Significance
Meiosis is essential for eukaryotic sexual reproduction, and meiotic recombination redistributes genetic variations among progeny. Homologous recombination is also important for DNA repair and genome stability. Current recombination models include DNA synthesis, particularly leading-strand synthesis, but its role has not been tested experimentally. Here, we showed that mutations in a DNA polymerase required for leading-strand elongation caused reduced fertility, defects in meiotic chromosome fragmentation and segregation, and reduction of interference-sensitive cross-overs (COs). Because the majority of meiotic COs are produced from the interference-sensitive pathway in budding yeast, mammals, and flowering plants, the discovery of leading-strand elongation in the differentiation of meiotic recombination pathways has important implications in understanding of human reproductive health and crop breeding.
Keywords: meiotic recombination, DNA synthesis, DNA polymerase-ε, Arabidopsis
Abstract
Meiosis halves diploid genomes to haploid and is essential for sexual reproduction in eukaryotes. Meiotic recombination ensures physical association of homologs and their subsequent accurate segregation and results in the redistribution of genetic variations among progeny. Most organisms have two classes of cross-overs (COs): interference-sensitive (type I) and -insensitive (type II) COs. DNA synthesis is essential for meiotic recombination, but whether DNA synthesis has a role in differentiating meiotic CO pathways is unknown. Here, we show that Arabidopsis POL2A, the homolog of the yeast DNA polymerase-ε (a leading-strand DNA polymerase), is required for plant fertility and meiosis. Mutations in POL2A cause reduced fertility and meiotic defects, including abnormal chromosome association, improper chromosome segregation, and fragmentation. Observation of prophase I cell distribution suggests that pol2a mutants likely delay progression of meiotic recombination. In addition, the residual COs in pol2a have reduced CO interference, and the double mutant of pol2a with mus81, which affects type II COs, displayed more severe defects than either single mutant, indicating that POL2A functions in the type I pathway. We hypothesize that sufficient leading-strand DNA elongation promotes formation of some type I COs. Given that meiotic recombination and DNA synthesis are conserved in divergent eukaryotes, this study and our previous study suggest a novel role for DNA synthesis in the differentiation of meiotic recombination pathways.
Eukaryotic sexual reproduction requires meiosis to produce haploid gametes from parents. Meiotic recombination not only ensures physical association of homologs to form bivalents but also, promotes their accurate segregation, and it is initiated by the formation of SPO11-programmed DNA double-strand breaks (DSBs) (1). Repair of meiotic DSBs is evolutionarily important, because it redistributes genetic variants in regions flanking the site of recombination [cross-overs (COs)] or gene conversion without flanking exchange [non-COs (NCOs)], resulting in offspring that are genetically distinct from parents and siblings (2, 3). Although DNA synthesis is thought to be essential for DSB repair (DSBR) (4), molecular genetic studies are quite limited. Recent genome-wide studies provide evidence that meiotic COs and NCOs may require different amounts of DNA synthesis (2, 3, 5–7), but the underlying mechanism remains unknown.
Premeiotic chromosome replication requires the same DNA replication machinery as that for mitotic DNA replication (8), which is carried out by coordinated activities of DNA polymerase-α (Pol-α), -δ, and -ε (9) as well as three ancillary complexes: proliferating cell nuclear antigen, replication factor C (RFC), and the minichromosome maintenance complex. Pol-α and -δ are primarily required for lagging-strand DNA synthesis (9, 10), whereas Pol-ε carries out leading-strand DNA synthesis (11). Given that DNA synthesis is essential for S-phase replication during the mitotic cell cycle, complete disruption of these genes causes lethality, which was observed in yeast, plants, and humans (11–14), making it difficult to study their functions in meiosis. In Saccharomyces cerevisiae, a partially defective mutation of Pol-δ causes no DNA replication defect but exhibits reduction in both the number of COs and the length of meiotic gene conversion tracts (15). In Arabidopsis, a partial loss of function of DNA replication factor C1 (RFC1) results in reduction of type I COs (16). These findings suggest that DNA synthesis likely affects the length of strand exchange intermediates and influences their resolution toward different CO pathways. Moreover, genome-wide analyses in both yeast and plants showed that meiotic gene conversion tracts vary in length, ranging from several hundreds to thousands of nucleotides, and that CO-associated tracts are usually longer than those of NCOs (2, 3, 5–7). This difference raises a question of whether there is any difference in DNA synthesis among CO-associated tracts with different lengths; however, this hypothesis has not been tested experimentally. Our previous research with RFC1 supports the idea that lagging-strand synthesis promotes type I COs (16). Here, we test the hypothesis that sufficient leading-strand synthesis is important for type I COs.
To investigate the role of leading-strand DNA elongation in meiotic recombination, we analyzed two partial loss-of-function alleles of the largest subunit of DNA Pol-ε (hereafter designated as POL2A). They have normal vegetative development but show defects in fertility and meiosis. Additional analyses show that POL2A has a role in the formation of normal bivalents and promoting formation of meiotic type I COs. Similar mutations in the yeast homolog showed reduced DNA replication efficiency in mitotic division (17–19), leading us to hypothesize that sufficient leading-strand DNA elongation could be a critical step for differentiation of types I and II COs. These observations provide a novel insight that meiotic types I and II COs are differentially dependent on leading-strand synthesis activity.
Results
POL2A Is Required for Normal Meiosis in Arabidopsis.
DNA Pol-ε is called POL2 or CDC20 in the budding and fission yeasts, respectively (20). Studies in yeast, flies, and humans showed that the POL2 N terminus is important for elongation and fidelity of DNA replication (19, 21), whereas the C-terminal region is essential for cell viability. Arabidopsis has two POL2 homologs (POL2A and POL2B), and POL2A was expressed in male meiocytes, whereas POL2B was almost undetectable (22), suggesting a potential function of POL2A in meiosis. To test this hypothesis, we obtained three pol2a mutants (SI Appendix, Fig. S1A). Among them, pol2a-1 (SALK_096341) is a transfer deoxyribonucleic acid (T-DNA) insertional allele in the 12th intron and has been shown to reduce abscisic acid (ABA) sensitivity and fertility (13) (Fig. 1A and SI Appendix, Fig. S1A), pol2a-2 (formally named til1-4) is a point mutation and displays a prolonged cell cycle during embryo development (12), and pol2a-3 (SALK_124217) also carries a T-DNA insertion affecting the conserved C-terminal domain (SI Appendix, Fig. S1 A and B) and exhibits aborted early embryo development (SI Appendix, Fig. S1F), consistent with an essential role of Pol-ε in DNA replication (19, 21). The lack of homozygous pol2a-3 plants precluded the use of this mutant to analyze possible meiotic defects.
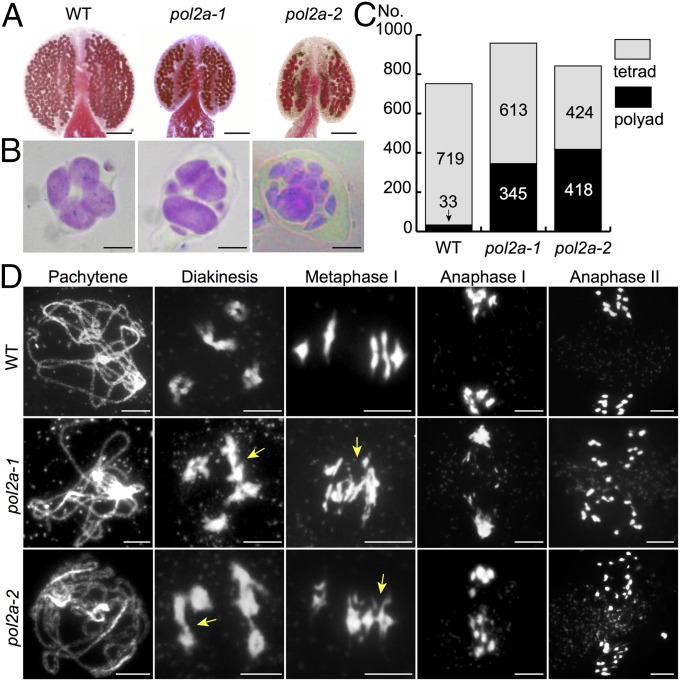
Fig. 1.
Mutations in POL2A cause reduced fertility and meiotic defects in Arabidopsis. (A) Alexander red staining of WT, pol2a-1, and pol2a-2 anthers. (B) Tetrad observation of the WT, pol2a-1, and pol2a-2. (C) A histogram of tetrads vs. polyads in the WT and mutants. (D) Chromosome morphologies of the WT, pol2a-1, and pol2a-2 by DAPI staining. Arrows indicate probable nonhomologous associations. (Scale bars: A, 100 μm; B, 10 μm; D, 5 μm.)
Both pol2a-1 and pol2a-2 plants had normal growth, including plant size and floral organ identity and number (SI Appendix, Fig. S1 C and E), indicating no obvious defects in their mitotic DNA replication. However, they had greatly reduced fertility (SI Appendix, Fig. S1D). Alexander staining of pollen viability showed that viable pollen grains were obviously reduced in pol2a-1 (294 ± 74; n = 16) and pol2a-2 (181 ± 73; n = 16) in contrast to the WT (542 ± 38; n = 29) (Fig. 1A). In addition, unlike WT tetrads with four microspores (Fig. 1B), male meiocytes of pol2a-1 and pol2a-2 produced polyads with variable microspore numbers ranging from five to eight with abnormality rates of 36.0% and 49.6%, respectively (Fig. 1 B and C), suggesting meiotic defects. Pollination of either pol2a mutant pistil with WT pollen grains also showed a defect in female fertility (SI Appendix, Table S1). We examined the POL2A expression level (SI Appendix, Fig. S1G) and found a significant reduction in pol2a-1, indicating that pol2a-1 is not a null mutant. In contrast, the mRNA level was slightly elevated in pol2a-2, indicating that the point mutation in the N-terminal region of POL2A likely diminishes its function, consistent with the observations in the budding and fission yeasts (19, 21).
POL2A Is Required for Normal Chromosome Interactions in Meiotic Prophase I.
The observation of polyads in pol2a suggests a meiotic defect; we, therefore, compared chromosome spreads of the WT and pol2a. Chromosome morphology showed no obvious differences up to the zygotene stage between the mutants and the WT (SI Appendix, Fig. S2). At pachytene, 73% (n = 100) pol2a meiocytes were similar to those of the WT (Fig. 1D), whereas the remaining 27% pachytene-like cells showed abnormal chromosome condensation and association (SI Appendix, Fig. S2). After the dissociation of the synaptonemal complex (SC) after the pachytene stage, WT meiocytes showed continued association at diakinesis of condensed homologs in five bivalents, which aligned along the equatorial plane at metaphase I, and subsequent separation of homologs at anaphase I. By contrast, both pol2a-1 and pol2a-2 meiocytes had entanglement between three or more chromosomes indicative of abnormal associations from diakinesis to metaphase I (Fig. 1D) and chromosome fragmentation at anaphase I and II, indicating the importance of POL2A for normal chromosome interactions. These findings also provide an explanation for the formation of polyads and reduced pollen viability.
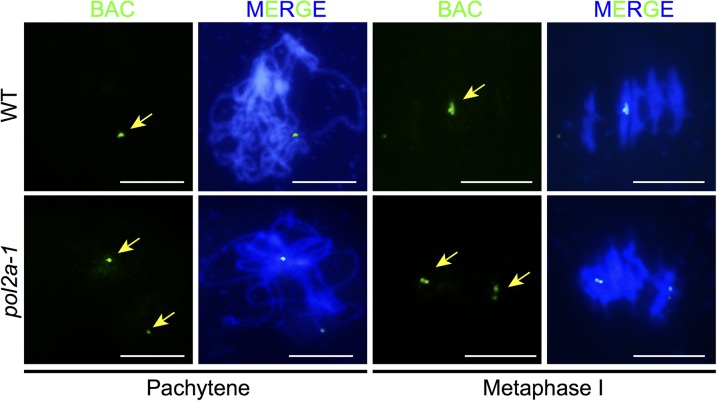
To test whether the associated chromosomes in pol2a-1 are homologs or nonhomologs, we performed chromosome-specific FISH using bacterial artificial chromosome (BAC) probe F1N21 for chromosome 1 (16). In the WT and pol2a-1, the F1N21 signals were similar at leptotene and zygotene (SI Appendix, Fig. S3). At pachytene, the WT had only one signal representing the closely synapsed homologous chromosome 1 arms (Fig. 2). In the pol2a-1 pachytene-like cells (n = 31), 77.4% had one signal similar to that in the WT, but 22.6% had two signals (Fig. 2), consistent with the results from DAPI-stained chromosome spreads described above. At metaphase I, 53.1% of pol2a-1 meiocytes (n = 32) had two signals that were on widely separate chromosomes; one of the signals appeared to be on a bivalent, whereas the other was often in a cluster of chromosomes in contrast to the one focus observed on one bivalent in the WT (Fig. 2). Similar results were also obtained using another BAC probe F4P13 located in the chromosome 3 left arm, with 13.2% of pachytene cells (n = 76) and 15.8% of metaphase I cells (n = 19) exhibiting abnormal separation of BAC signals (SI Appendix, Fig. S4). These results indicate that the two homologous chromosome 1 or 3 arms were sometimes separated in the mutant and suggest that they could associate with nonhomologs.
Fig. 2.
POL2A is required for homologous chromosome interactions in meiotic prophase I. BAC FISH with F1N21 probe at pachytene and metaphase I in the WT and pol2a-1 are shown. Arrows indicate the BAC probe signals. (Scale bar: 5 μm.)
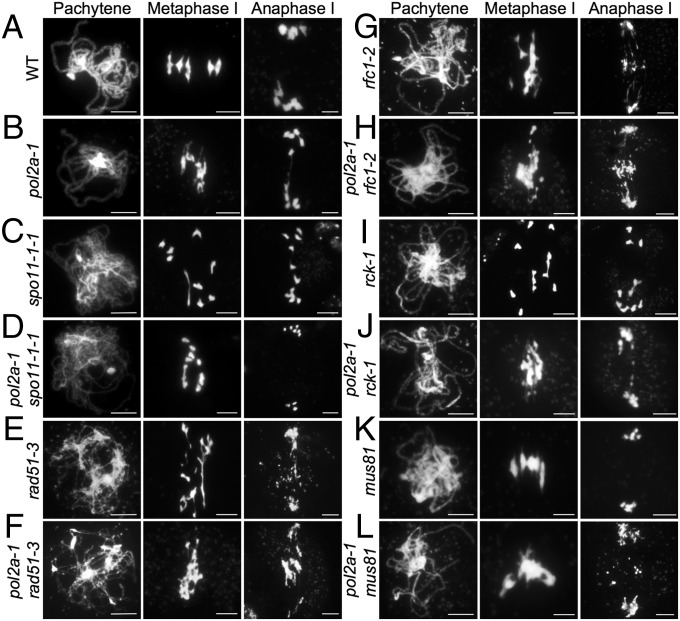
To determine whether the observed abnormal association and chromosome fragmentation in pol2a-1 are dependent on programmed meiotic DSBs, we tested for possible genetic interaction of POL2A with genes for DSB generation and repair. Arabidopsis SPO11-1 is required for DSB formation, chromosome pairing, and bivalent formation (Fig. 3C) (23). Repair of the DSBs requires RAD51 (24), and failed repair in atrad51 results in chromosome fragmentation (Fig. 3E), which is absent in the spo11-1 rad51 double mutant (24). Because meiotic DNA synthesis is proposed to occur after RAD51-dependent single-strand invasion, we hypothesized that chromosome fragmentation in pol2a-1 depends on SPO11-1 and that pol2a-1 would not enhance the defects of rad51. To test these hypotheses, we analyzed the double mutants with spo11-1 and rad51. As expected, the chromosome fragmentation in pol2a-1 was dependent on SPO11-1 (Fig. 3D), whereas the pol2a-1 rad51-3 double-mutant meiocytes resembled the rad51-3 single mutant, lacking distinctive pachytene chromosomes and bivalents (Fig. 3F). These results are consistent with the idea that POL2A acts downstream of RAD51.
Fig. 3.
Genetic analyses of POL2A with other meiotic recombination pathway genes. Chromosome morphologies shown by DAPI staining at pachytene, metaphase I, and anaphase I in (A) the WT, (B) pol2a-1, (C) spo11-1–1, (D) pol2a-1 spo11-1–1, (E) rad51-3, (F) pol2a-1 rad51-3, (G) rfc1-2, (H) pol2a-1 rfc1-2, (I) rck-1, (J) pol2a-1 rck-1, (K) mus81, and (L) pol2a-1 mus81. (Scale bar: 5 μm.)
Because the primary function of POL2A is DNA replication, several studies have shown that disruption of the DNA Pol-ε exonuclease domain causes reduced efficiency of mitotic DNA synthesis in yeast and plants (12, 13, 25). Thus, the pol2a-1 phenotype may be caused by abnormal premeiotic DNA replication. However, the normal vegetative morphology of pol2a-1 plant suggests that its mitotic DNA replication is unaffected (SI Appendix, Fig. S1C). Furthermore, analysis of meiotic chromosomes in pol2a-1 spo11-1–1 showed that premeiotic replication produces 20 chromatids (SI Appendix, Fig. S5), suggesting normal premeiotic DNA replication in pol2a-1. The lethality of pol2a-3 provides a means to test the phenotype of plants with possibly even less POL2A activity than pol2a-1 by generating the pol2a-1/pol2a-3 heteroallelic plants, which showed similar developmental morphology to pol2a-1 (SI Appendix, Fig. S6). Likewise, the pol2a-1/pol2a-3 meiocytes showed meiotic phenotypes similar to those of pol2a-1 and pol2a-2 (SI Appendix, Fig. S7). We also generated transgenic plants with a POL2A RNAi construct driven by the meiosis-specific DMC1 promoter and observed a meiotic defect similar to those of both single mutants (SI Appendix, Fig. S7). These results show that the meiotic phenotypes in pol2a are probably caused by loss of meiosis-specific function rather than partial loss of a general function.
POL2A Is Not Required for DSB Formation and Synapsis but Promotes Progression of Meiotic Recombination.
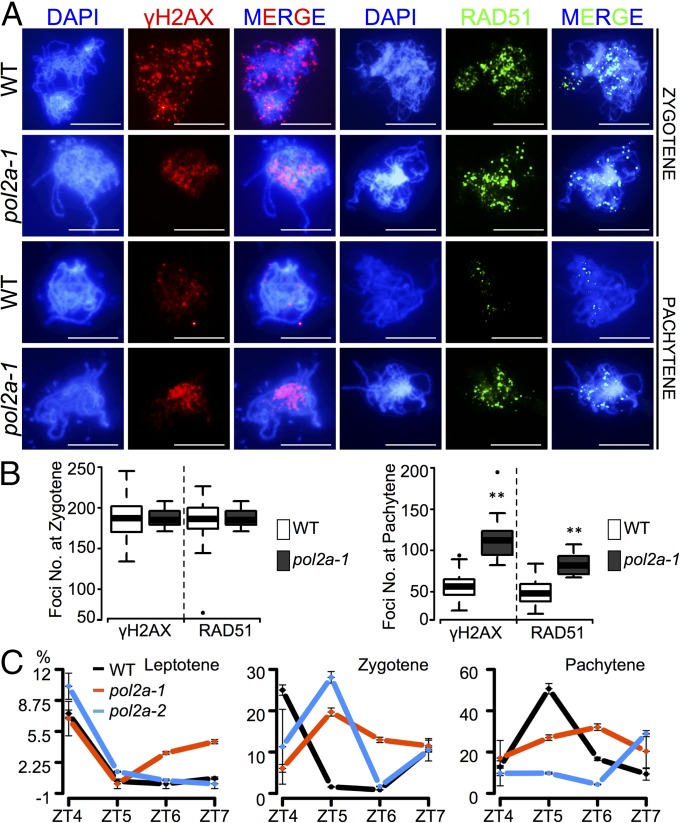
If these mutations in POL2A do not affect premeiotic DNA replication, another possible role for POL2A in meiosis is DNA synthesis for meiotic recombination (4). Defects in DNA synthesis could result in abnormal persistence of meiotic DSBs (16). To detect DSBs in pol2a-1, we examined the distribution of phosphorylated histone H2AX (γ-H2AX), which specifically marks DSBs (26). At zygotene, when DSBs are formed, no significant difference in γ-H2AX foci between the WT (189 ± 26; n = 39) and pol2a-1 (185 ± 16; n = 32) was observed (Fig. 4 A and B). We also examined the RAD51 recombinase, which mediates ssDNA invasion during recombination (24). No obvious difference was found between the WT (188 ± 24; n = 15) and pol2a-1 (187 ± 11; n = 14) at zygotene (Fig. 4 A and B). Together, the presence of γ-H2AX and RAD51 foci provides strong evidence that POL2A does not influence meiotic DSB formation. In contrast, at pachytene, the numbers of γ-H2AX and RAD51 foci in the WT decreased to 57 ± 12 (n = 55) and 51 ± 14 (n = 70), respectively, indicating completion of the majority of DSBR. At the equivalent stage, the pol2a -1 meiocytes maintained significantly more foci of γ-H2AX (113 ± 27; n = 19; P = 1.4E-08) and RAD51 (87 ± 15; n = 5; P = 0.003) (Fig. 4 A and B), suggesting delayed DSBR.
Fig. 4.
POL2A is not required for DSB formation but promotes progression of meiotic recombination. (A) Immunostaining with γ-H2AX (red), RAD51 (green), and DAPI counterstain (blue) at zygotene and pachytene in the WT and pol2a-1. (Scale bar: 5 μm.) (B) Variation of the γ-H2AX and RAD51 foci numbers above shown by boxplot in the WT and pol2a-1. **P ≤ 0.01 with a one-tailed Student t test. (C) Percentages of leptotene, zygotene, and pachytene cells at ZT4 (4 h after lights on), ZT5, ZT6, and ZT7, with error bars from analyses of three to five biological replicates in the WT, pol2a-1, and pol2a-2.
After formation of DSBs, synapsis is critical for stabilizing homolog interactions during meiotic prophase I (27). We examined the distributions of ASY1 and ZYP1, which mediate formation of the axial element and the SC, respectively (27). At zygotene, ASY1 showed linear signals with chromosomes in the WT and pol2a-1, and initiation of ZYP1 signals was also similar (SI Appendix, Fig. S8 A–H). At pachytene, the ASY1 signals in pol2a-1 seemed normal, and ZYP1 also had a linear distribution on the chromosomes in pol2a-1 (n = 128), similar to that of the WT (SI Appendix, Fig. S8 I–P). We conclude that ASY1 axis formation is normal in pol2a-1, and the normal localization of ZYP1 also suggests no obvious defect for SC formation in pol2a-1. However, the earlier described separate FISH signals for two BAC probes on different chromosomes suggest that sometimes synapsis occurred between nonhomologs.
Although there are no molecular markers for DNA synthesis during meiotic recombination, it presumably takes place in prophase I (5) soon after DSB formation. In Arabidopsis, DSBs are formed from leptotene to early zygotene, and their repair is completed by pachytene. We, therefore, compared the distribution of meiocytes from leptotene to pachytene at the four most relevant time points between the WT, pol2a-1, and pol2a-2 (Fig. 4C). Leptotene cells in the WT and mutants showed a higher level at zeitgeber time 4 (ZT4) (Fig. 4C). Zygotene cells in the WT also peaked at ZT4 and showed a gradual decrease to ZT5, whereas mutants displayed a gradual increase from ZT4 to reach the peak at ZT5 (Fig. 4C). In addition, unlike WT pachytene cells with a peak at ZT5, both mutant pachytene cells presented lower peaks with 1- or 2-h delays at ZT6 and ZT7, respectively (Fig. 4C). These results are consistent with the proposed delay in DSBR mentioned earlier and support a role of POL2A in progression of meiotic recombination.
POL2A Functions in Meiotic Interference-Sensitive CO Formation.
Similar to budding yeast and mammals, meiotic DSBR in Arabidopsis also yields NCOs and two types of COs (types I and II). The type I COs require ZMM proteins, such as MSH4 and MER3/RCK (27), and type II COs require the MUS81-MMS4 endonuclease (28, 29). The remaining fertility and bivalent-like cells in pol2a-1 (Fig. 1D) suggest formation of some COs. RFC1 (16) and MER3/RCK (30, 31) are required for formation of type I COs, whereas MUS81 (28) is important for type II COs. To determine which types of COs are formed in pol2a-1, we generated double mutants of pol2a-1 with rfc1-2 (16), rck-1 (30, 31), or mus81-1 (28) and compared their meiotic chromosome morphologies. The pol2a-1 rfc1-2 double mutant also displayed entangled chromosomes, resembling those found in either single mutant (being slightly more similar to those in rfc1-2) (Fig. 3H) and suggesting that they may act in the same pathway, whereas RFC1 has a greater role. In contrast to the rck-1 single mutant with one to two bivalents (Fig. 3I), the pol2a-1 rck-1 double mutant had entangled chromosomes (Fig. 3J), similar to those of the pol2a-1 single mutant (Figs. 1D and 3B), suggesting that POL2A acts upstream of RCK. In contrast, the pol2a-1 mus81-1 double mutant lacked typical bivalents but had more severely entangled chromosomes than those of the pol2a-1 single mutant, unlike the mus81-1 single mutant with normal chromosome morphology (Fig. 3 K and L). These data suggest that formation of bivalents in pol2a-1 depends on MUS81. Together, these results indicate that POL2A functions in the ZMM-dependent type I CO pathway, independent of the MUS81-dependent type II CO pathway.
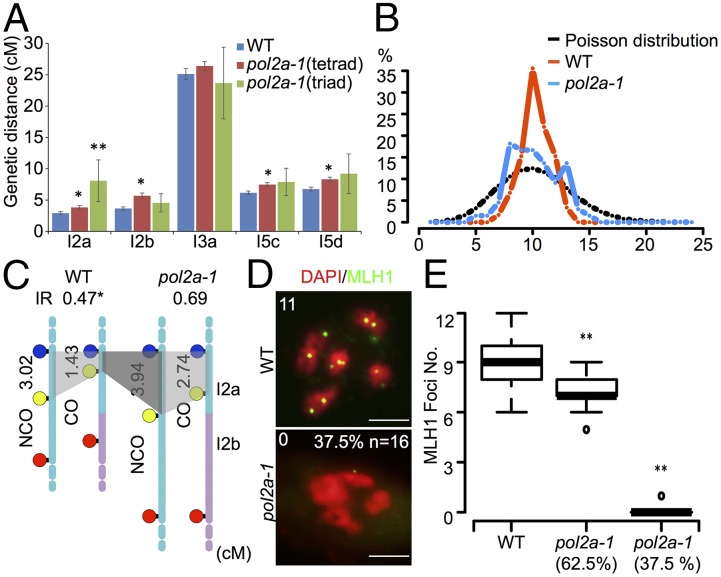
Because pol2a-1 plants produce some viable pollen grains, a fluorescence assay system was used to investigate the effect of the mutation on recombination frequency using three different fluorescent lines (SI Appendix, Fig. S9) (32). Four of five genetic intervals assayed showed significant increases in genetic distance (Fig. 5A and SI Appendix, Table S2). Because pol2a-1 had inviable pollen grains (Fig. 1A), to minimize biased estimation of recombination using viable pollen grains, we also counted triads with three viable pollen grains. The trend was in general agreement with the results from tetrad analysis (Fig. 5A and SI Appendix, Table S2). A possible explanation for the increased COs in pol2a-1 is that CO interference may be affected. To test this hypothesis, we estimated chiasma distributions in the WT and pol2a-1. The chiasma distribution in the WT deviated significantly from Poisson distribution (P = 3.1E-06; n = 59), indicative of interference, in contrast to that in pol2a-1 being statistically no different from Poisson (P = 0.16; n = 66) (Fig. 5B), suggesting reduced interference in pol2a-1. To verify the interference phenotype in pol2a-1, we calculated the genetic distance of interval I2a with or without CO in the adjacent I2b interval in the WT and pol2a-1. The WT interference ratio in I2a was 0.47 [P = 0.03 for hypothesis P(interference ratio = 1)], indicating a significant CO interference, whereas the interference ratio in pol2a-1 I2a was 0.69 [P = 0.15 for hypothesis P(interference ratio = 1)], indicating weaker CO interference (Fig. 5C). Together, these results provide evidence that POL2A functions in the type I CO pathway, and the residual COs in pol2a were type II, which depends on MUS81.
Fig. 5.
Estimation of meiotic recombination frequency and CO interference in pol2a. (A) Meiotic recombination frequency in five intervals from analyses of fluorescent tetrad or triad in the WT and pol2a-1. Positions of these intervals on chromosomes are indicated in SI Appendix, Fig. S9. Significant differences are shown by *P ≤ 0.05 and **P ≤ 0.01. (B) A graph of the chiasma frequency per nucleus in the WT and pol2a-1. Poisson distribution was based on 10,000 random samplings with an average of 10. (C) Genetic distance of interval I2a without or with CO in adjacent I2b in the WT and pol2a-1, respectively. Interference ratios are shown above with *P ≤ 0.05 against P(interference ratio = 1). (D) MLH1 foci (green) with DAPI (red) counterstain by immunostaining in the WT and pol2a-1. Numbers in the upper left indicate MLH1 foci numbers. (Scale bar: 5 μm.) (E) Statistics of MLH1 foci distribution in WT and pol2a-1 meiocytes. **P ≤ 0.01 with a one-tailed Student t test. IR, interference ratio.
To obtain independent validation that the observed COs in pol2a-1 were type II, we examined the MLH1 foci, which mark type I COs but not type II COs (33) at diakinesis in the WT and pol2a-1. As expected, pol2a-1 had significantly reduced MLH1 foci, with an average of 4.5 (P = 9.1E-05; n = 16) compared with the WT (an average of 9.0; n = 61) (Fig. 5D). Interestingly, the distribution of residual COs among meiocytes was bimodal: one group (62.5%) of pol2a-1 meiocytes had several (average of 7.2) MLH1 foci (n = 10), whereas another group (37.5%) of pol2a-1 meiocytes had one or no MLH1 foci (n = 6) (Fig. 5 D and E). Although the cause of bimodality is unknown, the clustering of COs in the second group of meiocytes is consistent with interference-insensitive type II COs.
Discussion
POL2A Plays a Role in DNA Synthesis During Meiotic Recombination.
DNA synthesis is required for both the mitotic cell cycle and meiosis. Because factors for DNA replication are essential for the mitotic S phase, null mutations in corresponding genes cause lethality in mammals, yeast, and plants (11–14, 34), making it difficult to study their functions. Nevertheless, characterization of weak mutations and/or generation of conditional mutations of DNA synthesis genes provide a feasible approach to study their functions, such as DNA Pol-α (35), Pol-δ (15, 36), Pol-ε (12, 13), and RFC1 (16) in multiple organisms. Genetic studies in yeast showed that the N-terminal region of Pol-ε is dispensable for DNA replication, DNA repair, and cell viability, whereas the noncatalytic C-terminal domain is essential (19–21). Consistently, both a previous study (12) and this study showed that disruption of the C-terminal region of POL2A also causes lethality in Arabidopsis, but a mutant affecting its N-terminal region is viable (SI Appendix, Fig. S1 C and F), consistent with previous observation in Drosophila (37) and humans (14), suggesting functional conservation of DNA Pol-ε among major eukaryotic groups (20).
Meiotic DSBR models invoke leading-strand DNA synthesis for yielding both COs and NCOs, but the role of DNA Pol-ε in meiosis has not been characterized. Here, we showed that two mutations in Arabidopsis POL2A and meiosis-specific knockdown of POL2 (pDMC1-POL2RNAi) cause similar defects in fertility and meiosis (Fig. 1 and SI Appendix, Fig. S7 K–O), supporting the idea that the observed meiotic defects in pol2a are caused by the POL2A mutation. Because POL2A is a DNA Pol, an obvious possibility of POL2A function in meiosis is in premeiotic DNA replication. Although we found no evidence for the mutants used here being defective in premeiotic DNA replication, we present several lines of evidence that support a crucial role of POL2A in DNA synthesis during meiotic recombination. First, pol2a mutants showed normal vegetative growth and produced 20 chromatids, indicating that these mutants could still replicate chromosomes. Second, observation of the delayed peak in pachytene cell distribution in mutant suggests that POL2A likely plays a role in a late stage of meiotic recombination (Fig. 4C). Third, the formation of DSBs seemed normal in pol2a (Fig. 4A). Fourth, the possible nonhomolog association and chromosome fragmentation in pol2a depend on SPO11-1–generated DSBs. Fifth, genetic analysis showed that POL2A acts downstream of RAD51 and upstream of RCK/MER3. Moreover, genetic studies of POL2A with RFC1 also support the idea that they function in a similar process in meiotic recombination. The weaker meiotic defect in pol2a than that in rfc1 suggests that lagging-strand synthesis is likely more important for double Holliday junction (dHJ) formation (Fig. 6), because Pol-δ, a DNA Pol required for lagging-strand elongation, can partially substitute the role for elongation in leading-strand synthesis (see below).
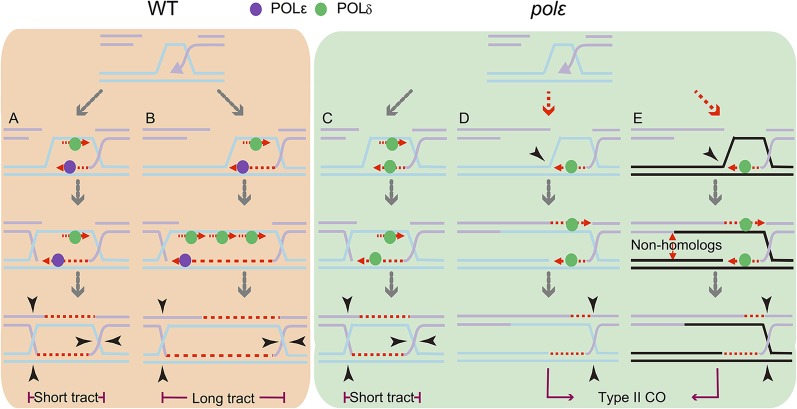
Fig. 6.
A model for POL-ε participation in meiotic type I COs. Our previous study presented that meiotic type I CO likely requires both leading- and lagging-strand DNA synthesis. In this model, we propose that, (A and B) in the WT, both short and long tracts involve POL-ε and POL-δ for leading- and lagging-strand synthesis, respectively. In the absence of POL-ε, for the COs with short tracts, POL-δ can replace POL-ε to carry out initiation and limited synthesis, such as (C) one Okazaki fragment, whereas for COs with long tracts, POL-δ cannot fully replace the elongation function of POL-ε. The failure of dHJ formation may result in the formation of MUS81-dependent type II COs between (D) homologs and (E) nonhomologs.
Elongation of DNA Synthesis Likely Promotes dHJ Formation.
Meiotic recombination is initiated from programmed DSBs (4), and DNA synthesis is required for DSBR during meiotic recombination. However, the underlying mechanism has not been characterized. Recently, we showed that formation of meiotic type I CO likely requires lagging-strand DNA synthesis (16) and proposed that lagging-strand synthesis promotes dHJ formation. This idea is consistent with the observation that more DNA synthesis accompanies CO formation than NCO formation in yeast (5). In mitotic DNA replication, POL-ε and POL-δ perform elongation of leading and lagging strands, respectively (8, 9). Unlike POL-ε for continuous elongation of the leading strand, POL-δ is only responsible for elongation of Okazaki fragments on the lagging strand (9). Studies in yeast showed that POL-δ can partially replace POL-ε for leading-strand synthesis (8, 9). This finding raises a question of whether there is a threshold of amount of DNA synthesis to differentiate the requirement for distinct DNA synthesis factors. In yeast mitosis, mutations in the N-terminal exonuclease domain of Pol-ε have no obvious defects in DNA replication initiation but greatly reduce efficiency of leading-strand elongation by 10- to 100-fold compared with the WT (17–19, 38), providing a possible means to study the role of elongation in meiosis. Here, we provided the following pieces of evidence to support the idea that POL2A participates in type I CO formation. First, the pol2a mutants have reduced type I COs, but the residual COs are more than those of rfc1 and the zmm mutants (27), indicating that the defect in elongation affects a portion of type I COs. Second, genetic analyses of pol2a with factors involved in the type I pathway, such as rfc1 and mer3/rck (Fig. 3 H and J), show similar chromosome morphologies to pol2a, whereas the pol2a double mutant with a mutation in the type II gene MUS81 has more severe phenotype than either single mutant (Fig. 3L). Third, CO interference in pol2a is significantly reduced (Fig. 5 B and C).
Why did POL2A only affect some type I COs? Recent genome-wide analyses showed that CO-associated tracts are usually longer than those of NCO in both yeast and Arabidopsis, ranging from several hundreds to thousands of base pairs (2, 6, 7, 39). In Arabidopsis, the mean length of CO-associated tracts is ∼500 bp (2, 7), whereas an Okazaki fragment for discontinuous lagging-strand synthesis is about 100–200 bp (40). It is reasonable to assume that only COs with long conversion tracts require elongation to fulfill the need for more extensive leading-strand synthesis. We, therefore, hypothesize that the CO-associated conversion tracts with >200 bp may require POL2A, providing an explanation for why POL2A affects some type I COs.
Previous models for meiotic recombination are not explicit regarding leading-strand elongation (4). The pol2a mutations that we analyzed here are similar to a mutation in yeast Pol-ε that showed no obvious defects in DNA replication initiation but caused greatly reduced leading-strand elongation efficiency (17–19, 38). The extensive phenotypic analyses presented here are consistent with these Arabidopsis mutations, affecting only elongation of leading strands but not initiation during meiotic recombination. Therefore, we present a revised model for describing the meiotic type I CO pathway (Fig. 6), incorporating both lagging-strand synthesis and leading-strand elongation. In this model, we suggest a threshold of DNA synthesis (say a length of 200 bp for discussion) to discriminate two different kinds of CO-associated conversion tracts: long tracts with >200 bp and short tracts with <200 bp. In the WT, both long and short tracts involve the functions of both Pol-δ and Pol-ε (Fig. 6 A and B). In the absence of POL2A (Pol-ε), for the COs with short tracts, Pol-δ can replace Pol-ε to carry out initiation and limited synthesis, similar to that for an Okazaki fragment (Fig. 6C), whereas for COs with long tracts, Pol-δ cannot perform the elongation function of Pol-ε. For these COs, a failure of dHJ formation in the pol2a mutation might result in formation of MUS81-dependent COs (Fig. 6D). Alternatively, the single invasion of the 3′ end may withdraw to find a nonhomolog to yield a CO as resolved by MUS81 (Fig. 6E). This model is further supported by the finding that a weak mutation of yeast Pol-δ causes a reduction in the length of meiotic gene conversion tracts and CO number (15, 36).
In summary, meiotic recombination is essential for sexual reproduction in eukaryotes, and most organisms have at least two distinct types I and II COs (41, 42), including the budding yeast, animals, and flowering plants; whereas DNA Pol is also highly conserved in eukaryotes (10), we hypothesize that discrimination of meiotic types I and II COs by the amount of associated DNA synthesis may be a general mechanism. However, the precise threshold length of the conversion tract that is required for factors to coordinate leading- and lagging-strand synthesis might be different in different organisms.
Materials and Methods
Materials used in this study are all Columbia (Col-0) background. Details of plant materials and growth conditions are described in SI Appendix, SI Materials and Methods. Assays of characterization of mutant phenotypes, like Alexander red staining, tetrad and chromosome morphology analyses, FISH and immunofluorescence observation, statistics of chiasmata distribution, and others are also described or follow the references listed in SI Appendix, SI Materials and Methods. Statistical analyses of t tests were performed in Microsoft Excel 2013 or Rstudio v0.98.501. Statistical significance was showed as P < 0.05 and P < 0.01.
Supplementary Material
Acknowledgments
We thank Prof. Raphaël Mercier at Institut National de la Recherche Agronomique (INRA) (France) for providing MLH1 antibody. This work was supported by Ministry of Science and Technology of China Grants 2011CB944603 and 2012CB910503, International Technology Cooperation Project of Shanghai Grant 13430720300, Shanghai “Post-Qi-Ming-Xing Plan” for Young Scientists Grant 13QA1400200, National Natural Science Foundation of China (NSFC) Grant 31370347, and funds from the State Key Laboratory of Genetic Engineering, Fudan University, and Rijk Zwaan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507165112/-/DCSupplemental.
References
- 1.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88(3):375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 2.Lu P, et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 2012;22(3):508–518. doi: 10.1101/gr.127522.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y, et al. Deep genome-wide measurement of meiotic gene conversion using tetrad analysis in Arabidopsis thaliana. PLoS Genet. 2012;8(10):e1002968. doi: 10.1371/journal.pgen.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 5.Terasawa M, et al. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proc Natl Acad Sci USA. 2007;104(14):5965–5970. doi: 10.1073/pnas.0611490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454(7203):479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijnker E, et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife. 2013;2:e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strich R. Meiotic DNA replication. Curr Top Dev Biol. 2004;61:29–60. doi: 10.1016/S0070-2153(04)61002-7. [DOI] [PubMed] [Google Scholar]
- 9.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7(12):e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317(5834):127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenik PD, Jurkuta RE, Barton MK. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell. 2005;17(12):3362–3377. doi: 10.1105/tpc.105.036889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H, et al. Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell. 2009;21(2):386–402. doi: 10.1105/tpc.108.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palles C, et al. CORGI Consortium WGS500 Consortium Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maloisel L, Bhargava J, Roeder GS. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics. 2004;167(3):1133–1142. doi: 10.1534/genetics.104.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, et al. The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 2012;8(11):e1003039. doi: 10.1371/journal.pgen.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′----5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shcherbakova PV, Pavlov YI. 3′-->5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142(3):717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3(5):679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 20.Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: An inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274(32):22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Lu P, Wang Y, Ma H. The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: The complexity and evolution of the meiotic process. Plant J. 2011;65(4):503–516. doi: 10.1111/j.1365-313X.2010.04439.x. [DOI] [PubMed] [Google Scholar]
- 23.Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20(3):589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA. 2004;101(29):10596–10601. doi: 10.1073/pnas.0404110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aksenova A, et al. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase ε. PLoS Genet. 2010;6(11):e1001209. doi: 10.1371/journal.pgen.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowndes NF, Toh GW. DNA repair: The importance of phosphorylating histone H2AX. Curr Biol. 2005;15(3):R99–R102. doi: 10.1016/j.cub.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 27.Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011;190(3):523–544. doi: 10.1111/j.1469-8137.2011.03665.x. [DOI] [PubMed] [Google Scholar]
- 28.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3(8):e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18(2):117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, Zhang W, Timofejeva L, Gerardin Y, Ma H. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J. 2005;43(3):321–334. doi: 10.1111/j.1365-313X.2005.02461.x. [DOI] [PubMed] [Google Scholar]
- 31.Mercier R, et al. Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3,whereas the other one is not. Curr Biol. 2005;15(8):692–701. doi: 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 32.Berchowitz LE, Copenhaver GP. Fluorescent Arabidopsis tetrads: A visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc. 2008;3(1):41–50. doi: 10.1038/nprot.2007.491. [DOI] [PubMed] [Google Scholar]
- 33.Jackson N, et al. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 2006;25(6):1315–1323. doi: 10.1038/sj.emboj.7600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albertson TM, et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci USA. 2009;106(40):17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, et al. Mutation in the catalytic subunit of DNA polymerase alpha influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 2010;61(1):36–45. doi: 10.1111/j.1365-313X.2009.04026.x. [DOI] [PubMed] [Google Scholar]
- 36.Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol Cell Biol. 2008;28(4):1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suyari O, et al. Differential requirement for the N-terminal catalytic domain of the DNA polymerase ε p255 subunit in the mitotic cell cycle and the endocycle. Gene. 2012;495(2):104–114. doi: 10.1016/j.gene.2011.12.056. [DOI] [PubMed] [Google Scholar]
- 38.Hogg M, Johansson E. DNA polymerase ε. Subcell Biochem. 2012;62:237–257. doi: 10.1007/978-94-007-4572-8_13. [DOI] [PubMed] [Google Scholar]
- 39.Qi J, et al. Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics. 2009;10:475. doi: 10.1186/1471-2164-10-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hübscher U, Seo YS. Replication of the lagging strand: A concert of at least 23 polypeptides. Mol Cells. 2001;12(2):149–157. [PubMed] [Google Scholar]
- 41.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: Localization and regulation. Nat Rev Genet. 2013;14(11):794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 42.Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.