Significance
The regulation of protein abundance of receptors and transporters at the plasma membrane is important for proper signaling in many biological pathways. The removal of plasma membrane proteins can occur via the endocytic protein degradation pathway, in which posttranslational modification by ubiquitin plays a key role. The activity of ubiquitinating and deubiquitinating enzymes can determine the ubiquitination status of a given target protein, and it has been shown that both classes of enzymes have important physiological roles. However, how these enzymes themselves are regulated at the molecular level has not yet been completely understood. In this study, we report a possible mechanism by which the deubiquitinating enzyme AMSH3 is regulated by its interacting protein, apoptosis-linked gene-2 interacting protein X (ALIX), in Arabidopsis.
Keywords: Arabidopsis, intracellular trafficking, ubiquitin, ESCRT-III
Abstract
Ubiquitination is a signal for various cellular processes, including for endocytic degradation of plasma membrane cargos. Ubiquitinating as well as deubiquitinating enzymes (DUBs) can regulate these processes by modifying the ubiquitination status of target protein. Although accumulating evidence points to the important regulatory role of DUBs, the molecular basis of their regulation is still not well understood. Associated molecule with the SH3 domain of signal transduction adaptor molecule (STAM) (AMSH) is a conserved metalloprotease DUB in eukaryotes. AMSH proteins interact with components of the endosomal sorting complex required for transport (ESCRT) and are implicated in intracellular trafficking. To investigate how the function of AMSH is regulated at the cellular level, we carried out an interaction screen for the Arabidopsis AMSH proteins and identified the Arabidopsis homolog of apoptosis-linked gene-2 interacting protein X (ALIX) as a protein interacting with AMSH3 in vitro and in vivo. Analysis of alix knockout mutants in Arabidopsis showed that ALIX is essential for plant growth and development and that ALIX is important for the biogenesis of the vacuole and multivesicular bodies (MVBs). Cell biological analysis revealed that ALIX and AMSH3 colocalize on late endosomes. Although ALIX did not stimulate AMSH3 activity in vitro, in the absence of ALIX, AMSH3 localization on endosomes was abolished. Taken together, our data indicate that ALIX could function as an important regulator for AMSH3 function at the late endosomes.
Ubiquitin-dependent protein degradation plays a pivotal role in almost all biological processes, as the timely and selective removal of regulatory proteins is essential in many signaling pathways (1, 2). Ubiquitin molecules can form topologically distinct ubiquitin chains that can serve as signals for different pathways (3). Among them, ubiquitin chains linked through lysine 63 (K63) have been associated with endocytosis and were shown to be required for the efficient endocytic degradation of plasma membrane cargos (4, 5). In eukaryotes, ubiquitinated membrane proteins are transported into the vacuole/lysosome for degradation by resident proteases, depending on the function of the endosomal sorting complex required for transport (ESCRT) machinery. Ubiquitinated cargos are recognized and transported to late endosomes through the function of ESCRT-0, ESCRT-I, and ESCRT-II, and are subsequently sequestered to the intraluminal vesicles (ILVs) of the multivesicular body (MVB) by ESCRT-III (6, 7). Plants lack homologs of ESCRT-0 (8, 9), and it is suggested that ubiquitin binding proteins such as the target of Myb (TOM)-LIKEs (TOLs) take over its function (10).
Ubiquitination of plasma membrane proteins depends on the activity of the ubiquitin conjugating machinery that creates an isopeptide bond between the C-terminal glycine of ubiquitin and a lysine of the substrate proteins or another ubiquitin molecule (11). Deubiquitinating enzymes (DUBs) can counteract the E3 ligase activity, in that they hydrolyze ubiquitin chains. In contrast to earlier assumptions that DUBs play merely a housekeeping role, it has been shown that DUBs can also be actively involved in the regulation of their target proteins (12, 13). The Arabidopsis genome encodes for at least 48 DUBs, although most of their molecular and biological functions are yet poorly understood (14). Whereas ubiquitinating enzymes interact specifically with their substrates (11), DUBs also can hydrolyze free ubiquitin chains unattached to target proteins (12, 15), and in most cases, do not require specific interactions with the substrate proteins. The elucidation of the spatiotemporal regulation of DUBs is therefore essential for a better understanding of the molecular mechanisms of DUB function.
Associated molecule with the SH3 domain of STAM (AMSH) is a metalloprotease DUB that was first identified as an interactor of the signaling molecule and ESCRT-0 component signal transduction adaptor molecule (STAM) in mammals (16). AMSH genes are conserved in higher eukaryotes and are essential for growth and development. Knockout of AMSH in mice causes postnatal lethality and neurodegenerative aberrations (17), and mutations in human AMSH were associated with an infant neurodegenerative disease (18), indicating its essential function in mammals. In our previous work, we conducted a genetic analysis of AMSH genes in Arabidopsis, named AMSH1, AMSH2, and AMSH3. We have shown that knockdown of AMSH1 causes altered pathogen response, and that the knockout of AMSH3 is lethal in plants, leading to growth arrest in the early stages of development (19–21).
AMSH proteins were shown to interact with ESCRT-III subunits and were implicated in endocytic protein degradation (21–26). Human AMSH and the Mpr1/Pad1 N-terminal (MPN)+ domain of Arabidopsis AMSH show specificity toward K63-linked ubiquitin chains (20–22), further supporting their function in endocytosis. Defects in AMSH function impair a number of intracellular trafficking events, including degradation of endocytosis cargos (22, 25, 27), vacuolar transport, and vacuole biogenesis (20), as well as defects in autophagic degradation (18, 20, 21). Although AMSH function in intracellular trafficking and protein degradation is well established, the molecular framework surrounding its function is not yet completely understood.
With the aim of elucidating the molecular basis of AMSH regulation, we screened for interactors of Arabidopsis AMSH proteins and found a homolog of human apoptosis-linked gene-2 interacting protein X (ALIX) and budding yeast bypass of C kinase 1 (BCK1)-like resistance to osmotic shock 1p (Bro1p) as a direct interactor of AMSH3. ALIX is a conserved protein in eukaryotes that was implicated in cytokinesis, ILV, and exosome biogenesis and endosomal sorting (28). Human ALIX was also suggested to play a role during viral infection and budding (29–31). Mammalian ALIX, yeast Bro1p, and their Arabidopsis homolog were all shown to interact with ESCRT-III via the charged multivesicular body protein 4/sucrose nonfermenting 7p (CHMP4/Snf7p) subunit (32–35). Bro1p was shown to interact also with the endosome-associated DUB degradation of alpha 4p (Doa4p) and to be essential for recruiting Doa4p to late endosomes (36). Doa4p belongs to the ubiquitin-specific protease (UBP) family of DUBs and is structurally unrelated to AMSH. The involvement of ALIX/Bro1p in the regulation of other DUBs during endosomal sorting has not yet been reported. Our data show that ALIX is essential in Arabidopsis and that it is important for the degradation of ubiquitinated proteins, vacuole, and MVB biogenesis, as well as for the localization of AMSH3 to endosomes.
Results
ALIX Can Directly Interact with AMSH3.
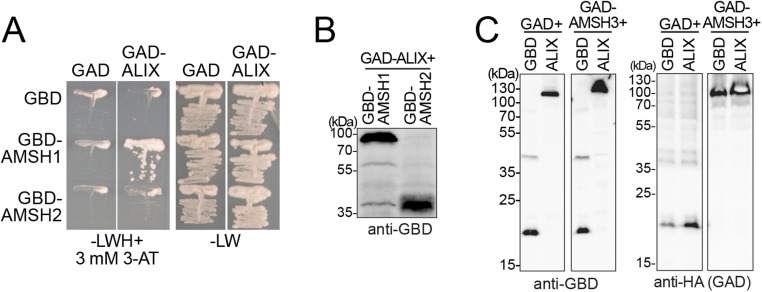
To identify interacting proteins or potential regulators of AMSH proteins, we carried out a yeast two-hybrid (YTH)-screen against 12,000 Arabidopsis ORFs, using AMSH1, AMSH2, and AMSH3 as bait. In this screen, we identified a Bro1 domain containing protein (AT1G15130) as a YTH interactor of AMSH1 and AMSH3, but not AMSH2 (Fig. 1A and Fig. S1 A–C). The identified protein showed highest homology to the yeast Bro1 domain containing proteins Bro1p (15.8% amino acid identities) and regulator of IME2 20 (Rim20p, 18%), as well as human ALIX (25.1%). We thus designated this protein Arabidopsis ALIX.
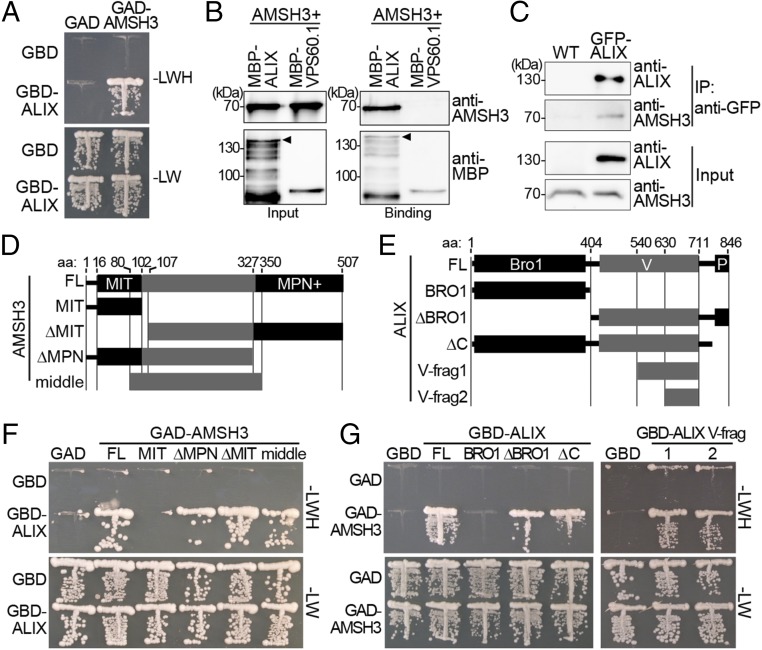
Fig. 1.
(A) ALIX interacts with AMSH3 in YTH. GBD and GBD-ALIX were transformed in yeast with either GAD- or GAD-AMSH3. Transformants were tested for their auxotrophic growth on synthetic complete medium lacking leucine and tryptophan (SC-LW) (-LW: SC-Leu-Trp) or synthetic complete medium lacking leucine, tryptophan, and histidine (SC-LWH) (-LWH: SC-Leu-Trp-His) media. (B) ALIX interacts directly with AMSH3 in vitro. Recombinant AMSH3 was incubated with either MBP-ALIX or MBP-VPS60.1 for 1 h at 4 °C and subjected to immunoblot analysis after extensive washing. Anti-AMSH3 and anti-MBP antibodies were used to detect beads-retained material. Arrowheads indicate MBP-ALIX. (C) ALIX and AMSH3 interact in planta. Immunoprecipitation (IP) was performed from total extracts of wild-type or GFP-ALIX-expressing seedlings using anti-GFP immobilized matrix. Immunoprecipitated material was subjected to immunoblot analysis. GFP-ALIX and endogenous AMSH3 were detected with anti-ALIX and anti-AMSH3 antibodies, respectively. (D and E) AMSH3- (D) and ALIX- (E) constructs used for YTH interaction studies in F and G. Bro1, Bro1-domain; FL, full-length; MIT, MIT domain; MPN+, MPN+ domain; P, Proline-rich domain; V, V-domain; V-frag1/2, V-domain fragment 1/2. (F) YTH analysis of AMSH3 domains responsible for the interaction with ALIX. GAD-fused AMSH3 constructs shown in D were cotransformed with either GBD or GBD-ALIX. Transformants were tested for their auxotrophic growth on SC-LW and SC-LWH media. (G) YTH analysis of ALIX domains responsible for the interaction with AMSH3. GBD-fused ALIX constructs shown in E were cotransformed with either GAD or GAD-AMSH3. Transformants were tested for their auxotrophic growth on SC-LW and SC-LWH media.
Fig. S1.
(A) ALIX interacts with AMSH1 in a YTH assay. Growth of representative colonies on SC-LW (-LW: SC-Leu-Trp) and SC-LWH (-LWH: SC-Leu-Trp-His) media supplemented with 3 mM 3-AT are shown. (B) Expression of the YTH clones shown in A. Yeast total extracts were prepared from transformants shown in A, and expression of GBD-AMSH1 and GBD-AMSH2 was verified using an anti-GBD antibody. (C) Expression of the YTH clones shown in Fig. 1A. Yeast total extracts were prepared from transformants shown in Fig. 1A and subjected to immunoblotting, using an anti-GBD antibody to detect GBD-fusion proteins and an anti-HA antibody to detect the GAD-HA-fusion proteins.
We first wanted to establish whether ALIX directly binds to AMSH proteins. Because we could not obtain enough recombinant AMSH1, we decided to focus our further studies on AMSH3. To analyze the interaction between ALIX and AMSH3, we purified recombinant maltose binding protein (MBP)-fused ALIX, an ESCRT-III subunit MBP-vacuolar protein sorting (VPS)60.1 that was shown not to interact with AMSH3 (26), and untagged AMSH3. In an in vitro binding assay, MBP-ALIX, but not MBP-VPS60.1, bound AMSH3 (Fig. 1B), indicating that ALIX and AMSH3 can directly interact with each other. Next, to test whether ALIX and AMSH3 interact in planta, we generated plants expressing a functional fusion protein GFP-ALIX driven by a native ALIX promoter fragment (Fig. S2 A and B). Immunoprecipitation of GFP-ALIX from total protein extracts prepared from wild-type or GFP-ALIX-expressing seedlings and subsequent immunoblotting revealed coimmunoprecipitation of endogenous AMSH3 with GFP-ALIX (Fig. 1C). This result indicates that AMSH3 and ALIX can interact in planta.
Fig. S2.

Complementation of alix-2 with ALIXpro:GFP-ALIX. (A) ALIXpro:GFP-ALIX complements seedling lethality of alix-2. Wild-type (WT), heterozygous alix-2 (hetero), and homozygous alix-2 plants expressing GFP-ALIX are shown. (Scale bar, 5 cm.) (B) Immunoblot analysis of total extracts from wild-type (WT), alix-2 with and without the ALIXpro:GFP-ALIX construct with an anti-ALIX antibody. Positions of endogenous ALIX and GFP-ALIX are indicated on the right of the panel. CDC2 was used as loading control.
To identify the binding domain of AMSH3 and ALIX, we conducted a directed YTH analysis. AMSH3 has an N-terminal microtubule interacting and trafficking (MIT) domain that interacts with two ESCRT-III subunits, 2.1 and VPS24.1 (26), and an MPN+ domain that comprises the catalytic center (20). Neither the MIT nor the MPN+ domain of AMSH3 was required for the interaction with ALIX, whereas the middle region was necessary and sufficient for the interaction (Fig. 1 D and F and Fig. S3A). A Pfam database search (37) showed that ALIX has an N-terminal Bro1 domain followed by a V-shaped domain (V-domain) and a C-terminal proline-rich domain (Fig. 1E). To identify the region in ALIX that is responsible for the binding to AMSH3, YTH interactions between different ALIX fragments and the full-length AMSH3 were tested. Both the Bro1- and the C-terminal region were dispensable for the interaction with AMSH3, whereas an 82-amino acid fragment of the ALIX V-domain was sufficient for the interaction (Fig. 1 E and G and Fig. S3 B and C).
Fig. S3.
Immunoblot analysis of YTH clones. (A–C) Expression of the YTH clones shown in Fig. 1. Yeast total extracts were prepared from transformants shown in Fig. 1F (A), Fig. 1G, Left (B), and Fig. 1G Right (C) and subjected to immunoblotting using an anti-GBD antibody to detect GBD-fusion proteins and an anti-HA antibody to detect the GAD-HA-fusion proteins.
alix Null Mutants Show Similar Phenotypes to amsh3.
AMSH proteins were previously shown to be essential for plant growth and development (20, 27). To investigate whether ALIX functions in the same biological pathway as AMSH3, we analyzed available T-DNA insertion lines of ALIX. Two lines, GABI 837H11 and SALK 063124, which we named alix-2 and alix-4, respectively, carried T-DNA insertions in exons (Fig. 2A). Homozygous alix-2 and alix-4 mutants showed reduced germination compared with the wild-type (Table 1), and even if germinated, did not develop beyond the seedling stage (Table 1 and Fig. 2B). This seedling lethal phenotype was very similar to the previously analyzed amsh3 mutant (Fig. S4A) (20), suggesting AMSH3 and ALIX could function in the same biological process. Immunoblotting with an anti-ALIX antibody showed that homozygous alix-2 and alix-4 mutants were null mutants (Fig. 2C).
Fig. 2.
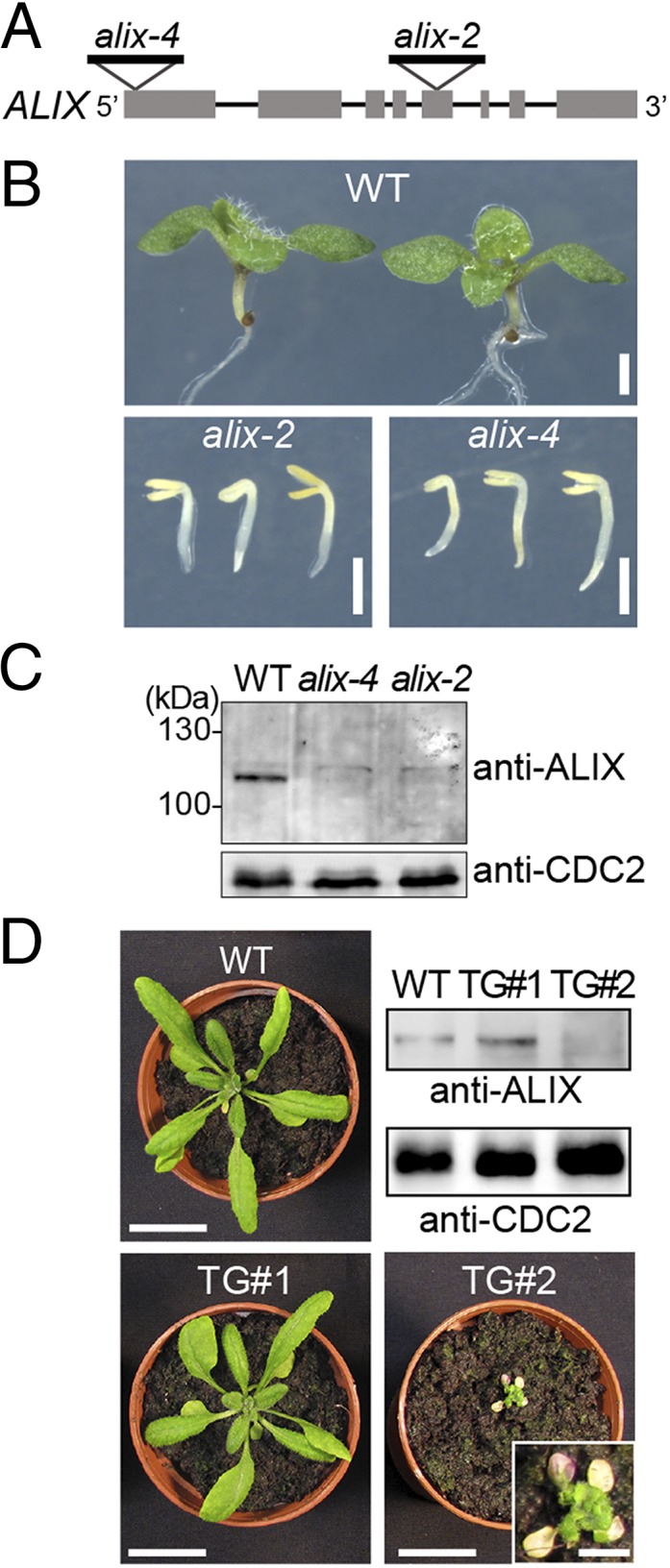
alix null mutants show seedling lethality. (A) Schematic presentation of the T-DNA insertion sites of alix-2 and alix-4. Lines indicate introns and gray boxes indicate exons. (B) Phenotypes of 9-d-old wild-type (WT), alix-2, and alix-4 seedlings. (Scale bars, 1 mm.) (C) alix-2 and alix-4 are null mutants. Total extracts from seedlings shown in B were subjected to immunoblot analysis, using an anti-ALIX antibody. CDC2 was used as loading control. (D) Photographs of 3-wk-old wild-type (WT) and two transgenic plants (TG#1 and TG#2) harboring 35Spro:ALIX. Total extracts from rosetta leaves were subjected to immunoblot analysis with an anti-ALIX antibody. CDC2 was used as loading control. Note that TG#2 with strongly reduced levels of ALIX (magnification in inset) shows severe growth defects and leaf chlorosis. (Scale bars, 2 cm; scale bar in inset, 0.5 cm.)
Table 1.
Germination assay of alix mutants
| Genotype of parental line | Phenotype of progeny | n | |||
| Wild type | Seedling lethal | Ungerminated | Seedling lethal+ ungerminated | ||
| WT (Columbia-0) | 100% | 0% | 0% | 0% | 424 |
| alix-2+/− (het) | 83.2% | 11.9% | 4.9% | 16.8% | 405 |
| alix-4+/− (het) | 77% | 9.5% | 13.5% | 23% | 400 |
Fig. S4.
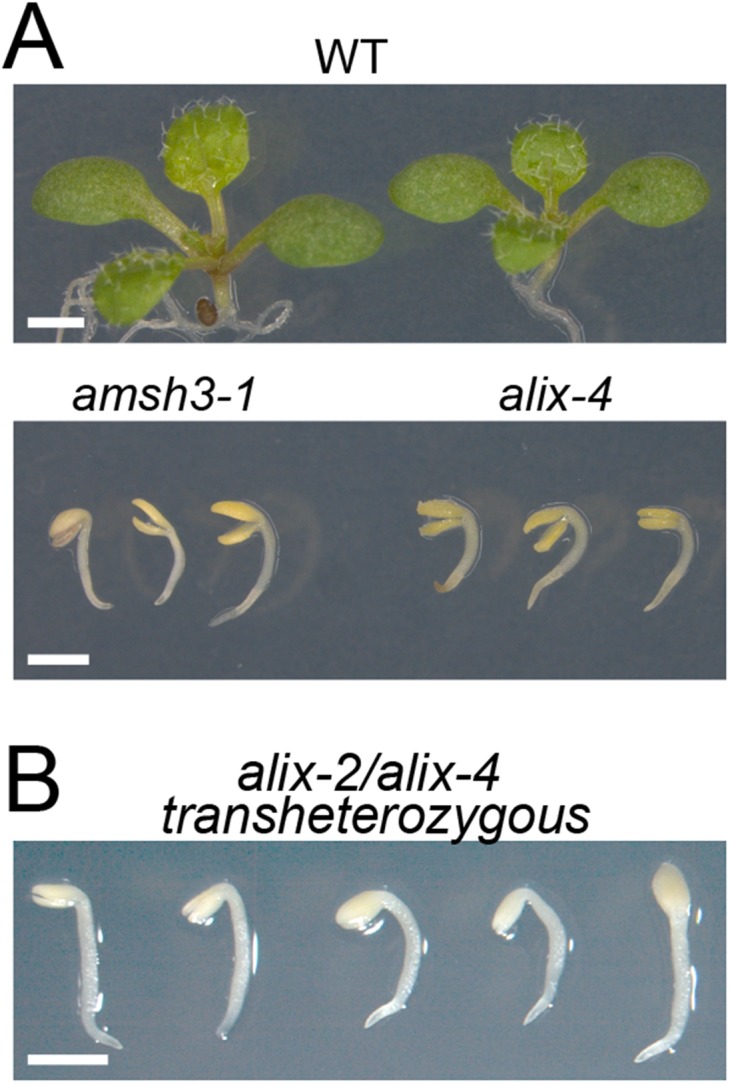
alix-2 and alix-4 are allelic to each other and resemble amsh3. (A) Comparison of the amsh3- and alix mutant phenotypes. Nine-day-old wild type (WT), amsh3-1, and alix-4 mutants are shown. (Scale bars, 1 cm.) (B) Phenotypes of alix-2/alix-4 transheterozygous seedlings. (Scale bar, 1 mm.)
To verify that the phenotype is caused by the loss of ALIX function, we crossed heterozygous alix-2 and alix-4 plants with each other and analyzed the F1 progeny. Transheterozygous (alix-2/alix-4) F1 progenies showed the same seedling phenotype as homozygous alix-2 or alix-4 mutants (Fig. S4B), indicating that alix-2 is allelic to alix-4. Moreover, seedling lethality of alix-2 could be complemented by ALIXpro:GFP-ALIX (Fig. S2 A and B), confirming that ALIX is indeed the causative gene for the alix mutant phenotype.
To investigate whether ALIX is also essential during plant growth, we tried to generate artificial microRNA lines, but were not able to identify lines with significantly reduced ALIX transcripts. We realized, however, that the introduction of the 35Spro:ALIX construct in a wild-type background leads to reduced protein levels of ALIX, probably because of cosuppression. Plants harboring the 35Spro:ALIX construct showed a range of phenotypes, but the severely growth-defective plants with chlorotic leaves had always reduced amount of ALIX in total protein extracts, whereas larger plants had at least wild-type levels of ALIX (Fig. 2D). These results suggest ALIX is indispensable during vegetative plant growth.
ALIX Is Required for Vacuole Biogenesis.
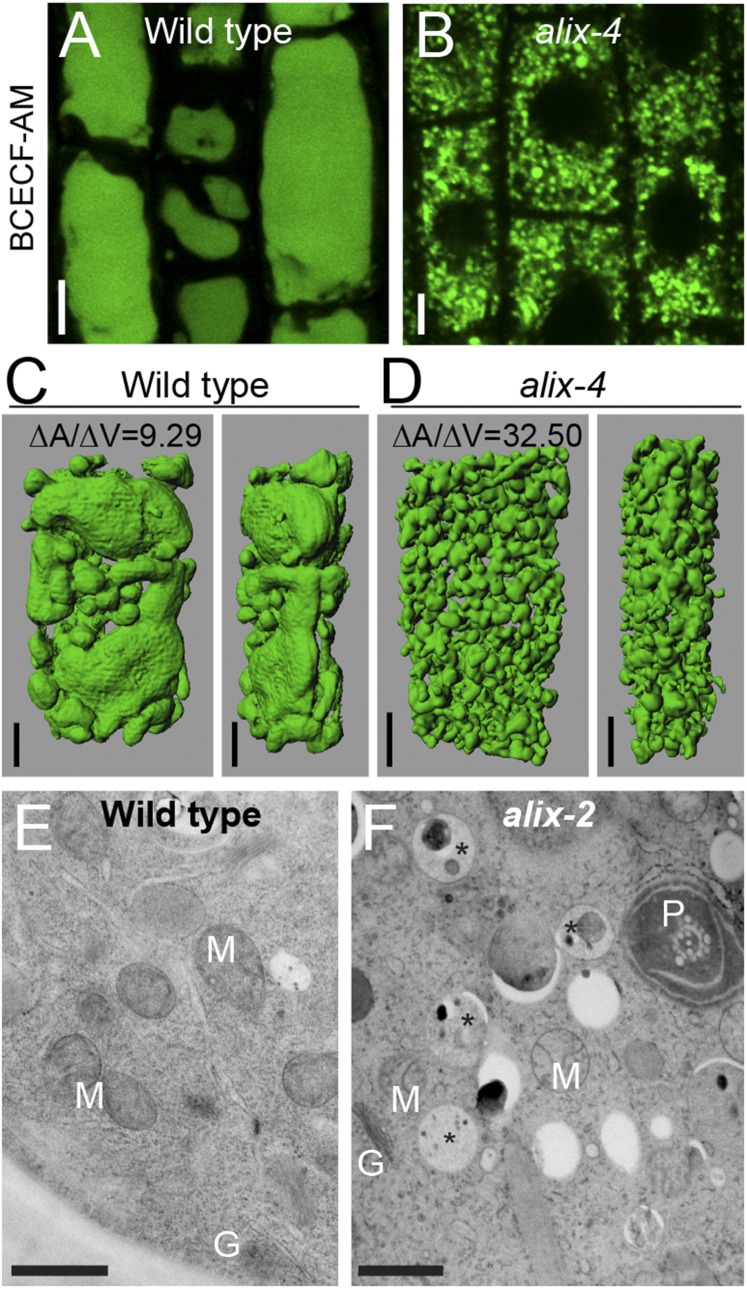
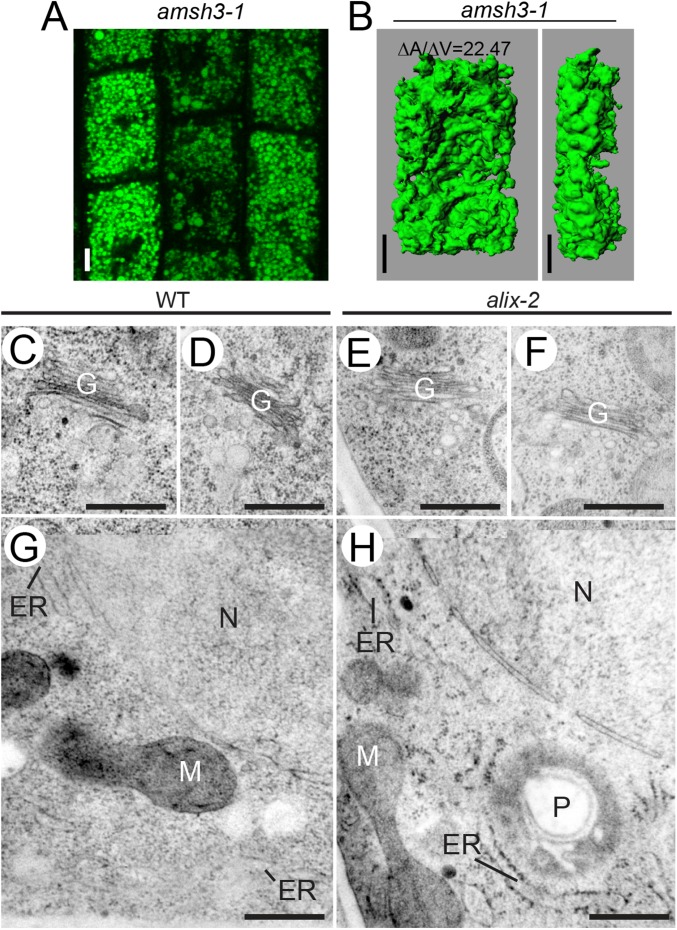
amsh3 mutants showed severe vacuole biogenesis defects and lacked large central vacuoles (20). Confocal microscope analysis of 2′,7′-bis-(2-carboxyethyl)-5-(and-6)- carboxyfluorescein (BCECF)-acetoxymethyl ester (AM)-stained wild-type, alix-4, and amsh3-1 vacuoles revealed that alix mutants also have abnormal vacuole morphology similar to amsh3-1, indicating that ALIX is necessary for proper vacuole biogenesis (Fig. 3 A and B and Fig. S5A). To analyze the vacuolar structure in more detail, we performed 3D reconstruction analysis on Z-stack images of BCECF-AM-stained wild-type, alix, and amsh3 vacuoles. Surface rendering revealed similar higher surface area-to-volume ratios for alix-4 and amsh3-1 in comparison with the wild type, reflecting the tubular interconnected vacuolar structures in these mutants (Fig. 3 C and D and Fig. S5B). In addition, the presence of 1 µm vacuolar or provacuolar profiles was confirmed by electron microscopy of high-pressure frozen/freeze substituted roots. These vacuolar structures contained membrane and cytoplasmic materials in their lumen and were abundant in alix root cells, but not in the wild-type (Fig. 3 E and F). In contrast, the morphology of other organelles such as the Golgi apparatus, endoplasmic reticulum (ER), nucleus, plastids, and mitochondria did not seem to be affected in alix-2 (Fig. 3 E and F and Fig. S5 C–H).
Fig. 3.
alix shows aberrant vacuole morphology. (A and B) Vacuole morphology of wild-type and alix mutants. Vacuoles of 2-d-old wild-type (A) and 7-d-old alix-4 (B) were stained with BCECF-AM and analyzed under a confocal microscope. Vacuoles in the root epidermis cells are shown. (Scale bars, 5 µm.) (C and D) 3D surface renderings of vacuoles from a representative root epidermal cell of 2-d-old wild-type (C) and 7-d-old alix-4 (D) seedlings. Z-stack images were processed to generate 3D reconstruction pictures of vacuoles. Views from the front (Left) and the side (Right) are shown. Note the tubular appearance of mutant vacuoles. (Scale bars, 5 µm.) ∆A/∆V: surface area to volume ratio. (E and F) Overview of WT (E) and alix-2 (F) root cells from 5-d-old seedlings. Note the presence of vacuolar/provacuolar compartments with membranes and cytoplasmic contents (asterisks). G, Golgi; M, mitochondria; P, plastid. (Scale bars, 1 µm.)
Fig. S5.
Vacuole morphology in amsh3-1 and organelle morphology in alix-2. (A) Vacuole morphology of amsh3-1. Vacuoles in 7-d-old amsh3-1 were stained with BCECF-AM and analyzed under a confocal microscope. (Scale bar, 5 µm.) (B) Z-stacks were processed to generate 3D reconstruction pictures of vacuoles in the root epidermis cell. Views from the front (Left) and the side (Right) are shown. (Scale bars, 5 µm.) (C–H) Morphology of organelles in alix-2 mutant cells. Morphology of the Golgi-apparatus in wild-type (C and D) and in the alix-2 (E and F), as well as the morphology of ER and mitochondria in the wild-type (G) and alix-2 (H), is indistinguishable. ER, endoplasmic reticulum; G, Golgi-apparatus; M, mitochondria; N, nucleus; P, plastid. (Scale bars, 500 nm.)
AMSH3 Shows Colocalization with Late Endosomal Markers.
Human ALIX and yeast Bro1p were reported to localize on endosomes and to be involved in intracellular trafficking (28). To investigate whether AMSH3 and ALIX localize on the same cellular compartment and function in intracellular transport, we expressed an AMSH3pro:AMSH3-YFP construct in plants. AMSH3pro:AMSH3-YFP complemented the seedling lethal phenotype of amsh3-1, indicating that AMSH3-YFP is a functional fusion protein (Fig. S6 A and B).
Fig. S6.
Complementation of amsh3-1 with AMSH3pro:AMSH3-YFP. (A) AMSH3pro:AMSH3-YFP complements seedling lethality of amsh3-1. Two independent transgenic lines (#1 and #2) along with wild-type (WT) plants are shown. (B) Expression of AMSH3-YFP in the transgenic lines and wild-type plants shown in A was verified by immunoblotting using an anti-AMSH3 antibody. Positions of the endogenous AMSH3 and AMSH3-YFP are indicated on the right of the panel, and the asterisk indicates an unspecific band that serves as loading control.
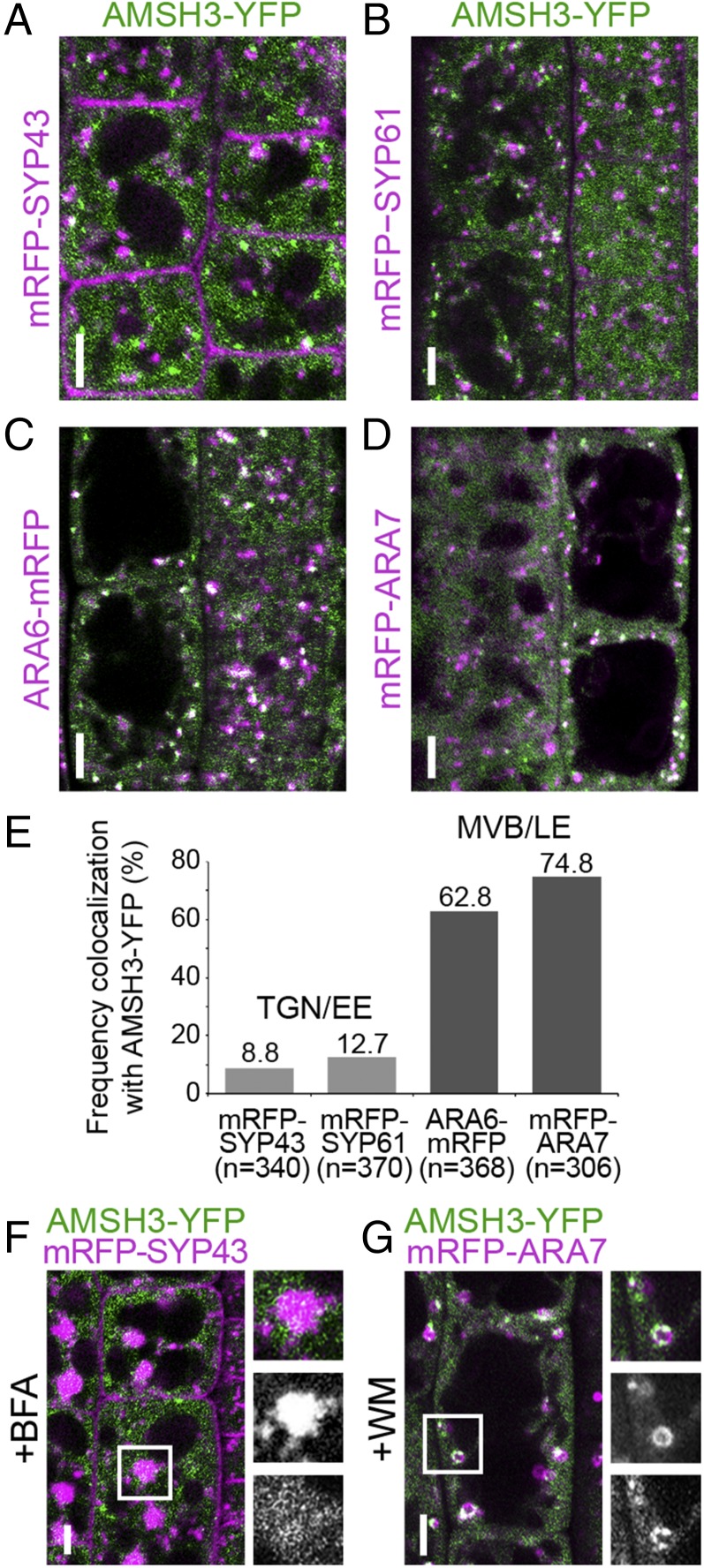
AMSH3-YFP showed strong cytosolic signals similar to the previously published N-terminal YFP fusion YFP-AMSH3 (26), but localized also on intracellular foci. To investigate the nature of these foci, we performed colocalization studies using trans-Golgi network (TGN)/early endosome (EE) markers monomeric red fluorescent protein (mRFP)-syntaxin of plants 43 (SYP43) and mRFP-SYP61 (38, 39), as well as late endosome (LE) markers arabidopsis rat sarcoma-related proteins in brain GTPase6 (ARA6)-mRFP and mRFP-ARA7 (40, 41). AMSH3-YFP signals did not show frequent colocalization with the TGN/EE markers (Fig. 4 A, B, and E). In contrast, 62.8% (n = 368) and 74.8% (n = 306) of AMSH3-YFP-positive vesicles showed colocalization with ARA6- and ARA7-labeled vesicles, respectively, indicating that the AMSH3-YFP foci represent mostly late endosomes (Fig. 4 C, D, and E).
Fig. 4.
AMSH3 localizes to ARA6- and ARA7-labeled late endosomes. (A to D) Seedlings expressing AMSH3pro:AMSH3-YFP were crossed with SYP43pro:mRFP-SYP43 (A), SYP61pro:mRFP-SYP61 (B), ARA6pro:ARA6-mRFP (C), and ARA7pro:mRFP-ARA7 (D) expressing plants and colocalization was examined under a confocal microscope. (Scale bars, 5 µm.) SYP43 and SYP61 are markers for TGN or EE, and ARA6 and ARA7 are markers for MVB or LE. (E) Numbers of AMSH3-YFP-positive vesicles that colocalize with the mRFP-tagged markers in A–D were counted to calculate the colocalization frequency. (F) An AMSH3-YFP and mRFP-SYP43 expressing seedling as in A was treated with 50 µM brefeldin A (BFA) for 60 min. Note that although mRFP-SYP43 relocalized to the BFA bodies, AMSH3-YFP did not. A magnification of the area indicated with the white rectangle is shown on the right side (from top to bottom: merged, mRFP-ARA7, and AMSH3-YFP). (Scale bars, 5 μm.) (G) An AMSH3-YFP and mRFP-ARA7 expressing seedling as in D was treated with 33 µM WM for 120 min. Both AMSH3-YFP and mRFP-ARA7 localize to WM-induced compartments. A magnification of the area indicated with the white rectangle is shown on the right side (from top to bottom: merged, mRFP-ARA7, and AMSH3-YFP). (Scale bars, 5 μm.)
To further establish the endosome localization of AMSH3, we treated the AMSH3-YFP/mRFP-SYP43 coexpressing line with the ADP ribosylation factor-GTP exchange factor (ARF-GEF) inhibitor brefeldin A (BFA) and the AMSH3-YFP/mRFP-ARA7 coexpressing line with the PI3K/PI4K inhibitor wortmannin (WM). Although mRFP-SYP43 localized to the so-called BFA bodies on 60-min BFA treatment, AMSH3-YFP signals were not found in the BFA-induced compartments (Fig. 4F). On treatment with WM, both mRFP-ARA7 and AMSH3-YFP localized to the surface of the swollen MVBs that appeared as ring-like structures (Fig. 4G). These results corroborate that AMSH3-YFP does not stably associate with TGN/EE but localizes together with LE markers on vesicles sensitive to WM.
AMSH3 and ALIX Colocalize on Late Endosomes.
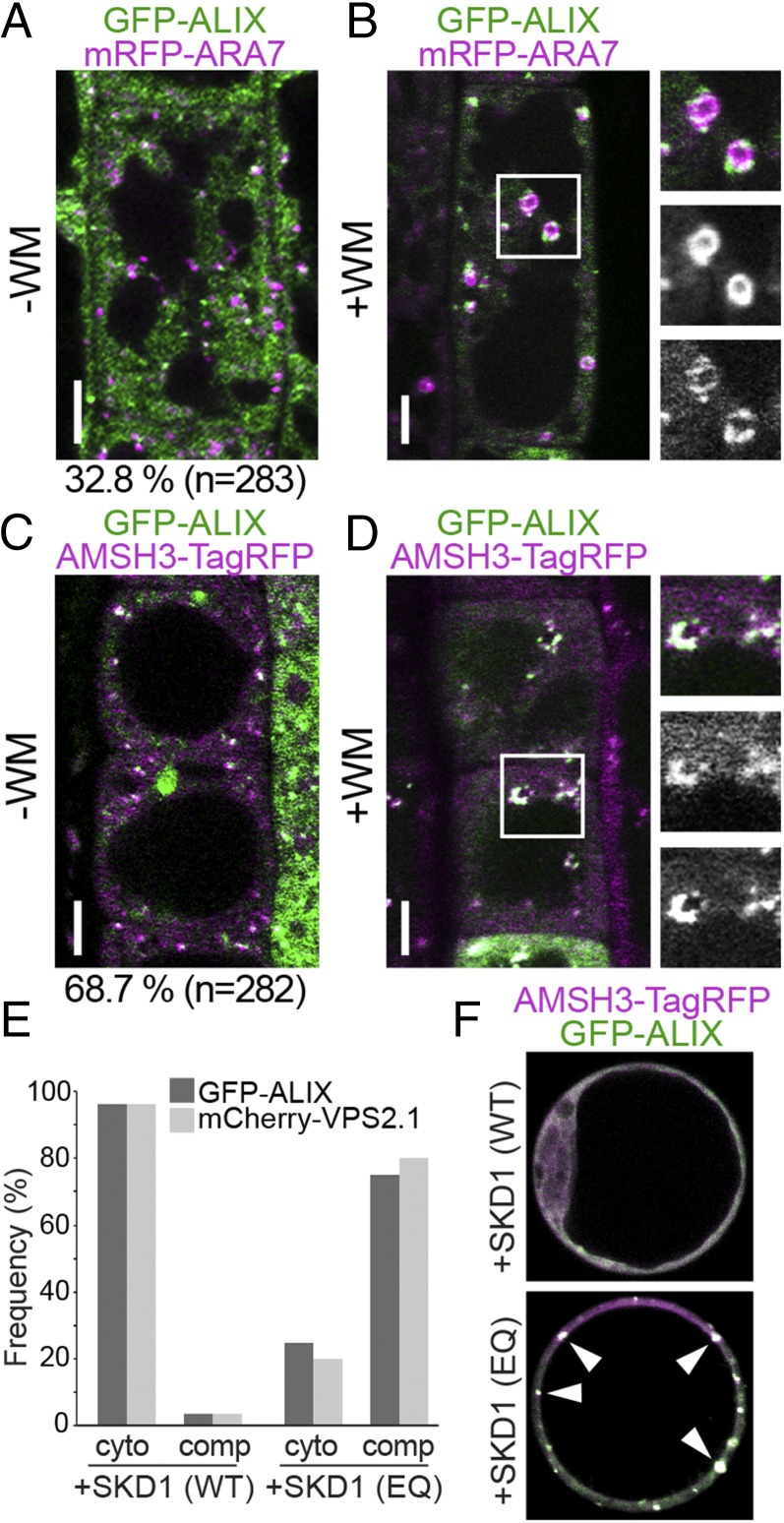
If ALIX functions together with AMSH3, they should show overlapping intracellular localizations. We thus examined the localization of GFP-ALIX with the late endosome marker mRFP-ARA7. Similar to AMSH3-YFP, GFP-ALIX signals showed cytosolic distribution but also localized on punctuate structures: 32.8% ALIX-positive vesicles (n = 283) colocalized with the LE-marker ARA7 (Fig. 5A), indicating that part of the ALIX pool localizes to late endosomes. Similar to AMSH3-YFP, GFP-ALIX relocalized to WM-induced compartments together with mRFP-ARA7 (Fig. 5B). Similar to AMSH3-YFP, GFP-ALIX-labeled vesicles did not efficiently colocalize with the TGN/EE-endosome marker mRFP-SYP43 (9.96%, n = 280) and did not react to BFA-treatment (Fig. S7 A and B). We next wanted to investigate whether AMSH3 and ALIX localize to the same cellular compartment and generated an RFP-fused AMSH3 construct, AMSH3pro:AMSH3-TagRFP. Similar to the YFP-tagged AMSH3, AMSH3-TagRFP showed both cytosolic and endosomal localization: 68.7% (n = 282) of AMSH3-positive compartments showed colocalization with GFP-ALIX (Fig. 5C). Compartments positive for both GFP-ALIX and AMSH3-TagRFP were sensitive to WM treatment (Fig. 5D), suggesting both proteins colocalize on late endosomes.
Fig. 5.
AMSH3 and ALIX colocalize on late endosomes. (A and B) ALIX localizes to ARA7-labeled late endosomes. Seedlings expressing ALIXpro:GFP-ALIX and ARA7pro:mRFP-ARA7 were examined with a confocal microscope without (A) or with (B) 120 min treatment with 33 µM WM. Percentage of GFP-ALIX vesicles colocalizing with mRFP-ARA7 vesicles is indicated below the panel in A. (Scale bars, 5 µm.) (C and D) Seedlings coexpressing ALIXpro:GFP-ALIX and AMSH3pro:AMSH3-TagRFP were analyzed without (C) or with 120 min treatment with 33 µM WM. (D) Percentage of GFP-ALIX vesicles colocalizing with AMSH3-TagRFP vesicles is indicated below the panel in C. (Scale bars, 5 µm.) (E) ALIXpro:GFP-ALIX was cotransformed with UBQ10pro:mCherry-VPS2.1 and either 35Spro:HA-SKD1(WT) or 35Spro:HA-SKD1(EQ) in Arabidopsis cell culture-derived protoplasts. Percentages of cells with cytosolic localization (cyto) or SKD1(EQ)-induced compartments (comp) of GFP-ALIX and mCherry-VPS2.1 are indicated. (F) AMSH3 and ALIX localize to SKD(EQ)-induced compartments. ALIXpro:GFP-ALIX and 35Spro:AMSH3-TagRFP were cotransformed with either 35Spro:HA-SKD1(WT) or 35Spro:HA-SKD1(EQ) in Arabidopsis cell culture-derived protoplasts. Representative cells are shown. Note that on coexpression of SKD1(EQ), both AMSH3-TagRFP and GFP-ALIX relocalize to enlarged endosomal compartments (arrowheads).
Fig. S7.
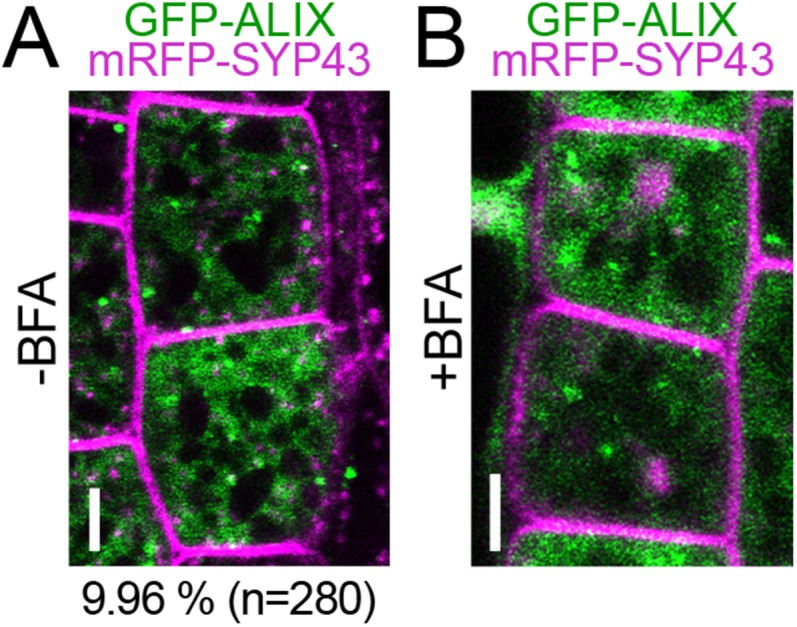
GFP-ALIX does not colocalize with the early endosome marker mRFP-SYP43. (A and B) Seedlings expressing ALIXpro:GFP-ALIX and SYP43pro:mRFP-SYP43 were analyzed with a confocal microscope before (A) or 60 min after (B) incubation with 50 µM brefeldin A (BFA). Percentage of GFP-ALIX vesicles colocalizing with mRFP-SYP43 vesicles is indicated below the panel in A. (Scale bars: 5 µm.)
A systematic YTH analysis of Arabidopsis ESCRT-related proteins has shown that ALIX interacts with the ESCRT-III subunit SNF7 (35). ESCRT-III components accumulate in aberrant late endosomal structures on inhibition of ESCRT-III disassembly by inactivation of the ATPases associated with diverse cellular activities (AAA)-ATPase suppressor of K+ transport growth defect 1(SKD1)/Vps4p (26, 41, 42). In Arabidopsis, the overexpression of ATPase-inactive SKD1(EQ) was shown to induce aggregation of endosomes (26, 41) that were labeled with the late endosome marker ARA7, but not those labeled with the early endosome marker SYP41 or clathrin light chain (43). In accordance with the previously reported interaction with SNF7 (35) and its partially late-endosomal localization, GFP-ALIX accumulated in the SKD1(EQ)-induced enlarged vesicular compartments together with the ESCRT-III subunit VPS2.1 and AMSH3-TagRFP (Fig. 5 E and F). Altogether these results show that ALIX and AMSH3 colocalize on late endosomal compartments that are sensitive to WM and SKD1 ATPase activity.
ALIX Binds Ubiquitin and Is Involved in Ubiquitin-Dependent Protein Degradation.
AMSH proteins were implicated in the degradation of ubiquitinated membrane proteins in both mammals and plants (22, 27). We reasoned that if ALIX functions in the same pathway as AMSH3, alix mutants would also show similar defects in the degradation of ubiquitinated cargos. Indeed, when we analyzed total extracts from seedlings using an anti-ubiquitin antibody, alix mutants showed high levels of high-molecular weight ubiquitinated proteins (Fig. 6A), suggesting ALIX plays a role in the removal of ubiquitin conjugates.
Fig. 6.
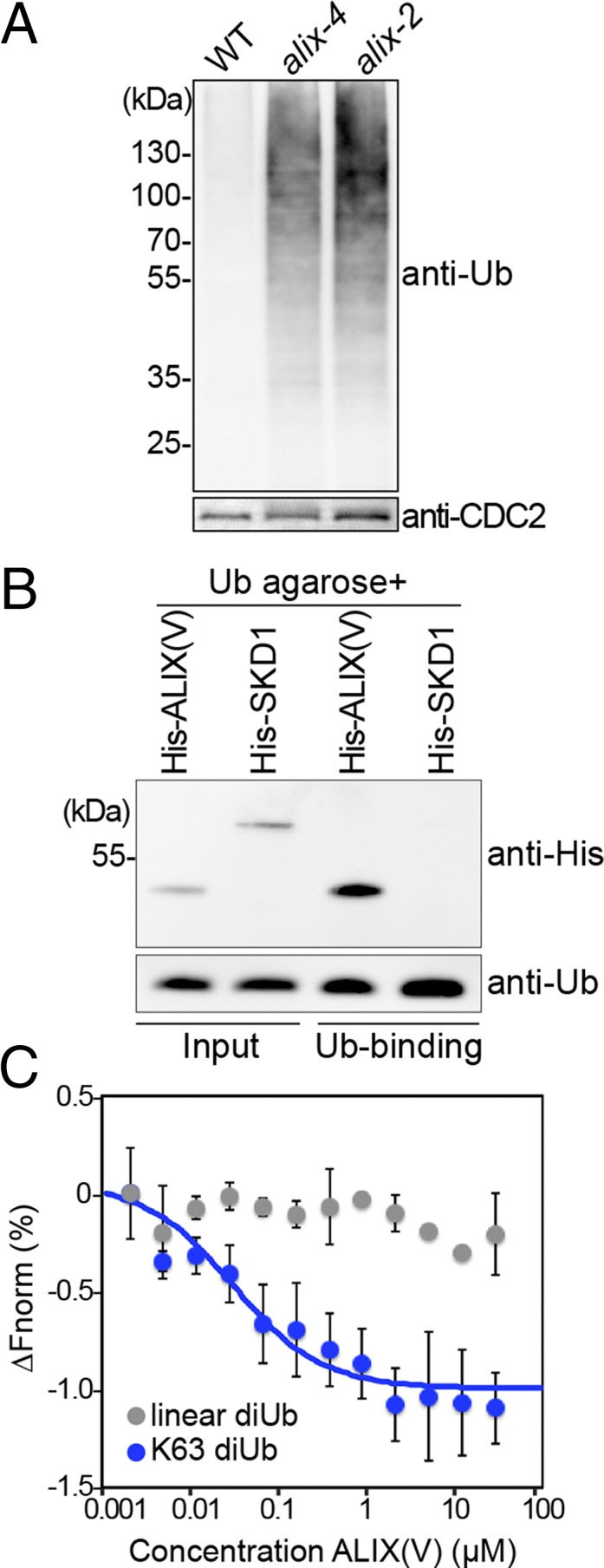
ALIX binds ubiquitin. (A) alix mutants accumulate ubiquitin conjugates. Total extracts of wild-type (WT), alix-2, and alix-4 were subjected to immunoblot analysis using an anti-ubiquitin antibody. CDC2 was used as loading control. (B) ALIX binds monoubiquitin. A recombinant His-ALIX fragment containing the V-domain (His-ALIX(V): amino acids 393–734), and His-SKD1 were incubated with ubiquitin agarose. After extensive washing, the eluate was analyzed by immunoblotting using an anti-His tag and an anti-Ub antibody. (C) MicroScale Thermophoresis assays show that recombinant His-ALIX(V) binds K63-linked diubiquitin, but not linear diubiquitin. The average of three experiments are shown. ∆Fnorm, normalized fluorescence. Blue, K63-lilnked diubiquitin; gray, linear diubiquitin. Error bars, SEM.
The V-domain of both Bro1p and human ALIX was shown to mediate the interaction with ubiquitin in intracellular trafficking (29, 31, 44). Because the amino acid sequence homology in the Arabidopsis ALIX V-domain is relatively moderate (25% amino acid identity with human ALIX and 17% with Bro1p), we tested whether it could bind ubiquitin. In a ubiquitin binding assay using ubiquitin agarose, recombinant His-ALIX(V) interacted with ubiquitin, whereas the negative control His-SKD1 did not (Fig. 6B), suggesting the ubiquitin-binding ability of the ALIX V-domain is conserved in Arabidopsis. Because K63-linked ubiquitin chains are implicated in endocytic degradation, we tested whether His-ALIX(V) can interact with this type of linkage. We analyzed the interaction of His-ALIX(V) with linear and K63-linked diubiquitin, using the MicroScale Thermophoresis technique that enables affinity measurements without the immobilization of either of the proteins (45). The results showed that His-ALIX(V) bound K63-linked diubiquitin, but not linear diubiquitin, implying that ALIX is involved in pathways mediated by K63-ubiquitin linkages (Fig. 6C).
DUB-interacting proteins can affect the activity of DUBs, as shown for the case of human AMSH and its binding partner STAM, which activates AMSH (46, 47). To establish whether ALIX can influence the enzymatic activity of AMSH3, we performed DUB assays with AMSH3 in the presence or absence of ALIX, using the fluorescent substrate diubiquitin-carboxytetramethylrhodamine (TAMRA). The cleavage of diubiquitin to monoubiquitin separates the quencher from the fluorophore, which can be measured as an increase in fluorescence. AMSH3 readily catalyzed this reaction; however, preincubation with equimolar amount of ALIX did not change the AMSH3 enzyme kinetics (Fig. S8), suggesting ALIX does not influence the DUB activity of AMSH3.
Fig. S8.
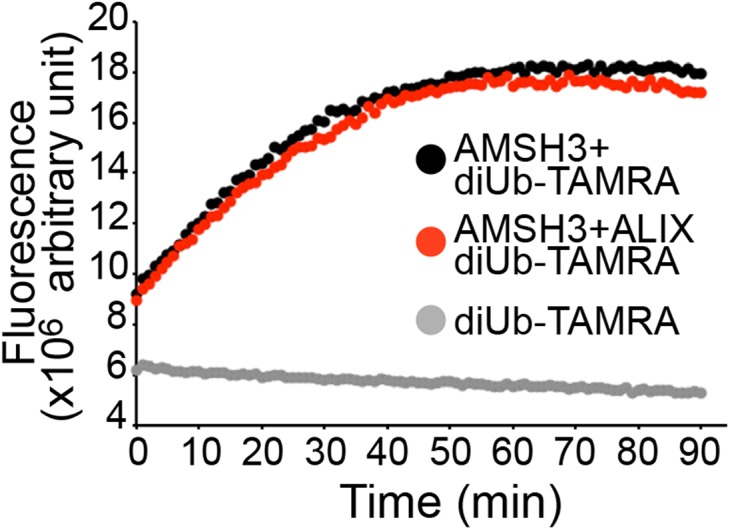
ALIX does not affect AMSH3 DUB activity. DUB assay with K63-linked diubiquitin-TAMRA. Recombinant AMSH3 was incubated with diubiquitin-TAMRA with or without preincubation with an equimolar amount of ALIX. Fluorescence was measured every minute. Black, diubiquitin-TAMRA with AMSH3; Red, diubiquitin-TAMRA with AMSH3 and ALIX; Gray, diubiquitin-TAMRA only.
ALIX Is Important for Localization of AMSH3 on ARA7-Labeled Vesicles.
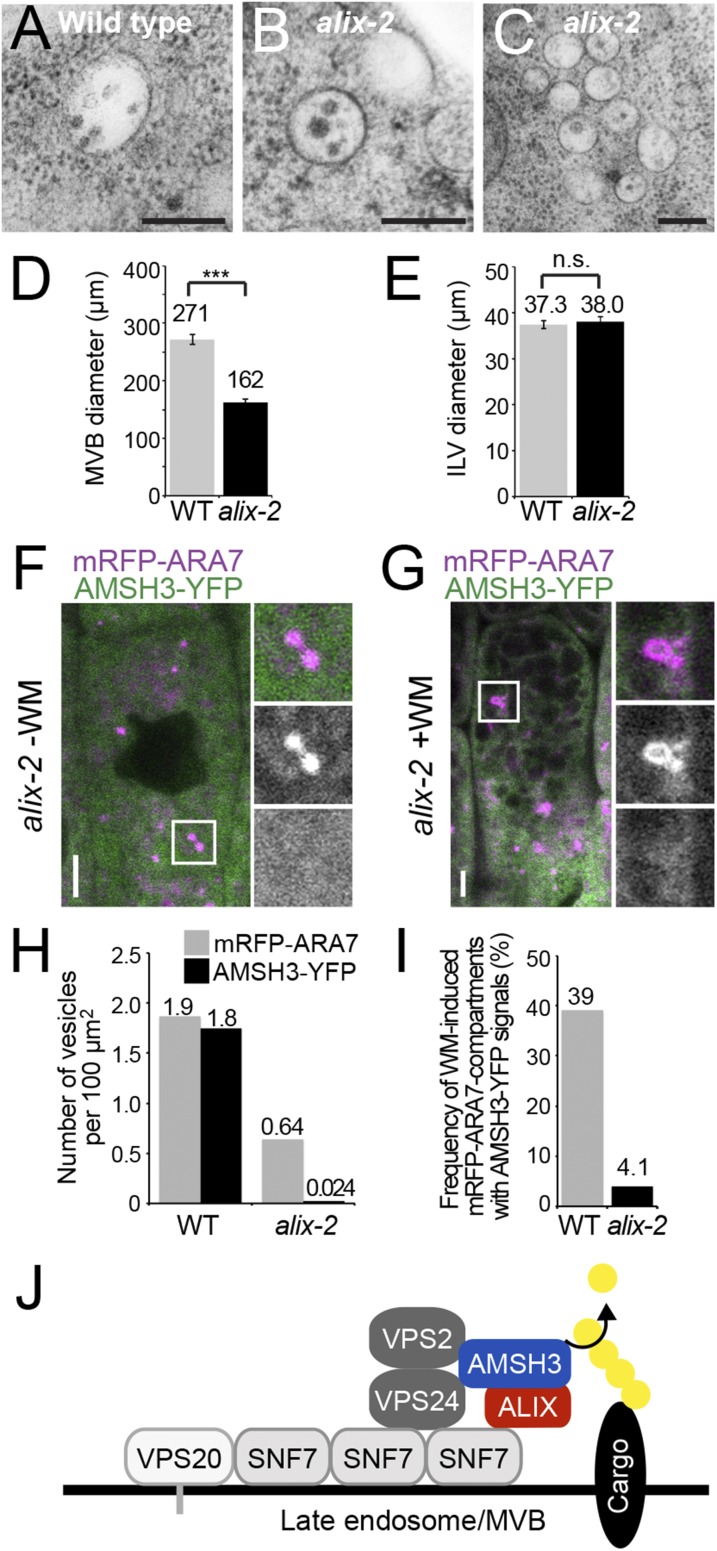
To investigate whether the ALIX null mutation affects MVB formation, we analyzed MVBs in wild-type and alix-2 roots by transmission electron microscopy. alix-2 mutants were able to form MVBs (Fig. 7 A and B); however, we also occasionally observed clustering of MVBs in alix-2 cells (Fig. 7C), implying a function of ALIX in MVB formation. The size of MVBs was significantly reduced in alix-2 cells compared with that of wild-type cells (Fig. 7D), supporting the idea that ALIX is involved in MVB formation. In contrast, there was no significant difference in the size of ILVs between wild-type and alix-2 (Fig. 7E).
Fig. 7.
AMSH3 does not localize on late ARA7-labeled vesicles in the alix mutant. (A and B) Electron micrographs of MVBs from wild-type (A) and alix-2 (B) cells. (Scale bars, 200 µm.) (C) Electron micrographs showing clustered MVBs in alix-2. (Scale bar, 200 µm.) (D) Diameter of MVBs in wild-type (light gray, n = 61) and alix-2 (dark gray, n = 50). Error bars, SEM. The difference between wild-type (WT) and alix-2 is highly significant, with P < 0.001 (***). (E) Diameter of ILVs in wild-type (WT; light gray, n = 28) and alix-2 (dark gray, n = 42). Error bars, SEM. The difference between WT and alix-2 is not significant (n.s.) (P > 0.05). (F and G) Localization of AMSH3-YFP and mRFP-ARA7 in root epidermis cells of alix-2 without (F) or with (G) 120 min WM treatment. Note that AMSH3-YFP localize neither on mRFP-ARA7-labeled vesicles nor on WM-enlarged late endosomes. Magnifications of the area indicated with white rectangles are shown on the right side (from top to bottom: merged, mRFP-ARA7, and AMSH3-YFP). (Scale bars, 5 µm.) (H) Number of mRFP-ARA7 and AMSH3-YFP vesicles per 100 µm2 in wild-type (WT) and alix-2. Note that reduction of the number of AMSH3-YFP vesicles is stronger than of mRFP-ARA7 vesicles in alix-2. (I) Number of WM-induced mRFP-ARA7 compartments containing AMSH3-YFP signals in wild-type (WT) and alix-2. (J) Predicted model for ALIX interaction with AMSH3 and ESCRT-III. AMSH3 associates with ESCRT-III subunits VPS2 and VPS24 through its MIT domain and with ALIX through the middle domain. ALIX binds to ESCRT-III component SNF7 via the BRO1 domain and to AMSH3 through the V-domain. ALIX does not influence AMSH3 DUB activity, but might function to stabilize the interaction of AMSH3 with ESCRT-III-positive MVBs.
Yeast Bro1p interacts with Doa4p, a UBP-class DUB unrelated to AMSH, and recruits it to endosomes (36, 48). To test whether loss of ALIX function affects association of AMSH3 to endosomes, we analyzed the localization of AMSH3-YFP together with the late-endosomal marker mRFP-ARA7 in alix-2. Whereas AMSH3-YFP and mRFP-ARA7 endosomes showed colocalization with high frequency in a wild-type background (Fig. 4 D and E), AMSH3-YFP signals were dispersed in the cytosol and did not colocalize with mRFP-ARA7 in alix-2 (Fig. 7F). The numbers of both mRFP-ARA7 and AMSH3-YFP-positive vesicles were decreased in the alix-2 mutant, among which AMSH-YFP signals were more affected (Fig. 7H). When treated with WM for 120 min, mRFP-ARA7 appeared in enlarged WM compartments, as it did in the wild type (Fig. 7G). In contrast, AMSH3-YFP showed reduced sensitivity toward WM and localized less frequently in ARA7-labeled WM compartments (structures with a diameter larger than 1.5 µm) in the alix-2 mutant background (Fig. 7 G and I). Together, these results indicate that the localization of AMSH3-YFP to mRFP-ARA7-labeled late endosomes requires intact ALIX function in planta (Fig. 7J).
Discussion
Our data show that Arabidopsis ALIX interacts directly with the late-endosome localized metalloprotease DUB AMSH3 and is important for its localization to late endosomes. Mutant analyses support a functional relationship between ALIX and AMSH3, as phenotypic alterations of amsh3 and alix mutants are strikingly similar in regard to seedling lethality, vacuole morphology, and accumulation of ubiquitinated proteins.
Class-E vps mutants in budding yeast, to which mutants of ESCRT-III and BRO1 belong, do not show apparent alterations in central vacuole morphology (49). In contrast, Arabidopsis alix mutants have abnormal vacuoles, suggesting that the function of ALIX homolog differs in this regard in yeast and plants. Vacuoles are essential organelles in plants (50), have developmental stage- or organelle-specific functions (51, 52), and as recently reported, can be also regulated by the phytohormone auxin (53). Thus, the underlying mechanisms of vacuole biogenesis might be more complex in plants than in yeast.
siRNA-mediated gene silencing of human ALIX in HeLa cells was shown to decrease the number of multivesicular endosomes (34). Our ultrastructural analysis of alix-2 mutant cells showed smaller MVBs that are occasionally found forming clusters, which indicates that the function of Arabidopsis ALIX is also important for proper MVB biogenesis. However, in contrast to the membranous aggregates that are observed in typical class-E mutants of budding yeast (54), defects in Arabidopsis class-E genes seem to affect MVB biogenesis in a different manner.
A Xenopus ALIX homolog was shown to interact with AMSH in cell lysates (55); however, neither the nature of this interaction nor the functional relationship between ALIX and AMSH has been established. Our analysis demonstrated that AMSH3 and ALIX interact directly in vitro and coimmunoprecipitate in vivo, suggesting these two proteins function together. They colocalize on WM-sensitive late endosomes, and ALIX is necessary for the endosomal localization of AMSH3. Furthermore, mutant analyses revealed phenotypic similarities between null mutants of ALIX and AMSH3, indicating they play a role in the same biological pathways.
Yeast Bro1p and a human ALIX-related protein HD-PTP were shown to recruit UBPs Doa4p and UBPY, respectively, to endosomes (36, 56). In contrast to Doa4p, and similarly to UBPY (57), AMSH3 interacts directly with ESCRT-III subunits through its MIT domain (26). Interaction between AMSH3 and ALIX alone is probably not enough for proper AMSH3 function, as the deletion of the MIT domain of AMSH3 renders it nonfunctional (27). Thus, ALIX may function to stabilize the interaction of AMSH3 with ESCRT-III. Alternatively, defects in degradation of ubiquitinated cargos or possible AMSH3 substrates in the alix mutants might also affect the localization of AMSH3 to endosomes.
In addition to the localization to vesicles, AMSH3-YFP signals were also observed dispersed in the cytosol. Differential regulation of AMSH3 activity in the cytosol and on endosomes could therefore be important to guarantee deconjugation of ubiquitin chains only on endosome-associated substrates. Human AMSH was shown to be activated by the binding of STAM (47). The Arabidopsis genome does not contain a sequence-homolog of STAM (9), and although Arabidopsis AMSH3 interacts with ESCRT-III subunits (26) and ALIX, none of them was shown to influence its DUB activity. Future research should identify factors that may inhibit AMSH3 activity before its recruitment to endosomes or enhance its activity on endosomal membranes.
Despite its relatively low conservation among the different kingdoms, Arabidopsis ALIX also interacted with ubiquitin through its V-domain, similar to its homologs (29, 31, 44). The V-domain was also reported to serve as a dimerization domain (58) and clathrin-binding domain (44). Whereas Bro1p interacts with the DUB Doa4p through its C-terminal proline-rich motif (59), our experiments identified a subregion in the V-domain of Arabidopsis ALIX to be sufficient for the interaction with AMSH3. How the binding of different proteins is coordinated within this domain is an intriguing question.
In addition to ALIX, the Arabidopsis genome encodes for four additional Bro1 domain-containing proteins (AT1G13310, AT1G17940, AT1G73990, and AT5g14020). These proteins are shorter than ALIX and lack other recognizable functional domains. Because the Bro1 domain is characterized as the interaction domain for ESCRT-III (60), these proteins might also play a role in ESCRT-mediated processes. The moderate frequency of colocalization of GFP-ALIX with both the early endosomal marker SYP43 and the late endosomal marker ARA7 suggests Arabidopsis ALIX, as its homologs, could play a broad role in membrane trafficking events, which will be an interesting topic for future studies.
Methods
Cloning Procedure.
Details of the cloning procedure and primers used for cloning are described in SI Methods and Table S1, respectively.
Table S1.
Primers used in this study
| Primer | Sequence |
| KK40 ALIX fw BamHI | AAGGGGATCCTTGGCTTCTTCTTCGCTCT |
| KK41 ALIX rv SalI | AAGGGTCGACTCATTGCCTGTAGTATCCTCC |
| KK72 alix-4 genotyping F | TGCTGTGCGGACAGA |
| KK73 alix-4 genotyping R | TGAATAGAACAGCTGCTTTCTC |
| KK77 ALIX fw NdeI | AAGGCATATGTTGGCTTCTTCTTCGCTC |
| KK78 ALIX rv BamHI | AAGGGGATCCTCATTGCCTGTAGTATCCTCC |
| KK89 ∆BRO1 fw BamHI | AAGGGGATCCGAGAGATTACAGCAAGCTAGC |
| KK94 alix-2 genotyping F | GCTGGCGAAAGTGATGT |
| KK95 alix-4 genotyping R | ACCATCGTATAGAATGCAATACA |
| KK100 ∆C rv SalI | AAGGGTCGACGCCGGAGCTTCTATGATC |
| KK125 ∆BRO1 fw NdeI | AAGGCATATGGAGAGATTACAGCAAGCTAGC |
| KK126 BRO1 rv BamHI | AAGGGGATCCTCACGCCTGAGTTCTAATTACATC |
| KK127 ∆C rv BamHI | AAGGGGATCCTCAAACTTCAATGTTTTGGGAAATATC |
| KK143 V-frag.1 fw NdeI | AAGGCATATGGAGCGATCAGTGAGAGAT |
| KK169 ALIX attB1_2 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTGATGGCTTCTTCTTCGCTCT |
| KK193 V-frag.2 fw NdeI | AAGGCATATGCTGAGGCAACTGGAAAATC |
| EI3 AMSH1 GW fw | AAAAAGCAGGCTTGGGGTCGTCTTTTGAGA |
| EI4 AMSH1 GW rv | AGAAAGCTGGGTCTATCTGAGATCAATGAC |
| EI5 AMSH2 GW fw | AAAAAGCAGGCTTGGTAACGCTCTCGTCTC |
| EI6 AMSH2 GW rv | AGAAAGCTGGGTTTAACGTAGATCAAAAAT |
| EI7 AMSH3 GW fw | AAAAAGCAGGCTTGAAGATTGATCTGAACA |
| EI8 AMSH3 GW rv | AGAAAGCTGGGTTTAGCGGAGATCGAGGAC |
| EI553 AMSH3 middle fw BamHI | AAGGGGATCCTTGGGAGTAGGAAGAGG |
| EI554 AMSH3 middle rv SalI | AAGGGTCGACTTATTCTGTGTTTGACCGAGC |
| TF1 ALIX GW fw | GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGCTTCTTCTTCGC |
| TF2 ALIX GW rv | GGGGACCACTTTGTACAAGAAAGCTGGGTTCATTGCCTGTAGTA |
| LBb1.3 (alix-4 genotyping) | ATTTTGCCGATTTCGGAAC |
| o8409 (alix-2 genotyping) | ATATTGACCATCATACTCATTGC |
Biological Material.
All experiments were performed with A. thaliana (Columbia-0 ecotype). The T-DNA insertion lines of ALIX were obtained from the Genomanalyse im biologischen System Pflanze-Kolner Arabidopsis T-DNA (GABI-Kat) collection (alix-2, GABI 83711) (61) and from the Nottingham Arabidopsis Stock Centre (alix-4, SALK 063124). alix-2 mutants were genotyped using primers KK94 and KK95 for wild-type and KK94 and o8409 for T-DNA insertion. alix-4 mutants were genotyped with primers KK72 and KK73 for the wild-type and KK72 and LBb1.3 for the T-DNA insertion.
Plant transformations were carried out using the floral dip method (62). Seedlings were grown in continuous light at 110 µmol m−2⋅s−1 light intensity. Standard Murashige & Skoog growth medium (Duchefa Biochemie) supplemented with 1% sucrose was used to grow seedlings. Adult plants were grown on soil. Arabidopsis root epidermis cell culture was used to produce protoplasts for transient transformation and localization analysis, as described previously (26).
Immunoblotting, Antibodies, and Immunoprecipitation.
To prepare total protein extracts from Arabidopsis seedlings, frozen seedlings were homogenized with the TissueLyser-II (Qiagen), using glass beads, and the pulverized tissue was mixed with the extraction buffer [50 mM Tris⋅HCl at pH 7.5, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitor mixture (Roche)]. The mixture was centrifuged 10 min at 13,000 × g, and the supernatant was used for further experiments. Immunoprecipitation of GFP-ALIX from plant total extracts was performed as described in ref. 63, using GFP-Trap_A (Chromotek). Protein purification from Escherichia coli and in vitro binding assays were performed as described previously (26). SDS/PAGE and immunoblotting were performed according to standard methods. An anti-ALIX antibody was raised in rabbits using a total amount of 800 µg of recombinant 6xHis-ALIX(V) as antigen (Eurogentec). The serum was used at a 1:1,000 dilution, and the specificity of the antibody was verified using total extracts from alix null mutants. Additional primary antibodies used were anti-AMSH3 (20), anti-CDC2 (Santa Cruz), anti-GAL4 DNA binding domain (Santa Cruz), anti-His (Sigma-Aldrich), anti-UB(P4D1) (Santa Cruz), and horseradish peroxidase conjugated anti-HA (Sigma-Aldrich).
Ub-Binding Assay.
The ub-binding assay was performed using ubiquitin agarose (Enzo Life Sciences), as described in ref. 64. For MicroScale Thermophoresis, His-ALIX(V) was labeled with Monolith Protein Labeling Kit RED-NHS (NanoTemper). Next, 50 nM labeled His-ALIX(V) was mixed with a twofold dilution series of K48- and K63- linked diubiquitin (Enzo Life Sciences), starting with a concentration of 37 µM. Themophoresis was measured in standard treated capillaries (NanoTemper) with a monolith NT.115 instrument (NanoTemper), as described in ref. 65.
Microscopy.
For the visualization of vacuoles in the root epidermis cells, seedlings were incubated in liquid Murashige and Skoog (MS) supplemented with 5 μM BCECF-AM (Molecular Probes) and 0.02% Pluronic F-127 (Molecular Probes, Invitrogen) for 1 h at room temperature in the dark. Brefeldin A (BFA) (Sigma Aldrich) and WM (Applichem) treatments were performed at a concentration of 50 and 33 µM, respectively, for 60 min for BFA and 120 min for WM at room temperature. GFP-, YFP-, or TagRFP/mRFP-fusion proteins as well as BCECF-AM staining were analyzed with an FV-1000/IX81 confocal laser scanning microscope (Olympus) equipped with GaAsP detectors (Olympus) and a UPlanSApo ×60/1.20 (Olympus) objective, using the 488-, 515-, and 559-nm laser line, respectively. Images were subsequently processed using FluoView (Olympus) and Photoshop CS6 (Adobe). For 3D-reconstruction and surface rendering of BCECF-stained vacuoles, 90 Z-stacks images with 0.2-µm step size were obtained and subsequently processed, using Imaris 8 (Bitplane) with the following parameters: surface area detail level, 0.12 µm; diameter of largest sphere, 1.00 µm; background subtraction, enabled. The area/volume ratio was calculated using the following formula: ∆A/∆V = area (µm2)/[volume (µm3)]^(1/3)^2. For the analysis of AMSH3-YFP signals on ARA7-positive WM compartments, ARA7-labeled structures with a minimal diameter of 1.5 µm were identified using Imaris 8 (Bitplane).
Transmission Electron Microscopy.
Root tips from 5-d-old wild-type and alix-2 seedlings were high-pressure frozen in a Baltec HP010, substituted in an AFS Leica device for 5 d at −90 °C in acetone containing 0.2% (wt/vol) uranyl acetate and 0.2% (vol/vol) glutaraldehyde, and embedded in Lowicryl HM20 resin (Electron Microscopy Sciences). Samples were sectioned and stained with 2% (wt/vol) uranyl acetate in 70% methanol and lead citrate [2.6% (wt/vol) lead nitrate and 3.5% (wt/vol) sodium citrate, pH12].
Note Added in Proof.
While this paper was in production, a paper by Cardona-López et al. entitled “ESCRT-III-associated protein AtALIX mediates high affinity phosphate transporter trafficking to maintain phosphate homeostasis in Arabidopsis” has been accepted for publication in The Plant Cell. In this paper, the authors also reported that ALIX is essential in Arabidopsis and involved in intracellular trafficking and vacuole biogenesis.
SI Methods
Cloning Procedures.
All primers used for cloning and subcloning are listed in Table S1. For YTH analysis, the ORFs of AMSH1 and AMSH2 were PCR-amplified using primer pairs EI3–EI4, EI5–EI6, and EI7–EI8, respectively, and cloned into pDest-DB (66) to obtain GBD-AMSH1, GBD-AMSH2, and GBD-AMSH3. The ORF of ALIX was amplified from cDNA, using primers KK77 and KK78, and cloned into pGBKT7 (Clontech), using the NdeI and BamHI sites. ALIX fragments, BRO1 domain alone, ∆BRO1, ∆C-term, V-domain fragment 1, and V-domain fragment 2 were amplified using primer pairs KK77–KK126, KK125–KK78, KK77–KK127, KK143–KK127, and KK193–KK127. All fragments were ligated into pGBKT7 between the NdeI-BamHI sites. The AD-fused full-length AMSH3 (pGADT7, Clontech), as well as AMSH3 fragments MIT(N154), deltaMIT, and deltaMPN, were described previously (26). The middle domain of AMSH3 was amplified with primers EI553–EI554 and cloned between the BamHI-SalI sites of pGADT7.
To generate His-ALIX (V), the region containing the V-domain of ALIX was amplified using primers KK89 and KK100 and cloned into pET21a (Merck Millipore) between the BamHI and SalI sites. For MBP-ALIX, the ORF of ALIX was amplified using primers KK40 and KK41 and cloned into pMAL-p2p (gift from Keiji Tanaka's laboratory) between BamHI and SalI sites. For MBP-VPS60.1, insert of a GST-VPS60.1 construct (26) was cut out from pGEX-6P-1 with BamHI and SalI and ligated into pMAL-p2p.
For AMSH3pro:AMSH3-YFP and AMSH3pro:AMSH3-TagRFP, the YFP moiety was cut out from a previously published AMSH3pro:YFP-AMSH3 construct (26). The stop codon was replaced with a NcoI site by PCR-based mutagenesis, and the coding sequence for YFP or TagRFP was cloned into the newly created NcoI site. ALIXpro::GFP-ALIX will be described elsewhere.
For overexpression in plants that resulted in reduced ALIX protein level, the ORF of ALIX was amplified using primers KK169 and TF2 and cloned into pDONR207 (Invitrogen), and subsequently into pFAST-R05 (67), to obtain 35Spro:ALIX. The presence of a stop codon at the end of the sequence caused the overexpression of an untagged protein.
Constructs used for expression in protoplasts, UBQ10pro:YFP-VPS24.1, 35Spro:HA-SKD1(WT), and 35Spro:HA-SKD1(EQ) were described previously (26).
DUB Assay.
For diubiquitin-TAMRA assay, 0.4 µM diubiquitin (K63-linked) FRET TAMRA Position 3 (R&D Systems) was incubated with 50 nM AMSH3 in the TAMRA DUB buffer [50 mM Tris⋅HCl at pH 7.5, 100 mM NaCl, 0.1% Pluronic F-127, 1 mM Tris(2-carboxyethyl)phosphine] in a volume of 100 µL. To examine the effect of ALIX on AMSH3 activity, AMSH3 was preincubated for 30 min with 50 nM of ALIX before the addition of di-Ub TAMRA. Fluorescence was measured with a Microplate reader (BioTek).
Acknowledgments
We thank Tomohiro Uemura, Takashi Ueda, and Akihiko Nakano (University of Tokyo) for kindly sharing published materials and for discussion; Yasushi Saeki, Yoko Kimura, and Keiji Tanaka for the pMal-p2p vector; and Genomanalyse im biologischen System Pflanze-Kolner Arabidopsis T-DNA and Nottingham Arabidopsis Stock Centre for providing alix mutant seeds. We are also grateful to Kamyar Hadian and Kenji Schorpp (Helmholtzzenrum München) for help with the MicroScale Thermophoresis measurement. This work was supported by grants from the Deutsche Forschungsgemeinschaft SFB924 (A10, to P.B.), the Spanish Ministry of Research, Development and Innovation BIO2013-46539-R (to V.R.), the National Science Foundation MCB1157824 (to M.S.O.), and the Deutsche Forschungsgemeinschaft IS221/4-1 and SFB924 (A06, to E.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510516112/-/DCSupplemental.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol. 2011;23(4):393–403. doi: 10.1016/j.ceb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125(Pt 2):265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 6.Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12(10):1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5(9):a016766. doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes FC, Buono R, Otegui MS. Plant endosomal trafficking pathways. Curr Opin Plant Biol. 2011;14(6):666–673. doi: 10.1016/j.pbi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Winter V, Hauser MT. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006;11(3):115–123. doi: 10.1016/j.tplants.2006.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korbei B, et al. Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr Biol. 2013;23(24):2500–2505. doi: 10.1016/j.cub.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 12.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 13.Clague MJ, Coulson JM, Urbé S. Cellular functions of the DUBs. J Cell Sci. 2012;125(Pt 2):277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 14.Isono E, Nagel MK. Deubiquitylating enzymes and their emerging role in plant biology. Front Plant Sci. 2014;5:56. doi: 10.3389/fpls.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695(1-3):189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka N, et al. Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J Biol Chem. 1999;274(27):19129–19135. doi: 10.1074/jbc.274.27.19129. [DOI] [PubMed] [Google Scholar]
- 17.Ishii N, et al. Loss of neurons in the hippocampus and cerebral cortex of AMSH-deficient mice. Mol Cell Biol. 2001;21(24):8626–8637. doi: 10.1128/MCB.21.24.8626-8637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonell LM, et al. FORGE Canada Consortium Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly-capillary malformation syndrome. Nat Genet. 2013;45(5):556–562. doi: 10.1038/ng.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isono E, et al. The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell. 2010;22(6):1826–1837. doi: 10.1105/tpc.110.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsiarimpa A, et al. The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell. 2013;25(6):2236–2252. doi: 10.1105/tpc.113.113399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullough J, Clague MJ, Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166(4):487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281(32):23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- 24.Kyuuma M, et al. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct Funct. 2007;31(2):159–172. doi: 10.1247/csf.06023. [DOI] [PubMed] [Google Scholar]
- 25.Ma YM, et al. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J Biol Chem. 2007;282(13):9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 26.Katsiarimpa A, et al. The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell. 2011;23(8):3026–3040. doi: 10.1105/tpc.111.087254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsiarimpa A, et al. The ESCRT-III-interacting deubiquitinating enzyme AMSH3 is essential for degradation of ubiquitinated membrane proteins in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(4):727–736. doi: 10.1093/pcp/pcu019. [DOI] [PubMed] [Google Scholar]
- 28.Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24(1):19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Dowlatshahi DP, et al. ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev Cell. 2012;23(6):1247–1254. doi: 10.1016/j.devcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bissig C, et al. Viral infection controlled by a calcium-dependent lipid-binding module in ALIX. Dev Cell. 2013;25(4):364–373. doi: 10.1016/j.devcel.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keren-Kaplan T, et al. Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. EMBO J. 2013;32(4):538–551. doi: 10.1038/emboj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikko E, Marini AM, André B. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J Biol Chem. 2003;278(50):50732–50743. doi: 10.1074/jbc.M306953200. [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, et al. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278(40):39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 34.Matsuo H, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303(5657):531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 35.Richardson LG, et al. Protein-Protein Interaction Network and Subcellular Localization of the Arabidopsis Thaliana ESCRT Machinery. Front Plant Sci. 2011;2:20. doi: 10.3389/fpls.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166(5):717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uemura T, et al. Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct Funct. 2004;29(2):49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- 39.Uemura T, et al. Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc Natl Acad Sci USA. 2012;109(5):1784–1789. doi: 10.1073/pnas.1115146109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueda T, Uemura T, Sato MH, Nakano A. Functional differentiation of endosomes in Arabidopsis cells. Plant J. 2004;40(5):783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- 41.Haas TJ, et al. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell. 2007;19(4):1295–1312. doi: 10.1105/tpc.106.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17(11):2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolb C, et al. FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 2015;167(4):1361–1373. doi: 10.1104/pp.114.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pashkova N, et al. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev Cell. 2013;25(5):520–533. doi: 10.1016/j.devcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 46.Kim MS, Kim JA, Song HK, Jeon H. STAM-AMSH interaction facilitates the deubiquitination activity in the C-terminal AMSH. Biochem Biophys Res Commun. 2006;351(3):612–618. doi: 10.1016/j.bbrc.2006.10.068. [DOI] [PubMed] [Google Scholar]
- 47.McCullough J, et al. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16(2):160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 48.Nikko E, André B. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: Application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot Cell. 2007;6(8):1266–1277. doi: 10.1128/EC.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3(12):1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell. 2001;1(2):303–310. doi: 10.1016/s1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- 51.Frigerio L, Hinz G, Robinson DG. Multiple vacuoles in plant cells: Rule or exception? Traffic. 2008;9(10):1564–1570. doi: 10.1111/j.1600-0854.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- 52.Zouhar J, Rojo E. Plant vacuoles: Where did they come from and where are they heading? Curr Opin Plant Biol. 2009;12(6):677–684. doi: 10.1016/j.pbi.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Löfke C, Dünser K, Scheuring D, Kleine-Vehn J. Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. Elife. 2015;4:e05868. doi: 10.7554/eLife.05868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3(2):271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 55.Dejournett RE, et al. Phosphorylation of the proline-rich domain of Xp95 modulates Xp95 interaction with partner proteins. Biochem J. 2007;401(2):521–531. doi: 10.1042/BJ20061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali N, et al. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol. 2013;23(6):453–461. doi: 10.1016/j.cub.2013.02.033. [DOI] [PubMed] [Google Scholar]
- 57.Row PE, et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282(42):30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 58.Pires R, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17(6):843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 2007;26(10):2454–2464. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, et al. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8(6):937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosso MG, et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53(1-2):247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 62.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 63.Isono E, Schwechheimer C. Co-immunoprecipitation and protein blots. Methods Mol Biol. 2010;655:377–387. doi: 10.1007/978-1-60761-765-5_25. [DOI] [PubMed] [Google Scholar]
- 64.Kalinowska K, Isono E. Analysis of global ubiquitylation and ubiquitin-binding domains involved in endosomal trafficking. Methods Mol Biol. 2014;1209:189–202. doi: 10.1007/978-1-4939-1420-3_15. [DOI] [PubMed] [Google Scholar]
- 65.Hadian K, et al. NF-κB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-κB activation. J Biol Chem. 2011;286(29):26107–26117. doi: 10.1074/jbc.M111.233163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dreze M, et al. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 67.Shimada TL, Shimada T, Hara-Nishimura I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010;61(3):519–528. doi: 10.1111/j.1365-313X.2009.04060.x. [DOI] [PubMed] [Google Scholar]