Significance
Tumor sequencing efforts have enabled the identification of cancer genes based on an excess of mutations in the gene or clustering of mutations along the (one-dimensional) DNA sequence of the gene. Here, we show that this approach can be extended to identify cancer genes based on clustering of mutations relative to the 3D structure of the protein product. By analyzing the PanCancer compendium of somatic mutations in nearly 5,000 tumors, we identified known cancer genes and previously unidentified candidates based on clustering of missense mutations in protein structures or at interfaces with binding partners. In addition, we found that 3D clustering is present in both oncoproteins and tumor suppressors—contrary to the view that such clustering is a hallmark of oncoproteins.
Keywords: cancer, cancer genetics, mutation clustering, protein structures, interaction interfaces
Abstract
Large-scale tumor sequencing projects enabled the identification of many new cancer gene candidates through computational approaches. Here, we describe a general method to detect cancer genes based on significant 3D clustering of mutations relative to the structure of the encoded protein products. The approach can also be used to search for proteins with an enrichment of mutations at binding interfaces with a protein, nucleic acid, or small molecule partner. We applied this approach to systematically analyze the PanCancer compendium of somatic mutations from 4,742 tumors relative to all known 3D structures of human proteins in the Protein Data Bank. We detected significant 3D clustering of missense mutations in several previously known oncoproteins including HRAS, EGFR, and PIK3CA. Although clustering of missense mutations is often regarded as a hallmark of oncoproteins, we observed that a number of tumor suppressors, including FBXW7, VHL, and STK11, also showed such clustering. Beside these known cases, we also identified significant 3D clustering of missense mutations in NUF2, which encodes a component of the kinetochore, that could affect chromosome segregation and lead to aneuploidy. Analysis of interaction interfaces revealed enrichment of mutations in the interfaces between FBXW7-CCNE1, HRAS-RASA1, CUL4B-CAND1, OGT-HCFC1, PPP2R1A-PPP2R5C/PPP2R2A, DICER1-Mg2+, MAX-DNA, SRSF2-RNA, and others. Together, our results indicate that systematic consideration of 3D structure can assist in the identification of cancer genes and in the understanding of the functional role of their mutations.
To elucidate the genetic basis of cancer, efforts have been initiated to sequence the exomes or genomes of many human tumors. Among them are large-scale efforts such as The Cancer Genome Atlas (TCGA) (1) and the International Cancer Genome Consortium (ICGC) (2), as well as many smaller-scale projects. These efforts have collectively found millions of somatic mutations in virtually all human genes (the vast majority of which are nonfunctional or “passenger” mutations) across thousands of tumor samples (3). The amounts of available cancer sequencing data are growing rapidly and will continue to grow in the foreseeable future. We and others have developed computational methods to detect cancer-associated genes and functional mutations from such data, based on a significant overall burden of mutations or on significant positional clustering of mutations in the one-dimensional (1D) gene sequences, corresponding to mutational hotspots (3–6).
For some cancer proteins, it has been observed that, although mutations may be distributed along the linear amino acid sequence, they tend to cluster in certain regions of the 3D structure, such as active sites. A clear example is KRAS, where particular missense mutations at the active site are positively selected in cancer because they disable the GTPase activity of the protein, locking it in its GTP-bound, active state, which promotes proliferation. As a result, recurrently mutated residues (e.g., G12, G13, I36, A59, Q61, K117, A146) tend to occur around the substrate-binding pocket of KRAS (Fig. 1). This and other individual examples of proteins showing 3D clustering of cancer missense mutations are sometimes used in the literature as supporting evidence for the involvement of those proteins in the disease or as a basis for functional hypotheses about the clustered mutations [e.g., EGFR (8), PIK3CA (9), DIS3 (10), SPOP (11), MRE11 (12), ERCC2 (13)]. Stehr et al. (14) and Ryslik et al. (15) assessed the structural clustering of missense mutations in 29 and 131 proteins, respectively, and demonstrated that taking into account 3D structural information can be helpful for identifying mutation hotspots in known cancer proteins or in new candidates.
Fig. 1.
Spatial mutation clustering in KRAS. (A) Protein sequence of KRAS (Isoform 2B; UniProt: P01116-2) with mutated residues from the PanCancer data set (3) shown in red. Recurrent mutations (at least three samples) are shown in larger font and are annotated with position and number of samples with such mutations. Gray arcs between such residues are shown if their centroids are located closer than 13 Å between each other in the protein structure; arc width and label show the spatial distance in these cases (wider arcs corresponding to shorter distances). The C-terminal part of the protein sequence not covered by the structure in B is shown in smaller, gray font. (B) 3D structure of KRAS (gray) with substrate GDP (blue) bound to its active site (PDB ID code 4LUC) (7). Mutated residues are shown in red (recurrent mutations: sticks, nonrecurrent mutations: thin lines) and color intensity scales with the number of mutations per residue.
Here, we seek to undertake comprehensive studies of 3D clustering of somatic missense mutations in cancer across all human proteins with available protein structures. Such integrative analysis may help to identify new cancer proteins that have been missed by other methods. In addition, it can help explain the functional roles of individual mutations based on their spatial location in the protein; for example, mutations that cluster at protein interaction interfaces may perturb key molecular interactions (16).
Results
To systematically discover genes in which somatic mutations show significant clustering in the 3D structure of the encoded protein, we analyzed the set of missense mutations identified by comprehensive (exome or genome) sequencing of 4,742 tumors from 21 cancer types (PanCancer compendium) (3) relative to the structural and cocomplex data available in the Protein Data Bank (PDB) (17) for >4,000 human proteins and >5,000 protein interactions, respectively (Fig. 2).
Fig. 2.
Schematic overview of the analyses in this study. (Upper) Triangles denote somatic missense mutations in individual samples; bars labeled s1–s5 represent PDB structures covering different parts of the protein sequence; the representative structures, s1 and s5, selected with our heuristic (Materials and Methods) are highlighted in black. (Lower) Mutated residues are shown as pins (red: recurrent, pink: nonrecurrent mutation).
CLUMPS Method.
We focused first on 3D mutation clustering within each protein, developing a statistical method called CLUMPS (clustering of mutations in protein structures) to assess the significance of mutational clustering in a given 3D structure. CLUMPS does not attempt to specify individual clusters but rather detects an overall enrichment of mutated residues that are spatially close to each other. The method uses a weighted average proximity (WAP) scoring function summarizing the pairwise Euclidean distances of all mutated residues in the structure, weighted by the normalized number of samples in which they are mutated (SI Appendix, Fig. S1 and Materials and Methods). We assess the significance of a given WAP score by comparing it to the null distribution obtained by randomly permuting the positions of the mutations across all residues in the structure (preserving the distribution of the number of samples mutated at a given residue) to obtain an empirical P value. CLUMPS is designed to be insensitive to other types of signals frequently used to discover cancer genes, such as overall mutational burden, clustering within the linear amino acid sequence, or mutation enrichment in evolutionarily conserved sites.
Using CLUMPS, we systematically looked for 3D mutation clustering in 4,062 human proteins, each of which had (i) somatic missense mutations in the PanCancer compendium and (ii) available structural information in the PDB. These proteins were represented by a total of 41,063 3D structures (after filtering out structures with less than three missense mutations). Because of the existence of multiple, often partially or completely overlapping structures for some proteins, we developed a heuristic method to select a set of minimally overlapping structures with maximal combined protein sequence coverage, to represent each protein (Materials and Methods). Using this heuristic, we selected 4,822 representative structures for the 4,062 proteins. To validate the P values generated with CLUMPS, we confirmed that the vast majority of data points were consistent with the null model and lay on the diagonal of the Q-Q plot (SI Appendix, Fig. S2).
Significant Mutation Clustering in Known Oncoproteins, Tumor Suppressors, and the Kinetochore Component NUF2.
Of the 4,062 human proteins tested, 10 showed significant 3D clustering of missense mutations at a false discovery rate (FDR) (Table 1 and Dataset S1). The list included four well-established oncoproteins (PIK3CA, PTPN11, BRAF, and HRAS), four well-established tumor suppressors (PTEN, TP53, FBXW7, and CDKN2A), and PPP2R1A, a central component of the protein phosphatase 2A (PP2A) complex that also functions as a tumor suppressor (19). All structures are shown in Dataset S15.
Table 1.
Proteins with significant 3D (i.e., spatial) mutation clustering identified with CLUMPS
| Protein | Spatial clustering | Positional clustering P | ||
| P | ||||
| Full analysis | ||||
| PTEN* | 1e-06 | 0.001 | 0.0001 | 8e-08 |
| PTPN11† | 1e-06 | 0.001 | 0.0001 | 0.0003 |
| PIK3CA† | 1e-06 | 0.001 | 0.0001 | 7e-08 |
| TP53* | 1e-06 | 0.001 | 0.0001 | 8e-08 |
| FBXW7* | 2e-05 | 0.02 | 0.002 | 6e-08 |
| BRAF† | 3e-05 | 0.02 | 0.002 | 5e-08 |
| PPP2R1A* | 6e-05 | 0.04 | 0.003 | 0.0003 |
| NUF2 | 9e-05 | 0.05 | — | 0.4 |
| HRAS† | 0.0001 | 0.05 | 0.005 | 9e-08 |
| CDKN2A* | 0.0001 | 0.05 | 0.005 | 6e-08 |
| Restricted analysis | ||||
| SPOP* | 0.001 | 0.4 | 0.04 | 4e-05 |
| STK11* | 0.001 | 0.4 | 0.05 | 0.9 |
| EGFR† | 0.001 | 0.5 | 0.05 | 6e-08 |
| VHL* | 0.002 | 0.5 | 0.05 | 1 |
| MTOR | 0.002 | 0.5 | 0.05 | 6e-08 |
| RAC1† | 0.002 | 0.5 | 0.06 | 1e-05 |
| FGFR3† | 0.003 | 0.6 | 0.07 | 0.02 |
qfull and qrestricted denote the FDR calculated in the full analysis of 4,062 human proteins represented in the PDB and in the restricted analysis of a subset of 311 previously implicated cancer proteins, respectively. Proteins in the bottom part of the table were significant in the restricted analysis but not in the full analysis. The P values in the “Positional clustering P” column were obtained with MutSig-CL (18) using mutations from the PanCancer compendium across the whole gene sequence (3).
Known tumor suppressor.
Known oncoprotein.
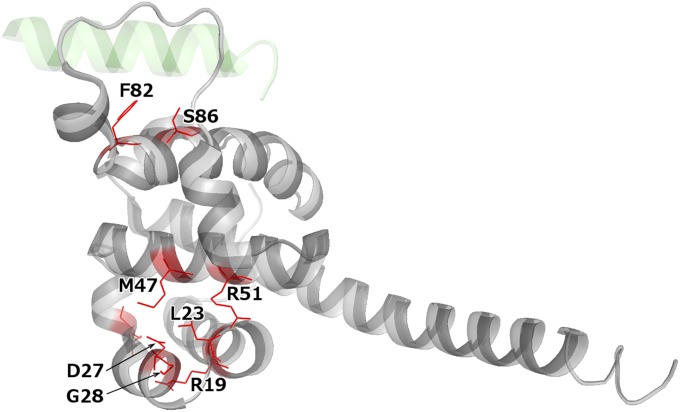
The final protein on the list was the kinetochore component NUF2 (, ). In the available protein structures (comprising the Nuf2 protein domain), missense mutations formed two clusters that involved six and two mutated residues, respectively (Fig. 3). The smaller cluster was located at the interaction interface with another kinetochore component, NDC80 (also called retinoblastoma-associated protein HEC), and was separated from the larger cluster (Fig. 3). Although one false positive among the 10 significant results might be expected given the threshold , the biological role of NUF2 (discussed below) supports the hypothesis that its mutations play a functional role in cancer. We also noted the presence of two likely mutational hotspots in portions of the protein not covered by available structures: an S340L missense mutation in three independent samples and a splice site mutation at the end of exon 8 in three separate samples (SI Appendix, Fig. S3).
Fig. 3.
Crystal structure of NUF2 (gray) bound to a fragment of NDC80 (pale green) (PDB ID code 3IZ0) (20). The structure covers protein domain Nuf2 (residues 4–156 of the reference NUF2 protein of 464 amino acids). Mutated residues are shown in red (all residues are mutated in a single tumor sample each with the exception of G28, which is mutated in two samples).
NUF2, also known as cell division associated 1 (CDCA1), is responsible for kinetochore-microtubule attachment and is hence pivotal for the proper segregation of sister chromatids during mitosis. A dysfunctional kinetochore may missegregate sister chromatids and cause chromosomal instability and aneuploidy, which often lead to cancer (21). In fission yeast, NUF2-null mutations indeed cause defects in chromosome segregation (22). Although NUF2 missense mutations have not been previously linked to human cancer, multiple levels of evidence implicate the gene in this disease. For example, elevated NUF2 gene expression has been found in a range of tumor types and cell lines (23–26) and is associated with poor outcome in cancer patients (23, 25). Furthermore, silencing of NUF2 inhibits tumor growth and leads to apoptosis in cancer cell lines (23–28), likely induced by the spindle checkpoint pathway (22). Experimental follow-up is required to ultimately understand the role of the clustered NUF2 mutations in cancer.
Restricting 3D Clustering Analysis to Known Cancer Proteins.
We also applied CLUMPS to a subset of 425 structures of 316 proteins that have previously been implicated in cancer [based on COSMIC Classic (29) or the Cancer Gene Census (30), or being significantly mutated (3); Dataset S2]. By focusing on this subset, the analysis restricts the number of statistical hypotheses tested and hence increases the statistical power.
The restricted analysis identified significant 3D mutation clustering () in seven additional proteins (Table 1 and Dataset S1): three tumor suppressors (SPOP, STK11, and VHL), three oncoproteins (EGFR, RAC1, and FGFR3), and the cancer-associated protein MTOR.
Spatial vs. Linear Patterns of Mutation Clustering.
We examined the relationship between 1D clustering (with respect to the linear DNA sequence, as calculated with MutSig-CL) (3) and 3D clustering (with respect to spatial structure of protein products, as calculated with CLUMPS) of missense mutations. Although some of the genes identified with CLUMPS as having significant 3D mutation clustering in the encoded protein structure also showed 1D clustering, others clearly did not (Table 1). For example, missense mutations in STK11 often affected residues at the substrate pocket of the protein product (SI Appendix, Fig. S4), but were nonrecurrent and nonadjacent in sequence, resulting in significant 3D clustering () but not 1D clustering ().
Overall, there was little correlation between the P values for 3D and 1D clustering for all tested proteins (Spearman’s ; SI Appendix, Fig. S5). This lack of correlation is not surprising, considering that the null model of CLUMPS preserves the distribution of the number of missense mutations per residue during the permutations. In contrast, the null distribution of MutSig-CL is created by permuting all mutations independently of each other (while preserving the mutational signatures of individual tumors) (3, 18); hence, MutSig-CL is by design highly sensitive to 1D mutation hotspots. To further ensure that the clustering signal captured by CLUMPS was not merely a consequence of mutations in consecutive residues (i.e., direct neighbors in the linear protein sequence), we repeated the full CLUMPS analysis after combining each uninterrupted sequence of mutated residues into a single meta-residue, represented by its centroid in three dimensions, and treating is as a single event during the permutations. This analysis yielded similar results (Dataset S3), indicating that spatially clustered residues were often not direct sequence neighbors but rather farther apart in the linear protein sequence.
Importantly, our clustering analysis found similar numbers of tumor suppressors and oncoproteins. On its face, this result might seem contrary to the frequent assumption that 3D (14) and 1D (5) clustering are hallmarks of oncogenes, as opposed to tumor suppressor genes (although mutational hotspots are known in some tumor suppressors such as TP53) (31). In fact, Yang et al. recently reported enrichment of missense mutations in particular protein domains of both tumor suppressors and oncoproteins; however, within individual domains, they reported mutation clustering for oncoproteins and uniform mutation distribution for tumor suppressors in 1D (32). 3D clustering of missense mutations in tumor suppressors, identified with CLUMPS, may reflect important properties of protein structure. Whereas most nonsense and frameshift mutations will suffice to abolish protein function, only a subset of single amino acid substitutions may suffice to abolish a tumor suppressor’s function, and these may be concentrated in particular regions of a protein critical to protein structure or protein interaction (see below).
We note that Stehr et al. did not observe a higher level of 3D clustering of mutations in tumor suppressors (including some identified here: PTEN, TP53, FBXW7, CDKN2A, STK11, VHL) than of common germ-line polymorphisms in the same proteins (14). Hence, they concluded that tumor suppressors, in contrast to oncoproteins, lack mutation clustering. The discrepancy with our results likely reflects major differences in methodology. Specifically, (i) we weighted mutations according to frequency of occurrence in a defined patient cohort, whereas Stehr et al. weighted all mutated residues equally and thus likely overweighted passenger mutations (which are expected to be scattered randomly across the protein) relative to driver mutations (which tend to recur across patients due to positive selection); (ii) Stehr et al. weighted small interresidue distances (up to 2 Å) very strongly compared with our score, which declines more slowly for distances up to 6–8 Å; and (iii) we studied mutations obtained through comprehensive sequencing of a defined set of tumors, whereas Stehr et al. examined mutations from COSMIC (which is subject to serious reporting bias).
CLUMPS aims to detect the tendency of mutated protein residues to cluster together, regardless of the number of clusters formed. As evident in Datasets S15 and S16, some proteins feature one mutation cluster (e.g., HRAS, CDKN2A, FGFR3), whereas in other proteins, more than one cluster is apparent (e.g., VHL, SPOP, EGFR, NUF2).
Spatial Patterns of Co-Occurring Mutations.
In our analyses above, for each protein, we collated missense mutations from all patients while ignoring the fact that, in some cases, a single patient may contribute more than one mutation. Mutations that co-occur in a patient and impact spatially proximal protein residues may act together, e.g., to change the binding affinity to another biomolecule beyond levels achievable through a specific single mutation. To explore such potential synergistic effects between co-occurring mutations, we searched for samples harboring pairs of spatially proximal mutations in the 17 significant protein structures identified with CLUMPS.
Overall, 167 patient–protein combinations had more than one mutation (Dataset S4). Of these, 23 had at least one spatially proximal ( 10 Å) pair of mutated residues. In 16 of these 23 cases (highlighted in Dataset S4), each of the residues in the pair were mutated in at least one other patient and hence were less likely to be passengers. The 16 pairs fell within a total of four proteins: EGFR, PTEN, PIK3CA, and TP53. Interestingly, the EGFR residue R108 located in the protein’s extracellular domain was affected by a missense mutation in a total of five glioblastoma multiforme patients, of which four had an additional missense mutation in the same protein domain. In two of these patients, the additional mutation affected the spatially proximal residue A289, whereas in two other patients, the distant residues P596 and G598 were affected, respectively. The co-occurrence of mutations in the extracellular domain of EGFR may be due to the complex mechanism of ligand-free, cancer-associated EGFR dimerization, which may require several simultaneous structural changes of EGFR (33).
Common and Tumor Type-Specific Mutation Clustering.
In our analysis above, we combined mutations from all tumor types because we were concerned that there would be insufficient statistical power to detect proteins with significant mutation clustering when considering individual tumor types separately (3). However, it is possible that there may be tissue-specific cancer mechanisms that are missed when merging all tumor types. We therefore applied the full CLUMPS analysis separately to each of the five tumor types with the largest patient cohorts (Dataset S5), omitting breast cancer because it is known to have multiple, very distinct subtypes. As we expected, the individual analyses revealed only small numbers (one to five) of significant proteins (Datasets S6–S10). In all but one case, the proteins were also detected in the combined analysis. The exception was GUSB, which showed significant mutation clustering only in kidney cancer.
We then focused on the proteins identified in the combined PanCancer analysis above (Table 1) and manually inspected the results to see if they showed specificity to particular tumor types. For example, it is well known that EGFR mutations found in lung adenocarcinoma and those found in glioblastoma multiforme cluster in different parts of the protein (intracellular protein kinase domain and extracellular region, respectively) (3, 32, 34). In fact, this difference is thought to be responsible for the differential sensitivity of these cancer types to EGFR kinase inhibitors (34). With CLUMPS, we were able to confirm the tissue-specific mutation clustering in EGFR (Datasets S7 and S10).
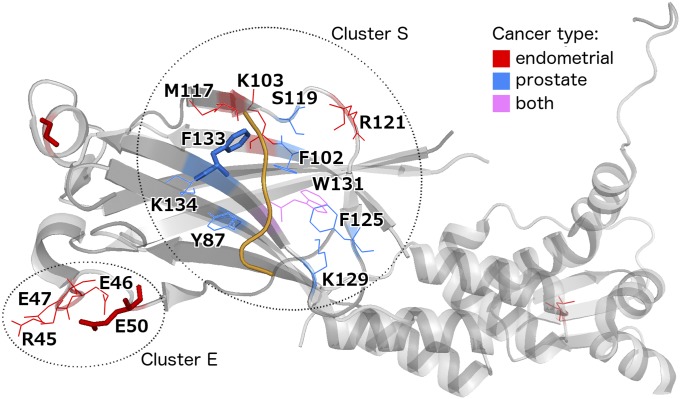
Interestingly, we also identified tumor type-specific clusters in SPOP, a substrate recognition component of an E3 ubiquitin-protein ligase complex that mediates the ubiquitination and subsequent proteasomal degradation of MAPK8, PTEN, and other cancer-related proteins. In Barbieri et al. (11), we described a 3D cluster of missense mutations from prostate tumors affecting mostly hydrophobic residues at the substrate-binding cleft of SPOP (called mutation cluster S for convenience). Here, we identified an additional, distant cluster (called cluster E), formed by mutations exclusively found in endometrial tumors and impacting four charged residues. The cluster was located in the same Math domain of the protein but spatially far from the substrate-binding pocket (Fig. 4 and SI Appendix, Fig. S6A). Residues forming cluster E were found to be mutated in a total of six samples; three samples had mutations at E50 (consistently changing this negatively charged residue to the positively charged lysine), whereas the rest of the residues (R45, E46, and E47) were mutated in a single sample each. Interestingly, using endometrial cancer mutation data that were recently generated by TCGA (https://tcga-data.nci.nih.gov) and were not part of our PanCancer dataset, we observed that the mutation incidence of all four residues from cluster E was increased in this data set (R45: two samples total, E46: two, E47: three, E50: four; the glutamate residues being altered to lysine in most cases), supporting the notion that cluster E contains driver mutations. To investigate the potential effects of SPOP mutations in clusters S and E on substrate protein levels, we analyzed TCGA reverse-phase protein array (RPPA) data from endometrial tumors (35). We expected mutations in cluster S (i.e., those affecting residues responsible for substrate interaction) to perturb the binding of SPOP with substrates and thus to dysregulate their ubiquitination and proteasomal degradation. Indeed, we found elevated levels of two known SPOP substrates, MAPK8 and PTEN, in endometrial tumors with mutations in cluster S relative to tumors with no mutations in SPOP (two-tailed t test, ). In contrast, the levels of MAPK8 and PTEN were not significantly changed in tumors with mutations in cluster E (SI Appendix, Fig. S6B). Theurillat et al. recently analyzed the protein levels of a novel SPOP substrate, DEK, in prostate tumor cell lines, where different SPOP variants were overexpressed (36). Consistent with our findings, they observed that DEK levels were significantly elevated in the case of missense variants in the substrate-binding pocket (cluster S) but not in the case of SPOP-E50K (which falls in our cluster E). Overall, our results suggest that the mutations in cluster E have a distinct, potentially endometrial-specific role, which remains to be elucidated experimentally.
Fig. 4.
Tissue-specific missense mutation clustering in SPOP (PDB ID code 3HU6) (37). All residues mutated in endometrial or prostate tumors are highlighted in color according to tissue of origin. Residues shown as thick sticks are recurrently mutated in the PanCancer data set (at least three samples). A substrate protein fragment is shown in orange. The distance between clusters S and E is 23 Å (average pairwise atomic distance); the corresponding within-cluster distances are 9 Å for cluster S and 6 Å for cluster E.
Mutation Clustering at Biomolecular Interaction Interfaces May Point to Interactions Potentially Perturbed in Cancer.
3D missense mutation clustering in some proteins may reflect selection for mutations that alter specific molecular interactions. Several studies have established that alteration of protein interactions plays a key role in many diseases (16, 38–42), including cancer (9, 16, 43). We tested for enrichment of missense mutations at known interfaces (inferred from structurally resolved complexes, such as cocrystals, from the PDB) of proteins, mediating interactions with other proteins, small molecule or ion ligands, DNA, and RNA. To quantify enrichment, we calculated the total number of samples that had a mutation at any residue (of either interaction partner) belonging to an interaction interface and compared it with a null distribution obtained by randomly scattering the mutations across all residues in the protein structure (preserving the number of samples per mutated residue and the number of mutations per structure). As in the CLUMPS analyses described above, we applied the FDR method to correct for multiple hypothesis testing and used a threshold to identify those interfaces that showed significant enrichment for mutations.
Enrichment of mutations at protein–protein interaction interfaces.
Among 1,145 heteromeric protein–protein interaction interfaces tested, 7 passed the significance threshold (Table 2 and Dataset S11). When we restricted the analysis to 304 protein–protein interfaces involving at least one known or candidate cancer protein (Dataset S2) to increase statistical power, we found 9 additional interfaces with significant clustering (Table 2 and Dataset S11). All structures are depicted in Dataset S15.
Table 2.
Molecular interactions showing enrichment of interface mutations
| Interactor A | Interactor B | Interface Mutations | P | ||
| Protein–protein | |||||
| Full analysis | |||||
| FBXW7 | CCNE1 | 66* | 1 | 1e-07 | 5.7e-05 | 1.5e-05 |
| PPP2R1A | PPP2R5C | 23* | 0 | 1e-07 | 5.7e-05 | 1.5e-05 |
| CAND1 | CUL4B | 6* | 9* | 9e-05 | 0.035 | 0.0092 |
| RASA1 | HRAS | 3 | 27* | 0.0002 | 0.044 | 0.012 |
| EXOSC7 | EXOSC2 | 2 | 4* | 0.0002 | 0.051 | – |
| OGT | HCFC1 | 8* | 3 | 0.0003 | 0.053 | – |
| PPP2R1A | PPP2R2A | 23* | 0 | 0.0003 | 0.053 | 0.02 |
| Restricted analysis | |||||
| ARHGEF25 | RHOA | 1 | 13* | 0.002 | 0.17 | 0.076 |
| RAC1 | NCF2 | 11* | 2 | 0.002 | 0.17 | 0.076 |
| PIK3CA | PIK3R1 | 299* | 21 | 0.002 | 0.17 | 0.076 |
| HLA-E | B2M | 3* | 4* | 0.002 | 0.17 | 0.076 |
| GRB14 | HRAS | 0 | 16* | 0.004 | 0.22 | 0.097 |
| FCGRT | B2M | 3 | 5* | 0.004 | 0.22 | 0.097 |
| ARHGEF11 | RHOA | 1 | 13* | 0.004 | 0.22 | 0.097 |
| TCEB1 | VHL | 4 | 23* | 0.004 | 0.22 | 0.097 |
| SMAD3 | SMAD4 | 6 | 20* | 0.004 | 0.22 | 0.097 |
| Protein–compound/ion | |||||
| Full analysis | |||||
| PTEN | L(+)-tartrate# | 76 | 9e-07 | 0.0034 | 0.00038 |
| HRAS | GTP | 30 | 9e-06 | 0.016 | 0.0018 |
| DICER1 | Mg2+ | 8 | 1e-05 | 0.016 | 0.0018 |
| FBXW7 | # | 52 | 3e-05 | 0.028 | 0.0031 |
| EP300 | Lys-CoA# | 16 | 6e-05 | 0.048 | 0.0054 |
| TP53 | 1,2-ethanediol# | 278 | 0.0002 | 0.098 | 0.011 |
| Restricted analysis | |||||
| HRAS | R,S,R-bisfuranol# | 21 | 0.0004 | 0.21 | 0.027 |
| KRAS | 20G# | 300 | 0.0006 | 0.23 | 0.03 |
| SETD2 | SAH# | 11 | 0.0009 | 0.25 | 0.042 |
| Protein–DNA/RNA | |||||
| Full analysis | |||||
| TP53 | DNA | 253 | 4e-05 | 0.0044 | 0.0013 |
| FOXO1 | DNA | 4 | 0.0005 | 0.022 | 0.0064 |
| WT1 | DNA | 10 | 0.0005 | 0.022 | 0.0064 |
| MAX | DNA | 9 | 0.0008 | 0.022 | 0.0065 |
| RUNX1 | DNA | 8 | 0.0009 | 0.022 | 0.0065 |
| TFAM | DNA | 6 | 0.001 | 0.027 | – |
| TDG | DNA | 4 | 0.002 | 0.027 | – |
| Restricted analysis | |||||
| SRSF2 | RNA | 5 | 0.02 | 0.25 | 0.059 |
The table shows all significant results both from a full analysis of all distinct interfaces in PDB and from a restricted analysis of interfaces involving at least one known cancer protein (as per Dataset S2). The corresponding FDR q-values are shown in columns and , respectively. A dash (–) in the column means that the corresponding interaction does not involve a known cancer protein. In the case of protein-protein interactions, the numbers of mutations at the interface of each partner are separated by the pipe () symbol; an asterisk (*) denotes that this number is individually significant (permutation test ) without considering mutations in the other partner. Protein–compound/ion interactions where a molecule other than the natural substrate occupies (or is near) the protein active site in the available PDB structure are marked with the hash symbol (#). SAH, S-adenosyl-l-homocysteine; 20G, N-1-[(2,4-dichlorophenoxy)acetyl]piperidin-4-yl-4-sulfanylbutanamide.
Most of the significant interfaces carried mutations in both interaction partners. In three cases (CAND1-CUL4B, PIK3CA-PIK3R1, and B2M-HLA-E), the number of interface mutations was significant () for each of the two partners individually, as well as for the combined number of mutations at the interface. In the other cases, only one of the interactors showed a significant number of mutations at the interface (in addition to the interface as a whole), perhaps because interface mutations in the other partner are deleterious for other reasons (such as essential interaction with a third partner). Below, we discuss some of the significant cases.
-
i)
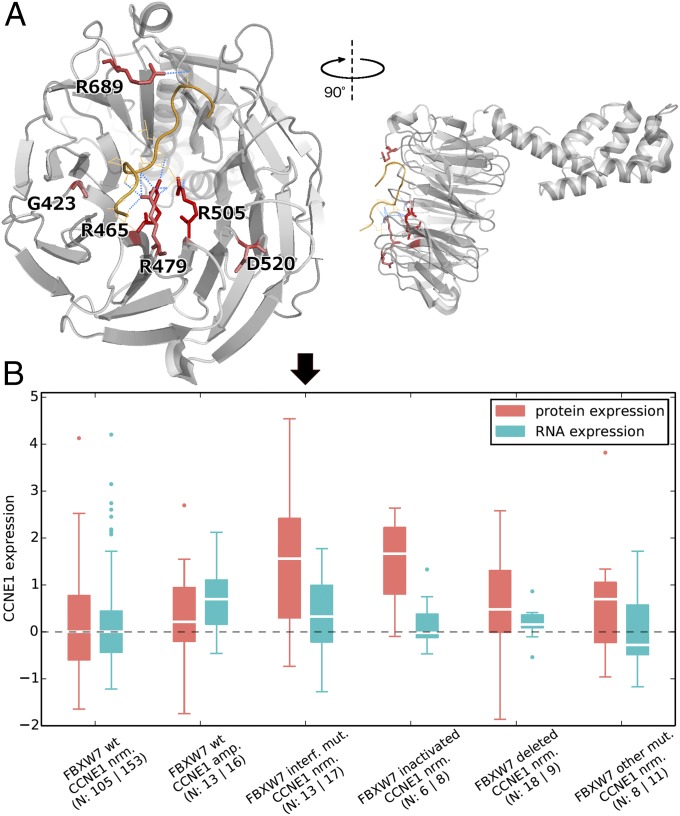
FBXW7-CCNE1. Cyclin E1 (CCNE1) is a critical cell cycle protein, which at abnormally high levels promotes premature cell division, genomic instability, and tumorigenesis. FBXW7 (F-box/WD repeat-containing protein 7) is a substrate recognition component of an E3 ubiquitin-protein ligase complex, mediating the ubiquitination and subsequent proteasomal degradation of CCNE1 and other cancer proteins like MYC and JUN. We found that all six recurrently mutated residues (found in at least three samples from our mutation dataset) of FBXW7 clustered together at the WD40 propeller domain of the protein product. Four of them, R465, R479, R505, and R689, interacted directly with the substrate CCNE1 through hydrogen bonds (Fig. 5A). Changes in these residues could perturb the interaction, causing insufficient ubiquitination/degradation of CCNE1 in tumor samples (as has been previously shown in model systems) (44, 46, 47).
-
ii)
PPP2R1A interaction with PPP2R5C or with PPP2R2A. PPP2R1A is a constant regulatory subunit of the heterotrimeric protein phosphatase 2 (PP2A) complex, serving as a scaffold for complex assembly. PP2A is a serine/threonine phosphatase, which controls numerous signaling pathways. It is involved in negative control of cell growth and division and has been implicated as a tumor suppressor (19). Perturbation of the PPP2R1A interactions with other PP2A subunits through mutations at the protein–protein binding interface (SI Appendix, Fig. S7) may disturb assembly of the complex and may hence abolish its tumor suppressor function.
-
iii)
CUL4B-CAND1. CUL4B (cullin 4B) serves as a scaffold for multiple Cullin-RING-based E3 ubiquitin-protein ligase complexes that mediate the ubiquitination of target proteins, followed by their proteasomal degradation. CUL4B is important for the regulation of cyclin E (48), members of the MTOR pathway (49) and multiple histones (50) and plays a role in DNA repair on damage from UV light (51). It has been implicated in cancer (52) and is significantly mutated in breast tumors (3). CAND1 (Cullin-associated NEDD8-dissociated 1) is a key regulator of cullin-based E3 ubiquitin ligases. It has been found to be transcriptionally deregulated in prostate cancer (53) and high-grade neuroendocrine lung tumors (54). Furthermore, targeted knockdown of CAND1 has been shown to promote proliferation of prostate carcinoma cells (55). Altogether, the enrichment of mutations at the CUL4B-CAND1 interface suggests positive selection of mutations that disrupt the interaction and hence potentially prevent the ubiquitination and degradation of cancer-related proteins. Moreover, mutations in CUL1 (a paralog of CUL4B), which are found frequently in prostate cancer, have been suggested to disturb the interaction with CAND1 and have been associated with aberrant centriole synthesis, which can lead to aneuploidy (53).
-
iv)
HRAS-RASA1. Mutations that disturb the interaction between HRAS and the GTPase activating protein RASA1 (SI Appendix, Fig. S8) would be expected to lead to constitutive activation of the HRAS oncoprotein.
-
v)
EXOSC2-EXOSC7. Both proteins are subunits of the RNA exosome complex responsible for the decay of all types of RNA. In Chapman et al. (10), we reported that multiple myeloma patients frequently harbor mutations in DIS3, the catalytic subunit of the RNA exosome. Based on their location at the enzymatic pocket of DIS3, and on prior experimental evidence from model organisms, those mutations were predicted to abolish the catalytic activity of the exosome (10). The enrichment of mutations at the EXOSC2-EXOSC7 interface found here may reflect selection for mutations disturbing the binding of these two subunits. Our findings thus support the potential causal role of the RNA exosome in cancer and suggest that exosome-mediated RNA decay may be disturbed in cancer in alternative ways: through mutations abolishing the enzymatic activity of the catalytic subunit or through mutations disabling exosome complex formation.
-
vi)
OGT-HCFC1. HCFC1 is involved in several processes important in cancer, including cell cycle control, positive regulation of proliferation, chromatin organization, histone acetylation, and transcriptional regulation. Notably, it is a major downstream effector of BRCA1-associated protein 1 (BAP1) (56), whose frequently observed mutations have a causal role in cancer (57). OGT is involved in the posttranslational modification and direct proteolysis of HCFC1 (58), thus influencing its activity and abundance. Interestingly, two of the three mutated interface residues of HCFC1 were threonines (SI Appendix, Fig. S9), which could be glycosylated by OGT (58). A plausible hypothesis based on the significant enrichment of mutations at the OGT-HCFC1 interface is that such mutations might disturb the regulation (through posttranslational modifications or cleavage) of HCFC1, leading to deregulation of cancer-related processes mentioned above.
-
vii)
RHOA-ARHGEF25. RHOA is a GTPase that controls cell contractility and motility and also promotes tumorigenesis through STAT3 activation. ARHGEF25 activates RHOA by exchanging GDP for GTP in its substrate pocket. In a recent publication (59), two mutation hotspots in RHOA, Y42 and D59, were identified in gastric adenocarcinoma that were predicted to localize at the interaction interface of RHOA with a downstream effector, ROCK1. This interface coincides with the binding interface or RHOA for ARHGEF25. In this study, the significant enrichment of missense mutations at the common protein binding interface of RHOA was mainly driven by a different positional hot-spot, E40 (reported in ref. 3). Mutations at the common interface of RHOA may act by preferentially modifying the affinity of RHOA to downstream effectors (like ROCK1), activators (like ARHGEF25), or perhaps to a third class of proteins, GTPase activating proteins (GAPs), that inactivate RHOA by promoting its GTP hydrolysis function.
-
viii)
PIK3CA-PIK3R1. The enrichment of mutations at the PIK3CA-PIK3R1 interaction interface is consistent with previous reports (9); it shows positive selection for mutations that disturb the negative regulation of the oncoprotein PIK3CA by PIK3R1, thus leading to constitutive activation of PIK3CA.
-
ix)
B2M-HLA-E. The formation of the HLA-E MHC class I, resulting from interaction with B2M, is important for cell recognition by natural killer cells and thus crucial for host immunity against cancer (60). Interface mutations in B2M or HLA-E that disrupt the interaction may result in failure of MHC complex formation and subsequently to immune system evasion, which is recognized as an emerging hallmark of cancer (61).
-
x)
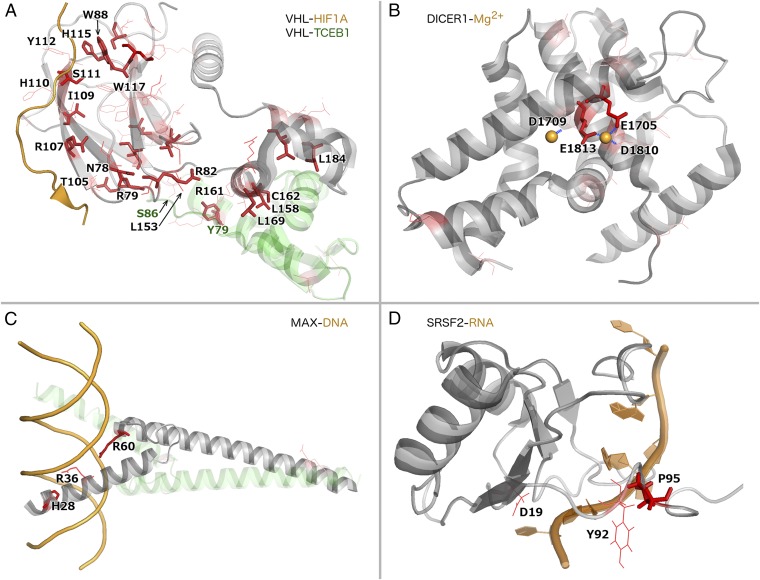
VHL-TCEB1. VHL is a well-known tumor suppressor that functions as the substrate recognition component of an E3 ubiquitin ligase complex also comprising TCEB1, TCEB2, and CUL2. It plays a central role in the ubiquitination and degradation of hypoxia-inducible transcription factors 1α (HIF1A) and 2α (HIF2A), which are important for tumor angiogenesis. Germ-line inactivating mutations in VHL cause the Von Hippel–Lindau cancer syndrome. Moreover, VHL is frequently affected by inactivating (nonsense, frameshift, or splice site) somatic mutations in sporadic kidney tumors. We found that somatic missense mutations in VHL are significantly enriched at the interface of the VHL protein product with another ubiquitin ligase component, TCEB1 () and at its interface with the substrate, HIF1A () (Fig. 6A). Such missense mutations likely lead to loss of VHL function through loss of interactions with either TCEB1 or HIF1A, which are essential for HIF1A regulation. Some of these missense mutations in VHL have been already demonstrated to cause interaction perturbations and/or HIF dysregulation (62, 66). Notably, TCEB1 was also mutated at its interface to VHL in four samples in our dataset, with three samples harboring mutations at TCEB1-Y79 (Fig. 6A). Mutations in TCEB1 could also abolish the interaction with VHL. Consistent with our hypothesis, alterations of this residue have been associated with HIF accumulation in cell lines (67).
Fig. 5.
Spatial location of FBXW7 mutation hotspots and their effect on CCNE1 protein levels. (A) Crystal structure of FBXW7 (gray) bound to the degron of CCNE1 (orange) (PDB ID code 2OVQ) (44). All recurrently mutated residues (at least three samples) of FBXW7 are shown as red sticks; color intensity scales with recurrence. Hydrogen bonds between such residues and CCNE1 residues are shown as blue dotted lines. (B) CCNE1 protein and RNA levels (as measured by RPPA and RNAseq, respectively) (45) in colorectal cancer samples with different FBXW7/CCNE1 genotypes (“nrm”: normal copy number; “amp.”: amplified, “interf. mut.”: mutation at interaction interface).
Fig. 6.
Examples for protein–protein, protein–ion, protein–DNA, and protein–RNA interaction interfaces significantly enriched with mutations. All mutated residues in each protein are shown in red, with color intensity scaling with the number of samples with mutations at the corresponding residue. Recurrently mutated residues are shown as sticks and residues mutated in one or two samples as thin lines. All mutated interface residues are labeled (in A, dark green labels correspond to TCEB1 residues). (A) Many mutations in VHL (gray) cluster at both its interfaces to substrate HIF1A (orange) and to cocomplex partner TCEB1 (pale green) (PDB ID code 1LM8) (62). (B) Mutations in the DICER1 C-terminal RNase III domain (gray) cluster at the interaction interface with its activator magnesium (orange sphere) (PDB ID code 2EB1) (63). (C) A heterodimer of MAX (gray) and MAD (pale green) bound to DNA (orange) (PDB ID code 1NLW) (64). (D) SRSF2 (gray) bound to RNA (orange) (PDB ID code 2LEB) (65).
Increased CCNE1 levels in colorectal tumors with FBXW7-CCNE1 interface mutations.
To follow up on results above concerning the FBXW7-CCNE1 interface, we sought experimental evidence. Because FBXW7 is known to regulate the degradation of CCNE1, we predicted that mutations at the interface would lead to abolished interaction and hence to increased levels of CCNE1 protein. By analyzing experimental RPPA and RNAseq data from TCGA (45), we found that primary colorectal tumors carrying FBXW7 mutations that affect interface residues indeed show normal levels of CCNE1 RNA but significantly elevated levels of CCNE1 protein (two-tailed t test, ; Fig. 5B) compared with colorectal tumors in which FBXW7 is nonmutant and CCNE1 has normal copy number. The same was true for samples with inactivating (i.e., nonsense, frameshift or splice-site) mutations in FBXW7 (two-tailed t test, ). Our results, based on human tumors, were consistent with previous studies in cell lines and model organisms (44, 46, 47). Interestingly, the RPPA data did not show significant protein-level changes in MYC (two-tailed t test, and for samples with interface and inactivating mutations in FBXW7, respectively) and JUN (analogously, and ), which are also substrates of FBXW7. Altogether, these results suggest that patients with either FBXW7 missense mutations at the substrate interface, or with FBXW7 inactivating mutations, may benefit from inhibitors of CCNE1 or of its downstream effector CDK2.
Enrichment of mutations at protein binding sites for small molecules or ions.
In addition to examining protein–protein interactions, we also looked for enrichment of mutations at binding interfaces with other types of biomolecules. We tested for enrichment at 3,759 unique protein interfaces for small molecules or metal ions, using an analogous statistical approach as for protein–protein interactions. We found six protein–compound/ion interaction interfaces with significant () mutation enrichment. When we restricted the analysis to 423 interfaces involving only cancer proteins, three additional hits were obtained (Table 2 and Dataset S12). Here we discuss two examples:
-
i)
HRAS-GTP. Mutations at the active site of HRAS, similarly to its paralog KRAS (Fig. 1), disturb the GTPase activity of the oncoprotein, locking it in its active, GTP-bound state (68).
-
ii)
DICER1-Mg2+. Mg2+ is required for the activity of DICER1 (69), which is pivotal in processing siRNAs that play important roles in posttranscriptional gene silencing. Germ-line mutations that inactivate DICER1 are known to predispose to a range of tumors (70), and DICER1 inhibition promotes metastasis (71). A significant fraction of missense mutations in DICER1 from our data affect negatively charged residues in direct contact with the positively charged magnesium ions, consistently altering these residues to positively or noncharged residues (Fig. 6B). The observed somatic mutations in the magnesium-binding residues likely abolish the DICER1–Mg2+ interaction, thereby interfering with DICER1 function and potentially leading to tumor formation, metastasis, or both.
In both examples above, perturbation of the protein–compound/ion interactions may drive cancer because they involve known cancer proteins and the ligands are required for the activity of those proteins. However, this might not be the case for some of the remaining seven significant interactions (marked with # in Table 2): For example, the significance of the FBXW7– interaction may reflect the fact that the sulfate ion is located near the CCNE1 binding interface of FBXW7. Similarly, missense mutations in PTEN cluster at its active site and most likely perturb its interaction with phosphatidylinositol trisphosphate; however, the available PDB structure has tartrate bound at the active site instead of this natural substrate (72).
Enrichment of mutations at protein interaction interfaces with DNA and RNA.
Finally, we analyzed protein–DNA and protein–RNA complexes from the PDB to look for enrichment of mutations affecting protein residues in direct contact with nucleic acids. We tested 124 protein–DNA and 51 protein–RNA interfaces and found 7 significant protein–DNA and no significant protein–RNA interfaces (Table 2 and Datasets S13 and S14). Five of the protein–DNA interactions involved known cancer proteins, including TP53 and MAX, whereas two interactions also implicated the proteins TFAM and TDG in cancer. A restricted analysis focusing only on known cancer proteins yielded one additional significant case, the SRSF2–RNA interface (Table 2 and Dataset S14). We discuss two examples below:
-
i)
MAX-DNA. A significant number of mutations in the MAX (Myc-associated factor X) transcription factor affected three positively charged residues in direct contact with the negatively charged DNA molecule (Fig. 6C). The overall mutation frequency of MAX in the PanCancer compendium was not significant (3), although an independent study has suggested that MAX is significantly mutated in pheochromocytoma and paraganglioma (73). MAX forms heterodimers with MYC, which is a classic cancer gene that is amplified in various cancers (74). Another heterodimerization partner of MAX, called MGA (MAX gene associated), was found significantly mutated in the PanCancer compendium (3).
-
ii)
SRSF2-RNA. SRSF2 is a pre-mRNA splicing factor and a component of the spliceosome. We and others found it to be significantly mutated in acute myeloid leukemia (3, 75), where it has been associated with adverse outcome (75). Here, we found that missense mutations in this protein clustered at the interaction interface with RNA, including the recurrently mutated residue P95 and two nonrecurrently mutated residues (Fig. 6D).
Although the examples all achieved statistical significance, several were supported by relatively few samples from the PanCancer compendium. For example, the TFAM–DNA and TDG–DNA interfaces were mutated in only four and six tumor samples, respectively (Table 2). Encouragingly, a potential association of these interactions with cancer makes good biological sense: truncating TFAM mutations that abolish interactions with DNA, frequently found in colorectal tumors, have been shown to cause resistance to apoptosis (76), and TDG plays important roles in DNA demethylation and damage repair (77). However, larger cohorts of sequenced tumors will be needed to draw robust conclusions, based solely on statistical methods, about the role of perturbing the interaction of these proteins with DNA in cancer.
Discussion
Using large-scale datasets of cancer somatic mutations and of 3D models of human proteins, we systematically searched for spatial clustering of missense mutations with respect to 3D protein structures. Such clustering likely results from positive selection for certain missense mutations in cancer. Overall, we identified 50 different proteins with clustering of mutations and/or enrichment of mutations at interaction interfaces (Tables 1 and 2). As anticipated, many of these proteins are known cancer drivers, including HRAS, FBXW7, and SPOP, whereas others, like NUF2, OGT, and HLA-E, represent previously unidentified candidates that require experimental follow-up. Our analyses not only identify candidate cancer genes, but also highlight specific alleles to be tested experimentally.
Interestingly, our analyses demonstrated that 3D clustering of somatic mutations is not only a characteristic of some oncoproteins but also of some tumor suppressors. It is broadly appreciated that missense mutations that constitutively activate oncoproteins: i.e., gain-of-function mutations tend to be localized to specific regions of the 3D structure of these oncoproteins (5, 9, 14, 15). Similarly, missense mutations capable of destroying the function of tumor suppressor proteins could also cluster spatially. They may occur preferentially at key residues in the 3D core of proteins, destabilizing them (14). Others may abolish specific molecular interactions and would tend to cluster at protein interaction interfaces (38, 78). Indeed, our analyses identified enrichment of missense mutations in interaction interfaces of known tumor suppressors with their substrates (e.g., in PTEN, FBXW7, SPOP, STK11, VHL), with essential cocomplex partners (e.g., in PPP2R1A, PIK3R1, VHL) and with DNA (e.g., in TP53). Furthermore, loss-of-interaction mutations of tumor suppressors that lead to loss of function may have similar effects on gene/protein expression to protein-destabilizing mutations. An example given above was FBXW7, where both inactivating (i.e., nonsense, frameshift, or splice-site) mutations throughout the protein and missense mutations located at the binding interface with CCNE1 showed the same effect on CCNE1 protein levels.
Discriminating driver from passenger missense mutations in the same gene is currently a central challenge in cancer genetics and has clear clinical implications. Although this problem was not within the scope of this study, our 3D mutation clustering approach may help prioritize potential driver mutations. More precisely, clustered mutations more likely reflect positive selection than their randomly scattered counterparts. For example, mutations clustered at molecular interaction interfaces would tend to disrupt important interactions. We are currently investigating the mutation discrimination potential of our approach and results will be published elsewhere.
Databases of somatic mutations in cancer and of protein structures are growing rapidly. Improved methods for 3D structure determination have led to unprecedented growth of the PDB (17). In addition, computational methods can help to infer 3D structures of as yet unresolved proteins and protein complexes, based on available structures of their homologs and/or on other types of experimental data (79). At the same time, dramatic decreases in sequencing costs enable the sequencing of many additional tumors. More extensive mutational and structural data will enable the discovery of 3D clustering of mutations in more proteins, in studies both within and across different tumor types. Such discoveries should lead to new insights into tissue-specific and general molecular mechanisms of cancer.
Materials and Methods
Mutation Data.
We used the somatic mutation dataset published in ref. 3. UniProt protein sequence coordinates for missense mutations were mapped using Oncotator (80).
Protein Structures.
We downloaded all human protein structures from PDB on 27 March 2014 and used SIFTS (81) to cross-map both protein identifiers and individual amino acid residues between PDB and UniProt. Structures with mutations in less than three tumors were filtered out, resulting in an input dataset of 41,063 structures (counting different PDB chains within the same PDB file separately) of 4,062 human proteins. Biomolecular interaction interface definitions were obtained from PDBsum (82) on 27 July 2014. PyMol (https://www.pymol.org) was used for structure visualization and residue distance calculations.
Selection of Representative Structures for Each Protein.
Many human proteins were represented by multiple PDB structures that often (i) covered only parts of the reference protein sequence (SI Appendix, Fig. S10) and (ii) overlapped partially or completely with each other (SI Appendix, Fig. S11). We developed a greedy algorithm to select a set of minimally overlapping, “representative” structures for each protein so that the set jointly covered a maximal part of the reference (UniProt) protein sequence. We built this set by consecutively adding the longest structure (i.e., that with largest protein sequence coverage) so that no pair of structures in the set overlapped with each other by more than 10% of the shorter structure. For groups of structures with comparable lengths but with high mutual overlap, we selected the structure with median CLUMPS P value. Although choosing the structure with the best P value might appear as a more intuitive choice, we reasoned that cancer proteins might tend to have more structures in PDB compared with their noncancer counterparts due to study bias. Thus, selecting the structure with the best P value would artificially reward cancer proteins, whereas selecting those with median P value would not. Our algorithm selected 4,822 (from the total of 41,063) representative structures corresponding to 4,062 human proteins. The joint protein sequence coverage of these representative structures is shown in SI Appendix, Fig. S12.
CLUMPS Methodology.
To identify significant clustering of mutations in proteins with available structural data, we first defined a WAP score summarizing all pairwise distances between mutated residues in a given 3D protein structure (SI Appendix, Fig. S1) as
| [1] |
where q and r () are protein residues; is the Euclidean distance (in Å) between the centroids of those residues; and (or ) is the number of samples where q (or r) is found mutated, normalized to the range [0,1] using the sigmoidal Hill function
| [2] |
Here, is the number of samples with a missense mutation impacting residue q of the protein; and and are parameters of the Hill function controlling the critical point (center) and steepness of the sigmoid function, respectively. The exponential function in Eq. 1 transforms the absolute spatial distance between residues to the interval [0,1] with shorter distances (relative to the parameter t that can be interpreted as a “soft” distance threshold and was set to Å) mapping to a value close to 1 and longer distances mapping to values near zero (SI Appendix, Fig. S13A). The absolute number of samples with mutations at a given residue was normalized as per Eq. 2 to avoid a disproportionally high influence of very frequently mutated residues (positional ultra-hotspots) compared with less frequently but still recurrently mutated ones. A sigmoidal function was chosen to down-weight residues mutated in only one sample while rewarding residues mutated in three or more samples (SI Appendix, Fig. S13B). We calculated a WAP score for each protein structure and assessed its significance using a null model assuming a uniform distribution of mutations across the protein residues covered by the given PDB structure, preserving the number of samples with mutations at a given residue. The null distribution was obtained through randomizations, and, if the resulting P value was less than 0.1, we extended the randomizations to . SI Appendix, Fig. S14 shows a comparison of P values of the top scoring 300 proteins against P values obtained with CLUMPS when mutated residues are weighed equally regardless of recurrence. All P values were corrected for multiple testing consistently throughout the manuscript using the FDR method (83), implemented in the p.adjust function in R (https://www.r-project.org/).
Additional method descriptions are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Andre Bernards, Emre Guney, Yves Jacob, Jakob Berg Jespersen, Ulrich Stelzl, and Ani Stoyanova for useful comments on the manuscript. A.K. is supported by the European Molecular Biology Organization long-term fellowship. G.G. is supported by National Institutes of Health Grant U24 CA143845. G.G. is the Paul C. Zamecnik, MD, Chair in Oncology at Massachusetts General Hospital.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516373112/-/DCSupplemental.
References
- 1.The Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29(18):2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porta-Pardo E, Godzik A. e-Driver: A novel method to identify protein regions driving cancer. Bioinformatics. 2014;30(21):3109–3114. doi: 10.1093/bioinformatics/btu499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 9.Huang CH, et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318(5857):1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 10.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: Understanding tumorigenic mutations. Structure. 2011;19(11):1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Van Allen EM, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4(10):1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stehr H, et al. The structural impact of cancer-associated missense mutations in oncogenes and tumor suppressors. Mol Cancer. 2011;10:54. doi: 10.1186/1476-4598-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryslik GA, et al. A spatial simulation approach to account for protein structure when identifying non-random somatic mutations. BMC Bioinformatics. 2014;15:231. doi: 10.1186/1471-2105-15-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, et al. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol. 2012;30(2):159–164. doi: 10.1038/nbt.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman HM, Kleywegt GJ, Nakamura H, Markley JL. The Protein Data Bank archive as an open data resource. J Comput Aided Mol Des. 2014;28(10):1009–1014. doi: 10.1007/s10822-014-9770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sablina AA, et al. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell. 2007;129(5):969–982. doi: 10.1016/j.cell.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467(7317):805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen KWY, Montpetit B, Hieter P. The kinetochore and cancer: What’s the connection? Curr Opin Cell Biol. 2005;17(6):576–582. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: A link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110(5):322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- 23.Hayama S, et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66(21):10339–10348. doi: 10.1158/0008-5472.CAN-06-2137. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko N, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390(4):1235–1240. doi: 10.1016/j.bbrc.2009.10.127. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi Y, et al. Cell division cycle-associated protein 1 overexpression is essential for the malignant potential of colorectal cancers. Int J Oncol. 2014;44(1):69–77. doi: 10.3892/ijo.2013.2177. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Dai SJ, Li H, Dong L, Peng YP. Silencing of NUF2 inhibits tumor growth and induces apoptosis in human hepatocellular carcinomas. Asian Pac J Cancer Prev. 2014;15(20):8623–8629. doi: 10.7314/apjcp.2014.15.20.8623. [DOI] [PubMed] [Google Scholar]
- 27.Sethi G, et al. An RNA interference lethality screen of the human druggable genome to identify molecular vulnerabilities in epithelial ovarian cancer. PLoS One. 2012;7(10):e47086. doi: 10.1371/journal.pone.0047086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu P, Chen X, Sun J, Bie P, Zhang L. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. Biosci Rep. 2015;35(1):e00170. doi: 10.1042/BSR20140124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Forbes SA, et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozturk M. p53 mutation in hepatocellular carcinoma after aflatoxin exposure. Lancet. 1991;338(8779):1356–1359. doi: 10.1016/0140-6736(91)92236-u. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, et al. Protein domain-level landscape of cancer-type-specific somatic mutations. PLOS Comput Biol. 2015;11(3):e1004147. doi: 10.1371/journal.pcbi.1004147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson JP, et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25(17):7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vivanco I, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theurillat JPP, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346(6205):85–89. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang M, et al. Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36(1):39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y, et al. Dissecting disease inheritance modes in a three-dimensional protein network challenges the “guilt-by-association” principle. Am J Hum Genet. 2013;93(1):78–89. doi: 10.1016/j.ajhg.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosca R, et al. dSysMap: Exploring the edgetic role of disease mutations. Nat Methods. 2015;12(3):167–168. doi: 10.1038/nmeth.3289. [DOI] [PubMed] [Google Scholar]
- 41.Ryan CJ, et al. High-resolution network biology: Connecting sequence with function. Nat Rev Genet. 2013;14(12):865–879. doi: 10.1038/nrg3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahni N, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161(3):647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishi H, et al. Cancer missense mutations alter binding properties of proteins and their interaction networks. PLoS One. 2013;8(6):e66273. doi: 10.1371/journal.pone.0066273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26(1):131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 45.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413(6853):311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 47.Akhoondi S, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67(19):9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 48.Zou Y, et al. Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem. 2009;284(48):33320–33332. doi: 10.1074/jbc.M109.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghosh P, Wu M, Zhang H, Sun H. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle. 2008;7(3):373–381. doi: 10.4161/cc.7.3.5267. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22(3):383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 51.Guerrero-Santoro J, et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008;68(13):5014–5022. doi: 10.1158/0008-5472.CAN-07-6162. [DOI] [PubMed] [Google Scholar]
- 52.Lee J, Zhou P. Cullins and cancer. Genes Cancer. 2010;1(7):690–699. doi: 10.1177/1947601910382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korzeniewski N, Hohenfellner M, Duensing S. CAND1 promotes PLK4-mediated centriole overduplication and is frequently disrupted in prostate cancer. Neoplasia. 2012;14(9):799–806. doi: 10.1593/neo.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salon C, et al. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: Relationships with CAND1 and cyclin E protein levels. J Pathol. 2007;213(3):303–310. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- 55.Murata T, et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010;13(4):356–361. doi: 10.1038/pcan.2010.32. [DOI] [PubMed] [Google Scholar]
- 56.Misaghi S, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29(8):2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carbone M, et al. BAP1 and cancer. Nat Rev Cancer. 2013;13(3):153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capotosti F, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144(3):376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 59.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng M, Chen Y, Xiao W, Sun R, Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol. 2013;10(3):230–252. doi: 10.1038/cmi.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 62.Min JH, et al. Structure of an HIF-1α -pVHL complex: Hydroxyproline recognition in signaling. Science. 2002;296(5574):1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 63.Takeshita D, et al. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374(1):106–120. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 64.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112(2):193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 65.Daubner GM, Cléry A, Jayne S, Stevenin J, Allain FHT. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31(1):162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rechsteiner MP, et al. VHL gene mutations and their effects on hypoxia inducible factor HIFα: Identification of potential driver and passenger mutations. Cancer Res. 2011;71(16):5500–5511. doi: 10.1158/0008-5472.CAN-11-0757. [DOI] [PubMed] [Google Scholar]
- 67.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 68.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Provost P, et al. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21(21):5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slade I, et al. DICER1 syndrome: Clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48(4):273–278. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- 71.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 72.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 73.Burnichon N, et al. MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clin Cancer Res. 2012;18(10):2828–2837. doi: 10.1158/1078-0432.CCR-12-0160. [DOI] [PubMed] [Google Scholar]
- 74.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang SJ, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119(19):4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo J, et al. Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer. Cancer Res. 2011;71(8):2978–2987. doi: 10.1158/0008-5472.CAN-10-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dalton SR, Bellacosa A. DNA demethylation by TDG. Epigenomics. 2012;4(4):459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Das J, et al. Elucidating common structural features of human pathogenic variations using large-scale atomic-resolution protein networks. Hum Mutat. 2014;35(5):585–593. doi: 10.1002/humu.22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alber F, Förster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 80.Ramos AH, et al. Oncotator: Cancer variant annotation tool. Hum Mutat. 2015;36(4):E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velankar S, et al. SIFTS: Structure Integration with Function, Taxonomy and Sequences resource. Nucleic Acids Res. 2013;41(Database issue):D483–D489. doi: 10.1093/nar/gks1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Beer TAP, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42(Database issue):D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat Soc. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.