Significance
Gastric cancer (GC) is one of the major causes of cancer-related deaths worldwide, but targeted therapy for GC is limited. Here, we identified two druggable targets from genomic alteration profiling of 103 patients with GC from Asia and validated the target suitability using patient-derived GC xenograft models, which recapitulate the tumor biology observed in patients. Combination therapy of irinotecan (standard treatment) with a BCL2L1 (BCL2-like 1)-targeted drug was effective in size reduction of GC tumors having amplification of the BCL2L1 gene, and genomic mutations of deleted in liver cancer 1 (DLC1) were associated with increased sensitivity to a ROCK inhibitor. Therefore, our study strongly suggests that BCL2L1 and DLC1 can serve as targets for novel GC therapies.
Keywords: gastric cancer, copy number alteration, whole-exome sequencing, patient-derived xenograft, druggable target
Abstract
Gastric cancer (GC) is the third leading cause of cancer-related deaths worldwide. Recent high-throughput analyses of genomic alterations revealed several driver genes and altered pathways in GC. However, therapeutic applications from genomic data are limited, largely as a result of the lack of druggable molecular targets and preclinical models for drug selection. To identify new therapeutic targets for GC, we performed array comparative genomic hybridization (aCGH) of DNA from 103 patients with GC for copy number alteration (CNA) analysis, and whole-exome sequencing from 55 GCs from the same patients for mutation profiling. Pathway analysis showed recurrent alterations in the Wnt signaling [APC, CTNNB1, and DLC1 (deleted in liver cancer 1)], ErbB signaling (ERBB2, PIK3CA, and KRAS), and p53 signaling/apoptosis [TP53 and BCL2L1 (BCL2-like 1)] pathways. In 18.4% of GC cases (19/103), amplification of the antiapoptotic gene BCL2L1 was observed, and subsequently a BCL2L1 inhibitor was shown to markedly decrease cell viability in BCL2L1-amplified cell lines and in similarly altered patient-derived GC xenografts, especially when combined with other chemotherapeutic agents. In 10.9% of cases (6/55), mutations in DLC1 were found and were also shown to confer a growth advantage for these cells via activation of Rho-ROCK signaling, rendering these cells more susceptible to a ROCK inhibitor. Taken together, our study implicates BCL2L1 and DLC1 as potential druggable targets for specific subsets of GC cases.
Gastric cancer (GC) is a highly prevalent malignancy and is the third leading cause of cancer-related deaths in the world (1). In unresectable and metastatic cases, the clinical outcome for this disease remains poor (median survival is 10–14 mo) (2), and other treatment options are often limited because of the lack of effective therapeutic approaches and molecular prognostic markers (2, 3). To date, with the exception of the application of trastuzumab [ERBB2 (ErbB2 receptor tyrosine kinase 2) antagonist] or ramucirumab [VEGFR2 (Vascular endothelial growth factor receptor 2) antagonist] for advanced GC cases (4, 5), drugs that target GC on a molecular level are limited.
Recent genomic studies have demonstrated the heterogeneous genomic characteristics of GC (6–10). In addition, previous studies of patients with GC by whole-genome and whole-exome sequencing (WES) have identified frequent somatic mutations in tumor suppressors such as TP53, ARID1A, APC, and FAT4, and oncogenes including PI3KCA, KRAS, and RHOA (6–10). However, these findings, although academically meaningful, are far from ready for clinical applications, largely because of the lack of identified druggable molecular targets and the availability of reliable preclinical models for validation of potential target inhibitors.
To identify novel therapeutic targets for GC, we explored genomic alterations in GC through an integrated genomic data set from WES and copy number alteration (CNA) analyses of tumors of patients with GC. Recurrent amplification of antiapoptotic gene BCL2L1 (BCL2-like 1) and recurrent mutation of DLC1 (deleted in liver cancer 1) were found and then strongly suggested as druggable candidate targets in subsets of GC tumors, using in vitro experiments and in vivo GC patient-derived xenograft (PDX) mouse models. We demonstrate that small molecules targeting these two genes have the potential to be effective treatments for subsets of GC harboring these genomic alterations.
Results
Genome-Wide Analyses of CNAs and Mutations in GC.
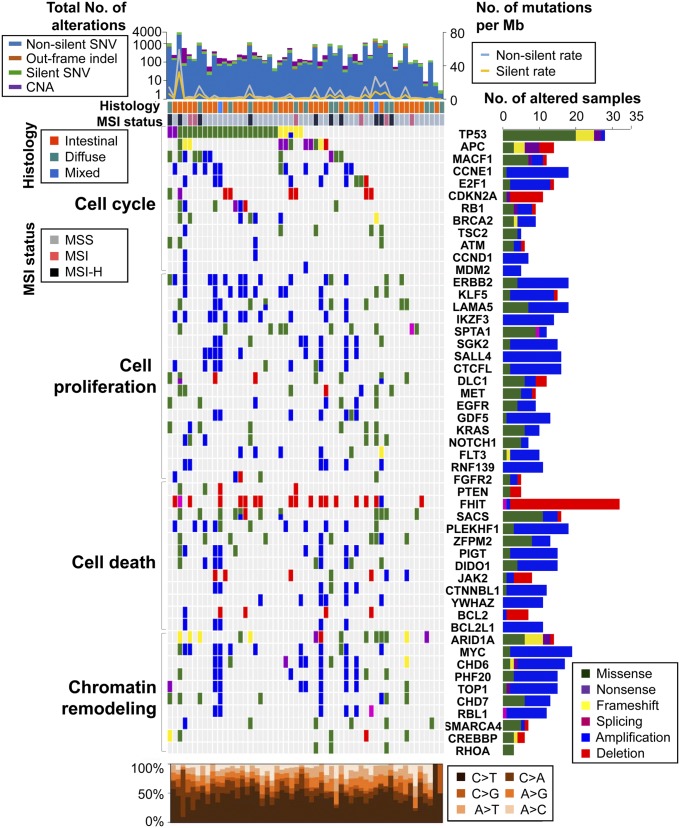
Using paired tumor and normal samples from 103 GC cases [containing at least 70% tumor cellularity by pathological examination (11); detailed clinical information is found in SI Appendix, Tables S1 and S2], we identified a total of 3,858 CNAs using a one million-feature genome-wide array comparative genomic hybridization (aCGH) platform (SI Appendix, Fig. S1A and Dataset S1). The average numbers of copy number gains and losses were 22.6 and 18.9 per patient, respectively. A previous study categorized GC into four subgroups: Epstein-Barr virus-positive, microsatellite instable (MSI), genomically stable, and chromosomal instability (6). In our study, the patients in the chromosomal instability subgroup (55.4 gains and 37.5 losses per patient) showed much higher CNA rates than the patients in the MSI (5.1 gains and 8.9 losses per patient) and genomically stable (4.4 gains and 5.0 losses per patient) subgroups. To validate the CNAs, we performed droplet digital PCR (ddPCR) experiments for 20 randomly chosen genes. Results of 140/178 of the ddPCR experiments correlated with our aCGH data, resulting in a predictive value of 78.7% (SI Appendix, Table S3).
We also conducted WES on 55 paired normal-tumor GC samples (SI Appendix, Fig. S1B). The average mapping percentages of reads were 99.8% and 99.9%, and the average coverage depths were 91.7x and 89.3x for normal and tumor samples, respectively (SI Appendix, Tables S4 and S5). From the WES data, we identified 61,926 somatic mutations in the 55 GC samples, of which 23,025 mutations occurred in protein-coding regions (14,060 were missense mutations, 748 were nonsense mutations, 6,563 were synonymous mutations, 328 mutations were at splice sites, 206 were unclassified mutations, and 1,120 were indels). The average nonsilent mutation rate (5.91 variants/Mb) was similar to that in previous reports (8, 12), and samples with a MSI-high status correspondingly showed higher mutation rates (19.58 variants/Mb; Fig. 1 and SI Appendix, Table S6) (10, 13). Sanger sequencing was used to validate mutations in 73 randomly selected regions, giving a positive prediction rate of 83.6% (SI Appendix, Table S7). However, Sanger sequencing can detect mutations down to 10–15% frequency, and accordingly, there must be some false negatives. Previous studies of GC mutational signatures showed high-frequency ratios of C > T, C > A, and A > G base changes (average: C > T, 45.1%; C > A, 14.9%; A > G, 14.4%) (6, 12), a signature pattern that was replicated in our study (average: C > T, 43.8%; C > A, 27.7%; A > G, 15.4%; Fig. 1). There was little difference between the mutation signatures according to Lauren histological classifications (diffuse, intestinal, and mixed types) and TCGA classifications (MSI, genomically stable, and chromosomal instability types; SI Appendix, Table S8).
Fig. 1.
Genomic alteration profiles for 103 patients with GC. (Top) Total number of genomic alterations (bar graph, left axis) and the rate of silent and nonsilent mutations (line graph, right axis) in 55 patients with GC analyzed by both whole-exome sequencing and aCGH. SNV, single-nucleotide variant. (Left) Matrix of significantly altered genes colored by types of mutations and CNAs. Each column stands for an individual cancer sample, and each row denotes a gene, except for the top two rows, which represent histological types and tumor MSI status. MSS, microsatellite stable; MSI-H, microsatellite instable-high. (Right) Number of samples with genomic alterations for each gene in 103 patients. (Bottom) Mutation spectrum of base changes in the samples shown in the matrix above.
Significantly Altered Pathways in GC.
According to the alteration frequencies of our data, gene lists of public cancer gene databases (14–16), and the Gene Ontology database (17), we selected 342 significantly altered genes (Dataset S2; a detailed description of the selection criteria can be found in SI Appendix, Materials and Methods) and annotated 51 of these genes in the Gene Ontology database for cancer-related biological processes (Fig. 1). The 342 significantly altered genes were grouped into two categories: predominantly mutated (61 genes) and predominantly copy number-altered (281 genes) genes. Of the 61 predominantly mutated genes, nine (TP53, ARID1A, RNF43, APC, PIK3CA, REV3L, DCLK1, KRAS, and RIMS2) were significantly altered (P < 0.05) by MutSig analysis (Dataset S2) (18). Among the 342 significantly altered genes, we also found previously known critical genetic drivers including TP53, ARID1A, PIK3CA, KRAS, MYC, FAT4, CTNNB1, and ERBB2 (SI Appendix, Table S9) (6, 19). Using the JAX Cancer Treatment Profile analysis pipeline, we also identified clinically actionable mutations in 144 genes as candidate targets for clinical application (JAX-CTP; Dataset S3) (20).
GC is classified into two histological types: intestinal and diffuse types. A previous study reported the enrichment of RHOA mutations in diffuse-type GC (9). However, there was no similar association of RHOA mutations with histological type and differentiation status in our study (Fig. 1 and SI Appendix, Table S10). In our data, DCBLD2, PER1, RNF207, DYNC1H1, and TCHH were significantly mutated in diffuse-type GC compared with intestinal-type GC, with a P value of less than 0.01 (SI Appendix, Table S11).
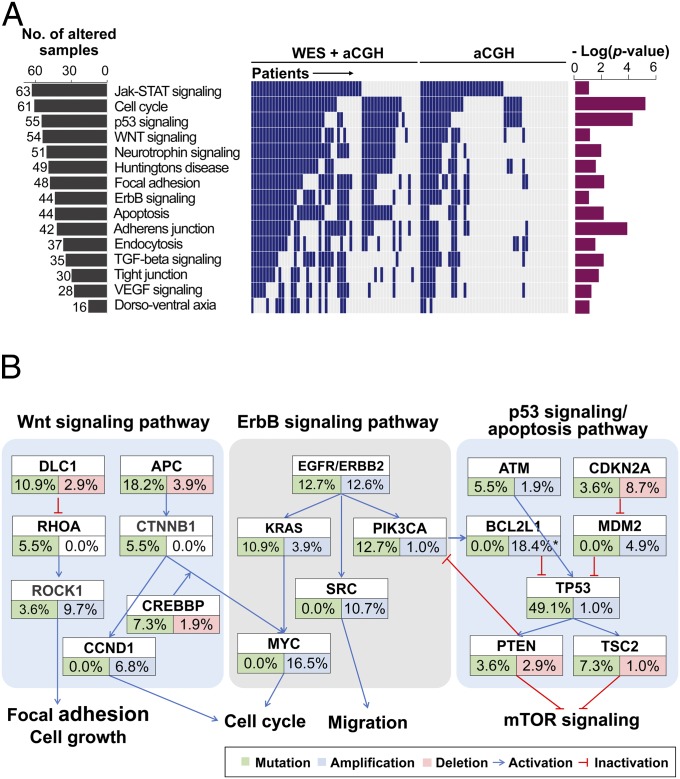
We performed pathway analysis on the 342 significantly altered genes, using the Kyoto Encyclopedia of Genes and Genomes analysis tool (21) in the DAVID bioinformatics resources (https://david.ncifcrf.gov) (22). Several important pathways, including the Wnt signaling, ErbB signaling, and p53 signaling/apoptosis pathways, were altered at the genomic level (Fig. 2 A and B and SI Appendix, Table S12). Alterations in the Wnt pathway were detected in 52.4% of the 103 GC cases. We also found mutations in genes in both the canonical Wnt pathway [APC (18.2%) and CTNNB1 (5.5%)] and the planar cell polarity pathway [DLC1 (10.9%), RHOA (5.5%), and ROCK1 (3.6%)]. Genes in the p53 signaling and apoptosis pathway were mutated in 65.5% of patients with GC and amplified or deleted in 38.8% of cases; some of the alterations include TP53 mutations (49.1%), TSC2 mutations (7.3%), and BCL2L1 amplification (18.4%). Moreover, ErbB signaling pathway components, including ERBB2, KRAS, PIK3CA, and MYC, were recurrently mutated (combined rate of 23.6%) or amplified (combined rate of 31.1%; Fig. 2B). Given that the ErbB signaling pathway is already being used as a molecular target for GC with the drug trastuzumab (4), we further investigated the other two pathways (Wnt pathway, p53 signaling and apoptosis pathway) for novel drug targets for GC.
Fig. 2.
Significantly altered pathways in gastric cancer. (A) List of somatically altered pathways analyzed by the Kyoto Encyclopedia of Genes and Genomes database (P < 0.1). (Left) Number of affected patients (of 103 patients) for each pathway. (Middle) Alteration events in each patient (column). (Right) P values for pathway analyses using the DAVID bioinformatics resources. (B) Somatically altered genes in the Wnt signaling, ErbB signaling, and p53 signaling/apoptosis pathways. Alteration frequencies are expressed as a percentage of cases (total number of mutation cases = 55; total number of CNA cases = 103). The amplification rate for BCL2L1 was determined by droplet digital PCR (indicated with an asterisk).
Amplification of BCL2L1 as a Molecular Target for Combination Therapy in a Subset of GC.
BCL2L1 is a component of the p53 signaling and apoptosis pathway and a member of the antiapoptotic BCL2 family (23, 24). Increased expression and amplification of BCL2L1 has been reported in several hematologic and solid malignancies and has been linked to both poor prognosis and resistance to conventional forms of therapies (25–28). Amplification of BCL2L1 was detected in 11 (10.7%) of the 103 GC cases analyzed by aCGH (SI Appendix, Fig. S2A and Table S13). In the TCGA database, the putative amplification rate of BCL2L1 using the GISTIC algorithm was 2.7% (6/220) for stomach adenocarcinoma cases (www.cbioportal.org) (29, 30). However, targeted ddPCR analyses in our GC cases showed amplification of the BCL2L1 gene in 18.4% (19/103) of the patients with GC studied (more than or equal to three copies; SI Appendix, Fig. S2B and Table S13) and in 35.0% (7/20) of available GC cell lines (more than or equal to three copies; SI Appendix, Fig. 2C). Because ddPCR is a more precise method for determination of gene copy number compared with aCGH, this suggests that the amplification of BCL2L1 in GC is much more prevalent than indicated in our aCGH analysis and in the TCGA dataset. The difference in BCL2L1 amplification frequency between the TCGA dataset and our data are not attributed to the ethnicity effect, because we found no significant difference between Asian and non-Asian samples with respect to copy number of BCL2L1 in TCGA dataset (P = 0.6862; SI Appendix, Table S14). In addition, the expressed amount of BCL2L1 protein in GC cells correlated well with the copy number of the BCL2L1 gene determined by ddPCR (P = 0.0026; SI Appendix, Fig. S2D). However, the mRNA expression levels of HM13 and COX4I2, which are located in same amplicon with BCL2L1, did not show significant correlations with copy numbers of these genes in GC cells (P = 0.7231 for HM13 and P = 0.0528 for COX4I2; SI Appendix, Fig. S2E), indicating a distinctive correlation between CNAs and gene expressions of BCL2L1 compared with the neighboring genes. APC and TP53 were significantly mutated in BCL2L1-amplified cases (P < 0.05; Dataset S4).
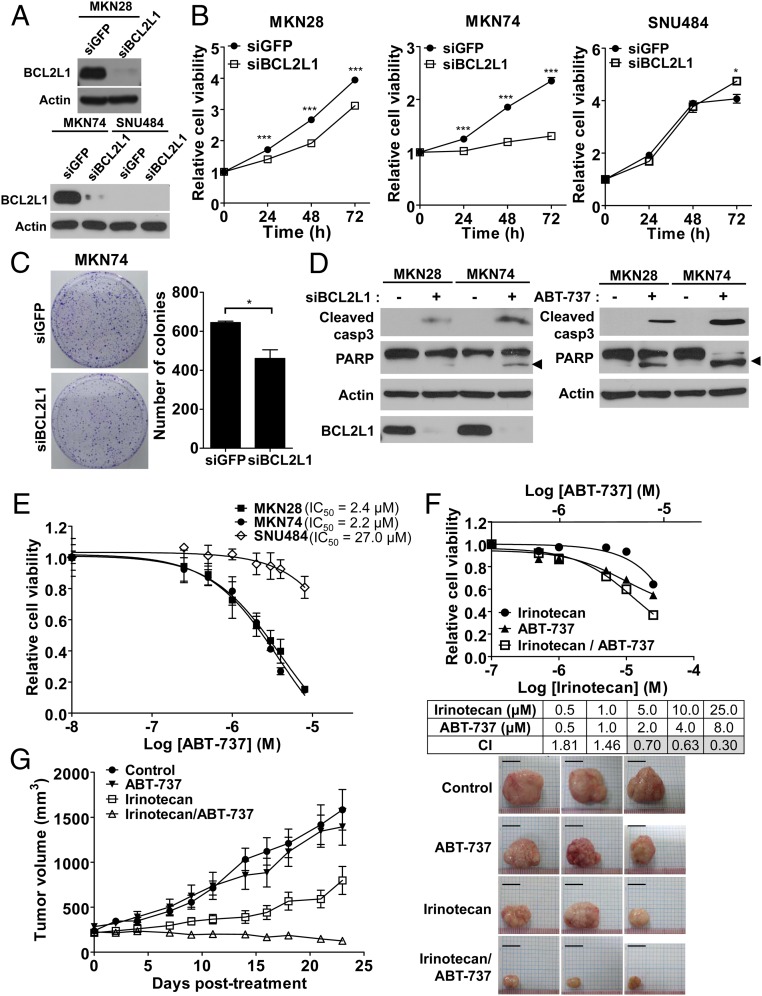
The functional role of BCL2L1 amplification was further evaluated, using siRNA for BCL2L1 in cells with and without BCL2L1 amplification (Fig. 3A). Down-regulation of BCL2L1 using siRNA in GC cells reduced the proliferation rates only in cell lines with increased copy numbers of BCL2L1, including MKN28 (three copies) and MKN74 (six copies), but not in SNU484, which had a normal copy number (two copies) of BCL2L1 and low levels of BCL2L1 protein expression (Fig. 3B). Knock-down of BCL2L1 also reduced colony formation of BCL2L1-amplified cells in clonogenic assays (Fig. 3C). These effects were partly attributed to increased apoptosis estimated by the cleavage of caspase 3 and poly ADP ribose polymerase proteins (Fig. 3D) (31, 32). The BH3 mimetic ABT-737, an inhibitor of BCL2L1 (33, 34), showed a greater antiproliferative effect by increasing apoptosis on BCL2L1-amplified cells (IC50 = 2.4 μM for MKN28 and IC50 = 2.2 μM for MKN74) compared with on BCL2L1 nonamplified cells (IC50 = 27.0 μM for SNU484; Fig. 3 D and E).
Fig. 3.
Increased drug sensitivity via inhibition of BCL2L1 in vitro and in vivo. (A) Knock-down effects of BCL2L1 siRNA were validated using Western blot analysis in MKN28 (BCL2L1-amplified cells; three copies), MKN74 (BCL2L1-amplified cells; six copies), and SNU484 (BCL2L1-nonamplified cells; two copies) cells. (B) BCL2L1 knock-down effect on cell proliferation. After BCL2L1 was knocked-down using siRNA, viable cells were monitored for 72 h using WST cell viability assay in MKN28, MKN74, and SNU484 cells. (C) The BCL2L1 knock-down effect on colony formation in MKN74 cells. The colonies were visualized using crystal violet staining and counted by ImageJ software. Asterisks indicate statistically significant differences (*P < 0.05; ***P < 0.001) compared with control siRNA-treated cells. (D) The effect of BCL2L1 knock-down (Left) and the BCL2L1 inhibitor ABT-737 (8 μM; Right) on the cleavage of caspase 3 (Cleaved casp3) and poly ADP ribose polymerase (PARP). The arrowhead indicates cleaved PARP. (E) ABT-737 dose-inhibition curve for cell viability of MKN28, MKN74, and SNU484 cells. IC50 values for ABT-737 are given. (F) Combination cytotoxicity of irinotecan and ABT-737. WST assays were used to examine the cell growth inhibitory effect in MKN74 cells. (Bottom) Calculated combination index values at applied concentrations. Gray boxes represent synergistic effect of the two drugs (combination index < 0.9). (G) In vivo efficacy of irinotecan and ABT-737 in a GC patient-derived xenograft model. BCL2L1-amplified tumor tissues from patients with GC (11 copies) were injected into the flanks of NOD/SCID/IL-2γ-receptor null (NSG) mice. The tumors were treated with irinotecan (50 mg/kg/week), ABT-737 (100 mg/kg/day), or the combination of the two drugs for 23 d (n = 6). Average tumor sizes for each group are plotted (Left) and representative tumors after treatment were shown (Right). (Scale bar, 10 mm.)
Next, we investigated whether the BCL2L1 inhibitor potentiates sensitivity of GC cells to conventional and other targeted chemotherapeutic drugs. Inhibition of the BCL2L1 protein using ABT-737 exhibited synergistic interactions with conventional chemotherapeutic drugs, including irinotecan, cisplatin, and paclitaxel, as indicated by cell viability in in vitro cell culture systems (Fig. 3F and SI Appendix, Fig. S3 A and B). ERBB2 amplification is a molecular target for GC (4), and afatinib is a second-generation ERBB2 inhibitor with high potency (35, 36) that is currently in phase 2 clinical trials for ERBB2-amplified GC cases (https://clinicaltrials.gov, NCT01522768). Among ERBB2-amplified GC cells, BCL2L1-amplified cells [including those in the GC cell lines SNU216 (five copies) and NCI-N87 (three copies)] were more resistant to afatinib treatment compared with ERBB2-amplified, BCL2L1-nonamplified cells, including those in the GC cell lines SNU19 (two copies) and SNU484 (two copies; SI Appendix, Fig. S4A). Importantly, targeting BCL2L1 synergistically with ABT-737 enhanced sensitivity to afatinib in ERBB2-amplified, BCL2L1-amplified GC cells (SI Appendix, Fig. S4 B and C).
We further established PDX mice as avatar models of GC. PDX mice are highly immune-deficient mice in which patient tumors have been transplanted; thus, they recapitulate the genomic characteristics and drug responsiveness observed in patients (37). Synergistic effects of ABT-737 with the conventional drug irinotecan were also verified via decreased tumor volume in both BCL2L1-amplified GC PDX models (11 copies; Fig. 3G) and GC cell line xenografts (MKN74; SI Appendix, Fig. S5), consistent with the notion that this combination therapy could be applied to patients with GC with increased genomic copy numbers of BCL2L1 (or possibly even in patients with dysregulation of BCL2L1 leading to increased levels of transcriptional products of BCL2L1).
Association of Mutations of DLC1 with Sensitivity to a ROCK Inhibitor.
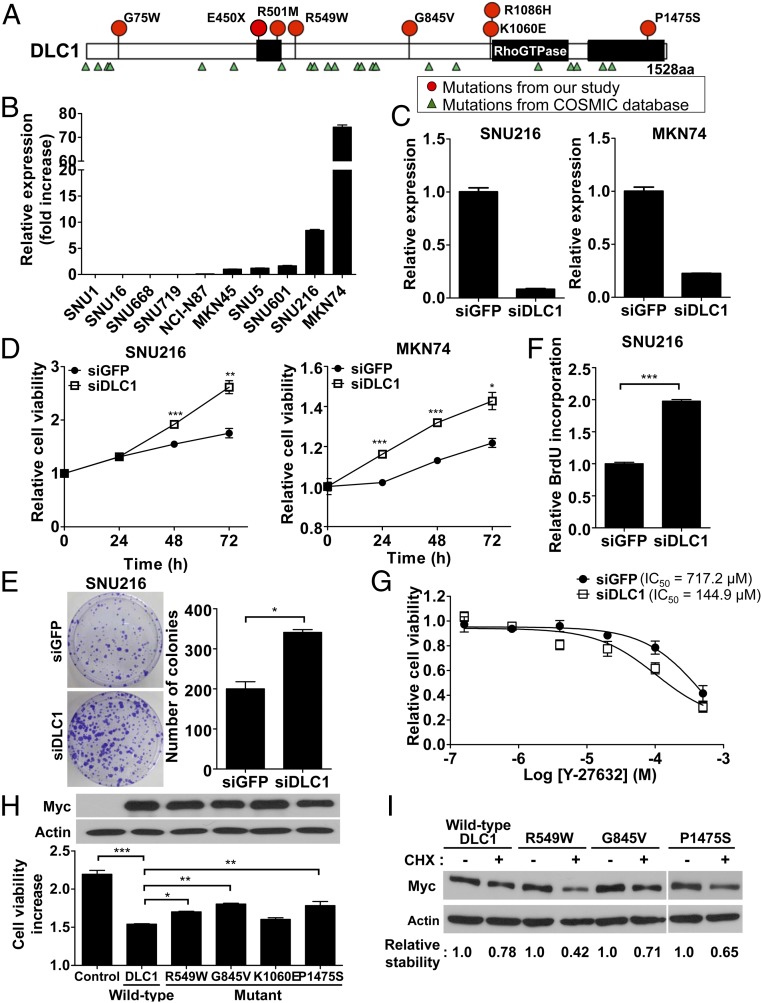
DLC1 is a Wnt pathway component and a GTPase activating protein (GAP) for Rho proteins, including RhoA and Cdc42 (38). DLC1 negatively regulates Rho proteins via its GAP functions and has been suggested as a candidate tumor suppressor gene (38). DLC1 mutations were detected in 10.9% (6/55) of GC cases sequenced, and DLC1 deletions were found in 2.9% (3/103) of the patient samples studied (Figs. 1 and 4A and Dataset S1). We detected a total of eight mutations of DLC1 in six patients (Fig. 4A) and found that one patient (S477) had three mutations of DLC1 (G75W, E450X, and R501M), suggesting the possibility of mutations in both alleles of this patient. In the TCGA database, DLC1 mutations and deletions were detected in 9.1% (20/220) and 2.3% (5/220), respectively, of stomach adenocarcinoma cases (www.cbioportal.org) (29, 30). In addition, DLC1 has been shown to have low expression levels in many human solid tumors (39), and the epigenetic silencing of DLC1 via methylation of CpG islands has also been reported in GC cell lines and tissues (40). Correspondingly, expression of DLC1 was undetectable in several of our GC cell lines (Fig. 4B). Finally, DLC1 mutations tended to co-occur with mutations of MYC, TOP1, CDKN2A, ARID1A, CHD7, and CREBBP (P < 0.01; SI Appendix, Fig. S6).
Fig. 4.
Association of mutations of DLC1 with gastric cancer growth and protein stability. (A) The locations of somatic mutations of DLC1 in 55 GC samples (red circles) and the COSMIC database (green triangles). (B) The expression of DLC1 mRNA in 10 GC cell lines. The relative expression levels were calculated compared with the expression of MKN45 cells. (C) Knock-down effects of DLC1 siRNA were validated using real-time PCR assays in DLC1-expressing SNU216 and MKN74 cells. (D) DLC1 knock-down effect on cell proliferation. After DLC1 was knocked-down using siRNA, viable cells were monitored for 72 h using the WST cell viability assay in SNU216 and MKN74 cells. (E) DLC1 knock-down effect on colony formation in SNU216 cells. The colonies were visualized using crystal violet staining and counted by ImageJ software. (F) DLC1 knock-down effect on DNA synthesis, which was estimated by BrdU incorporation assays in SNU216 cells. (G) DLC1 knock-down effect on sensitivity to a ROCK inhibitor, Y-27632, in SNU216 cells. IC50 values for Y-27632 are given. (H) The overexpression effect of wild-type and mutant DLC1 on cell proliferation. (Upper) Protein levels for wild-type and mutant DLC1-expressing cells. Asterisks indicate statistically significant differences (*P < 0.05; **P < 0.01; ***P < 0.001) compared with control siRNA-treated cells (D–F) or wild-type DLC1-transfected cells (H). (I) Protein stability of wild-type and mutant DLC1. The protein levels of each mutant DLC1 were evaluated after a 3-h treatment with cycloheximide (CHX; 100 μg/mL). The density of each band was quantitated and compared with CHX-untreated cells.
To better estimate the DLC1 mutation rate in GC, we performed WES in an additional 27 Korean GC samples (cohort 2). We found DLC1 mutations in three samples of the 27 patients [11.1% (3/27); SI Appendix, Fig. S7]. The amino acid changes of these mutations were A4T, R502H, and L1480F. This result is compatible with that in our original cohort, in which the mutation rate of DLC1 was 10.9% (6/55; SI Appendix, Fig. S7). We also determined the mutation rates of genes associated with the RhoA pathway in cohorts 1 and 2 and found that three genes (DLC1, AKAP13, and ARHGAP35) were mutated in more than 5% of a total of 82 samples (55 samples in cohort 1 and 27 samples in cohort 2; SI Appendix, Fig. S7). In addition, in the 82 samples from the two cohorts, DLC1 showed significant co-occurrence with ARHGAP35 in the RhoA pathway (P < 0.05; SI Appendix, Table S15).
The function of DLC1 was investigated using siRNA against DLC1 in DLC1-expressing GC cells (GC cell lines SNU216 and MKN74; Fig. 4C). Knock-down of DLC1 expression in GC cells increased cell proliferation and colony formation, as shown in clonogenic assays and measurement of DNA synthesis (Fig. 4 D–F). Given that DLC1 inhibits the Rho-ROCK signaling pathway through its Rho-GAP activity (38), down-regulation of DLC1 was anticipated to activate this signaling pathway, which rendered the cells more susceptible to the ROCK inhibitor Y-27632 (Fig. 4G and SI Appendix, Fig. S8 A and B) (41). Correspondingly, overexpression of wild-type DLC1 reduced cell proliferation (Fig. 4H). Among four randomly selected patients with GC with DLC1 mutations, we found that three cases (R549W, G845V, P1475S) exhibited less inhibition of proliferation compared with the wild-type DLC1 after overexpression of the wild-type or mutant DLC1 (Fig. 4H), suggesting that the mutant DLC1 proteins detected in patients with GC have functional deficits. In cells treated with cycloheximide, which blocks protein synthesis, the three mutants (R549W, G845V, P1475S) had decreased DLC1 protein stability compared with wild-type DLC1 protein (Fig. 4I). Computational modeling showed a decrease in protein stability with a high reliability index (≥3) in six DLC1 mutants, including these three mutants (SI Appendix, Table S16) (42). In addition, using three kinds of bioinformatics algorithms [SIFT (43), PolyPhen-2 (44), and MutationTaster (45)], we found that seven mutations showed “damaging” functional changes, as indicated in multiple analytic algorithms (SI Appendix, Table S17). Taken together, these results suggest that deletions and mutations of the DLC1 gene influence GC development, and that DLC1 is therefore potentially a druggable target for therapy.
Discussion
In this study, genetic analyses of somatic mutations and CNAs in 103 GC genomes have provided two new candidate druggable targets for therapy. Functional and inhibitor assays implicate BCL2L1 as a novel oncogenic driver in GC progression and DLC1 as a novel tumor suppressor gene. Furthermore, chemotherapy and targeted therapy combinations appeared to be an effective treatment strategy in patients with GC with BCL2L1 CNAs.
Antiapoptotic BCL2 family proteins including BCL2 and BCL2L1 are inhibited by BH3-only proteins, which have only the BH3 domain (46). Therefore, BH3 mimetics, which mimic the BH3-only proteins by interacting with the hydrophobic groove of antiapoptotic BCL2 family proteins, have been proposed as apoptosis-inducing agents and candidate drugs for cancer treatment (46). In this study, we propose the application of irinotecan with BH3 mimetics as a second-line therapy in this particular subset of GC cases.
The RhoA signaling pathway is recurrently altered in patients with GC (9, 10), and DLC1 has been suggested as a tumor suppressor that inhibits the Rho-ROCK pathway (38, 39). Inactivation of DLC1 via promoter hypermethylation, heterozygous deletion, and somatic mutation has been found in various types of cancer (38). In our analysis, mutations and deletions of DLC1 were detected mainly in intestinal-type patients [83.3% (5/6) of cases for mutations; 100% (3/3) of cases for deletions]. Most DLC1 mutants showed functional defects in growth inhibitory activity, most likely as a result of decreased protein stability (Fig. 4 H and I). DLC1-inactivated cells were more sensitive to a ROCK inhibitor (Fig. 4G), suggesting that activation of the RhoA pathway via mutation of DLC1 has the potential to yield novel therapeutic targets.
In summary, integration of genomic molecular profiling and PDX mouse models provides a valuable platform for novel drug target discovery and validation. Our study has validated that approach, revealing two potential novel molecular mechanisms for the treatment of subsets of GC cases: suppression of BCL2L1 amplifications in patients within increased copy numbers of BCL1L1, and suppression of the Rho-ROCK pathway for DLC1-mutated cases.
Materials and Methods
Additional details for materials and methods can be found in SI Appendix, Materials and Methods.
Array Comparative Genomic Hybridization.
We designed a copy number variation-targeted aCGH platform using the 1M format on SurePrint G3 Human CGH Microarrays (Agilent Technologies). We conducted aCGH experiments according to the manufacturer’s instructions (Agilent Technologies) and analyzed data as depicted in SI Appendix, Fig. S1A.
Exome Capture, Library Construction, and Sequencing.
For all 110 samples (55 normal and 55 tumor samples), 1 μg of DNA per sample was sheared with a Covaris SS Ultrasonicator. Exome capture was performed with Agilent SureSelect Human All Exon Kit V5 (Agilent Technologies). Each sample was sequenced on an Illumina HiSeq 2000 instrument using a read length of 2 × 101 bp. Image analysis and base calling were performed using the Illumina pipeline with default settings.
Animal Experiments.
All animal procedures were approved by the Institutional Animal Care and Use Committee at Seoul National University (IACUC approval SNU-14–0016-C0A0). The surgically resected GC tissues or GC cells were injected into the flanks of 4-wk-old NOD/SCID/IL-2γ-receptor null female mice. Drug treatments began after tumors reached ∼200 mm3.
Supplementary Material
Acknowledgments
We thank J. H. Park for supporting this international collaboration, J. W. Ping Yam for providing the expression construct of Myc-tagged DLC1, and S. Sampson and the Scientific Program Development of The Jackson Laboratory for critical comments on this manuscript. This work was supported by grants from the Seoul National University Invitation Program for Nobel Laureates (to C.L.); the Ewha Womans University Research Grant of 2015; the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (Grant HI13C2148); and the Seoul National University Hospital Research fund supported by Heung Soon Park (Grant 30-2012-0200).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Nucleotide Archive (accession no. PRJEB10531).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507491112/-/DCSupplemental.
References
- 1.McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11(11):664–674. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- 2.Yang W, Raufi A, Klempner SJ. Targeted therapy for gastric cancer: Molecular pathways and ongoing investigations. Biochim Biophys Acta. 2014;1846(1):232–237. doi: 10.1016/j.bbcan.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 3.De Vita F, et al. Clinical management of advanced gastric cancer: The role of new molecular drugs. World J Gastroenterol. 2014;20(40):14537–14558. doi: 10.3748/wjg.v20.i40.14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang YJ, et al. ToGA Trial Investigators Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs CS, et al. REGARD Trial Investigators Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383(9911):31–39. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 8.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 9.Kakiuchi M, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46(6):583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 11.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative ICGC Breast Cancer Consortium ICGC MMML-Seq Consortium ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmermann B, et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One. 2010;5(12):e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Allen EM, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20(6):682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ananda G, et al. Development and validation of the JAX Cancer Treatment Profile™ for detection of clinically actionable mutations in solid tumors. Exp Mol Pathol. 2015;98(1):106–112. doi: 10.1016/j.yexmp.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao X, et al. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corcoran RB, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23(1):121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011;278(3):403–413. doi: 10.1111/j.1742-4658.2010.07965.x. [DOI] [PubMed] [Google Scholar]
- 25.Straten Pt, Andersen MH. The anti-apoptotic members of the Bcl-2 family are attractive tumor-associated antigens. Oncotarget. 2010;1(4):239–245. doi: 10.18632/oncotarget.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tonon G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102(27):9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith LT, et al. 20q11.1 amplification in giant-cell tumor of bone: Array CGH, FISH, and association with outcome. Genes Chromosomes Cancer. 2006;45(10):957–966. doi: 10.1002/gcc.20354. [DOI] [PubMed] [Google Scholar]
- 29.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakhani SA, et al. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science. 2006;311(5762):847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decker P, Isenberg D, Muller S. Inhibition of caspase-3-mediated poly(ADP-ribose) polymerase (PARP) apoptotic cleavage by human PARP autoantibodies and effect on cells undergoing apoptosis. J Biol Chem. 2000;275(12):9043–9046. doi: 10.1074/jbc.275.12.9043. [DOI] [PubMed] [Google Scholar]
- 33.van Delft MF, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10(5):389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konopleva M, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10(5):375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Janjigian YY, et al. Monitoring afatinib treatment in HER2-positive gastric cancer with 18F-FDG and 89Zr-trastuzumab PET. J Nucl Med. 2013;54(6):936–943. doi: 10.2967/jnumed.112.110239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solca F, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther. 2012;343(2):342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 37.Tentler JJ, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko FC, Ping Yam JW. Regulation of deleted in liver cancer 1 tumor suppressor by protein-protein interactions and phosphorylation. Int J Cancer. 2014;135(2):264–269. doi: 10.1002/ijc.28505. [DOI] [PubMed] [Google Scholar]
- 39.Liao YC, Lo SH. Deleted in liver cancer-1 (DLC-1): A tumor suppressor not just for liver. Int J Biochem Cell Biol. 2008;40(5):843–847. doi: 10.1016/j.biocel.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TY, et al. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003;22(25):3943–3951. doi: 10.1038/sj.onc.1206573. [DOI] [PubMed] [Google Scholar]
- 41.Ishizaki T, et al. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol Pharmacol. 2000;57(5):976–983. [PubMed] [Google Scholar]
- 42.Bava KA, Gromiha MM, Uedaira H, Kitajima K, Sarai A. ProTherm, version 4.0: Thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32(Database issue):D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 46.Billard C. BH3 mimetics: Status of the field and new developments. Mol Cancer Ther. 2013;12(9):1691–1700. doi: 10.1158/1535-7163.MCT-13-0058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.