prostaglandin synthesis in C. parapsilosis, we generated OLE2 deletion mutants using a previously described gene disruption technique.26 However, we found that there was no difference between the PGE2 secretion of WT and OLE2 KO C. parapsilosis cells. The fact that OLE2 is not required for PG synthesis in C. parapsilosis is unexpected, but can be explained considering that C. albicans and C. parapsilosis have crucial differences in their fatty acid metabolism. Interestingly, we also found that OLE2 mutants had a different fatty acid composition compared to WT cells, containing higher amounts of palmitoleic and oleic acids. These UFAs are produced from palmitic and stearic acids, respectively, by Δ9-desaturases and are further converted to PUFAs via Δ12-desaturases.27 The accumulation of palmitoleic and oleic acids in OLE2 KO cells argues against the role of OLE2 as an exclusive Δ9-desaturase and suggests that it may have other enzymatic activities as well. These results are in line with the findings of Krishnamurthy et al., who demonstrated that heterologous expression of CaOLE2 could not reconstitute the phenotype of S. cerevisiae OLE1 (Δ9-desaturase) mutants, implicating that Ole2 is not a Δ9-desaturase.21 Moreover, UFA auxotrophic OLE1 C. parapsilosis mutants are unable to grow in SFA-supplemented media without the presence of UFA, indicating that Ole1 is the only enzyme in C. parapsilosis responsible for the synthesis of these essential fatty acids.17 Further supporting these results, we found that Ole2 proteins from different Candida species (including C. parapsilosis and C. albicans) form a phylogenetically distinct group which is far from both Δ9− and Δ12− fungal desaturases (Fig. S6).
Interestingly, we also found that OLE2 KO C. parapsilosis cells were phagocytosed and killed more efficiently by human macrophages compared to WT cells. One potential explanation for this decreased virulence is that the accumulated UFAs are toxic to yeast cells; however, we found no difference between the growth of OLE2 KO and WT strains in medium conditions, making this possibility unlikely. On the other hand, as it has been shown with OLE1 KO C. parapsilosis cells,17 OLE2 mutants could become hypersensitive to cellular stress. However, we found that the lack of OLE2 did not impact the capacity of C. parapsilosis to cope with extracellular stressors such as H2O2 or SDS (data not shown). In addition to increased phagocytosis and killing, OLE2 mutants also stimulated higher IL-10 production in macrophages compared to WT cells. IL-10 has been shown to increase the phagocytic capacity of macrophages28; therefore, suppression of IL-10 secretion may also be beneficial for the pathogen during infection.
There is an increasing amount of evidence showing that the integrity of fatty acid biosynthesis pathways in pathogenic fungi is essential for growth and virulence. For example, the downregulation of fatty acid synthase genes FAS1 and FAS2 substantially affects the virulence of C. neoformans in a murine model of pulmonary cryptococcosis.29 Similarly, fatty acid synthase 2 (FAS2) mutant C. parapsilosis cells are defective in biofilm formation, are highly sensitive to human serum and have decreased virulence in vivo compared to WT cells.30 Moreover, the stearyl-CoA desaturase Ole1 regulates the stress response and cellular morphology of C. albicans and C. parapsilosis, and it plays a major role in the pathobiology of both species.17,22 In accordance with these findings, our results further support the role of fatty acid homeostasis in the virulence of pathogenic fungi.
Taken together, we have shown here that C. parapsilosis is capable of producing prostaglandins similarly to other Candida spp. Furthermore, we generated a OLE2 KO C. parapsilosis strain to assess the role of this gene in prostaglandin synthesis. Although we found that OLE2 is not required for PGE2 synthesis in C. parapsilosis, we show that OLE2 deficient C. parapsilosis cells have altered fatty acid composition, which is accompanied by decreased virulence in vitro. Although further investigation is needed to reveal the exact function of OLE2 during infection, our results contribute to the better understanding of fatty acid biosynthesis pathways in Candida parapsilosis.
Materials and Methods
Strains and growth conditions
Unless otherwise specified, C. albicans SC5314, Candida parapsilosis strains (GA1 wild type [WT]31 and OLE2 mutant [ΔCpole2/ΔCpole2::FRT]) were grown overnight in liquid YPD medium (1% yeast extract, 2% bactopepton, 2% glucose) at 30°C. Cells were harvested by centrifugation, washed twice with PBS (phosphate-buffered saline; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4; pH 7.4) and counted in a Bürker-chamber prior to experiments.
Generation of the disruption constructs pSFS2Ole2 and pSFS2NewOle2
The pSFS2Ole2 and pSFS2NewOle2 plasmids were used to disrupt the open reading frame of the OLE2 gene, consisting of 1 653 nucleotides. In order to construct the pSFS2Ole2 vector, a 599bp upstream and a 594bp downstream fragment was amplified from C. parapsilosis GA1 genomic DNA. For the amplification of the upstream PCR product “CpOLE2 upstKpnIfrw” (5′-ttttttggtaccGAAAAATGAAGCTAAATCCTCCAAGGGAC-3′; [KpnI site underlined]) and “CpOLE2 upstXhoI rev” (5′-ttttttctcgagATAGGAAAAGTAGTTGAAAATGCGCACAC-3′; [XhoI]) primers were used. For the amplification of the downstream PCR product “CpOLE2 down NotIfrw” (5′-ttttttgcggccgcGCGTTGTTTCTCATCCATTTGAGGAAATTC-3′; [NotI]) and “CpOLE2 down SacI rev” (5′-ttttttgagctcGGTTTATACTCAAATGGATGAATAGTCTTCTG-3′; [SacI]) primers were used. To construct the pSFS2NewOle2 vector, only the upstream region was changed for easier detection of the elimination of the second allele. For the amplification of the upstream 417 bp PCR product “CpOLE2 NewupstKpnIfrw” (5′-ttttttggtaccCTATTTACACTAAGCAGCTTCGGCAG-3′; [KpnI]) and “CpOLE2 NewupstXhoI rev” (5′-ttttttctcgagGGCACACCAATATCTAAACTACCATTG-3′; [XhoI]) primers were used. The PCR fragments were ligated into pSFS232 appropriate cloning sites (KpnI/XhoI and NotI/SacI) to generate the pSFS2Ole2 and pSFS2NewOle2 disruption plasmids.
Candida parapsilosis transformation and generation of CpOle2 deletion mutants
C. parapsilosis WT cells were transformed by electroporation as previously described,31 with 5 μg KpnI-SacI digested and purified fragment from the pSFS2Ole2 plasmid. For the disruption of the second allele, heterozygous mutants (OLE2/Δ ole2 :: FRT) were transformed with 5 μg KpnI-SacI digested and purified fragment from the pSFS2NewOle2 plasmid. Transformants were regenerated and analyzed by Southern blotting (Fig. Supplementary 1). As our primary focus was to measure the production of prostaglandins and we did not find any difference between the WT and OLE2 strain, we did not include the reconstituted strain in our study.
Determination of prostaglandin profile by HPLC-MS
For prostaglandin measurements, 100 ml PBS (control) or 100 ml PBS+AA (500 μM AA [Sigma, A9673] in PBS) was inoculated with 2 × 109 yeast cells and incubated for 24 h at 30°C. Subsequently, cells were harvested by centrifugation and the supernatants were collected by sterile filtration. Samples were analyzed with HPLC-MS technique using an Agilent 1100 liquid chromatograph and an Agilent 6410 triple quadrupole mass spectrometer (Palo Alto, USA). For the HPLC separation, a YMC Pack ProC18 (150 mm × 2.1 mm, 5 µm) (Dinslaken, Germany) column was applied with a guard column (20 mm × 2.1 mm, 5 µm). The A eluent was water, while the B eluent was acetonitrile/methanol (95:5) and both of them were supplemented with 0.005% acetic acid. The flow rate of the mobile phase was 0.3 ml/min with the gradient program progressing from 25% to 40% at 10 minutes and 50% at 20 minutes, and then reduced to 25% for an additional 15 minutes. For ionization, an ESI ion source was used with negative ionization at the following mass spectrometric parameters: nitrogen drying gas temperature, 325°C; drying gas flow, 12 l/min; capillary voltage, 3800 V; fragmentation voltage, 110 V. The analyzer operated in the multiple reaction monitoring (MRM) mode using 2 transitions for each components with the following transition and collosion energy (CE) parameters: 13,14-dihydro-15-keto-prostaglandin F2α (DKPGF2α, 353.2→309.1-20V, 291.2-20V), 11β-Prostaglandin F2α (11βPGF2α, 353.2→309.1-20V, 291.2-20V), Prostaglandin E1 (PGE1, 353.2→309.1-20V, 273.3-20V), Prostaglandin F1α (PGF1α, 355.5→311.2-20V, 293.2-20V), Prostaglandin H2 (PGH2, 351.3→271.2-15V, 180.1-15V), Prostaglandin E2 (PGE2, 351.3→271.2-15V, 315.1-15V), Prostaglandin F2α (PGF2α, 353.2→317.2-20V, 273.3-20V), Prostaglandin D2 (PGD2, 351.3→271.2-15V, 189.1-15V), Prostaglandin F2β (PGF2β, 353.2→291.2-20V, 317.2-20V), Prostaglandin A2 (PGA2, 333.3→271.2-20V, 189.2-20V), Prostaglandin J2 (PGJ2, 333.3→271.2-20V, 189.2-20V), Prostaglandin B2 (PGB2, 333.3→235.0-20V, 175.0-20V). For method development and quantifications, the Prostaglandin HPLC mixture (10002), Cyclopentenone Prostaglandin HPLC mixture (10000), Prostaglandin Metabolite HPLC mixture (10005) and Prostaglandin H2 (17020, all from Cayman Chemical) were used. All measurements were carried out in triplicates.
Determination of fatty acid composition of C. parapsilosis strains by gas chromatography
For fatty acid measurements, strains were grown as described in 1 l YPD medium. After washing with PBS, yeast cells were lyophilized. The sample preparation was carried out according to Wei et al.33 with minor modifications. Approximately 100 mg of a lyophilized sample was supplemented with 100 µg heptadecanoic acid (Sigma, H3500) as internal standard (1000 μg/ml). Samples were suspended in 5 ml of 5% methanolic solution of potassium hydroxide and saponified at 70°C for 1 hour. Subsequently, the pH of the mixture was adjusted to 2 with HCl. A mixture of 4 ml water and 2 ml chloroform was added and the samples were vigorously shaken. The chloroform phase was collected after centrifugation (4000 rpm, 4°C, 15 min) and the extraction was repeated again from the upper phase with 1 ml chloroform. The collected chloroform phases were pooled and evaporated under a stream of nitrogen. Four ml of 14% methanolic solution of boron-trifluoride was added to the saponified samples, and incubated at 70°C for 1.5 hour. The fatty acid methyl-esters (FAMEs) were partitioned with hexane, which was evaporated to dryness under nitrogen. Finally, the derivatized samples were reconstituted in 50 µl hexane before chromatographic analysis. The samples were analyzed with an Agilent 6890N (Palo Alto, USA) gas chromatograph (GC) and fatty acid methyl esters were identified by comparison of their retention times with those of standards (methyl palmitate, Sigma, P5177; methyl palmitoleate, Sigma, P9667; 9(Z),12(Z)-hexadecadienioc-acid methyl ester, Larodan AB, 20-1620-4; methyl stearate, Sigma, S5376; methyl oleate, Sigma, 311111; methyl linoleate, Sigma, L1876; methyl linolenate, Sigma, L2626). The instrument was equipped with an FID detector and an HP-Innowax (60 m*0.25 mm*0.5 µm) (Hewlett-Packard, USA) column. The injected volume was 1 µl of sample in split mode (split ratio was 50:1) and the injector and detector temperatures were 250°C. The temperature gradient was 50°C for 2 min, then increased to 200°C by 20°C/min, and then followed by increases of 3°C/min to reach 240°C where it was maintained for 50 min. For the separations, constant pressure mode (32 psi) was used and the flow of the detector gases were 30 ml/min, 300 ml/min and 20 ml/min for the hydrogen, air and nitrogen, respectively. The samples were analyzed under the same conditions at least 3 times.
Isolation of human peripheral blood mononuclear cells (PBMCs) and differentiation of monocyte-derived macrophages (MDM) (under approval of the institutional ethical review board of Szeged University)
PBMC isolation and differentiation was carried out as described34 with minor modifications. Following isolation, PBMCs were plated on different cell culture plates: 96-well for killing assay (5 × 105cell/well); tissue culture coverslips (13mm, SARSTEDT, 83.1840.002) for acridine orange staining (5 × 105cell/coverslip) or 12-well for cytokine measurement (107cell/well). To differentiate macrophages, isolated monocytes were cultured for 7 d in X-VIVO 15 medium (Lonza, 04-744Q) supplemented with 10 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF, Sigma-Aldrich, SRP3050) and 1% 100ċ penicillin–streptomycin solution (Sigma-Aldrich, P0781).
Fluorescent microscopy
MDMs were cultured on tissue culture coverslips (13 mm, SARSTEDT) in 12-well cell culture plates. The in vitro infection of the macrophages was carried out at a 1:5 macrophage:yeast ratio. After 3 h incubation at 37°C, the samples were stained with 0.01% acridine orange dye (Sigma, 235474) as described.34 This method allows differentiation between live and dead yeast cells. When acridine orange binds to intact (ds) DNA, it emits green fluorescence, while binding to damaged (ss) DNA it shows red fluorescence, indicating dead cells. For quenching the fluorescence of non-phagocytosed yeast cells, 0.05% crystal violet (Sigma, C3886) dye was used. The killing efficiency was calculated as follows: (number of dead yeast cells / number of engulfed yeast cells) × 100. Phagocytosis% was calculated as follows: (phagocytosing macrophages / all macrophages) × 100. In every experiment, 10 fields in each well were counted, and approximately 1000 MDMs were analyzed in each well.
Killing assay
Human macrophages were cultured on 96-well tissue culture plates. The in vitro infection of the macrophages was carried out at a 1:5 macrophage:yeast ratio. As a control, the same number of yeast cells were incubated in cell culture medium without macrophages. The killing assays were carried out as described.34
Cytokine measurements
Human macrophages were cultured on 12-well tissue culture plates. The in vitro infection of the macrophages was carried out at a 1:5 macrophage:yeast ratio. Cell culture supernatants were collected after 24 h and stored at −20°C until assayed for cytokine production. The concentration of cytokines in cell culture supernatants was determined by DuoSet ELISA Kits (R&D Systems; TNFα, DY210; IL-1β, DY201; IL-6, DY206; IL-10, DY217B) according to the manufacturer's instructions.
PGE2 EIA
For PGE2 measurement, the strains were grown as described above. Subsequently, 100 ml PBS (control) or 100 ml PBS + AA (500 μM AA [Sigma] in PBS) was inoculated with 2 × 109 yeast cells and incubated for 24 h at 30°C. Cells were harvested by centrifugation and the supernatants were sterile filtered. Samples were purified on a PGE2 Affinity Column (Cayman Chemical, 400056) and analyzed with a Prostaglandin E2 EIA Kit – Monoclonal (Cayman Chemical, 514010) according to the manufacturer's instructions.
Statistical analysis
GraphPad Prism 5 software was used for statistical analysis. Data were analyzed by paired or unpaired t-test (see Figure legends for details) and differences were considered statistically significant at P < 0.05. All experiments were performed at least twice (see Results and figure legends for details).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
We thank László G Nagy for his help with the phylogenetic analysis.
Funding
This research was supported by the European Union and the State of Hungary, co-financed by the European Social Fund in the framework of TÁMOP 4.2.4. A/2-11-1-2012-0001 “National Excellence Program.” AG is supported by OTKA NN100374, NF84006 and by EMBO Installation Grant 1813. AG is also supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.van Asbeck EC, Clemons KV, Stevens DA. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit Rev Microbiol 2009; 35:283-309; PMID:19821642; http://dx.doi.org/10.3109/10408410903213393 [DOI] [PubMed] [Google Scholar]
- 2.Kreusch A, Karstaedt AS. Candidemia among adults in Soweto, South Africa, 1990–2007. Int J Infect Dis 2013; 17:e621-3; PMID:23535300; http://dx.doi.org/10.1016/j.ijid.2013.02.010 [DOI] [PubMed] [Google Scholar]
- 3.Nucci M, Queiroz-Telles F, Alvarado-Matute T, Tiraboschi IN, Cortes J, Zurita J, et al. . Epidemiology of candidemia in Latin America: a laboratory-based survey. PLOS One 2013; 8:e59373; PMID:23527176; http://dx.doi.org/10.1371/journal.pone.0059373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 2008; 21:606-25; PMID:18854483; http://dx.doi.org/10.1128/CMR.00013-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 2001; 294:1871-5; PMID:11729303; http://dx.doi.org/10.1126/science.294.5548.1871 [DOI] [PubMed] [Google Scholar]
- 6.Murakami M, Kudo I. Recent advances in molecular biology and physiology of the prostaglandin E2-biosynthetic pathway. Prog Lipid Res 2004; 43:3-35; PMID:14636669; http://dx.doi.org/10.1016/S0163-7827(03)00037-7 [DOI] [PubMed] [Google Scholar]
- 7.Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun 2001; 69:2957-63; PMID:11292712; http://dx.doi.org/10.1128/IAI.69.5.2957-2963.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris SG, Padilla J, Koumas L, Ray D, Phipps RP. Prostaglandins as modulators of immunity. Trends Immunol 2002; 23:144-50; PMID:11864843; http://dx.doi.org/10.1016/S1471-4906(01)02154-8 [DOI] [PubMed] [Google Scholar]
- 9.Noverr MC, Toews GB, Huffnagle GB. Production of prostaglandins and leukotrienes by pathogenic fungi. Infect Immun 2002; 70:400-2; PMID:11748207; http://dx.doi.org/10.1128/IAI.70.1.400-402.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erb-Downward JR, Noverr MC. Characterization of prostaglandin E2 production by Candida albicans. Infect Immun 2007; 75:3498-505; PMID:17470538; http://dx.doi.org/10.1128/IAI.00232-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta 2007; 1771:255-70; PMID:16950653; http://dx.doi.org/10.1016/j.bbalip.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 12.Nielsen J. Systems biology of lipid metabolism: from yeast to human. FEBS Lett 2009; 583:3905-13; PMID:19854183; http://dx.doi.org/10.1016/j.febslet.2009.10.054 [DOI] [PubMed] [Google Scholar]
- 13.Noverr MC, Huffnagle GB. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect Immun 2004; 72:6206-10; PMID:15501745; http://dx.doi.org/10.1128/IAI.72.11.6206-6210.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, Sillaots S, et al. . Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A 2010; 107:22044-9; PMID:21135205; http://dx.doi.org/10.1073/pnas.1009845107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen LN, Gacser A, Nosanchuk JD. Secreted lipases supply fatty acids for yeast growth in the absence of de novo fatty acid synthesis. Virulence 2011; 2:538-41; PMID:22030857; http://dx.doi.org/10.4161/viru.2.6.18244 [DOI] [PubMed] [Google Scholar]
- 16.Schneiter R, Guerra CE, Lampl M, Gogg G, Kohlwein SD, Klein HL. The Saccharomyces cerevisiae hyperrecombination mutant hpr1 Delta is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol Cell Biol 1999; 19:3415-22; PMID:10207065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen LN, Gacser A, Nosanchuk JD. The stearoyl-coenzyme A desaturase 1 is essential for virulence and membrane stress in Candida parapsilosis through unsaturated fatty acid production. Infect Immun 2011; 79:136-45; PMID:20974817; http://dx.doi.org/10.1128/IAI.00753-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweizer E, Hofmann J. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev 2004; 68:501-17, table of contents; PMID:15353567; http://dx.doi.org/10.1128/MMBR.68.3.501-517.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneiter R, Tatzer V, Gogg G, Leitner E, Kohlwein SD. Elo1p-dependent carboxy-terminal elongation of C14:1Delta(9) to C16:1Delta(11) fatty acids in Saccharomycescerevisiae. J Bacteriol 2000; 182:3655-60; PMID:10850979; http://dx.doi.org/10.1128/JB.182.13.3655-3660.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin CE, Oh CS, Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta 2007; 1771:271-85; PMID:16920014; http://dx.doi.org/10.1016/j.bbalip.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy S, Plaine A, Albert J, Prasad T, Prasad R, Ernst JF. Dosage-dependent functions of fatty acid desaturase Ole1p in growth and morphogenesis of Candidaalbicans. Microbiology 2004; 150:1991-2003; PMID:15184585; http://dx.doi.org/10.1099/mic.0.27029-0 [DOI] [PubMed] [Google Scholar]
- 22.Xu D, Sillaots S, Davison J, Hu W, Jiang B, Kauffman S, et al. . Chemical genetic profiling and characterization of small-molecule compounds that affect the biosynthesis of unsaturated fatty acids in Candidaalbicans. J Biol Chem 2009; 284:19754-64; PMID:19487691; http://dx.doi.org/10.1074/jbc.M109.019877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ells R, Kock JL, Albertyn J, Pohl CH. Arachidonic acid metabolites in pathogenic yeasts. Lipids Health Dis 2012; 11:100; PMID:22873782; http://dx.doi.org/10.1186/1476-511X-11-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiraki Y, Ishibashi Y, Hiruma M, Nishikawa A, Ikeda S. Candidaalbicans abrogates the expression of interferon-gamma-inducible protein-10 in human keratinocytes. FEMS Immunol Med Microbiol 2008; 54:122-8; PMID:18647352; http://dx.doi.org/10.1111/j.1574-695X.2008.00457.x [DOI] [PubMed] [Google Scholar]
- 25.Joo M, Sadikot RT. PGD synthase and PGD2 in immune resposne. Mediators Inflamm 2012; 2012:503128; PMID:22791937; http://dx.doi.org/10.1155/2012/503128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Attila Gácser DT, Schäfer W, Joshua D. Nosanchuk JD. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest 2007; 117:3049-58; PMID:17853941; http://dx.doi.org/10.1172/JCI32294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uemura H. Synthesis and production of unsaturated and polyunsaturated fatty acids in yeast: current state and perspectives. Appl Microbiol Biotechnol 2012; 95:1-12; PMID:22562166; http://dx.doi.org/10.1007/s00253-012-4105-1 [DOI] [PubMed] [Google Scholar]
- 28.Capsoni F, Minonzio F, Ongari AM, Carbonelli V, Galli A, Zanussi C. IL-10 up-regulates human monocyte phagocytosis in the presence of IL-4 and IFN-gamma. J Leukoc Biol 1995; 58:351-8; PMID:7665991 [DOI] [PubMed] [Google Scholar]
- 29.Chayakulkeeree M, Rude TH, Toffaletti DL, Perfect JR. Fatty acid synthesis is essential for survival of Cryptococcus neoformans and a potential fungicidal target. Antimicrob Agents Chemother 2007; 51:3537-45; PMID:17698629; http://dx.doi.org/10.1128/AAC.00442-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen LN, Trofa D, Nosanchuk JD. Fatty acid synthase impacts the pathobiology of Candida parapsilosis in vitro and during mammalian infection. PLOS One 2009; 4:e8421; PMID:20027295; http://dx.doi.org/10.1371/journal.pone.0008421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gacser A, Trofa D, Schafer W, Nosanchuk JD. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest 2007; 117:3049-58; PMID:17853941; http://dx.doi.org/10.1172/JCI32294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004; 341:119-27; PMID:15474295; http://dx.doi.org/10.1016/j.gene.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 33.Wei DS, Zhang YH, Xing LJ, Li MC. Agrobacterium rhizogenes-mediated transformation of a high oil-producing filamentous fungus Umbelopsis isabellina. J Appl Genet 2010; 51:225-32; PMID:20453313; http://dx.doi.org/10.1007/BF03195734 [DOI] [PubMed] [Google Scholar]
- 34.Toth A, Nemeth T, Csonka K, Horvath P, Vagvolgyi C, Vizler C, et al. . Secreted Candida parapsilosis lipase modulates the immune response of primary human macrophages. Virulence 2014; 5:555-62; PMID:24626151; http://dx.doi.org/10.4161/viru.28509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.