Significance
Bacteria living in seawater must cope with low-sodium environments that they may encounter. Here we show an unexpected finding that remodeling of the Sec protein export machinery plays a pivotal role in this adaptation. Vibrio alginolyticus possesses alternative SecDF1 and SecDF2 homologs that use the transmembrane gradient of Na+ and that of H+, respectively, to enhance protein export by cooperating with the SecYEG translocon. The synthesis of SecDF2 is induced in low-Na+ environments, and this induction is essential for the bacterium to survive low salinity. Remarkably, the Vibrio species use a nascent polypeptide, VemP, to monitor the functional state of the Sec pathway and to up-regulate translation of SecDF2 when activity of the SecDF1-containing Sec machinery declines.
Keywords: protein export, proton-motive force, ribosome, SecD, translation arrest
Abstract
SecDF interacts with the SecYEG translocon in bacteria and enhances protein export in a proton-motive-force-dependent manner. Vibrio alginolyticus, a marine-estuarine bacterium, contains two SecDF paralogs, V.SecDF1 and V.SecDF2. Here, we show that the export-enhancing function of V.SecDF1 requires Na+ instead of H+, whereas V.SecDF2 is Na+-independent, presumably requiring H+. In accord with the cation-preference difference, V.SecDF2 was only expressed under limited Na+ concentrations whereas V.SecDF1 was constitutive. However, it is not the decreased concentration of Na+ per se that the bacterium senses to up-regulate the V.SecDF2 expression, because marked up-regulation of the V.SecDF2 synthesis was observed irrespective of Na+ concentrations under certain genetic/physiological conditions: (i) when the secDF1VA gene was deleted and (ii) whenever the Sec export machinery was inhibited. VemP (Vibrio export monitoring polypeptide), a secretory polypeptide encoded by the upstream ORF of secDF2VA, plays the primary role in this regulation by undergoing regulated translational elongation arrest, which leads to unfolding of the Shine–Dalgarno sequence for translation of secDF2VA. Genetic analysis of V. alginolyticus established that the VemP-mediated regulation of SecDF2 is essential for the survival of this marine bacterium in low-salinity environments. These results reveal that a class of marine bacteria exploits nascent-chain ribosome interactions to optimize their protein export pathways to propagate efficiently under different ionic environments that they face in their life cycles.
Cells must export specific proteins to their surfaces for growth, reproduction, and interaction with the environment. In bacteria, the SecA ATPase and the SecYEG translocon play central roles in translocation of secretory proteins across the cytoplasmic membrane (1, 2). In addition, efficient operation of protein export in bacteria requires SecDF, a membrane protein complex having 12 transmembrane segments and large periplasmic domains of functional importance (3). SecDF is encoded either by the separate secD and secF genes (4) or as a single polypeptide (5). The crystal structures of Thermus thermophilus SecDF (TSecDF) and structure-instructed functional studies of the Escherichia coli ortholog suggest that SecDF drives polypeptide translocation by capturing a substrate that is emerging from the translocon into the periplasmic space and undergoing proton-motive-force-dependent cycles of conformational changes (5, 6).
In the present study, we address the functions and expression regulation of the two SecDF paralogs in a halophilic, marine-estuarine bacterium, Vibrio alginolyticus. The Vibrio group of gram-negative bacteria such as Vibrio parahaemolyticus are important for human health because many of them cause severe foodborne illness and wound infections (7, 8). V. alginolyticus was reported to be involved in toxin production in infected sea animals (9). These bacteria living in seawater, including estuary areas, encounter, migrate into, or are brought by associated animals to different salinity environments. Upon infection to animals, they are also exposed to intrabody and even intracellular fluid (7, 10), whose Na+ concentrations may be different from those of the previous environments. Therefore, it is conceivable that these bacteria are equipped with mechanisms that allow them to cope with changing concentrations of sodium, a major solute of seawater.
These bacteria have two circular chromosomes, large (chromosome 1) and small (chromosome 2), both of which are indispensable for viability (11). Whereas chromosome 1 carries many essential genes involved in protein synthesis and housekeeping, chromosome 2 contains a number of genes encoding transporters and transcription regulators involved in environmental adaptation (12). The genomic information reveals that Vibrio species possess a single copy of the secA, secY, secE, and secG genes on chromosome 1. Because each of the SecA (13), SecY (14), and SecE (15) homologs of Vibrio can function when introduced into E. coli cells, the fundamental makeup of the Sec machinery may be common among these bacterial species. However, protein translocation into the Vibrio membrane vesicles in vitro was enhanced by the Na+-motive force, instead of the proton-motive force (16). Furthermore, protein export in E. coli cells expressing the V. alginolyticus SecDF1 (V.SecDF1) protein (discussed below) was enhanced by Na+ (5). Whereas the sodium dependence of the Vibrio SecDF is in accord with their marine origin, V. alginolyticus and other Vibrio species actually have two sets of secDF genes, which we designate secDF1 and secDF2, on chromosomes 1 and 2, respectively. The present study addresses whether these SecDF homologs play different roles in the bacterial physiology and how their expression is regulated.
A remarkable mechanism is known about regulation of protein targeting/translocation machineries, in which specific monitoring substrates directly monitor the activities of the respective systems; SecM monitors the Sec protein export activity in E. coli (17, 18) and MifM monitors the YidC protein insertion activity in Bacillus subtilis (19). They up-regulate translation of secA and yidC2, respectively, in response to defects in the respective protein localization pathways, thereby contributing to the homeostasis of cellular protein localization processes (20–23). SecM and MifM, called regulatory nascent polypeptides, undergo translation arrest in a regulated manner such that the arrest is released upon engagement of their nascent chains in the localization processes (17–19, 24–26). The arrested ribosome disrupts the secondary structure of the mRNA to expose the ribosome-binding (Shine–Dalgarno) sequence of the target gene, thus allowing the free ribosomes of the cell to initiate translation of the target gene. Here, we report that the Vibrio species of bacteria use a regulatory nascent polypeptide, VemP, to up-regulate translation of secDF2 upon encountering a low-Na+ environment, in which V.SecDF1 cannot function. Our results reveal that translation arrest contributes as an essential mechanism to the environmental adaptation in marine bacteria.
Results
V. alginolyticus Possesses Na+-Dependent and Na+-Independent SecDF Paralogs.
Although SecD1, SecF1, SecD2, and SecF2 are encoded by separate cistrons in Vibrio, we call the SecD-SecF complexes collectively V.SecDF1 and V.SecDF2 and their genes secDF1 (secD1-secF1 on chromosome 1) and secDF2 (secD2-secF2 on chromosome 2), respectively. In V. alginolyticus, secDF1VA is more closely related than secDF2VA to the E. coli secD-secF genes in the amino acid sequences of the encoded proteins (Fig. S1) as well as in the yajC-secD1VA-secF1VA arrangement, presumably forming an operon (Fig. 1A). The secD2VA and secF2VA genes on chromosome 2 are preceded by an ORF (termed vemP in this work, as shown below), which is conserved among Vibrio species but unrelated to yajC. To elucidate functions of these V.SecDF paralogs, we expressed them in E. coli cells carrying the secD1 (Cs) mutation that virtually eliminated the endogenous SecDF expression and impaired protein export severely (Fig. 1B, lane 9). When V.SecDF1 was expressed, export of outer membrane protein A (OmpA) and maltose-binding protein (MBP) was recovered markedly provided that the medium was supplemented with 300 mM NaCl (Fig. 1B, compare lanes 1 and 3) but not KCl (lane 2) (5). Such Na+ dependence was not observed when the mutant cells were complemented with V.SecDF2 or E. coli SecDF, which fully corrected the defect even without added NaCl (Fig. 1B, lanes 4–7). These results suggest that V.SecDF1 is unable to use the proton gradient and can instead use the Na+ gradient to execute its protein export function, whereas V.SecDF2 is independent of Na+ and presumably H+-dependent, as has been shown for the E. coli SecDF protein (5). Thus, V.SecDF1 has been optimized to the primary habitat, seawater, of the Vibrio strain whereas V.SecDF2 has retained or gained (27) the proton dependence.
Fig. S1.
Sequence alignment of E. coli SecDF and Vibrio SecDF paralogs. (A) Amino acid sequences of E. coli SecD (E.SecD), V. alginolyticus SecD1 (V.SecD1), and V. alginolyticus SecD2 (V. SecD2) are aligned by the crystal W program. Boldface and asterisks at the bottom indicate residues 100% conserved among the three proteins, red shows residues of 2/3 conservation, and green shows residues of high similarity. Transmembrane regions predicted from the TtSecDF structure (5) are indicated by blue lines at the top. Amino acid sequences that were used as peptide antigens for preparation of the V.SecD antibodies are underlined. (B) A sequence alignment of the SecF paralogs is similarly presented. Amino acid sequences that were used as peptide antigens for preparation of the V.SecF antibodies are underlined. The functionally essential Arg residue in TM 11 (Fig. S3C) is indicated by a red asterisk. (C) Identity/similarity scores are summarized and shown in red and blue, respectively.
Fig. 1.
Cation dependence of the V.SecDF paralogs expressed in E. coli: V.SecDF1 requires Na+. (A) Gene organization of the secDF loci in E. coli and V. alginolyticus. ORFs are shown by arrows. (B) Protein export-enhancing activities of V.SecDF1 and V.SecDF2 assessed using the E. coli secD1(Cs) mutant. E. coli secD1 cells expressing the indicated SecDF protein were grown in M63 medium supplemented with 300 mM Na+ or K+, as indicated, at 37 °C until an early log phase, shifted to 20 °C, and cultivated for additional 30 min. Cells were pulse-labeled with [35S]methionine for 1 min. Immunoprecipitated precursor (p) and mature (m) forms of labeled MBP and OmpA were separated by 10% Laemmli gel and detected by phosphorimaging (Upper). Export efficiencies (proportions of mature form) of MBP (gray) and OmpA (black) are shown with SD (n ≥ 3) (Lower). See also Fig. S1.
SecDF2 Is Inducible upon Na+ Deprivation and Crucial for the Life of V. alginolyticus in Low-Na+ Environments.
To examine Na+ dependence of V. alginolyticus growth, we used M63-amino acids medium supplemented with a total of 300 mM of NaCl–KCl combinations. A secDF1VA+ secDF2VA+ strain VIO5 (28), called WT strain hereafter, was able to grow in the presence of as low as 13 mM of Na+, albeit at a slightly reduced rate than in the presence of higher concentrations of Na+ (Fig. 2A, Left and Fig. S2C). Thus, this halophilic V. alginolyticus strain can adapt to limited-Na+ environments as long as the total ionic strength of the medium is maintained. We constructed V. alginolyticus mutants deleted for secDF1VA (ΔsecDF1) or secDF2VA (ΔsecDF2) (SI Materials and Methods and Fig. S2D). The ΔsecDF1 mutant exhibited Na+-independent growth similarly to the WT strain (Fig. 2A, Middle). By contrast, the ΔsecDF2 mutant depended heavily on the Na+ concentrations; it showed maximum growth rate in the presence of 300 mM Na+, which was progressively declined at lower Na+ concentrations (Fig. 2A, Right). Thus, V.SecDF2 is essential for growth in the absence of sufficient levels of Na+.
Fig. 2.
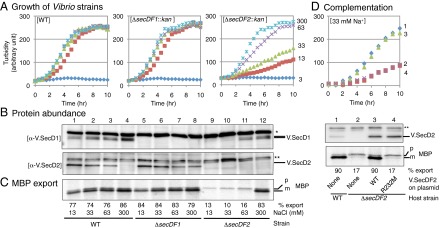
Physiological roles of V.SecDF1 and V.SecDF2 in V. alginolyticus. (A) V.SecDF2 is important for growth under Na+-limited conditions. WT and the indicated deletion mutants of V. alginolyticus were grown at 30 °C in M63 medium supplemented with a total of 300 mM of NaCl and KCl, in which Na+ concentrations were 3 (blue), 13 (brown), 33 (pale green), 63 (purple), and 300 (pale blue) mM. Cell densities were monitored by turbidity measurements. (B) Cellular abundance of V.SecD2 correlates inversely with Na+ concentrations and the presence of V.SecDF1. V. alginlyticus strains indicated at the bottom of C were grown at 30 °C until a mid log phase in the media supplemented with the indicated concentrations of Na+. Total cellular proteins of equivalent amounts were separated by Laemmli SDS/PAGE, followed by immunodetection of V.SecD1 (Upper) and V.SecD2 (Lower). Asterisks show nonspecific background bands. (C) Severe protein export defects in the absence of V.SecDF2 and sufficient Na+ levels. The same Vibrio strains used in B were grown at 30 °C in M63 medium supplemented with the indicated concentrations of Na+ and pulse-labeled for 30 s. Extracts of equivalent radioactivities were used for immunoprecipitation of MBP. Upon electrophoresis, precursor (p) and mature (m) bands of MBP were quantified to give the efficiencies of processing (export) shown at the bottom. (D) Activities of V.SecDF2 to support growth and protein export. (Upper) Growth at 30 °C in M63 medium supplemented with 33 mM NaCl and 10 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) was followed for the following V. alginolyticus strains: 1, WT (VIO5) carrying empty vector and 2–4, ΔsecDF2 carrying the vector (2), carrying the V.SecDF2-expressing plasmid (3), and carrying the V.SecDF2(R232M)-expressing plasmid (4). (Middle) Cellular abundance of V.SecD2 was examined by anti-V.SecD2 immunoblotting. Asterisks show nonspecific background signals. (Lower) Export of MBP was followed for the above strains by [35S]methionine pulse-labeling for 1 min at 30 °C. Processing efficiencies (percent) are shown at the bottom. See also Figs. S2 and S3.
Fig. S2.
Evaluation of antibodies and construction of the Vibrio mutant cells used in this study. (A) Specificity of the antibodies against V.SecD and V.SecF paralogs. Equal amounts of total proteins prepared from E. coli cells expressing either V.SecDF1 (lane 1), V. HA-SecDF2 (lane 2), or E.SecDF (lane 3) or E. coli cells with vector (lane 4) were separated by 10% (for V.SecD antibodies) and 15% (for V.SecF antibodies) Laemmli gels, followed by immunoblotting using the antibodies indicated. Each antibody specifically recognizes the corresponding protein. (B) Reactivity of the V.SecD antibodies. A V.SecD1 variant containing two closely arranged His10 stretches in the P1 domain was purified with Talon affinity chromatography in buffer containing 0.1% SDS from a membrane protein mixture, solubilized with 1% SDS, of a strain overexpressing the V.SecDF1 derivative. Similarly, V. HA-SecD2 protein with an N-terminal HA tag was purified with HA-antibody conjugated agarose beads in buffer containing 1% Triton X-100 from a membrane protein mixture, solubilized with 1% SDS, of a strain overexpressing the SecDF2 derivative. Detailed purification procedures are described in SI Materials and Methods. The indicated amounts of the purified proteins (lanes 1–5 for V.SecD1; lanes 6–10 for V.SecD2) were applied to 10% Laemmli gels and detected by CBB staining (the upper gel) and immunostaining with respective antibodies (the lower gels). Both antibodies have roughly equivalent reactivities against respective proteins. (C) Changes in accumulation levels of the V.SecDF paralogs upon a low-to-high Na+ concentration shift. (Left) Growth. WT V. alginolyticus 138-2 strain (the lateral flagellate, parental strain of VIO5) was first grown in M63 medium containing 300 mM NaCl and then inoculated (inoculum size 1/100) into M63 medium supplemented with 10 mM NaCl and 290 mM KCl. Cultivation was continued at 30 °C with rotational shaking of 180 rpm. At a mid log phase indicated by the dotted line, 1/100 volume was inoculated into fresh M63 medium supplemented with 300 mM NaCl and cultivation continued under the same physical conditions. Cell densities were monitored by turbidity measurement. (Right) Immunodetection of V.SecD1 and V.SecD2. Equivalent amounts of whole-cell proteins prepared from cells at the numbered sampling points (Fig. S2C, Left) were separated by 10% SDS/PAGE, and their V.SecD paralogs were detected with anti-V.SecD1 (upper panel) and anti-V.SecD2 (lower panel) immunoblotting. Samples prepared from E. coli cells expressing either V.SecD1-His6-F1 (lane 11) or HA-V.SecD2-F2 (lane 12) or E. coli cells with vector (lane 13) were also analyzed as a control. Nonspecific background signals are indicated by single and double asterisks. Relative intensities of the signals are shown below each lane. (D) PCR analysis of the Vibrio mutants constructed in this study. (Left) Schematic representations of the secDF1VA locus on the chromosome 1 (the upper two lanes) and the secDF2VA locus on the chromosome 2 (the lower two lanes) of WT and mutant strains. Predicted sizes of DNA fragments that should be amplified with the primers shown in the figure are presented in parentheses. (Right) Genomic DNAs of Vibrio strains VIO5 (WT), NR23 (ΔsecDF1), and HM3740 (ΔsecDF2), as indicated, were used as a template for PCR reactions primed by the primers shown at the top. The amplified DNAs were electrophoresed on 1% agarose gel and stained with Gel-Red. Asterisks indicate nonspecific products. All of the amplified fragments exhibited electrophoretic mobilities that agreed well with the predicted values.
The differential roles played by the two paralogs of SecDF were corroborated by immunologically detecting them (Fig. S2 A and B). V.SecD1, detected as a doublet band, persisted in the WT V. alginolyticus strain grown at any concentrations of Na+, with a slight decrease at lower Na+ concentrations (Fig. 2B, Upper, lanes 1–4). By contrast, V.SecD2 was undetectable in the presence of 300 mM Na+ (Fig. 2B, Lower, lane 4 and Fig. S2C, Lower, lanes 6–10). It became detectable at 63 mM or lower concentrations of Na+, in an inversely concentration-dependent manner (Fig. 2B, Lower, lanes 1–3 and Fig. S2C, Lower, lanes 1–5). We eliminated the possibility that the apparent induction of V.SecD2 at low Na+ concentrations was due to its selective degradation at higher Na+ concentrations by pulse-labeling and immunoprecipitation experiments. The synthesis rate of V.SecD2, assessed by brief pulse-labeling of V. alginolyticus cells with [35S]methionine, was indeed repressed at 300 mM Na+ (Fig. 3A, lane 1) and induced at lower Na+ concentrations (Fig. 3A, lanes 2–5). We also found that the synthesis of V.SecF2 was regulated almost identically to V.SecD2 (Fig. S3A, Right, lanes 1 and 4).
Fig. 3.
The synthesis rate of V.SecD2 is affected not only by Na+ but also by the absence of V.SecDF1 or the export-inhibiting azide. (A) WT V. alginolyticus grown in M63 medium supplemented with the indicated concentrations of Na+ were pulse-labeled for 1 min with [35S]methionine, followed by immunoprecipitation of V.SecD2. Asterisks indicate nonspecific background bands. (B) WT (lane 1 and lanes 4–9), ΔsecDF1 (lane 2), and ΔsecDF2 (lane 3) strains of V. alginolyticus grown in M63-300 mM Na+ medium at 30 °C were pulse-labeled for 1 min without NaN3 treatment (lanes 1, 2, 3, 8, and 9) or at the indicated time points after addition of NaN3. Samples of equivalent radioactivities were subjected to immunoprecipitation with antibodies against V.SecD1 (Upper) and V.SecD2 (Lower). Asterisks indicate nonspecific background bands. The numbers at the bottoms show intensities of V.SecD1 relative to that in lane 1 (Upper) and those of V.SecD2 relative to that in lane 2 (Lower). (C) To verify that azide inhibits protein export in V. alginolyticus, cells were pulse-labeled for 0.5 min with (lane 1) or without (lane 2) 1 min NaN3 pretreatment. Processing efficiencies of MBP are shown.
Fig. S3.
In vivo phenotypes of the Vibrio mutant cells. (A) Cellular levels of V.SecF paralogs are regulated in parallel with V.SecD paralogs. Total membrane proteins were prepared from WT (lanes 1 and 4), ΔsecDF1 (lanes 2 and 5), and ΔsecDF2 (lanes 3 and 6) cells grown in M63 medium supplemented with 300 mM (lanes 1–3) or 33 mM (lanes 4–6) Na+, separated by 10% Laemmli gel (Upper) or 10% wide-range gel (Lower), followed by immunodetection of V.SecD1 and V.SecD2 (Upper) as well as V.SecF1 and V.SecF2 (Lower). (B) Export of MBP in the Vibrio ΔsecDF2 mutant. The Vibrio ΔsecDF2 mutant cells were grown at 30 °C in M63 medium supplemented with the indicated concentrations of NaCl until an early log phase and pulse-labeled for 30 s. Cell extracts of equivalent amounts of radioactivities were immunoprecipitated with antibodies against E. coli MBP, followed by SDS/PAGE and phosphor imaging. Export efficiencies (proportions of mature form) of MBP are plotted against the medium NaCl concentrations. Relative intensities of immunoprecipitated MBP species (both the precursor and the mature forms) are also presented below each lane. (C) Functionality of the V.SecDF2 variants. (Upper Left) Growth complementation. A highly conserved Arg232 residue in TM5 of V.SecF2 was replaced with Met, on the basis of our previous findings that a substitution of methionine for the corresponding Arg247 of E.SecF and Arg240 of V.SecF1 (Fig. S1B) resulted in a complete loss of SecDF function (5). The E. coli secD1 mutant cells carrying a plasmid expressing V.SecDF2 (1), V.SecDF2(R232M) (2), E.SecDF (3), or no cloned gene (vector) (4) were grown until a mid log phase, diluted 103-fold, and spotted onto L-broth-Amp-arabinose (0.02%) agar plates and incubated at 37 °C for 18 h (Upper) or 20 °C for 52 h (Lower). (Lower Left) Protein export complementation. The secD1 mutant cells described in A were grown at 37 °C in M9 medium supplemented with 0.4% glycerol, 0.2% maltose, and 18 amino acids (except Met and Cys) until an early log phase. Then the cultures were sifted to 20 °C and cultivated for additional 30 min. Cells were pulse-labeled with [35S]methionine for 1 min. The labeled MBP and OmpA proteins were immunoprecipitated using respective antibodies. Their precursor (p) and mature (m) forms were separated by 10% Laemmli gel and analyzed by phosphor imaging. Export efficiencies (proportions of the mature form) of MBP and OmpA are shown below each lane. (Right) Accumulation of V.SecDF2 mutant proteins. Equivalent amounts of whole proteins prepared from the cells shown in A were separated by 10% Laemmli gel (the upper gel) or 10% wide range gel (the lower gel), followed by immunodetection of V.SecD2 (Upper) and V.SecF2 (Lower). The V.SecF2 antibodies cross-reacted with two bands, presumably the full-length V.SecF2 and its partial degradation product. Although the Arg232Met mutation seems to partially destabilize V.SecF2 protein, normal accumulation of the V.SecD2 was observed in the E. coli secD1 (Cs) cells. These results suggest that the Arg232 residue in V.SecF2 is crucial for its function.
We assessed protein export abilities of the V. alginolyticus strains by pulse-labeling MBP. Rapid export of MBP was observed with WT and the ΔsecDF1 cells, as shown by preferential labeling of the mature form, irrespective of the Na+ concentrations (Fig. 2C, lanes 1–8). By contrast, MBP export in the ΔsecDF2 cells was severely impaired, as judged from the exclusive labeling of its precursor form (Fig. 2C, lanes 9–11), unless cells were grown in the Na+-rich (300 mM) medium (Fig. 2C, lane 12). The efficiencies of MBP export correlated well with the concentrations of Na+ (Fig. S3B), although we observed that labeling of the MBP species was reduced under the export-compromised conditions for unknown reasons. The growth and export defects of the Na+-limited ΔsecDF2 cells were rescued when functional V.SecDF2 was supplied in trans (Fig. 2D and Fig. S3C), excluding the possibility that the chromosomal deletion mutation affected expression of some other growth/export-related genes. These results establish that Vibrio cells adapt to sodium-limited environments by inducing V.SecDF2 capable of using the proton-motive force.
We noted that the cellular abundance of V.SecD1 and V.SecD2 exhibited some interdependence. First, V.SecD2 (Fig. 2B, lanes 5–8) and V.SecF2 (Fig. S3A, Right, lanes 2 and 5) were markedly induced in the ΔsecDF1 mutant cells, as if they compensated for the loss of V.SecDF1. Second, the cellular abundance of V.SecD1 decreased when ΔsecDF2 mutant cells were grown in the presence of 33 mM or lower concentrations of Na+ (Fig. 2B, lanes 9 and 10).
Vibrio Cells Sense Lowered Functionality of the Sec Protein Export Pathway, Instead of Directly Sensing the Decreased Na+ Concentrations, to Up-Regulate V.SecDF2 Production.
Vibrio cells did not seem to sense extracellular Na+ concentrations for regulating secD2VA, because V.SecDF2 was induced markedly in the ΔsecDF1 cells even in the Na+-rich medium (Fig. 2B, lane 8). Pulse-labeling experiments confirmed that this induction took place at the level of the synthesis of V.SecD2 (Fig. 3B, Lower, lane 2). We then considered a possibility that the bacterium senses lowered functionality of the SecA-SecYEG-SecDF1 machinery. As a simple method to address this possibility, we examined effects of NaN3, an inhibitor of SecA, on the synthesis rate of V.SecDF2 (Fig. 3B). We first confirmed that NaN3 is a potent inhibitor of protein export in V. alginolyticus by its ability to inhibit signal peptide processing of pulse-labeled pre-MBP molecules (Fig. 3C). We then demonstrated that the synthesis of V.SecD2 was up-regulated markedly after addition of NaN3 (Fig. 3B, Lower, lanes 4–7). The V.SecD1 synthesis was unaffected (Fig. 3B, Upper). Thus, an impairment of the Sec protein export process results in specific up-regulation of V.SecDF2 synthesis.
VemP, Encoded by the Upstream ORF of secD2VA, Is a Secretory Protein That Undergoes Translational Elongation Arrest in Response to a Failure in Efficient Export.
The upstream ORF of the secD2VA gene consists of 159 codons. The deduced amino acid sequence shows that its N-terminal region contains a segment characteristic of a signal sequence for protein export; it is enriched with hydrophobic amino acids, which are preceded by two positively charged amino acids. We also note that the C-terminal ∼25-residue segment of the ORF is well conserved among Vibrio species (Table S1). We name this gene vemP, for Vibrio protein export monitoring polypeptide. Remarkably, VemP executes the function of monitoring the Sec activity and controlling the expression of secDF2VA by undergoing regulated translational elongation arrest, as discussed below.
Table S1.
Conserved features of VemP homologs among Vibrio species
| Vibrio species | Hydrophobic | C-terminal sequence of VemP | Residues |
| V. alginolyticus 138-2 | Yes | FYHFTSDHRI SGWKETNAMY VALNSQFSA | 159 |
| V. sp. (Ex25) | Yes | FYHFTSDHRI SGWKETNAMY VALNSQFSA | 159 |
| V. parahaemolyticus 10329 | Yes | FYHFTSDHRI SGWKETNAMY VALNSQFSA | 159 |
| V. alginolyticus 12G01 | Yes | FYHFTSDHRI SGWKETNAMY VALNSQFSA | 159 |
| V. harveyi 1DA3 | No | FNHFVSDHRL SGWKETNAMY VALNSQFPA | 144 |
| V. sp. EJY3 | Yes | FHHFTSDYRL SGWKETNAMY VALNSQFSA | 159 |
| V. harveyi (BBA-1116) | Yes (weak) | FNHFVSDHRL SGWKETNAMY VALNSQFSA | 159 |
| V. harveyi HY01 | Yes (weak) | FNHFVSNHRL SGWKETNAMY VALNSQFSA | 159 |
| V. vulnificus (YJ016) | Yes | QYHYVANHRL AGWKETNAMY VALNSQFS | 175 |
| V. tubiashii ATCC 19109 | Yes (weak) | YNSPYRI SGWKETNALY VALNGQYSVI S | 160 |
| V. brasiliensis LMG 20546 | Yes (weak) | YTSPHRI AGWKESNALY VALNAHYNSI F | 160 |
| V. furnissii (DSM 14383) | Yes | FHPSYRL SGWKETNAMY VALNSQYFH | 151 |
| V. coralliilyticus BAA-450 | Yes | FTSNHRI SGWKESNALY VALNAQFV | 158 |
| V. mimicus MB451 (SX-4) | Yes | FTSTHRL AGWKESNAMY VALNSQYQVS TFKI | 156 |
| V. sinaloensis DSM 21326 | Yes | FTANHRI SGWKDSNALY VALNSQFA | 159 |
| V. anguillarum (68554) | Yes | SNHRI SGWKETNAMY VALNSQY | 167 |
| V. orientalis CIP 102891 | Yes | RV SGWKESNTLY VALNGHF | 155 |
| V. cholerae HC-21A1 | Yes | FTSSHRL AGWKESNAMY VALNSQF | 155 |
We compared the VemP homologs among 18 Vibrio species. Most of the deduced VemP amino acid sequences were predicted by the TMHMM program to have a putative signal sequence at the N-terminal region, as indicated by “Yes” (with “weak” indicating relatively low hydrophobicity), whereas V. harvevi 1DA3 may lack the signal sequence. The third column shows the C-terminal segments of VemP, which are highly conserved among Vibrio species. The residues that are identical to those of V. alginolyticus 138-2 are highlighted in boldface.
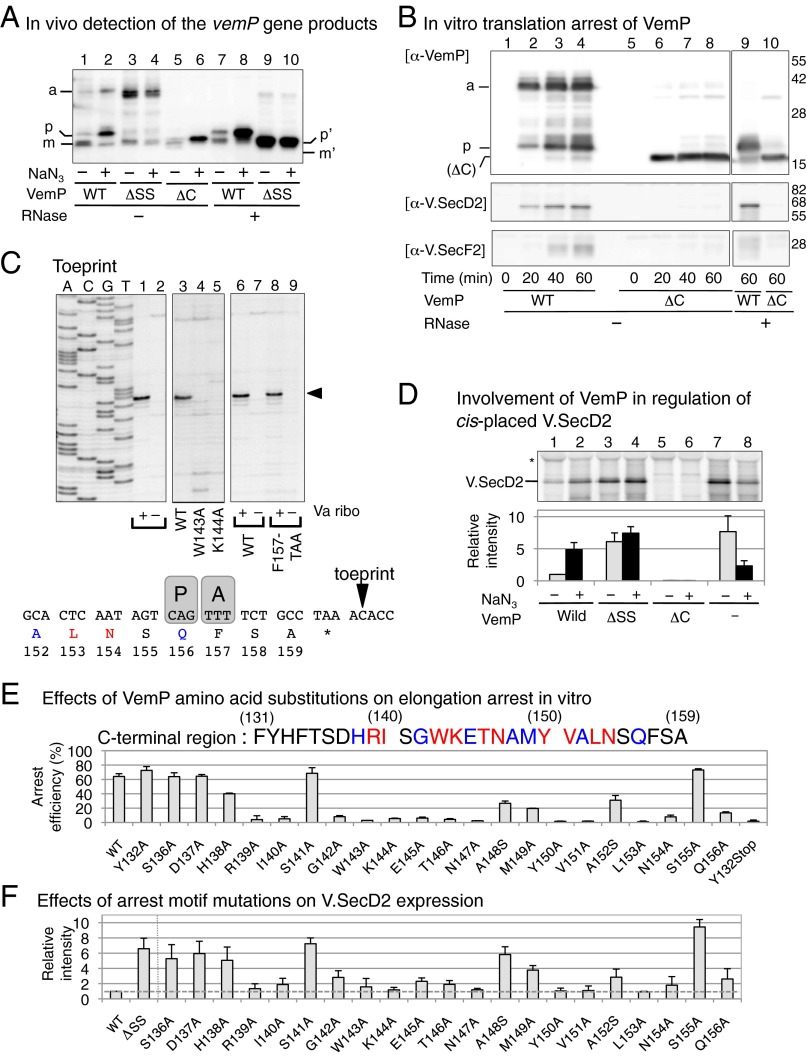
To characterize vemP, we expressed the plasmid-cloned vemP-secDF2VA genes in E. coli. Accumulation of the vemP-encoded protein was examined by immunoblotting. We used neutral pH SDS/PAGE to resolve protein species, which proved to include peptidyl-tRNA forms. Also, we focused on the effects of NaN3, a Sec inhibitor, because VemP has the putative signal sequence for export across the membrane. Indeed, VemP was detected as different molecular forms in response to administration of NaN3. Without NaN3, a majority of VemP was electrophoresed at the position indicated as “m” (Fig. 4A, lane 1) expected for the signal sequence-processed molecular species, along with two faint bands, indicated as “p” (for precursor form) and “a” (for arrest product; discussed below) of slower migration. When cells were treated with NaN3 for 30 min, the VemP species at the positions of “a” and “p” increased strikingly, whereas the “m” material diminished (Fig. 4A, lane 2). We reasoned that the large electrophoretic retardation of the “a” band was due to the presence of a covalently linked tRNA (26, 29); namely, translation elongation of VemP was arrested near the stop codon. In support of this possibility, treatment of the sample with RNase A converted the band “a” product to a material that migrated at the position “p” (Fig. 4A, lane 8). We note that VemP′-tRNA that gave band “a” was transferred poorly upon immunoblotting, explaining the intensity increase observed at position “p” after the RNase treatment (Fig. 4A, lanes 7 and 8). These results reveal two properties of VemP. First, it is indeed a secretory protein, which is handled by the SecA-SecYEG export pathway. Second, translation of VemP is subject to elongation arrest before termination, especially when its export is blocked. The materials that migrated at “p” may have represented either the polypeptide moiety of the arrested VemP′-tRNA produced by the ester bond hydrolysis in vivo or in vitro or, alternatively, translation-completed but signal peptide-unprocessed molecules.
Fig. 4.
Characterization of VemP as a Sec-monitoring substrate undergoing regulated translational arrest. (A) Detection of the vemP gene products. E. coli cells with plasmid carrying the vemP-secDF2VA genes containing the indicated vemP mutations were grown at 30 °C in M9-amino acids medium until a mid log phase. For the final 30 min of growth, they received 500 μM IPTG and 3 mM NaN3 (lanes 2, 4, 6, 8, and 10). Total proteins of the cultures of equivalent amounts, with or without RNase A treatment (40 μg/mL for 30 min at 37 °C) as indicated, were separated by wide-range gel and detected with immunodetection of VemP. p′ and m′ indicate precursor and mature forms of VemP(ΔC), respectively. (B) In vitro translation of the vemP-secD2VA-F2VA gene complex. DNA fragments of vemP-secDF2VA (WT) and vemP(Y132amber)-secDF2VA (ΔC) were used as a template for PURE System in vitro transcription–translation. At the indicated time points, samples, with or without RNase A treatment, were analyzed by neutral SDS/PAGE and immunoblotting detection of VemP (Upper), V.SecD2 (Middle), and V.SecF2 (Lower). (C) Ribosome stalling on the vemP mRNA studied by toeprinting. The vemP gene, with a mutation when indicated, was subjected to the PURE System reaction that lacked the E. coli ribosome (−) or that was supplemented with the ribosome prepared from V. alginolyticus (Va ribo) (+). The translation complexes were then used as a template for the reverse transcriptase reaction that was primed by a downstream primer (SI Materials and Methods). The cDNA products were electrophoretically separated along with the dideoxy sequencing ladders (left four lanes). Arrowhead indicates the signal generated by the ribosome stalling on vemP, which is schematically depicted below the toeprint gel on the basis of the comparison with the control reactions presented in Fig. S4A. P and A indicate the ribosomal P-site and A-site. (D) VemP controls secDF2VA expression in vivo. The plasmid-carried gene complex of vemP-secD2VA-F2VA (lanes 1–6) with a vemP mutation (ΔSS, signal sequence deletion; ΔC, Y132amber) as indicated, as well as secD2VA-F2VA DNA fragment directly placed under the control of the lac promoter on the vector (lanes 7 and 8) were induced in E. coli with 500 μM IPTG for 15 min at 30 °C. Cells were pulse-labeled with [35S]methionine for 1 min with (+, black) or without (−, gray) 5 min NaN3 (3 mM) pretreatment. Cell extracts of equivalent radioactivities were subjected to immunoprecipitation of V.SecD2. Intensities of the V.SecD2 band relative to that of the lane 1 sample are presented below the gel pattern (n ≥ 3). (E) Effects of VemP amino acid substitutions on translation arrest in vitro. WT and mutant forms of vemP having the indicated amino acid substitution mutations were translated in vitro at 37 °C for 45 min in the presence of [35S]methionine. Products were separated by neutral SDS/PAGE (Fig. S4D), and proportions (percent) of the arrested VemP-tRNA band in the sum of the tRNA-less VemP and the VemP-tRNA bands are presented (n ≥ 2). Amino acid residues, whose mutations decreased the translation arrest to below 10% peptidyl-tRNA (red) and to 10–40% peptidyl-tRNA (blue) are highlighted at the top. (F) Effects of VemP amino acid substitutions on in vivo expression of V.SecD2. WT and mutant forms of Δss-vemP-secD2VA-F2VA having the indicated amino acid substitution mutations were induced with 0.006% arabinose for 15 min in E. coli growing at 37 °C. Cells were then pulse-labeled for 1 min. Cell extracts of equivalent radioactivities were used for immunoprecipitation of V.SecD2, which was quantified after SDS/PAGE separation. Intensities of V.SecD2 relative to that of the WT sample are presented (n ≥ 3). See also Figs. S4 and S5.
The increased mobility of “m” in comparison with “p” is ascribable to the removal of the signal peptide, because the position of “m” was virtually identical to the position of the signal sequence-deleted construct of VemP (ΔSS) that was treated with RNase A (Fig. 4A, compare lanes 1 and 9/10). Strikingly, the results without RNase treatment indicate that the ΔSS variant of VemP was produced as the peptidyl-tRNA form that was electrophoresed at the position close to “a” whether or not the cells were treated with NaN3 (Fig. 4A, lanes 3 and 4). Thus, the WT product “m” was produced upon release from the arrested state, in which restored elongation (for the last three amino acid residues of VemP) accompanied the export-coupled signal peptide cleavage (discussed below). By contrast, no arrest release took place for the ΔSS construct. Taken together with the NaN3 effects observed with WT VemP, the constitutive elongation arrest seen with ΔSS VemP supports the notion that elongation arrest of VemP takes place strongly when VemP does not receive an effective export reaction.
To further characterize translation of VemP, we then used the in vitro translation system with the purified translation factors of E. coli and the ribosomes from either E. coli or V. alginolyticus, which behaved almost identically in translating vemP (Fig. S4 B and D). In the in vitro reactions, VemP was synthesized mainly as the peptidyl-tRNA form (Fig. 4B, lanes 2–4), which was converted by RNase treatment to the lower-molecular-weight product with an electrophoretic mobility similar to that of the in vivo product “p” (Fig. 4B, lane 9). Thus, in vitro translation of vemP is arrested near the C terminus. A greater proportion of the products was at the peptidyl-tRNA position at earlier time points than after prolonged reaction (Fig. 4B, compare lanes 2 and 4), suggesting that slow and spontaneous arrest release took place in vitro even in the absence of the Sec factors. Primer extension interference (toeprint) experiments verified that the ribosome indeed stalls at a specific position on the vemP mRNA in vitro, producing a specific length of reverse transcript (toeprint signal) (Fig. 4C, lane 1). Its alignment with the patterns of dideoxy sequencing using the same primer as well as with toeprint signals of control samples (Fig. S4A) indicates that translation of vemP is arrested as the P-site of the ribosome encounters the Gln156 codon (CAG) (Fig. 4C). Northern blot analysis of the translation product demonstrated that the arrested VemP'-tRNA carried a tRNAGln moiety (Fig. S4B), indicating that it resided in the P-site. Thus, the VemP-translating ribosome fails in peptidyl transfer reaction from the VemP1–156-tRNAGln to the A-site Phe-tRNAPhe.
Fig. S4.
In vitro analyses of VemP protein. (A) Calibration of the VemP toeprint signal. The vemP gene without (lanes 1 and 2) or with (lane 3) a missense mutation or a nonsense mutation as indicated (lanes 4–9) was subjected to the PURE system reaction that lacked the E. coli ribosome (−) (lane 2) or that was supplemented with ribosome prepared from V. alginolyticus (Va ribo) (+) (lanes 1 and 3–9). Additionally, the release factors 1, 2, and 3 were omitted for lanes 4–9 to make the ribosome stall at the defined stop codons. The translation complexes were then used as a template for the reverse transcriptase reaction that was primed by a downstream primer as described in SI Materials and Methods. The cDNA fragments were separated by 6% polyacrylamide–7 M urea gel along with the dideoxy sequencing ladders (left four lanes). Comparison of the signal produced from WT vemP (asterisk, lane 1) with those of the control samples (lanes 4–9) allowed us to identify the point of VemP translation arrest, where the Phe157 codon resides in the A site of the ribosome. (B) Western and Northern blotting characterization of VemP′-tRNAs. The flag-vemP fusion gene, encoding the Flag epitope tag followed by residues 26–159 of VemP (lanes 1 and 5), and flag-vemP with the F157stop mutation (lanes 4 and 8) were subjected to in vitro translation reaction using the Vibrio hybrid PURE System (lanes 1–4) or E. coli PURE System (lanes 5–8) in the presence of release factors. As controls we also used flag-vemP mRNA truncated after the 156th codon (lanes 2 and 6) and its Q156F version (lanes 3 and 7) as a template. The products were separated by 10% Nu-PAGE and the regions, where VemP′-tRNA molecules migrated, were analyzed by anti-FLAG immunoblotting (Upper) and Northern blotting using specific probes for tRNAGln (Middle) and tRNAPhe (Lower). WT VemP′-tRNA was hybridized with the tRNAGln probe (lane 1), indicating that the arrested nascent product contained a tRNAGln moiety. Thus, VemP translation undergoes an arrest at the step of peptidyl transfer from VemP1–156-tRNAGln to the A-site Phe-tRNAPhe. The product from the vemP(F157Stop) mutant was also hybridized with the same probe, indicating that the nascent VemP can arrest the termination step of translation as well. (C) Determination of the C-terminal end required for the VemP translation arrest. PCR products of vemP with a stop codon placed at the positions indicated were used as a template for in vitro translation in the presence of [35S]methionine for 45 min at 37 °C. The samples, treated without (lanes 1–7) or with (lanes 8–14) RNase A, were separated by 10% neutral SDS/PAGE, followed by phosphor imaging. (D) Effects of VemP amino acid substitutions on efficiency of translation arrest. WT and mutant forms of vemP having the indicated amino acid substitution mutations were translated using the Vibrio hybrid PURE system (the upper gel) and E. coli PURE system (the lower gel) in the presence of [35S]methionine at 37 °C for 45 min. Synthesized proteins were separated by neutral 10% SDS/PAGE. Proportions (percent) of the arrested VemP-tRNA band in the sum of the tRNA-less VemP and the VemP-tRNA bands are presented with SD in Fig. 4E (n ≥ 2 in the upper panel). Amino acid residues whose mutations compromised the translation arrest strongly and less strongly are highlighted in red and blue, respectively, at the top.
Determinants of the VemP Elongation Arrest.
A variant of VemP that is chain terminated by an amber mutation at the 132nd codon, termed VemPΔC hereafter, did not exhibit elongation arrest in vivo (Fig. 4A, lanes 5 and 6) or in vitro (Fig. 4B, lanes 6–8), indicating that the C-terminal 27 residues are required for the elongation arrest. We determined the C-terminal end of this requirement by a stop codon scanning. The in vitro elongation arrest withstood a stop-codon placement at the 157th position or its downstream (Fig. S4C, lanes 1–3) but not at position 156 or its upstream (Fig. S4C, lanes 4–7). Thus, “arrest sequence” of VemP ends at Gln156. Toeprint experiments showed that the stop codon at position 157 stalled the ribosome even though the full complement of release factors were included in the reaction (Fig. 4C, lane 8), indicating that the amino acid identity at position 157 is not essential for the ribosomal dysfunction.
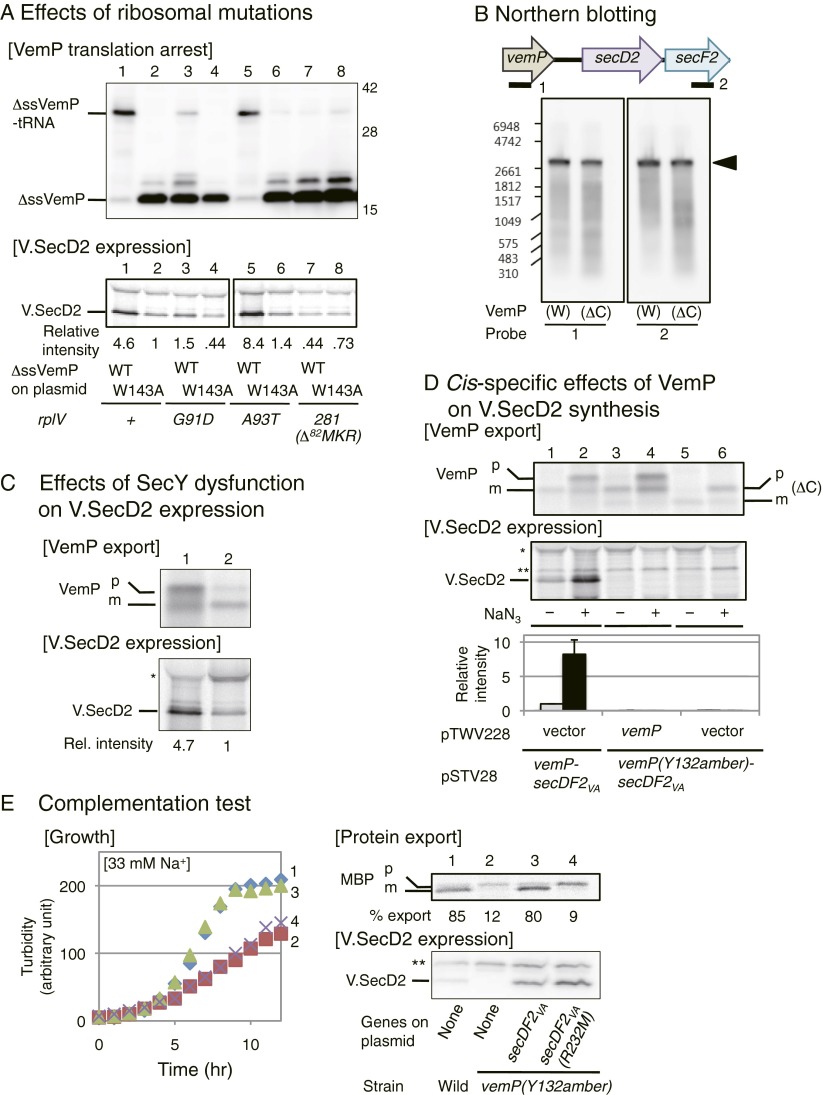
To further identify VemP elements required for the elongation arrest, we carried out alanine (serine when original residue was alanine) scanning mutagenesis of the C-terminal conserved segment. In vitro translation allowed us to determine the arrest-required amino acid residues, whose mutations are color-coded for strong (red) and moderate to weak (blue) defects in the arrest (Fig. 4E and Fig. S4D). Toeprint experiments confirmed defective ribosome stalling caused by the W143A and the K144A mutations (Fig. 4C, lanes 4 and 5). Thus, of the 21 amino acid residues of the segment examined, as many as 17 contribute to the translation halt of VemP. We also examined effects of the three mutations of the ribosomal protein L22 that were shown previously to compromise the SecM elongation arrest in E. coli (18) on elongation arrest of VemPΔSS expressed in E. coli. L22 (rplV) mutations, G91D and ΔM82KR, but not A93T, impaired the elongation arrest of VemPΔSS (Fig. S5A). Thus, the ribosome participates actively in the VemP-based regulation, but nascent chain–ribosome interactions are not identical for VemP and SecM, which differ totally in the primary sequences (see Fig. 7B). These results show that a specific amino acid sequence in the nascent VemP polypeptide is responsible for the elongation arrest through its interaction with the ribosomal exit tunnel.
Fig. S5.
Effects of VemP elongation arrest on V.SecDF2 expression. (A) (Upper) Effects of ribosomal mutations on VemP translational arrest. Plasmids pBAD24-ΔssvemP-secDF2VA (WT, odd lanes) and pBAD24-ΔssvemP(W143A)-secDF2VA (W143A, even lanes) were introduced into cells of WT E. coli (lanes 1 and 2) or E. coli with a ribosomal mutation, rplV(G91D) (lanes 3 and 4), rplV(A93T) (lanes 5 and 6), or rplV281 (lanes 7 and 8). Transformed cells were grown at 37 °C until a mid log phase (lanes 1–8) and induced with 0.02% arabinose for 1 h. Equivalent amounts of proteins were separated by 10% wide-range gel, followed by anti-VemP immunoblotting. (Lower) Effects of ribosome mutations on V.SecD2 expression. The WT and the ribosomal mutants of E. coli, carrying the plasmid indicated, were grown at 37 °C in M9 medium until an early log phase, induced with 0.02% arabinose for 10 min and then pulse-labeled with [35S]methionine for 1 min. Cellular proteins of equivalent radioactivities were used for immunoprecipitation of V.SecD2, which was separated by 10% SDS/PAGE and detected by phosphor imaging. Whereas the signal sequence-less VemP, which shows constitutive elongation arrest, renders the V.SecD2 synthesis constitutive, the ribosomal mutations that compromised the arrest reduced the synthesis levels of V.SecD2 (lanes 3 and 7). (B) Northern blotting analysis of the in vitro vemP operon transcript. PCR products of vemP-secDF2VA (W) and vemP(Y132amber)-secDF2VA (ΔC) were used as a template for PURE System transcription/translation reactions in vitro, using the E. coli ribosomes. Total RNAs (1 μg) were extracted from the in vitro samples and separated by 0.9% agarose gel electrophoresis and detected by Northern blotting using probes 1 and 2, which were complementary to the indicated parts of the vemP and secF2VA, respectively. (C) Protein export defect-dependent up-regulation of V.SecD2 synthesis. The secY24 (Ts) mutant of E. coli carrying psyd and pTV119N-vemP-secDF2VA (lane 1) and WT E. coli carrying pSTV29 and pTV119N-vemP-secDF2VA (lane 2) were grown at 30 °C in M9 medium until an early log phase. Syd, which inhibits the altered translocon, and VemP-V.SecDF2 were induced with 1 mM IPTG and 2 mM cAMP for 30 min. Then, cells were pulse-labeled with [35S]methionine for 1 min. VemP and V.SecD2 were immunoprecipitated from total cellular proteins of equal radioactivities. Under the pH conditions used in this experiment (pH 8.1), the peptidyl-tRNA ester bond had been hydrolyzed. Labeled proteins were separated by 10% wide-range gel for VemP (upper gel) and 10% Laemmli gel for V.SecD2 (lower gel). Overproduction of Syd in the secY24 strain led to preferential labeling of the precursor form of VemP, indicative of Sec dysfunction. Under such conditions, labeling of V.SecD2 was enhanced evidently (lane 1) compared with that in the WT cells. Asterisk shows a nonspecific background. (D) Cis-specific effect of vemP on V.SecD2 synthesis. WT E. coli cells containing the two plasmids indicated at the bottom were grown at 37 °C in M9 medium until an early log phase. The cultures were then treated with (even lanes) or without (odd lanes) 3 mM NaN3 for 5 min, followed by pulse-labeling with [35S]methionine for 1 min. Labeled VemP (Upper) and V.SecD2 (Middle) were immunoprecipitated, separated by 10% wide-range gel and 10% Laemmli gel, respectively, and analyzed by phosphor imaging. The notations of p indicates the polypeptide part of the elongation arrested VemP and/or the precursor form of VemP, and m indicates the mature form of VemP (left side) and VemP (ΔC) (right side). Single and double asterisks indicate nonspecific backgrounds. (Lower) Relative intensities of V.SecD2, taking that of lane 1 as unity with SD (n = 3). For up-regulated synthesis of V.SecD2 in response to a protein export defect, the cis placement of vemP is essential. (E) Defects of the Na+-limited Vibrio vemP(Y132amber) mutant are rescued by expression of functional V.SecDF2. (Left) Growth at 30 °C in M63 medium supplemented with 33 mM NaCl and 10 μM IPTG was followed for the following V. alginolyticus strains. 1, WT (VIO5) carrying the empty vector; 2–4, vemP(Y132amber) mutant carrying vector (2), carrying the V.SecDF2-expressing plasmid (3), and carrying the V.SecDF2(R232M)-expressing plasmid (4). Cell densities were monitored by turbidity measurement. (Upper Right) Export of MBP was followed for the above strains by [35S]methionine pulse-labeling for 30 s at 30 °C, followed by immunoprecipitation using cells extracts of equivalent radioactivities and anti-E. coli MBP. MBP species were separated by SDS/PAGE and detected by phosphor imaging. Processing efficiencies (percent) of MBP are shown at the bottom. (Lower Right) Cellular abundance of V.SecD2 was examined for the above strains, using equivalent numbers of cells grown as above, followed by anti-V.SecD2 immunoblotting. Asterisks show a nonspecific background signal.
Fig. 7.
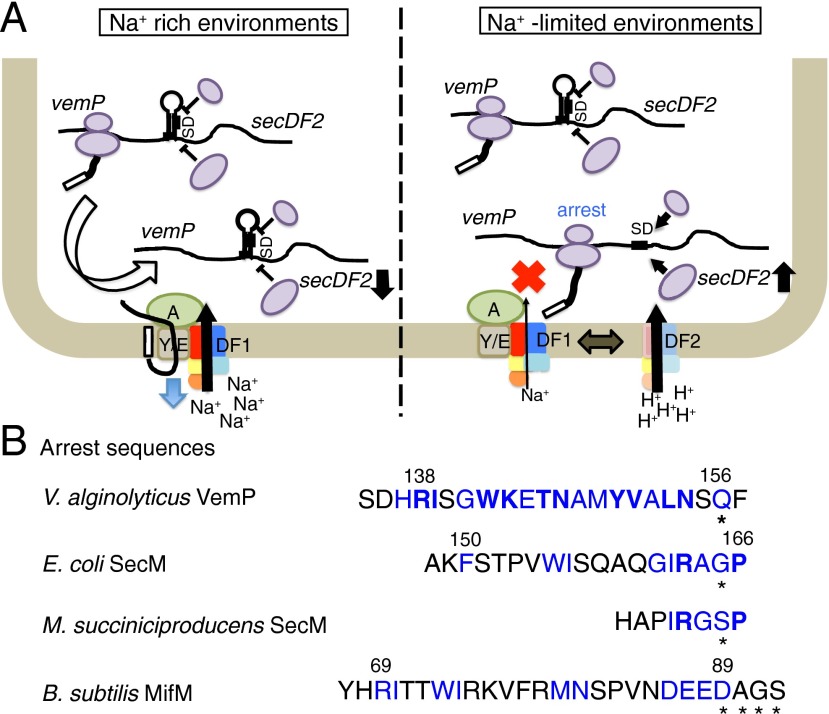
(A) Adaptation of Vibrio species to salinity changes through export-controlled translation arrest of VemP. (Left) Repression of V.SecDF2 synthesis in Na+-rich environments. Because the VemP export occurs efficiently by the V.SecDF1-containing Sec machinery capable of using the Na+-motive force, the VemP elongation arrest is transient. In this situation, the duration of the arrest, in which the Shine–Dalgarno sequence of V.SecD2 is available, is short, leading to low-level synthesis of V.SecD2. (Right) Up-regulation of V.SecDF2 synthesis in Na+-limited environments. In the absence of sufficient concentrations of Na+, activity of the V.SecDF1-containing Sec machinery will decline, leading to retarded export and prolonged elongation arrest of VemP. This in turn results in higher rates of initiation of secDF2VA translation. Newly synthesized V.SecDF2 molecules will substitute for V.SecDF1 such that the bacteria can now use the proton-motive force to effectively localize their secretory proteins. (B) Diversity in the intrinsic class of arrest sequences. Shown are the amino acid sequences that induce translation arrest without any cofactor, which have been elucidated for VemP, SecM (18, 52), and MifM (19). Residues whose identities contribute to the elongation arrest are highlighted in blue; particularly important residues are shown in boldface. Asterisks indicate the last residue of the arrested peptidyl-tRNA, which resides in the P-site of the ribosome. See also Fig. S6.
VemP Regulates Translation of V.SecDF2 and Is Essential for the Vibrio’s Adaptation to Environments of Different Sodium Levels.
To study roles of VemP in the synthesis of V.SecDF2, we expressed the vemP-secDF2VA gene complex in E. coli. Pulse-labeling experiments showed that the V.SecD2 synthesis was up-regulated markedly by NaN3 treatment (Fig. 4D, lanes 1 and 2). The V.SecD2 synthesis-enhancing effect was not specific for the chemical NaN3, because an independent, genetic means to impair the Sec function also led to the up-regulated synthesis of V.SecDF2 in an E. coli secY mutant (Fig. S5C). By contrast, a direct connection of secDF2VA to the lac promoter resulted in NaN3-independent production of V.SecD2, which was rather lowered by the drug, presumably due to general inhibition of metabolism (Fig. 4D, lanes 7 and 8). Thus, VemP is required for the regulation. The amber mutation at the 132nd codon of vemP, which produced the arrest-defective VemPΔC, prevented the V.SecD2 synthesis even when cells were treated with NaN3 (Fig. 4D, lanes 5 and 6). The VemP function to support the NaN3-dependent up-regulation of V.SecDF2 synthesis was cis-specific, because coexpression of WT VemP singly from a compatible plasmid did not restore V.SecDF2 synthesis from vemP(Y132amber)-secDF2VA (Fig. S5D, compare lanes 2 and 4). In contrast to the arrest-defective mutation, deletion of the VemP signal sequence, which caused constitutive arrest, allowed high-level and NaN3-independent synthesis of V.SecD2 (Fig. 4D, lanes 3 and 4). Good correlations were also observed between the extents of VemP arrest and the levels of V.SecDF2 expression among the VemP and the rplV (L22) missense mutations that affected the arrest efficiencies (Fig. 4, compare E and F, and Fig. S5A). The effects of the vemP and the rplV missense mutations rule out any polarity effects on the expression of the downstream target, secDF2VA. We then recapitulated this regulation in vitro. In the in vitro coupled transcription–translation system, vemP, secD2VA, and secF2VA were transcribed into a single mRNA (Fig. S5B), from which both V.SecD2 and V.SecF2 were translated (Fig. 4B, lower panels, lanes 2–4). However, arrest-defective VemPΔC did not allow the synthesis of V.SecD2 or V.SecF2 (Fig. 4B, Lower, lanes 6–8) without affecting the mRNA production. From these results, taken together, we conclude that the cis placement of vemP upstream of secDF2VA is required for the regulated translation of V.SecD2.
To address whether VemP plays crucial roles in the V.SecD2 production and consequent adaptation to low-sodium environments in V. alginolyticus, we introduced the elongation arrest-impairing Tyr132 amber mutation into the vemP gene on chromosome 2. Remarkably, the mutant V. alginolyticus cells did not contain any detectable amount of the V.SecD2 protein (Fig. 5C, lane 4). Their ability to export MBP was markedly impaired as well (Fig. 5B, lane 4). Furthermore, the mutant cells exhibited marked growth retardation in medium containing 33 mM Na+ (Fig. 5A, Right, growth curve 4), but not in the presence of 300 mM Na+ (Fig. 5A, Left). The observed defects in protein export and growth were similar to those observed with the ΔsecDF2 mutant (Fig. 5, strain 3) and, in fact, complemented by the forced expression of WT V.SecDF2 from a plasmid (Fig. S5E, lane 3). A nonfunctional SecDF2 variant was incompetent in this complementation (Fig. S5E, lane 4). These results establish that the VemP nascent polypeptide has an essential role in the adaptation of the marine Vibrio to low-sodium environments through its regulated elongation arrest that enables the efficient translation of the secD2VA-secF2VA genes.
Fig. 5.
VemP with the normal ability to arrest its own translation is essential for V. alginolyticus to grow rapidly and induce V.SecDF2. (A) Growth of V. alginolyticus strains with mutations as indicated by numbers (1, WT, squares; 2, ΔsecDF1, diamonds; 3, ΔsecDF2, triangles; 4, Y132amber, crosses) was followed at 30 °C in M63 minimum medium supplemented with either 300 mM Na+ (Left) or 33 mM Na+ (Right). (B and C) The Vibrio strains used in A were grown in the M63 medium supplemented with 33 mM Na+ until an early log phase and then pulse-labeled with [35S]methionine for 30 s. Labeled MBP and V.SecD2 were immunoprecipitated and separated by 10% SDS/PAGE, followed by phosphor imaging. See also Fig. S5.
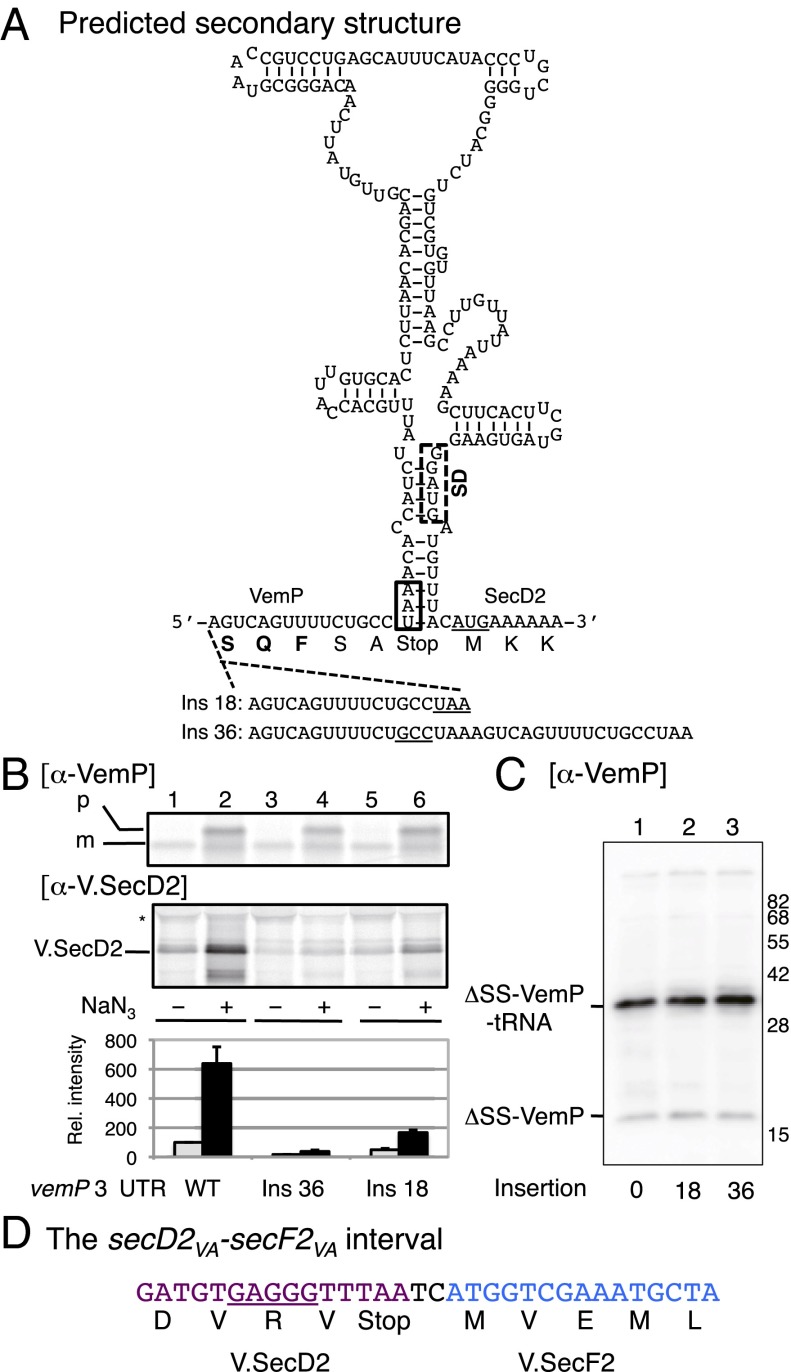
Finally, we address the importance of the elements in the intergenic regions of the vemP-secD2-secF2VA mRNA that support the VemP-based regulation. A secondary structure model by CentroidHomfold (www.ncrna.org/centroidhomfold/) shows that the ribosome-binding (Shine–Dalgarno) sequence of secD2VA is likely to be sequestered in a stem structure (Fig. 6A). The ribosome stalling at the arrest point in vemP, close to the stem-forming region, is then expected to interfere with the stem-loop formation, and, therefore, to liberate the Shine–Dalgarno sequence for initiation of the secD2VA translation by the free ribosomes. To experimentally validate the above model, we increased the distance between the arrest point and the secondary structure-forming region, by an insertion of one or two units of an 18-nucleotide sequence (Fig. 6A). Such a manipulation resulted in a severely impaired induction of the V.SecD2 synthesis upon export inhibition (Fig. 6B), without affecting the VemP translation arrest per se (Fig. 6C). Thus, the regulatory significance of the ribosome stalling requires its occurrence in the close vicinity of the secondary structure-forming unit of the mRNA. The fact that V.SecF also receives this regulation may rest on the short secD2VA-secF2VA intergenic region, having only two nucleotides (Fig. 6D). Here, the translational coupling mechanism (30, 31) may allow coordinated regulation of secD2VA and secF2VA by VemP.
Fig. 6.
Regulatory importance of the vemP-secD2-secF2 operon arrangement. (A) Predicted secondary structure of mRNA at the vemP-secD2VA intergenic region. The RNA sequence from the fifth last codon of vemP to the third codon of secD2VA are shown with the secondary structure predicted by CentroidHomfold. The putative SD sequence and the start codon of secD2VA are indicated by box and underline, respectively. The P site and the A site of the VemP-stalled ribosome (Fig. 4C) are shown schematically. To separate the VemP arrest point and the secondary structure-forming region, we inserted one or two copies of the 18 nucleotides encoding the last five amino acid residues of VemP followed by the termination codon, shown at the bottom. (B) Separation of the arrest point and the stem-loop-forming region impairs the regulation. E. coli cells carrying indicated vemP-secDF2VA plasmids (with or without the insertion mutations shown in A) were induced with IPTG for 15 min at 37 °C and treated with (+) or without (−) 3 mM NaN3 for 5 min. Cells were then pulse-labeled for 1 min. Cell extracts of equivalent radioactivities were subjected to anti-VemP (upper gel) and anti-V.SecD2 (lower gel) immunoprecipitation. In the upper gel, “p” indicates the signal peptide unprocessed form of VemP, which included the polypeptide part of the elongation arrested VemP, whereas “m” indicates the signal peptide-processed mature form. Relative intensities of V.SecD2 are shown in the bottom graph, taking the value of lane 1 as unity (n = 3). Gray and black bars represent results obtained without and with NaN3 treatment, respectively. (C) The insertion mutations do not affect translation arrest of VemP. E. coli cells carrying pBAD24-Δss-vemP-V.secDF2 with or without the insertion mutations were induced with 0.02% arabinose for 1 h at 37 °C. The same amounts of proteins were separated by 10% wide-range gel electrophoresis, followed by immunodetection of VemP. (D) The secD2VA-secF2VA interval. The nucleotide sequence from the fourth last codon of secD2VA to the fifth codon of secF2VA is shown. The predicted SD sequence of secF2VA is underlined.
SI Materials and Methods
Media.
L broth rich medium (53) and M9 synthetic medium supplemented with maltose (final 0.2%), glycerol (final 0.4%), 18 amino acids (except Met and Cys; final concentration of 20 μg/mL each) (53) were used for cultivation of E. coli strains. VC rich medium (54) and M63 synthetic medium (55) supplemented with maltose (final 0.2%), glycerol (final 0.4%), the 18 amino acids, and a total of 300 mM of NaCl–KCl combinations were used for cultivation of Vibrio cells. Note that the minimal-amino acids media are called simply M9 and M63 media in this paper. For E. coli, antibiotics were used at the following concentrations: ampicillin, 50 μg/mL; chloramphenicol, 20 μg/mL; kanamycin, 25 μg/mL; and tetracycline, 25 μg/mL. For V. alginolyticus, antibiotics were used at the following concentrations: chloramphenicol, 2.5 μg/mL and kanamycin, 100 μg/mL.
Chemicals.
Super-high-grade chemical reagents purchased from Sigma-Aldrich or Nacalai Tesque were used for all of the experiments. Restriction endonucleases (New England BioLabs) and the Ligation high Ver. 2 DNA ligation kit (Toyobo Co., Ltd.) were used for cloning experiments. Prime Star max DNA polymerase (Takara Shuzo Co., Ltd.) was used for polymerase chain reactions. In-Fusion HD cloning kit (Takara Shuzo Co., Ltd.) was used along with the classic cloning protocols for plasmid construction.
Antibodies.
For preparation of antibodies against V.SecD and V.SecF paralogs and VemP, the two synthetic oligopeptides listed in Table S4 were used for production of each antibody. The two antigenic polypeptides were mixed and conjugated with a carrier protein, keyhole limpet hemocyanin, via the Cys residue attached at their N termini and used to raise antibodies in rabbits. We used both the antisera against V.SecD1 and V.SecD2 for immunoblotting and immunoprecipitation experiments, whereas the anti-V.SecF1 and anti-SecF2 antisera were only usable for immunoblotting. We used the unfractionated anti-VemP serum for immunoprecipitation and its affinity-purified IgG fraction for immunoblotting. Anti-E. coli MBP antibodies (50) and anti-E. coli OmpA antibodies, gifts from the late Shoji Mizusihima, Tokyo University of Pharmacy and Life Sciences, Hachioji, Japan, were used for immunoprecipitation of pulse-labeled species of these proteins to assess efficiencies of their export from E. coli and Vibrio cells.
Table S4.
Synthetic peptides used for preparation of antisera in this study
| Names | No. | Peptide sequences | Corresponding residue numbers |
| V.SecD1 | 1 | C-QEEAFRSELREAKI | 138–151 |
| 2 | C-GRAPAGSEIKMDRD | 287–300 | |
| V.SecD2 | 1 | C-GNRIAVNSESDTAL | 199–212 |
| 2 | C-SGQDLVLKDNDGRT | 317–330 | |
| V.SecF1 | 1 | C-GFEKPANLEKIRT | 55–67 |
| 2 | C-VRLRPRDDVSGETL | 90–103 | |
| V.SecF2 | 1 | C-VKVDKSLTNHDLLD | 50–63 |
| 2 | C-YSAPEEGQTLPSVE | 88–101 | |
| VemP | 1 | C-VSEETTQSPISESHA | 59–73 |
| 2 | C-VSHLREGLDDEHVD | 87–100 |
Strains and Plasmids.
The E. coli and Vibrio strains used in this study are listed in Table S2. The E. coli strains newly prepared in this study were constructed by P1 transduction using appropriate strains and selective markers. The Vibrio strains were constructed as described below. The plasmids used in this study are listed in Table S3. Nucleotide sequences were determined by PRISM3130 DNA analyzer (Applied Biosystems Co., Ltd.). We confirmed all of the relevant DNA segments that were subjected to in vitro replication by sequencing the final constructs.
Table S2.
Strains used in this study
| Names | Genotypes | Source |
| E. coli | ||
| MC4100 | F−, araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 62 |
| KI297 | MC4100 rpsE zhd-33::Tn10 sY24 /F' lacIqlacPL8 lacZ+Y+A+pro+ | 63 |
| AD16 | Δprolac thi ara+/F' lacIqlacZΔM15Y+A+pro+ | 64 |
| AD202 | MC4100 ompT::kan | 65 |
| HM3314 | AD202 ara+ secD1 Tn10 (linked to secDF) | 5 |
| THE521 | AD202 secD1 Tn10 (linked to secDF) | Laboratory stock |
| HM1752 | MC4100 ara+ F'lacIq lacZΔM15Y+A+pro+ | 66 |
| β3914 | β2163 gyrA462 zei-298::Tn10 | 60 |
| HM1246 | AD202 ara+ | 67 |
| NH493 | AD16 rplV281 zhd-33::Tn10 | 18 |
| NH553 | AD16 rplV(G91D) zhd-33::Tn10 | 18 |
| NH555 | AD16 rplV(A93T) zhd-33::Tn10 | 18 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpirRK6 | 68 |
| HM4025 | AD202 ara+ rplV+ zhd-33::Tn10 | This study |
| HM4023 | AD202 ara+ rplV(G91D) zhd-33::Tn10 | This study |
| HM4026 | AD202 ara+ rplV281 zhd-33::Tn10 | This study |
| HM4028 | AD202 ara+ rplV(A93T) zhd-33::Tn10 | This study |
| V. alginolyticus | ||
| 138-2 | WT | 69 |
| VIO5 | 138–2 Rifr Pof+ Laf− | 28 |
| NR23 | VIO5 ΔsecDF1::kan | This study |
| HM3740 | VIO5 ΔsecDF2::kan | This study |
| EI308 | VIO5 vemP(Y132amber) | This study |
Table S3.
Plasmids used in this study
| Names | Vectors | Genes on plasmid | Relevant experiment | Source |
| pHM810 | pBAD24 | vemP-secDF2VA | Figs. 4 B, C, E, F, S4 C, D | This study |
| pHM813 | pBAD24 | Δss-vemP-secDF2VA | Figs. 4F, S5A | This study |
| pHM814 | pBAD24 | vemP(Y132amber)-secDF2VA | Fig. 4 B, E | This study |
| pHM829 | pBAD24 | vemP(S158stop)-secDF2VA | Fig. S4 A, C | This study |
| pHM830 | pBAD24 | vemP(F157stop)-secDF2VA | Fig. S4 A, C | This study |
| pHM832 | pBAD24 | vemP(Q156stop)-secDF2VA | Fig. S4 A, C | This study |
| pHM834 | pBAD24 | vemP(S155stop)-secDF2VA | Fig. S4 A, C | This study |
| pHM836 | pBAD24 | vemP(N154stop)-secDF2VA | Fig. S4 A, C | This study |
| pHM838 | pBAD24 | vemP(L153stop)-secDF2VA | Fig. S4 A, C | This study |
| pHM846 | pBAD24 | vemP(W143A)-secDF2VA | Figs. 4C, S4 A, D | This study |
| pHM848 | pBAD24 | vemP(K144A)-secDF2VA | Figs. 4C, S4D | This study |
| pHM915 | pBAD24 | Δss-vemP(132A)-secDF2VA | Fig. 4F | This study |
| pHM934 | pBAD24 | Δss-vemP(S136A)-secDF2VA | Fig. 4F | This study |
| pHM918 | pBAD24 | Δss-vemP(D137A)-secDF2VA | Fig. 4F | This study |
| pHM935 | pBAD24 | Δss-vemP(H138A)-secDF2VA | Fig. 4F | This study |
| pHM899 | pBAD24 | Δss-vemP(R139A)-secDF2VA | Fig. 4F | This study |
| pHM919 | pBAD24 | Δss-vemP(I140A)-secDF2VA | Fig. 4F | This study |
| pHM900 | pBAD24 | Δss-vemP(S141A)-secDF2VA | Fig. 4F | This study |
| pHM920 | pBAD24 | Δss-vemP(G142A)-secDF2VA | Fig. 4F | This study |
| pHM902 | pBAD24 | Δss-vemP(W143A)-secDF2VA | Fig. 4F | This study |
| pHM904 | pBAD24 | Δss-vemP(K144A)-secDF2VA | Fig. 4F | This study |
| pHM921 | pBAD24 | Δss-vemP(E145A)-secDF2VA | Fig. 4F | This study |
| pHM923 | pBAD24 | Δss-vemP(T146A)-secDF2VA | Fig. 4F | This study |
| pHM906 | pBAD24 | Δss-vemP(N147A)-secDF2VA | Fig. 4F | This study |
| pHM925 | pBAD24 | Δss-vemP(A148S)-secDF2VA | Fig. 4F | This study |
| pHM927 | pBAD24 | Δss-vemP(M149A)-secDF2VA | Fig. 4F | This study |
| pHM908 | pBAD24 | Δss-vemP(Y150A)-secDF2VA | Fig. 4F | This study |
| pHM910 | pBAD24 | Δss-vemP(V151A)-secDF2VA | Fig. 4F | This study |
| pHM929 | pBAD24 | Δss-vemP(A152S)-secDF2VA | Fig. 4F | This study |
| pHM912 | pBAD24 | Δss-vemP(L153A)-secDF2VA | Fig. 4F | This study |
| pHM937 | pBAD24 | Δss-vemP(N154A)-secDF2VA | Fig. 4F | This study |
| pHM931 | pBAD24 | Δss-vemP(S155A)-secDF2VA | Fig. 4F | This study |
| pHM939 | pBAD24 | Δss-vemP(Q156A)-secDF2VA | Fig. 4F | This study |
| pHM997 | pTV119N | vemP-secDF2VA | Fig. 4 A, D | This study |
| pHM1023 | pTV119N | Δss-vemP-secDF2VA | Fig. 4 A, D | This study |
| pHM1002 | pTV119N | vemP(Y132amber)-secDF2VA | Fig. 4 A, D | This study |
| pHM1001 | pTV119N | secDF2VA | Fig. 4 A, D | This study |
| pHM1025 | pSTV28 | vemP-secDF2VA | Fig. S5D | This study |
| pHM1027 | pSTV28 | vemP(Y132amber)-secDF2VA | Fig. S5D | This study |
| pHM1033 | pTWV228 | vemP | Fig. S5D | This study |
| pHM1054 | pSTV28 | vemP-(36 bases ins.)-secDF2VA | Fig. 6B | This study |
| pHM1055 | pSTV28 | vemP-(18 bases ins.)-secDF2VA | Fig. 6B | This study |
| pHM1057 | pBAD24 | Δss-vemP-(18 bases ins.)-secDF2VA | Fig. 6C | This study |
| pHM1058 | pBAD24 | Δss-vemP-(36 bases ins.)-secDF2VA | Fig. 6C | This study |
| pHM754 | pBAD24 | secD1VA(his)6 | Figs. 1B, S2A | 5 |
| pHM756 | pBAD33 | secF1VA | Figs. 1B, S2A | 5 |
| pHM753 | pSTV28 | ha-.secDF2VA | Figs. 1B, S2 A, B | This study |
| pGK02 | pSTV28 | ha-secDFEC | Figs. 1B, S2A | 5 |
| pHM802 | pBAD24 | secD1VA internal (his)10-GT-(his)10 | Fig. S2B | This study |
| pHM769 | pBAD24 | secDF2VA | Fig. S3C | This study |
| pHM1014 | pBAD24 | secDF2(R232M)VA | Fig. S3C | This study |
| pHM748 | pBAD24 | secDFEC(his)6 | Fig. S3C | This study |
| pST30 | pACYC | syd | Fig. S5C | 49 |
| pEI7 | pMMB206 | secDF2VA | Figs. 2D, S5E | This study |
| pEI8 | pMMB206 | secDF2(R232M)VA | Figs. 2D, S5E | This study |
| pHM775 | pTH100 | secD2VA 5′-kan-secF2VA 3' | Fig. S2D | This study |
| pNR5 | pTH100 | secD1VA 5′-kan-secF1VA 3' | Fig. S2D | This study |
| pEI9 | pSW7848 | vemP(Y132amber)-secD2VA | Fig. 5 | This study |
| pBAD24 | 70 | |||
| pBAD33 | 70 | |||
| pTV119N | Takara Bio | |||
| pTWV228 | Takara Bio | |||
| pSTV28 | Takara Bio | |||
| pFLAG-CTC | lacIq | Sigma-Aldrich | ||
| pCH85 | pTWV228 | ha-tag | 56 | |
| pTH100 | 54 | |||
| pSW7848 | 57 | |||
| pMMB206 | 71 |
Discussion
SecDF uses the proton-motive force across the membrane to enhance protein export that has been initiated by the SecA-SecYEG core Sec components (5). It is important for the normal physiology of E. coli, because its depletion leads to severe defects in protein export and cell growth, especially at low temperatures (32). However, V.SecDF1 and V.SecDF2 are not simply redundant-essential factors in Vibrios but differentiated with respect to the cations that energize the export-enhancing functions. In order for the Vibrio strain to cope with and survive the environments of changing salinity, they must use a proper V.SecDF paralog. We have presented pieces of evidence that support the above conclusions, including the following: (i) V.SecDF1 uses the sodium-motive force, whereas V.SecDF2 uses the proton-motive force; (ii) production of V.SecDF2 is inversely correlated with sodium concentrations of media; (iii) in this regulation, the Sec protein export activity is directly monitored by VemP, which undergoes regulated translation arrest to stimulate translation of V.SecDF2; and (iv) V. alginolyticus requires the VemP-mediated up-regulation of V.SecD2 production to grow and propagate rapidly under low-sodium conditions.
The constitutive expression of the sodium-dependent paralog, V.SecDF1, in Vibrio cells seems to be in accordance with the primary habitat, marine and estuarine water, of this species. The location of secDF1VA on the “housekeeping” chromosome 1 and the similarity of its organization to that of the E. coli yajC-secD-secF operon could suggest that the V.SecDF1 protein has evolved from the proton-using prototype during marine bacteria’s adaptation to seawater (33). However, an alternative view that the sodium motive force was the primordial form of energy (27, 34) suggests that the proton-using SecDF homologs, including V.SecDF2, had evolved from a V.SecDF1-like ancestor. Whatever the evolutionary scenarios may be, the V.SecDF2 protein in Vibrios today supports their life in such low-sodium environments as brackish water and intrabody or intracellular fluid of host animals. The regulated expression of SecDF2 should contribute to the prevalence of this group of bacteria in marine–estuarine interfaces.
The existence of the two V.SecDF paralogs with different ion selectivity is reminiscent of the dual flagella systems, Na+- vs. H+-driven, in V. alginolyticus (35). However, the H+-dependent lateral flagella is induced in response to an increase in external viscosity (36). V. alginolyticus uses a distinct mechanism to maintain high protein export activity in different ionic environments by exchanging the SecDF paralogs (Fig. 7A). The secretory protein VemP possesses an “arrest sequence” that prevents the ribosome from continuing elongation beyond the Gln156 codon. The secD2VA translation remains switched on as long as the ribosome stalling continues, and the stalled ribosome liberates the Shine–Dalgarno sequence of the secD2VA gene (Fig. 6A). VemP, which may be called a “Sec-monitoring substrate,” can give rapid, real-time on/off outputs for V.SecD2 translation in response to fluctuations of the cellular protein export activity. Moreover, this regulation is directly coupled to the translation of the other subunit, V.SecF2, of the complex through the translational coupling mechanism (Fig. 6D). We envisage that any other mechanisms that use a secondary signaling molecule, if any, will only be able to give greatly delayed outputs compared with the Sec-monitoring substrate, which can register the primary event in real time.
High-Na+ environments (∼600 mM in seawater) allow Vibrio to depend on the Na+-driven V.SecDF1, which is abundantly present in the cell (Fig. 7A, Left). By contrast, low-Na+ environments prevent V.SecDF1 from functioning, and the resulting decline in protein export is sensed by VemP, which immediately activates translation of V.SecDF2 to enable cells to use the H+ gradient for protein export. A recent transcriptome analysis of V. parahaemolyticus suggests that SecDF2 is a member of the low-salt stimulon (37). However, it remains to be determined whether the observed increase in the secDF2 message under low-salt conditions was generated by transcriptional regulation or secondarily by the VemP-mediated translational regulation.
Importantly, the rapid up-regulation of the V.SecDF2 production will only be effective if the synthesized V.SecDF2 molecules can collaborate with the SecYEG translocon. Rapid subunit exchange and/or preferential translocon association of V.SecDF2 will help the immediate translocon remodeling. We suggest that a quick Sec machinery remodeling upon a cell’s encounter with a low-salinity environment may be aided by preferential degradation of V.SecDF1 in the absence of sufficient levels of both V.SecDF2 and sodium ions (Fig. 2B). Decreased V.SecDF1 concentrations will then facilitate the formation of the SecDF2-containing export machinery that can use protons. Inversely, upon transition from a low-sodium to a high-sodium medium, the autogenously controlled V.SecDF2 synthesis will be terminated eventually due to cross-talks with now-activated V.SecDF1. In fact, the cellular abundance of V.SecD2 became negligible already at the time point of 3 h after the medium change (Fig. S2C), pointing to the possibility of active degradation. Possible involvement of a membrane protein quality control system (38) in Vibrio’s bidirectional adaptation to salinity changes will be an interesting research subject in the future.
Vibrio species lack the secM gene and instead have acquired vemP. It is intriguing that the Sec monitoring substrate in E. coli (SecM) and in V. alginolyticus (VemP) regulates the expression of different Sec components, SecA and SecDF, respectively. Probably, the Vibrio species chose V.SecDF2 as the target of regulation because SecDF2 carries out an environment-sensitive part of the export processes in this organism. The use of VemP seems to be advantageous because it can sense any conditions that disturb the protein export pathway to up-regulate V.SecDF2, which can use protons available ubiquitously. The rapid response mechanism by the monitoring substrate may be advantageous for Vibrio bacteria to infect animals successfully . A possible pathogenic importance of the V.SecDF2 paralog of Vibrio has been suggested by global transcriptome analyses, revealing secDF2 induction upon infection (39, 40).
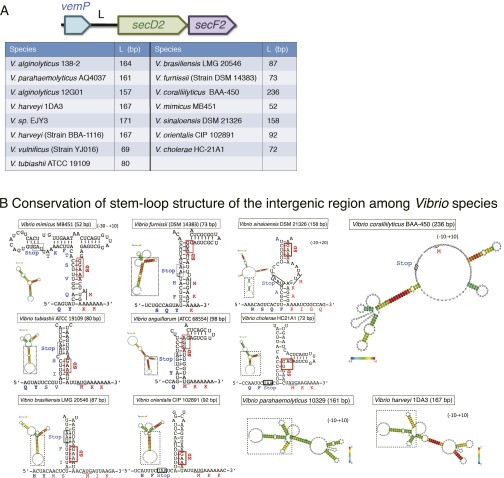
VemP adds to the concept, already discussed in the literature (19, 25, 41, 42), that each of the regulatory nascent polypeptides might have been acquired late in evolution by a specific group of species in phylogenetic lineages. Indeed, VemP is unique to Vibrio (Fig. S6A) and possibly other quite limited species. The high degree of conservation of the vemP-secDF2 gene complexes (Fig. S6A) and the VemP arrest motifs (Table S1) as well as the stem-loop-forming potential of the vemP-secDF2 intergenic regions (Fig. S6B) suggest that Vibrio species generally use VemP for regulation of secDF2, although the lengths of the intergenic regions are variable (Fig. S6A). In agreement with the late evolution theory, VemP has the arrest motif of unique amino acid sequence, which features as many as 17 amino acid residues that contribute to the temporary dysfunction of the translating ribosome. Because mutations of each one of these residues impair the arrest, the VemP elongation arrest must be a manifestation of their cooperative intramolecular interaction as well as interactions with ribosomal components. In line with the notion that ribosome arrest peptides interact with the ribosome in idiosyncratic ways (25, 43), ribosomal mutations affect the VemP-mediated arrest with a spectrum different from that observed for the SecM-mediated arrest. Furthermore, unlike the elongation arrest in SecM, which requires prolyl-tRNA in the A-site, that of VemP does not require a specific A-site amino acid; the P-site VemP1–156-tRNA did not even receive efficient termination reaction when the A-site was programmed by a stop codon. Although the ribosome-tethered VemP1–156-tRNA likely interferes with the ribosomal peptidyl transferase activity, the mode of its action is also different from that used by MifM, which induces multisite stalling of the B. subtilis ribosome, but not the E. coli ribosome (24, 44). We showed that VemP can stall the ribosomes from V. alginolyticus and E. coli, both being gram-negative, equally well. Species specificity of nascent polypeptide–ribosome interactions is an interesting subject to be explored.
Fig. S6.
Conservation of the stem-loop structures of vemP-secD2 intergenic regions. (A) The vemP-secD2 intergenic regions of different Vibrio strains. Lengths of the intergenic regions between vemP and secD2 from Vibrio species are listed in the table. (B) Predicted secondary structures formed in the intergenic regions. RNA sequences from 10–30 bases upstream of the VemP stop codon to 10–20 bases downstream of the V.SecD2 start codon of various Vibrio species were analyzed by the CentroidHomfold program. The boxed regions of the modeled RNA structures are enlarged to show the stem structures, in which the putative SD sequences of secD2 are highlighted by red boxes. The initiation codons of secD2 are underlined. In the case of V. parahaemolyticus and Vibrio harveyi, only the whole structures are presented, because their core stem structures are almost identical to that from V. alginolyticus (Fig. 6A). Vibrio coralliilyticus BAA-450 is exceptional in having no stem structure in front of the secD2 start codon. In the other strains, we observed several common features in the structures: (i) The start points of the stem structure are very close to the arrest points (a Gln codon near the stop) determined in this study, and (ii) the putative SD sequences of V.SecD2 are engaging in the stem structures. These features are in good agreement with our model of regulation of V.SecDF2 translation.
It should be noted that VemP represents the third member of the “intrinsic” class of regulatory nascent polypeptides (45) that arrests translation without involving a small-molecular-weight cofactor (see refs. 46 and 47 for the latter examples). Instead, their translation-arrested states are subject to release by the nascent chain engagement in protein localization reactions (Fig. 7B) (18, 19, 25). The unique arrest sequence of VemP indicates that living organisms have evolved ribosome arrest sequences in the course of genetic information diversification. However, they must have exploited the regulatory potential of the arrest sequences by arranging them in a convergent way into a regulatory unit such as the vemP-secD2VA-secF2VA, the secM-secA, and the mifM-yidC2 operons. The involvement of the large number of VemP amino acid residues in arrest will give a unique opportunity to study how the ribosome recognizes nascent polypeptides and how the dynamics of the nascent chain affects the translation progression (48). Whereas these molecular mechanisms deserve focused and combined approaches of biochemistry, genetics, and structural biology, we should also understand how living organisms integrate such mechanisms to survive changing environments. Microorganisms in marine–estuarine interfaces may continue to provide unique insights into environmental adaptation.
Materials and Methods
Strains and Plasmids.
Strains and plasmids used in this study are listed in Tables S2 and S3, respectively and details of the constructions are described in SI Materials and Methods.
SDS/PAGE.
Proteins were separated by 10% Laemmli SDS/PAGE for analysis of MBP, OmpA, and V.SecD paralogs and by 10% neutral pH SDS/PAGE, either wide-range gel (Nacalai Tesque) or Nu-PAGE (Life Technologies), for analysis of VemP, VemP-tRNA, and V.SecF paralogs (26, 29).
Immunoblotting.
Immunoblotting analysis was carried out according to the standard protocols described previously (49), using antisera against V.SecD1 (1/3,000), V.SecD2 (1/1,000), and VemP (after affinity-purified, 1/1,000), which were prepared as described in SI Materials and Methods and Table S4. Antisera against V.SecF1 and V.SecF2 were diluted 103-fold with PBS containing 5% (wt/vol) skim milk and 0.1% Tween 20 and pretreated with crude lysates prepared from the Vibrio ΔsecDF1 and ΔsecDF2 mutant strains, respectively, at 4 °C for 1 h to reduce background signals.
Pulse-Labeling Experiments.
E. coli and Vibrio were cultivated in the specified minimal medium (SI Materials and Methods, Media) until an early log phase at 37 °C or 30 °C, respectively, and pulse-labeled with [35S]methionine (ARS 0104A, 37 TBq/mmol) for 30 s or 1 min. The labeled cultures were immediately precipitated with ice cold TCA [final 5% (vol/vol)]. Immunoprecipitation was performed as described previously (50).
Purification of the V. alginolyticus Ribosomes.
Purification of Vibrio ribosome was carried out according to the procedures described by Ohashi et al. (51) with modifications presented in SI Materials and Methods.
In Vitro Translation.
In vitro transcription/translation reaction was carried out using PUREfrex System (Gene Frontier Co., Ltd) in combination with either the Vibrio or the E. coli ribosomes, according to the manufacturer’s instructions, as detailed in SI Materials and Methods.
Toeprint and Northern Blotting Dissection of the Ribosome-Nascent Chain Complexes on the vemP mRNA.
The ribosomes from the Vibrio strain were combined with PUREfrex System for in vitro translation/translation of the vemP gene. DNA templates for transcription/translation reactions were prepared by two successive PCR reactions as detailed in SI Materials and Methods. In vitro synthesized polypeptidyl-tRNAs of VemP derivatives were analyzed by Northern blotting using biotinylated oligodeoxynucleotide probes complementary to either tRNAGln or tRNAPhe as described previously (24). Translation complexes were also subjected to toeprint analysis as described in SI Materials and Methods.
Acknowledgments
We thank D. Mazel for providing the plasmid pSW7848 and the strain β3914; Y. Hizukuri and H. Terashima for helpful suggestions; S. Zhu for unpublished information about the chromosome engineering; and M. Sano for technical support. This work was supported by Grants-in-Aid for Scientific Research provided by Ministry of Education, Culture, Sports, Science and Technology and Japan Society for the Promotion of Science Grants 24117003 and 22370070 (to H.M.), Grant 15H01532 (to Y.A.), and Grants 26116008 and 25291006 (to S.C.), as well as by the Uehara Memorial Foundation (H.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513001112/-/DCSupplemental.
References
- 1.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78(5):835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 2.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427(6969):36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 3.Nouwen N, Piwowarek M, Berrelkamp G, Driessen AJ. The large first periplasmic loop of SecD and SecF plays an important role in SecDF functioning. J Bacteriol. 2005;187(16):5857–5860. doi: 10.1128/JB.187.16.5857-5860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pogliano KJ, Beckwith J. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol. 1994;176(3):804–814. doi: 10.1128/jb.176.3.804-814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474(7350):235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mio K, et al. Conformational variation of the translocon enhancing chaperone SecDF. J Struct Funct Genomics. 2014;15(3):107–115. doi: 10.1007/s10969-013-9168-4. [DOI] [PubMed] [Google Scholar]
- 7.Reilly GD, Reilly CA, Smith EG, Baker-Austin C. 2011. Vibrio alginolyticus-associated wound infection acquired in British waters, Guernsey, July 2011. Euro surveill 16(42):19994.
- 8.Zhang L, Orth K. Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol. 2013;16(1):70–77. doi: 10.1016/j.mib.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Noguchi T, et al. Vibrio alginolyticus, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu vermicularis vermicularis. Mar Biol. 1987;94(4):625–630. [Google Scholar]
- 10.Zhang L, et al. Type III effector VopC mediates invasion for Vibrio species. Cell Reports. 2012;1(5):453–460. doi: 10.1016/j.celrep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino K, et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V. cholerae. Lancet. 2003;361(9359):743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 12.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunioka E, Matsuyama S, Tokuda H. Cloning and expression of the secA gene of a marine bacterium, Vibrio alginolyticus, and analysis of its function in Escherichia coli. Gene. 1998;216(2):303–309. doi: 10.1016/s0378-1119(98)00343-6. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya D, Das J. The secY gene of V. cholerae: Identification, cloning and characterization. Gene. 1997;196(1-2):261–266. doi: 10.1016/s0378-1119(97)00238-2. [DOI] [PubMed] [Google Scholar]
- 15.Nishiyama K, Furuta M, Tokuda H. Molecular cloning and functional characterization of SecE of a marine bacterium, Vibrio alginolyticus. Biochem Biophys Res Commun. 1998;251(3):894–897. doi: 10.1006/bbrc.1998.9573. [DOI] [PubMed] [Google Scholar]
- 16.Tokuda H, Kim YJ, Mizushima S. In vitro protein translocation into inverted membrane vesicles prepared from Vibrio alginolyticus is stimulated by the electrochemical potential of Na+ in the presence of Escherichia coli SecA. FEBS Lett. 1990;264(1):10–12. doi: 10.1016/0014-5793(90)80751-4. [DOI] [PubMed] [Google Scholar]
- 17.Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001;7(1):185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 18.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108(5):629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 19.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28(22):3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiba S, Ito K. MifM monitors total YidC activities of Bacillus subtilis, including that of YidC2, the target of regulation. J Bacteriol. 2015;197(1):99–107. doi: 10.1128/JB.02074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami A, Nakatogawa H, Ito K. Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc Natl Acad Sci USA. 2004;101(33):12330–12335. doi: 10.1073/pnas.0404907101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver DB, Beckwith J. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell. 1982;30(1):311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- 23.Pogliano KJ, Beckwith J. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133(4):763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba S, Ito K. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol Cell. 2012;47(6):863–872. doi: 10.1016/j.molcel.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Chiba S. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 26.Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell. 2006;22(4):545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 27.Mulkidjanian AY, Dibrov P, Galperin MY. The past and present of sodium energetics: May the sodium-motive force be with you. Biochim Biophys Acta. 2008;1777(7-8):985–992. doi: 10.1016/j.bbabio.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178(8):2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito K, et al. Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One. 2011;6(12):e28413. doi: 10.1371/journal.pone.0028413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoner BE, Belagaje RM, Schoner RG. Translation of a synthetic two-cistron mRNA in Escherichia coli. Proc Natl Acad Sci USA. 1986;83(22):8506–8510. doi: 10.1073/pnas.83.22.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spanjaard RA, van Duin J. Translational reinitiation in the presence and absence of a Shine and Dalgarno sequence. Nucleic Acids Res. 1989;17(14):5501–5507. doi: 10.1093/nar/17.14.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13(3):554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deamer DW. The first living systems: A bioenergetic perspective. Microbiol Mol Biol Rev. 1997;61(2):239–261. doi: 10.1128/mmbr.61.2.239-261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takekawa N, et al. Sodium-driven energy conversion for flagellar rotation of the earliest divergent hyperthermophilic bacterium. Sci Rep. 2015;5:12711. doi: 10.1038/srep12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355(6356):182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 36.McCarter L, Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990;4(7):1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, et al. The low-salt stimulon in Vibrio parahaemolyticus. Int J Food Microbiol. 2010;137(1):49–54. doi: 10.1016/j.ijfoodmicro.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 39.Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA. 2003;100(3):1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bisharat N, Bronstein M, Korner M, Schnitzer T, Koton Y. Transcriptome profiling analysis of Vibrio vulnificus during human infection. Microbiology. 2013;159(Pt 9):1878–1887. doi: 10.1099/mic.0.067900-0. [DOI] [PubMed] [Google Scholar]
- 41.Cruz-Vera LR, Yanofsky C. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of Tna operon expression. J Bacteriol. 2008;190(14):4791–4797. doi: 10.1128/JB.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Sluis EO, Driessen AJ. Stepwise evolution of the Sec machinery in Proteobacteria. Trends Microbiol. 2006;14(3):105–108. doi: 10.1016/j.tim.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol. 2011;21(2):274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Sohmen D, et al. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat Commun. 2015;6:6941. doi: 10.1038/ncomms7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito K, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun. 2010;393(1):1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong F, Ito K, Nakamura Y, Yanofsky C. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro) Proc Natl Acad Sci USA. 2001;98(16):8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71(4):811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 48.Goldman DH, et al. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348(6233):457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimoike T, et al. Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J Biol Chem. 1995;270(10):5519–5526. doi: 10.1074/jbc.270.10.5519. [DOI] [PubMed] [Google Scholar]
- 50.Baba T, et al. Characterization of cold-sensitive secY mutants of Escherichia coli. J Bacteriol. 1990;172(12):7005–7010. doi: 10.1128/jb.172.12.7005-7010.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem Biophys Res Commun. 2007;352(1):270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 52.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009;34(2):201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori H, Akiyama Y, Ito K. A SecE mutation that modulates SecY-SecE translocase assembly, identified as a specific suppressor of SecY defects. J Bacteriol. 2003;185(3):948–956. doi: 10.1128/JB.185.3.948-956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terashima H, Koike M, Kojima S, Homma M. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol. 2010;192(21):5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 56.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16(16):2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. Genome engineering in Vibrio cholerae: A feasible approach to address biological issues. PLoS Genet. 2012;8(1):e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kusumoto A, et al. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem. 2006;139(1):113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- 59.Zhu S, Kumar A, Kojima S, Homma M. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol. 2015;23 doi: 10.1111/mmi.13103. [DOI] [PubMed] [Google Scholar]
- 60.Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol. 2007;73(3):777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- 62.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1984. [Google Scholar]
- 63.Ito K, Akiyama Y. In vivo analysis of integration of membrane proteins in Escherichia coli. Mol Microbiol. 1991;5(9):2243–2253. doi: 10.1111/j.1365-2958.1991.tb02154.x. [DOI] [PubMed] [Google Scholar]
- 64.Kihara A, Akiyama Y, Ito K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc Natl Acad Sci USA. 1995;92(10):4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem Biophys Res Commun. 1990;167(2):711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 66.Mori H, Ito K. The long alpha-helix of SecA is important for the ATPase coupling of translocation. J Biol Chem. 2006;281(47):36249–36256. doi: 10.1074/jbc.M606906200. [DOI] [PubMed] [Google Scholar]
- 67.Nakatogawa H, Murakami A, Mori H, Ito K. SecM facilitates translocase function of SecA by localizing its biosynthesis. Genes Dev. 2005;19(4):436–444. doi: 10.1101/gad.1259505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tokuda H, Nakamura T, Unemoto T. Potassium ion is required for the generation of pH-dependent membrane potential and delta pH by the marine bacterium Vibrio alginolyticus. Biochemistry. 1981;20(14):4198–4203. doi: 10.1021/bi00517a038. [DOI] [PubMed] [Google Scholar]
- 70.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morales VM, Bäckman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97(1):39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]