Significance
J paramyxovirus represents a group of unclassified paramyxoviruses that encode an integral membrane protein, transmembrane (TM), that is not found in other classified paramyxoviruses. In this work we have found that TM expression is involved in cell-to-cell fusion when coexpressed with the viral fusion protein F or coexpressed with F and the receptor-binding protein G. This work may define a separate class of viral proteins that promote cell-to-cell fusion.
Keywords: J paramyxovirus, TM, fusion, syncytia
Abstract
Paramyxoviruses include many important animal and human pathogens. Most paramyxoviruses have two integral membrane proteins: fusion protein (F) and attachment proteins hemagglutinin, hemagglutinin–neuraminidase, or glycoprotein (G), which are critical for viral entry into cells. J paramyxovirus (JPV) encodes four integral membrane proteins: F, G, SH, and transmembrane (TM). The function of TM is not known. In this work, we have generated a viable JPV lacking TM (JPV∆TM). JPV∆TM formed opaque plaques compared with JPV. Quantitative syncytia assays showed that JPV∆TM was defective in promoting cell-to-cell fusion (i.e., syncytia formation) compared with JPV. Furthermore, cells separately expressing F, G, TM, or F plus G did not form syncytia whereas cells expressing F plus TM formed some syncytia. However, syncytia formation was much greater with coexpression of F, G, and TM. Biochemical analysis indicates that F, G, and TM interact with each other. A small hydrophobic region in the TM ectodomain from amino acid residues 118 to 132, the hydrophobic loop (HL), was important for syncytial promotion, suggesting that the TM HL region plays a critical role in cell-to-cell fusion.
The Paramyxoviridae contains many important human and animal pathogens including measles virus, parainfluenza viruses 1–5, mumps virus, respiratory syncytial virus (RSV), human metapneumovirus (hMPV), and Hendra and Nipah viruses. J paramyxovirus (JPV) was isolated from moribund wild mice (Mus musculus) in Australia in 1972 (1, 2). J virus was identified as a paramyxovirus based on morphological studies. The full-length genome of JPV was determined in 2005, and it contains 18,954 nucleotides (3). The JPV genome contains eight genes in the order of 3′-N-P/V/C-M-F-SH-TM-G-L-5′. Its unique features do not match any known paramyxovirus genes, and currently it is an unclassified paramyxovirus. The unedited P mRNA is predicted to encode the phosphoprotein, and an edited mRNA molecule created by the addition of one or two nontemplated G residues at the editing site is predicted to encode the V or W protein, respectively. An alternative ORF present in both the unedited and edited mRNA molecules encodes the putative C protein. The JPV fusion (F) protein is predicted to be a type I membrane protein. The highly hydrophobic signal peptide sequence is predicted to be in the first 18 aa residues, the predicted transmembrane domain is located between amino acid residue 486 and 516, leaving a cytoplasmic tail of 28-aa residue. The predicted F cleavage site, GVPGVR, is monobasic and does not conform to the consensus motif for cleavage by furin, R-X-K/R-R (4), conserved in the majority of the Paramyxoviridae except Sendai virus F protein and F of avirulent strains of NDV, where the cleavage site contains a monobasic residue. The G gene encodes a putative 709-aa glycoprotein and distally contains a 2,115-nt second ORF, termed “ORF X.” A protein corresponding to this ORF has not been detected in virus-infected cells. Nucleotide probes specific to the G protein-coding region and ORF X both identified a mRNA species corresponding to the predicted length of the G gene (5). JPV lacking the SH ORF gene (JPV∆SH) induces more apoptosis in tissue culture cells than JPV, and the virus-induced apoptosis was inhibited by neutralizing antibody against TNF-α, suggesting that SH inhibits TNF-α production and signaling in tissue culture cells (6). J virus encodes a fourth integral membrane protein, TM, of 258 aa. Transmembrane (TM) is predicted to be a type II integral membrane protein (5). No significant amino acid sequence homology with any known protein in the Swiss Prot database was identified.
There are two known strains of JPV: JPV-LW and JPV-BH (7). JPV-LW strain is not pathogenic in mice, and JPV-BH strain is pathogenic in mice. There are five nucleotide sequence differences between JPV-BH and JPV-LW—one in the leader sequence, one in the G gene, and three in the L gene. JPV-BH-L-LW, a recombinant JPV-BH containing the L gene from JPV-LW, was attenuated in mice, suggesting that the nucleotide sequence differences in the L gene play a critical role in JPV pathogenesis (7).
To enter cells, the Paramyxoviridae fuse their envelope at neutral pH (except some isolates of hMPV) (8, 9) with the plasma membrane of the target cells and then enter cells and release the viral genomes into the cytoplasm. Membrane fusion is mediated by two viral glycoproteins, F protein, and its cognate attachment protein [hemagglutinin–neuraminidase (HN), hemagglutinin (H), or glycoprotein (G) depending on the genus] except that Hendra and Nipah viruses F and G are interchangeable (10). It is generally believed that membrane fusion by F is triggered after the binding of the attachment protein to the receptor: sialic acid residues or cell-surface proteins. The metastable prefusion form of F refolds into a highly stable postfusion form and does the work of bringing the viral membrane and cellular membrane together (11, 12). The requirement for a cognate attachment protein for fusion promotion suggests that there is a specific interaction between the two viral glycoproteins, and interactions of F protein with the membrane proximal stalk domains of attachment proteins have been detected. Mutations in the stalk domains have been shown to block F activation, mostly by blocking F–HN (H or G) interactions (13–18).
In addition to JPV, two other viruses Beilong (BeiPV) and Tailam (TlmPV) viruses, isolated in rats in 2005 (19) and 2011 (20), respectively, contain the same genome organization. Recently, JPV-like viruses have been identified in rodents in Africa (21) and in bats in Europe (22). They all contain the previously unidentified TM gene that does not exist in other paramyxoviruses.
Using biochemical assays and the reverse genetics system, we investigated the function of TM in this work.
Results
TM Is an Integral Membrane Protein Oriented as a Type II Membrane Protein.
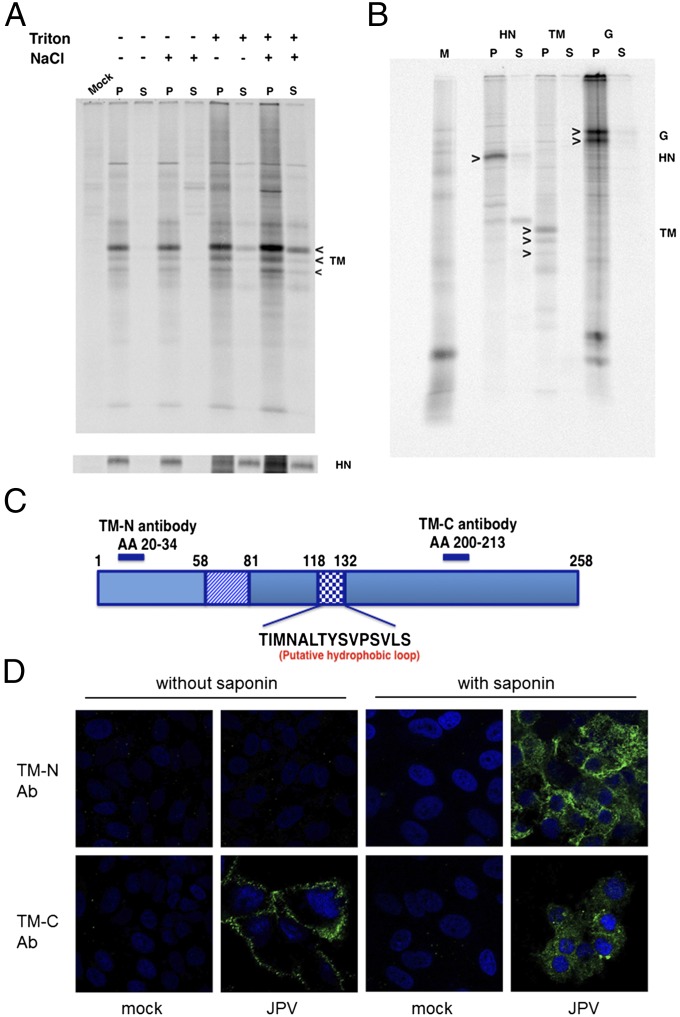
To provide evidence that TM is an integral membrane protein, detergent/high-salt solubility and insolubility to alkali (pH 11) extraction of TM-transfected cells were tested. PIV5 HN was used as a control membrane protein. As shown in Fig. 1, TM was solubilized by 2% (vol/vol) Triton X-100 treatment and was insoluble to alkali (pH 11) extraction from microsomes. G protein consistently migrated on SDS/PAGE as two species for unknown reasons. TM migrated as three species due to glycosylation heterogeneity (5). To further confirm that TM is a type II transmembrane protein, site-specific immunofluorescence staining (IF) was performed on virus-infected cells (Fig. 1 C and D). When cells were treated with saponin, which permeabilized the cell membrane, immunofluorescence was detected using cells stained with TM-N and TM-C antisera (Fig. 1D). However, when Vero cells were not treated with saponin, fluorescence was detected only from TM-C antibody-stained cells (Fig. 1D), suggesting that TM is a type II transmembrane protein. This is consistent with a previous report that TM is a type II membrane protein (5).
Fig. 1.
TM has properties of an integral membrane and type II transmembrane protein. (A) TM is solubilized by 0.5 M NaCl and 2% Triton X-100. Metabolically labeled transfected Vero cells expressing TM (or PIV5 HN as a control) were treated with 0.5 M NaCl and/or 2% Triton X-100 as described previously (37). Proteins were immunoprecipitated with α-FLAG or anti-HN sera, and polypeptides were analyzed by SDS/PAGE. P, pellet; S, supernatant. (B) The TM protein is resistant to extraction from microsomal membranes with alkali (pH 11.0). Microsomes from metabolically labeled Vero cells expressing TM, G, or PIV5 HN (as a control) were obtained and treatment of microsomal membranes with alkali (pH 11) was performed as described previously (35). TM, G, or HN was immunoprecipitated with α-FLAG, α-HA, and α-HN sera, respectively, and polypeptides were analyzed by SDS/PAGE. (C) Schematic representation of TM. The putative TM membrane spanning domain is from residues 58 to 81. The HL is from residues 118–132. The peptides used to generate N-terminal and C-terminal site-specific antibodies are indicated. (D) Detection of TM by specific antisera and immunofluorescence staining. Vero cells were mock-infected or infected with JPV at an MOI of 0.1. Cells were treated with or without saponin and stained with polyclonal sera TM-N or TM-C and FITC-conjugated goat anti-rabbit secondary antibody.
Recovery of Recombinant Virus JPV∆TM with TM Gene Deleted.
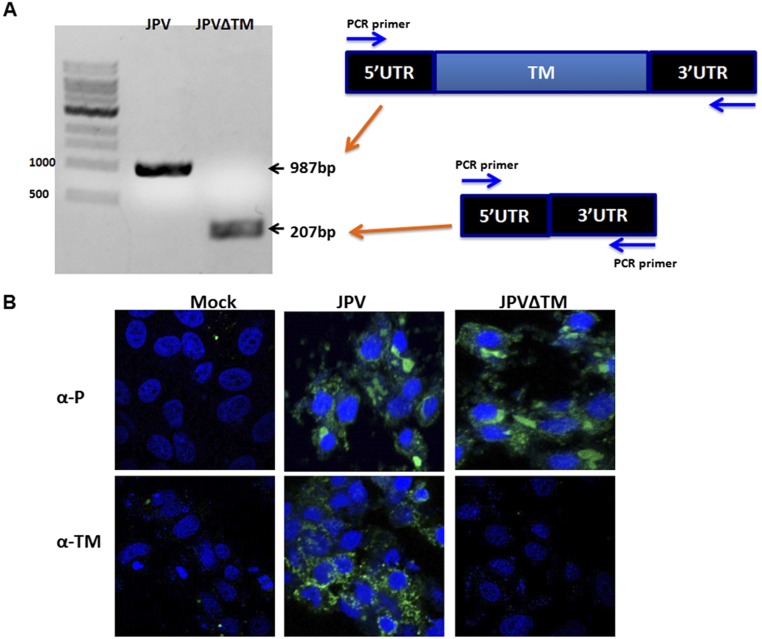
To study the function of TM, a plasmid containing the full-length genome of JPV-BH lacking TM (JPV∆TM) was constructed. This plasmid, together with three helper plasmids encoding N, P, and L proteins and a plasmid expressing T7 RNA polymerase, were cotransfected into HEK293T cells as described previously (7). After obtaining the rescued virus, the entire genome sequence of JPV∆TM was confirmed by RT-PCR and nucleotide sequencing using primers specific for the JPV sequence as described previously (6). That the mutant virus lacks the expression of TM was further confirmed using RT-PCR and an IF assay (Fig. S1).
Fig. S1.
Confirmation that JPV∆TM lacks expression of TM. (A) RT-PCR confirmation of JPV∆TM. cDNA from JPV or JPV∆TM virus-infected cells was amplified using primers derived from the 5′ and 3′ UTR of TM. The size of targets is shown. (B) Examination for TM protein expression in JPV and JPV∆TM-infected Vero cells. Vero cells were mock-infected or infected with JPV or JPV∆TM. Cells were stained with polyclonal sera to P or TM rabbit sera and FITC-conjugated goat anti-rabbit secondary antibody.
Analysis of JPV∆TM Growth Kinetics in Vero Cells.
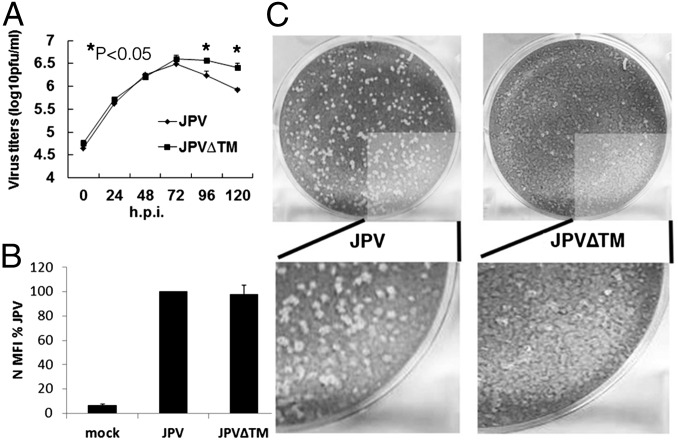
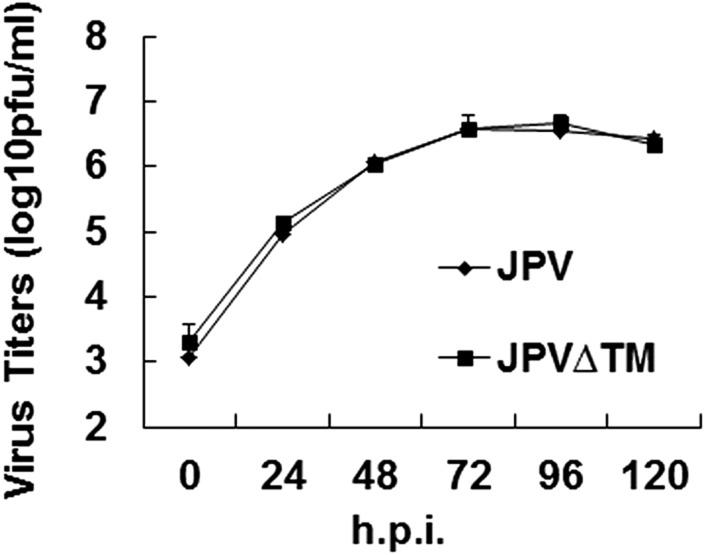
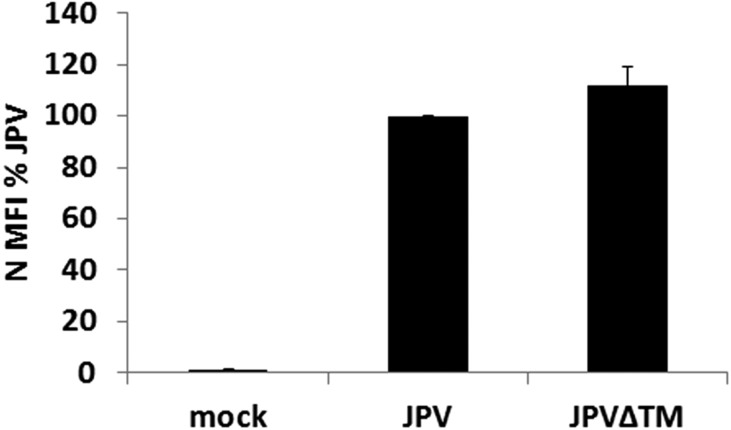
To compare the growth kinetics of WT JPV and JPV∆TM, single- and multiple-cycle growth curves were determined in Vero cells. JPV∆TM replicated similarly to JPV at the early stage and seemingly better at the later stage of a single-cycle growth curve (Fig. 2A). The higher replication titers of JPV∆TM in Fig. 2A may likely be due to more cells left in JPV∆TM-infected cells than in JPV-infected cells. JPV∆TM replicated similarly to JPV in a multiple-cycle growth curve (Fig. S2). To examine whether there was a difference in JPV viral protein expression levels, the mean fluorescence intensity (MFI) of the nucleocapsid (N) protein in virus-infected cells was examined. On staining with anti-N sera, JPV and JPV∆TM produced similar levels of N protein (Fig. 2B). Plaques formed by JPV or JPV∆TM were stained, and it was observed that JPV formed clear plaques; however, JPV∆TM formed opaque plaques (Fig. 2C).
Fig. 2.
Analysis of JPV∆TM in Vero cells. (A) Single-cycle growth curves of JPV and JPV∆TM. Vero cells were infected with JPV or JPV∆TM at an MOI of 5. The media were collected at 24-h intervals. Virus titers were determined by plaque assay on Vero cells. Error bars represent SD. Three independent experiments were performed. (B) Protein expression of JPV and JPV∆TM. Vero cells were mock-infected or infected with JPV or JPV∆TM at an MOI of 5. At 2 d.p.i., the cells were collected, and the MFI of the nucleocapsid protein (N) was determined by flow cytometry. MFI is shown as a percentage of WT JPV. Error bars represent SD. Three independent experiments were performed. (C) JPV∆TM formed opaque plaques on Vero cells. The cells were stained with Giemsa and photographed.
Fig. S2.
Multiple-cycle growth curves of JPV and JPV∆TM. Vero cells were infected with JPV or JPV∆TM at an MOI of 0.1. The media were collected at 24-h intervals. Virus titers were determined by plaque assay on Vero cells. Error bars represent SD. Three independent experiments were performed.
TM Plays a Role in Cell-to-Cell Fusion.
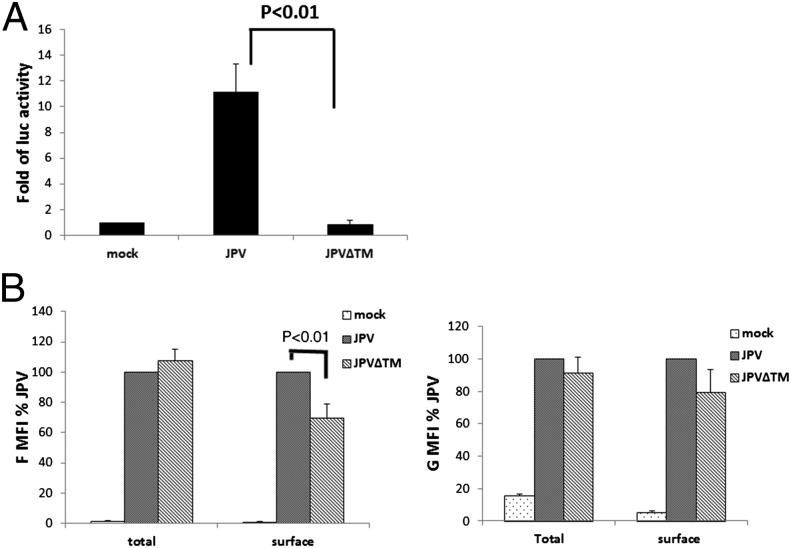
Virus can spread from cell to cell by infecting a fresh cell through virus-to-cell fusion or by promoting cell-to-cell fusion. We speculated that the opaque plaque phenotype was due to a lack of cell-to-cell fusion in JPV∆TM-infected cells. This was confirmed by using a quantitative fusion assay. The results showed that JPV induced higher levels of cell-to-cell (i.e., syncytia) activity than JPV∆TM (Fig. 3A). As the surface expression levels of F affect fusion activity, the surface expression of F and G in JPV∆TM-infected cells were examined (Fig. 3B). The F surface expression level in JPV∆TM-infected cells was lower than in JPV-infected cells, whereas total F expression levels were similar. It is possible that TM enhances F surface expression by stabilizing F on cell surface and/or by preventing internalization of F. However, there were no significant differences in G surface and total expression between JPV∆TM- and JPV-infected cells. As a control, the internal N protein expression levels were examined and found to be similar between cells infected with the mutant and WT viruses (Fig. S3).
Fig. 3.
JPV∆TM is defective in promoting cell-to-cell fusion. (A). Luciferase reporter assay for cell fusion was performed in virus-infected Vero cells. (B) F and G protein expression levels in JPV or JPV∆TM virus-infected cells. Vero cells were mock-infected or infected with JPV and JPV∆TM at an MOI of 2. At 2 d.p.i., the cells were collected, and F and G surface or total protein expression levels were determined by flow cytometry. Error bars represent SD. Three independent experiments were performed.
Fig. S3.
Expression levels of N in JPV or JPV∆TM virus-infected cells. Infected Vero cells (MOI = 2) were subjected to flow cytometry as described in the legend to Fig. 3. Error bars represent SD. Three independent experiments were performed.
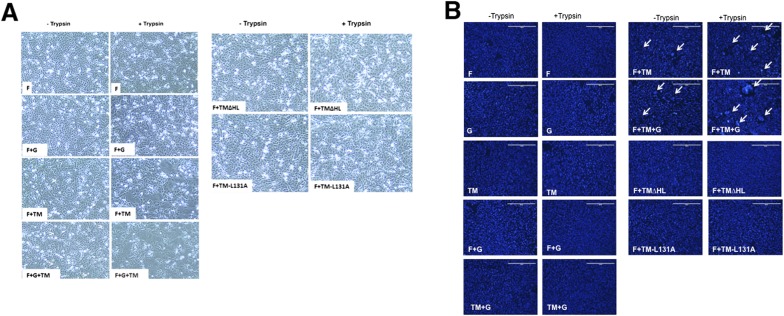
To determine whether promotion of syncytia by TM is infection-dependent, cells were transfected with plasmids expressing F, G, and/or TM. In the plasmid-transfected Vero cells, expression of F, G, TM, or G+TM did not cause formation of syncytia. Unexpectedly, no syncytia were observed in the F+G transfected cells. However, syncytia formed in cells expressing F+TM, and more syncytia formed in cells expressing F+G+TM (Fig. S4). As the JPV F cleavage site contained only a single basic residue, it was predicted that, like the Sendai virus F protein (23), JPV F would be cleaved to the biologically active form of F1 and F2 by the addition of trypsin (1 μg/mL). It was observed that addition of trypsin to the media greatly enhanced syncytia formation (Fig. S4).
Fig. S4.
TM coexpression with F or F and G promotes cell fusion. (A) Live cell images. Vero cells were transfected with F alone or in combination with G and/or TM or TM mutants. At 24 h.p.t., cells were incubated with or without trypsin (1 μg/mL) for 30 min. After 3 h of further incubation, the cells were photographed. (B) DAPI staining. The cells in A were stained with DAPI and photographed. Clustered nuclei are indicated by white arrows.
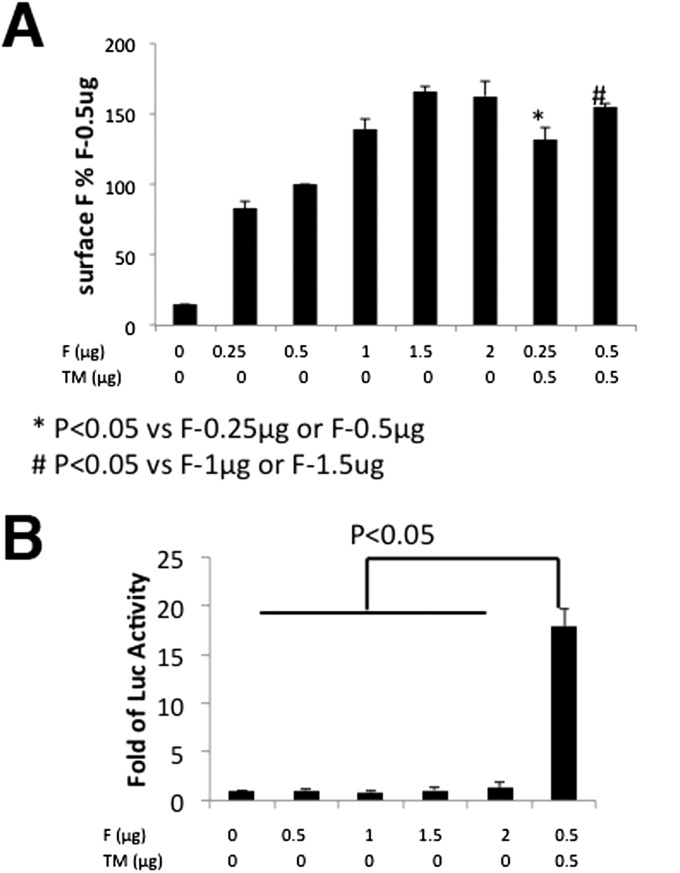
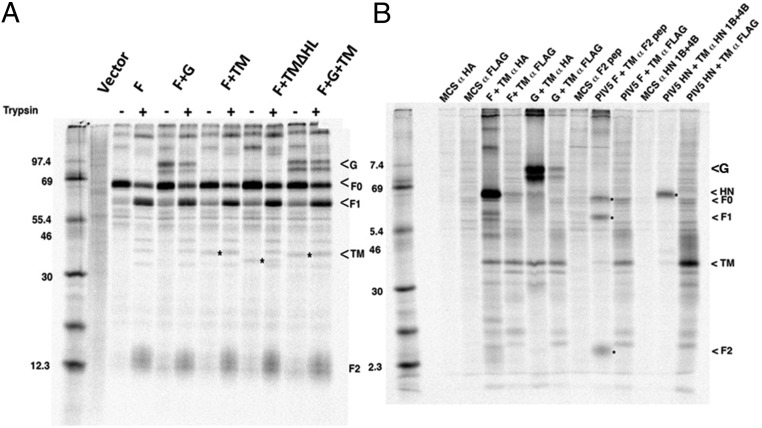
Quantification of fusion activity using a luciferase assay was performed with cells expressing combinations of F, G, and TM proteins and treated with and without trypsin. As shown in Fig. 4, cells expressing F and TM proteins showed some luciferase activity that was increased on trypsin treatment of cells. However, a large increase in luciferase activity was observed on coexpression of F, G, and TM that was increased on trypsin treatment of cells. Expression of F alone, even at a level higher than F+TM, did not promote fusion (Fig. S5), suggesting that TM is required for cell-to-cell fusion. To show that trypsin treatment cleaved F0 to F1 and F2, transfected cells expressing combinations of F, G, and TM were metabolically labeled with [35S] label for 30 min and incubated in unlabeled medium for 180 min (Chase). F proteins were immunoprecipitated using anti-F serum. As shown in Fig. 5A, although some F1 and F2 could be detected without trypsin treatment, the amount of F1 and F2 increased considerably on trypsin treatment. Cleavage of F0 was not complete after a 3-h incubation, suggesting very slow transport of F0 to the cell surface, although this has not been investigated further. Slow transport of F to the cell surface would explain the level of increase in fusion observed on F0 cleavage (Fig. 4). In Fig. 5A, it can also be seen that expressed G and/or TM coprecipitated with F. To investigate the interaction further, when F or G epitope-tagged with HA were immunoprecipitated with αHA, TM was coimmunoprecipitated (Fig. 5B). TM epitope-tagged with FLAG was immunoprecipitated with α-FLAG, and F or G were coimmunoprecipitated (Fig. 5B). To rule out the possibility that the coimmunoprecipitation was due to the formation of mixed micelles, TM was coexpressed with PIV5 F or HN and immunoprecipitated with α-FLAG sera for FLAG-tagged TM or anti-F sera or anti-HN sera for F and HN, respectively. No coimmunoprecipitation of TM with PIV5 F or HN was observed, indicating that the coimmunoprecipitation of TM and JPV F or HN was not artifactual (Fig. 5B) and suggesting that JPV F, G, and TM form a complex.
Fig. 4.
F plus TM promote cell fusion. Quantitative fusion assay on cells expressing combinations of F, G, TM, and TM mutants. Vero cells were transfected with combinations of F, G, TM, or TM mutants. At 24 h.p.t., the cells were incubated with or without trypsin (1 μg/mL) for 30 min. After 3 h of further incubations, the cells were overlaid with BSR cells, and a luciferase assay was performed. Data from four independent experiments were normalized, setting luciferase activity from expression of F and G and TM (plus trypsin) at 100%. The F and G and TM (no trypsin) luciferase activity was 73% of the values for plus trypsin. All other “no trypsin” values were normalized by resetting 73–100%. MCS, vector of expression plasmid; TM del, plasmid expressing TM∆HL; TM L-A, plasmid expressing TM HL L131A.
Fig. S5.
Expression of F did not promote cell-to-cell fusion. (A) F surface expression levels in transfected cells. Vero cells were transfected with F or F plus TM at indicated amounts. F surface protein expression levels were determined by flow cytometry as described in the legend to Fig. 3. (B) Quantification of cell-to-cell fusion. A quantitative fusion assay was performed as in Fig. 4. Error bars represent SD. Three independent experiments were performed.
Fig. 5.
Interactions among TM, F, and G. (A). TM, F, and G coimmunoprecipitate: F0 can be cleaved to F1 and F2. TM, F, and G were expressed in 293T cells in various combinations. At 24 h.p.t., cells were metabolically labeled for 1 h and treated (+) or untreated (−) with trypsin for 3 h. Cell lysates were immunoprecipitated with anti-F sera, and polypeptides were analyzed by SDS/PAGE. (B) 293T cells expressing combinations of F, G, and TM or PIV5 F, HN, and JPV TM were metabolically labeled as described above, and polypeptides immunoprecipitated. Antibodies were α-FLAG for TM, α-HA for JPV F and G, α-F2 pep for PIV5 F, and monoclonal antibodies HN 1b and HN 4b. F and G were tagged with HA epitope, and TM and TM mutants were tagged with FLAG epitope. MCS, vector of expression plasmid.
The TM putative hydrophobic loop.
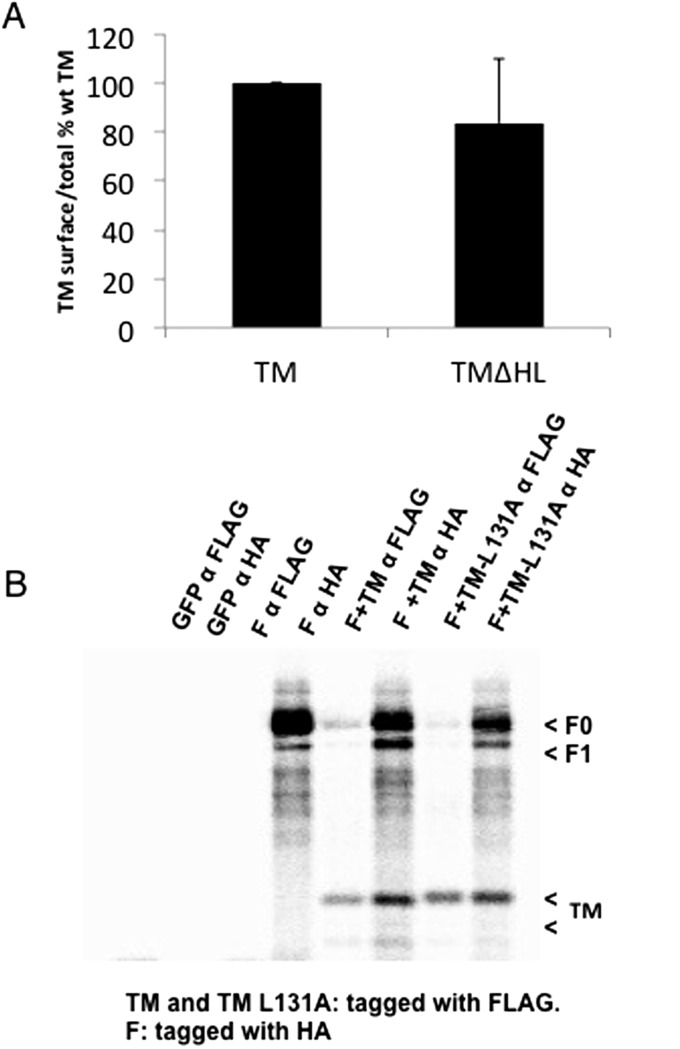
The DAS-transmembrane prediction server (www.sbc.su.se/∼miklos/DAS/) (24) predicted that TM contained a small hydrophobic region in the ectodomain from amino acid residue 118 to 132. This small hydrophobic region was designated as the hydrophobic loop (HL). The HL was deleted from TM (TM∆HL), and this deletion did not affect expression of TM (Fig. S6A). Expression of TM∆HL also did not affect coimmunoprecipitation of F (Fig. 5A). Interestingly, no syncytia were observed in Vero cells when F+TM∆HL were coexpressed with or without trypsin treatment (Fig. S4). A point mutation, L131A, was introduced into the TM HL, and coexpression of TM HL L131A with F with or without coexpression of G and with or without trypsin treatment abolished fusion activity (Fig. 4 and Fig. S4). TM HL L131A coexpression with F did not affect coimmunoprecipitation of TM with F (Fig. S6B).
Fig. S6.
Expression levels of TM mutants and interaction between F and TM HL L131A. (A) Surface expression of TM∆HL. Transfected cells were subjected to flow cytometry as described in the legend to Fig. 3. Error bars represent SD. Three independent experiments were performed. (B) TM HL L131A interacted with F. Vero cells were cotransfected with F and WT TM or TM HL L131A. At 24 h.p.t. cells were metabolically labeled for 2 h. Cell lysates were immunoprecipitated with anti-FLAG or anti-HA antibody as indicated. The immunoprecipitated proteins were resolved in SDS/PAGE.
JPV TM HL in Recovered Virus.
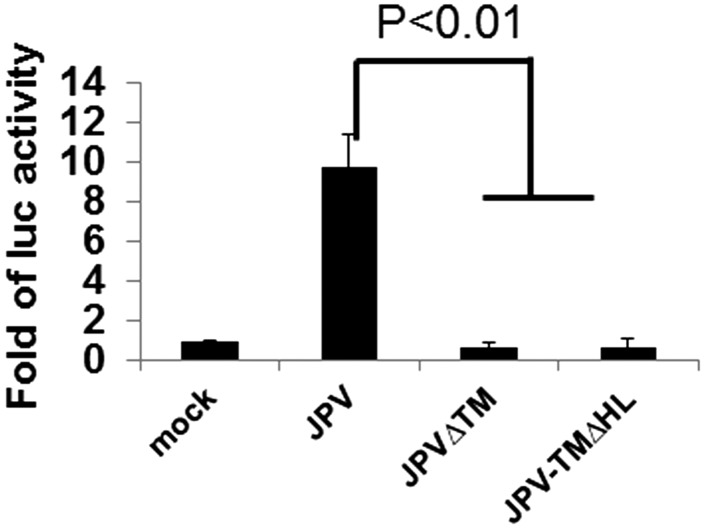
A recombinant virus with TM HL deleted (JPV-TM∆HL) was recovered. JPV-TM∆HL formed opaque plaques like those formed by JPV∆TM. The F surface expression level in virus-infected cells was examined by flow cytometry. JPV∆TM and JPV-TM∆HL had lower F surface expression than WT JPV (Fig. S7A). However, the total F expression levels were similar in virus-infected cells (Fig. S7B). JPV-TM∆FL was defective in promoting cell-to-cell fusion in a quantitative fusion assay like JPV∆TM (Fig. 6), indicating that the HL region of TM is critical for its ability to promote cell-to-cell fusion.
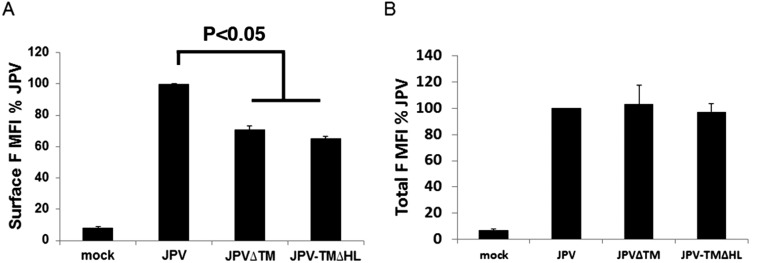
Fig. S7.
Expression levels of F in JPV∆TM and JPV-TM∆FL-infected cells. (A) Surface and (B) total expression of F in JPV∆TM and JPV-TM∆HL–infected Vero cells (MOI = 2). Infected Vero cells were subjected to flow cytometry as described in the legend to Fig. 3. Error bars represent SD. Three independent experiments were performed.
Fig. 6.
The involvement of the putative TM HL in fusion activity. Vero cells were infected with JPV viruses expressing WT TM, ∆TM, or TM∆HL at an MOI of 2 and at 2 d.p.i. fusion was quantified by a luciferase assay. Error bars represent SD. Three independent experiments were performed.
Discussion
For most paramyxoviruses the expression of the F protein with its cognate receptor-binding protein (HN, H, or G) is required for membrane fusion. It is generally thought that, upon receptor binding, a conformational change occurs in HN (H or G) that in turn activates the F protein to begin refolding from a prefusion metastable state to a postfusion stable state. In doing so, the F protein does the work of bringing two membranes together such that they can fuse. The precise mechanism by which HN (H or G) activates F is unclear. Paramyxovirus F proteins can promote cell-to-cell fusion (syncytia formation) as well as virus-to-cell fusion (infection).
Here we report that JPV, which expresses in addition to F and G an integral membrane protein designated TM, is required with F and G for efficient cell-to-cell fusion. However, TM is not a required gene to replicate JPV in tissue culture cells. The JPV∆TM virus forms opaque plaques, suggesting a defect in cell-to-cell fusion. Expression of cDNAs encoding F, G, or TM or coexpression of F+G did not cause cell-to-cell fusion (syncytia formation) or fusion in a quantitative luciferase assay whereas coexpression of F+TM caused a small amount of fusion and coexpression of F+G+TM caused extensive fusion. For many other paramyxoviruses, F+HN (H or G) coimmunoprecipitate, suggesting that they form a complex in which they function together. Similarly, on coexpression of JPV F+G+TM proteins, the proteins were found to coimmunoprecipitate, suggesting that they form a complex that may function together.
The JPV F protein appears to have the hallmarks of other paramyxovirus F proteins. It contains a cleavage site adjacent to the hydrophobic fusion peptide that can be cleaved by the addition of exogenous trypsin, and, as expected, cleavage of F increases fusion activity. The cysteine residues in F of JPV align in an amino acid sequence alignment with other paramyxovirus F proteins, suggesting an atomic structure similar to that of PIV5 and RSV F (25, 26).
The mechanism of triggering of F by HN (H or G) is not clear. The requirement for expression of a homotypic attachment protein for fusion triggering suggested a specific interaction between F and attachment proteins. This physical association has been well documented for a number of paramyxoviruses, including NDV, PIV2, PIV3, Nipah, and Hendra viruses (16, 27–29). It is also known for PIV5, mumps virus, NDV, Nipah virus, measles virus, and CDV that the HN (H or G) stalk region can function in triggering without expression of the receptor-binding globular head, and a region in the middle of the stalk is thought to interact directly with the F protein, suggesting a conserved core mechanism of activation among paramyxovirus F proteins (13, 14, 30–32). The F and HN interaction is thought to lower the energy of the activation barrier for triggering metastable F.
The mechanism by which JPV TM protein triggers cell-to-cell fusion on expression of F+TM and greatly enhances fusion on expression of F+G+TM is not known. Among many hypotheses, TM could affect the conformation of F to induce fusion. In this case, TM itself is not a fusion protein, but it plays an indirect role in membrane fusion. TM, instead of G, may trigger F for cell-to-cell fusion. This is consistent with the observation that there was an interaction between F and TM in the absence of G. However, the observation that TM∆HL and single-point mutation TM-L131A coimmunoprecipitate with F but do not promote cell-to-cell fusion could be used to argue against TM serving as a trigger for an F conformation change. Another possibility for a role of TM in fusion is that it plays a direct role in membrane fusion. It was not expected that a point mutation (L to A in the ectodomain at residue 131) would ablate activation of fusion. The point mutation in TM is in a hydrophobic region that is exposed extracellularly. The mutation may block an activity of TM while maintaining its ability to form a complex with F and G. At present, we cannot exclude the remote possibility that TM promotes fusion through an intermediate with TM interacting with a host protein that in turn triggers cell-to-cell fusion with F.
It is interesting that JPV encodes an additional protein to promote cell-to-cell fusion, especially as JPV has the largest genome of any known member of the paramyxovirus family. Perhaps the virus has divided functions differently among three integral membrane proteins to mediate virus-to-cell fusion and cell-to-cell fusion, allowing the virus to use different proteins in a complex to cause cell-to-cell fusion or virus-to-cell fusion depending on circumstances. As the deletion of TM did not affect replication of JPV in tissue culture cells, it is possible that the deletion of TM will result in attenuation in animals due to its inability to spread from cell to cell. Further studies of TM-promoted fusion will lead to new knowledge on membrane fusion as well as to determining the role of TM in fusion. Comparative studies of JPV and JPV∆TM pathogenesis will be helpful in elucidating the functions of the TM.
Materials and Methods
Cells and Antibodies.
Monolayer cultures of HEK293T and Vero cells were maintained in DMEM containing 10% FBS, 100 IU/mL penicillin, and 100 µg/mL streptomycin. All cells were incubated at 37 °C, 5% CO2. Virus-infected cells were grown in DMEM containing 2% FBS. Plaque assays were performed using Vero cells. Polyclonal anti-P protein C-terminal serum was used as described previously (6). TM polyclonal antibodies specific for N (TM-N Ab) or C terminus (TM-C Ab), F polyclonal antibody, and G polyclonal antibody were raised in rabbits (Genscript USA, Inc.). F or G monoclonal antibodies specific for the TM ectodomain or α-HA (12CA5) antibody were prepared from hybridoma cell-culture supernatants. PIV5 F2 peptide sera and PIV5 HN 1b and 4b monoclonal antibodies have been described previously (33). α-FLAG antibody was obtained from Sigma.
Virus Rescue and Nucleotide Sequencing.
In this work, we used exclusively JPV-BH and its genes. A plasmid containing the full-length genome of JPV lacking TM gene (pJPV∆TM) was constructed using standard molecular biology techniques. The recombinant virus JPV∆TM was rescued as described previously (7) with some modifications. A plasmid expressing T7 polymerase (pT7P); three plasmids, pJPV-N, pJPV-P, and pJPV-L, encoding the N, P, and L proteins, respectively; and pJPV∆TM were cotransfected into HEK293T cells at 95% confluence in 6-cm plates with Jetprime (Polyplus-Transfection, Inc.). The amounts of plasmids used were as follows: 3 µg pJPV∆TM, 1 μg pT7, 0.1 µg pJPV-N, 0.05 µg pJPV-P, and 3.75 µg pJPV-L. Two days posttransfection, 1/10 of the HEK293T cells were cocultured with 1 × 106 Vero cells. The mixed cells were cocultured 1–2 wk, the media were harvested, and cell debris were pelleted by low-speed centrifugation (3,000 × g, 10 min). Plaque assays on Vero cells were used to obtain single clones of recombinant viruses.
The full-length genome of plaque-purified JPV∆TM was sequenced. Total RNAs from JPV∆TM-infected Vero cells were purified using the RNeasy kit (Qiagen Inc.). cDNAs were prepared using random hexamers and aliquots of the cDNA were then amplified in PCR reactions using appropriate oligonucleotide primer pairs as described previously (6). The improved RACE PCR was used to amplify the leader and trailer sequences (34). Sequences of all primers for sequencing of the complete genome of JPV∆TM are available on request. DNA sequences were determined using an Applied Biosystem sequencer (ABI).
Growth Kinetics of JPV and JPV∆TM.
Vero cells in six-well plates were infected with JPV or JPV∆TM at an MOI of 5 or 0.1. The cells were then washed with PBS and maintained in DMEM–2% FBS. Media were collected at 24-h intervals. The titers of viruses were determined by plaque assay on Vero cells.
Detection of Viral Protein Expression.
To investigate if TM is an integral membrane protein, Triton X-100 solubilization and insolubility to alkali (pH 11) were performed as described previously (35). An immunofluorescence assay was used to confirm that TM is oriented in membranes as a type II transmembrane protein. Vero cells in 24-well plates were mock-infected or infected with JPV at an MOI of 0.1. At 2 d postinfection (d.p.i.), the cells were washed with PBS and then fixed in 0.5% formaldehyde. The cells were permeabilized in PBS–0.1% saponin solution for total protein staining or directly incubated for 30 min with polyclonal TM-N Ab or TM-C Ab for cell-surface protein staining. The cells were washed with PBS–1% BSA and then incubated with fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit antibody. The cells were incubated for 30 min, washed, and photographed using a confocal microscope (Nikon Eclipse Ti).
To confirm the lack of expression of TM in JPV∆TM-infected cells, Vero cells in 24-well plates were mock-infected or infected with JPV or JPV∆TM at an MOI of 0.1. At 2 d.p.i., the cells were treated as described above. The cells were incubated with polyclonal anti-P antibody or TM-N Ab. The cells were photographed using a confocal microscope.
Immunoprecipitation of Polypeptides.
Vero cells were transfected with expression plasmids for F, G, TM, TM∆HL, or TML131A alone or in combination. Trypsin (1 μg/mL) was added into the media. At 2 d.p.i., the cells were labeled for 2 h with [35S]-Met/Cys Promix (100 μCi/mL). The cells were lysed in RIPA buffer (20 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 0.2% Triton-X100, 0.1% SDS, 5 mM iodoacetanide containing protease inhibitor mixture.), and aliquots were immunoprecipitated using α-FLAG, α- HA (12CA5), PIV5 F, or PIV5 HN and JPV F polyclonal antibodies. Proteins were analyzed on a 15% SDS–PAGE and radioactivity detected by autoradiography using a Storm phosphorimager (Molecular Dynamics, Inc.).
Flow Cytometry.
The cell-surface and total expression of F or G and the total expression of N were quantified by flow cytometry using a flow cytometer (Becton Dickinson LSR II) as described previously (13, 36). For cell-surface expression, the virus-infected or plasmid-transfected cells were detached from dishes using 10 mM EDTA. After blocking in PBS-1% BSA, the cells were incubated with anti-F or G serum (1:100) for 1 h at 4 °C. The cells were stained with anti-mouse antibody labeled with phycoerythrin for 1 h at 4 °C in the dark. After washing, the cells were resuspended in PBS containing 0.5% formaldehyde. Sodium azide (0.02%) was added to buffers to prevent internalization of surface protein. The mean fluorescence intensity was measured using a flow cytometer. For total protein expression, the cells were collected, resuspended in FBS–DMEM (50:50), and permeabilized in 70% ethanol overnight, and then the permeabilized cells were treated as described above.
Cell Fusion Luciferase Assay.
Vero cells in six-well plates were transfected with expression plasmids as follows: 750 ng pT7-Luc and 750 ng of pCAGGS JV F, G, or TM with the balance made up with pCAGGS vector for a total of 3 μg per well. Transfection was carried out using Lipofectamine and Plus Reagent according to the manufacturer’s protocol (Life Technologies). At 20 h posttransfection (h.p.i.), the transfected cells were incubated for 1 h in the presence or absence of 1 μg/mL N-acetyl trypsin, overlaid with BSR-T7 [BSR derived from baby hamster kidney (BHK) cells that express T7 RNA polymerase] cells, and incubated until 30–50% of the monolayer was fused.
The luciferase activity was analyzed using the Bright-Go luciferase assay system (Promega) and a SpectraMax M5 microplate reader (Molecular Devices).
Vero cells in 12-well plates were mock-infected or infected with JPV or JPV∆TM at an MOI of 2. At 2 h postinfection, media were changed into DMEM–2% FBS (vol/vol). The 0.5 μg T7-Luc and 5 ng RL-TK plasmids were transfected into Vero cells. Vero cells in 10-cm plates were transfected with 10 μg T7P. At 2 d postransfection, Vero cells in 12-well plates were overlaid with the Vero cells transfected by plasmid encoding T7 RNA polymerase in 10-cm plates. At 1 d post-overlay, the luciferase activity was determined as prescribed above.
Statistical Analysis.
All statistical analysis was calculated by using a t test. P values < 0.05 were considered significant.
Acknowledgments
We appreciate helpful discussion and technical assistance from all the members of the B.H. and R.A.L. laboratories. This work was supported by the endowment of the Fred C. Davison Distinguished University Chair in Veterinary Medicine (B.H.). R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509476112/-/DCSupplemental.
References
- 1.Mesina JE, Campbell RS, Glazebrook JS, Copeman DB, Johnson RH. The pathology of feral rodents in North Queensland. Tropenmed Parasitol. 1974;25(1):116–127. [PubMed] [Google Scholar]
- 2.Jun MH, Karabatsos N, Johnson RH. A new mouse paramyxovirus (J virus) Aust J Exp Biol Med Sci. 1977;55(6):645–647. doi: 10.1038/icb.1977.63. [DOI] [PubMed] [Google Scholar]
- 3.Jack PJ, Boyle DB, Eaton BT, Wang LF. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J Virol. 2005;79(16):10690–10700. doi: 10.1128/JVI.79.16.10690-10700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosaka M, et al. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266(19):12127–12130. [PubMed] [Google Scholar]
- 5.Jack PJ, et al. Expression of novel genes encoded by the paramyxovirus J virus. J Gen Virol. 2008;89(Pt 6):1434–1441. doi: 10.1099/vir.0.83638-0. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, et al. Function of the small hydrophobic protein of J paramyxovirus. J Virol. 2011;85(1):32–42. doi: 10.1128/JVI.01673-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, et al. The L gene of J paramyxovirus plays a critical role in viral pathogenesis. J Virol. 2013;87(23):12990–12998. doi: 10.1128/JVI.02039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. Low-pH triggering of human metapneumovirus fusion: Essential residues and importance in entry. J Virol. 2009;83(3):1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herfst S, et al. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J Virol. 2008;82(17):8891–8895. doi: 10.1128/JVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossart KN, et al. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J Virol. 2002;76(22):11186–11198. doi: 10.1128/JVI.76.22.11186-11198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102(26):9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bose S, et al. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 2011;85(24):12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ader N, et al. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem. 2012;287(20):16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 2006;80(2):623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone-Hulslander J, Morrison TG. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J Virol. 1997;71(9):6287–6295. doi: 10.1128/jvi.71.9.6287-6295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bose S, Jardetzky TS, Lamb RA. 2015. Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479–480:518–531.
- 18.Lee JK, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283(24):16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, et al. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology. 2006;346(1):219–228. doi: 10.1016/j.virol.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Woo PC, et al. Complete genome sequence of a novel paramyxovirus, Tailam virus, discovered in Sikkim rats. J Virol. 2011;85(24):13473–13474. doi: 10.1128/JVI.06356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki M, et al. Molecular epidemiology of paramyxoviruses in Zambian wild rodents and shrews. J Gen Virol. 2014;95(Pt 2):325–330. doi: 10.1099/vir.0.058404-0. [DOI] [PubMed] [Google Scholar]
- 22.Kurth A, et al. Novel paramyxoviruses in free-ranging European bats. PLoS One. 2012;7(6):e38688. doi: 10.1371/journal.pone.0038688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu M, Choppin PW. Analysis of Sendai virus mRNAs with cDNA clones of viral genes and sequences of biologically important regions of the fusion protein. Proc Natl Acad Sci USA. 1984;81(24):7732–7736. doi: 10.1073/pnas.81.24.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cserzö M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: The dense alignment surface method. Protein Eng. 1997;10(6):673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 25.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci USA. 2012;109(41):16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci USA. 2011;108(23):9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilar HC, et al. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol. 2007;81(9):4520–4532. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop KA, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81(11):5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao Q, Hu X, Compans RW. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J Virol. 1997;71(1):650–656. doi: 10.1128/jvi.71.1.650-656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bose S, Song AS, Jardetzky TS, Lamb RA. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol. 2014;88(8):3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bose S, et al. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci USA. 2012;109(39):E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng R, et al. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology. 1999;253(1):43–54. doi: 10.1006/viro.1998.9501. [DOI] [PubMed] [Google Scholar]
- 33.Paterson RG, Hiebert SW, Lamb RA. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc Natl Acad Sci USA. 1985;82(22):7520–7524. doi: 10.1073/pnas.82.22.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Yu M, Zhang H, Wang HY, Wang LF. Improved rapid amplification of cDNA ends (RACE) for mapping both the 5′ and 3′ terminal sequences of paramyxovirus genomes. J Virol Methods. 2005;130(1-2):154–156. doi: 10.1016/j.jviromet.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Paterson RG, Lamb RA. Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell. 1987;48(3):441–452. doi: 10.1016/0092-8674(87)90195-4. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, et al. Single-dose vaccination of a recombinant parainfluenza virus 5 expressing NP from H5N1 virus provides broad immunity against influenza A viruses. J Virol. 2013;87(10):5985–5993. doi: 10.1128/JVI.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paterson RG, Takeda M, Ohigashi Y, Pinto LH, Lamb RA. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306(1):7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]