Significance
Pancreas differentiation is of interest as an example of organogenesis and for its medical implications. We report here that regulation of protein stability is a player in the differentiation of certain cell lineages in the early zebrafish pancreas. The related E3 ubiquitin ligases Ligand of Numb protein-X (Lnx)2a and Lnx2b affect differentiation of exocrine cells, apparently by destabilizing Numb in exocrine progenitor cells. Numb is an inhibitor of Notch, a key regulator of pancreatic development. We suggest that Lnx2a/b destabilize Numb to derepress Notch, allowing precursors to proliferate and support subsequent differentiation. This study also highlights the fact that detailed analysis of morpholino versus mutant phenotypes may be required for a full understanding of their relationship.
Keywords: pancreas, Numb, Notch, morpholino, TALEN
Abstract
The gene encoding the E3 ubiquitin ligase Ligand of Numb protein-X (Lnx)2a is expressed in the ventral-anterior pancreatic bud of zebrafish embryos in addition to its expression in the brain. Knockdown of Lnx2a by using an exon 2/intron 2 splice morpholino resulted in specific inhibition of the differentiation of ventral bud derived exocrine cell types, with little effect on endocrine cell types. A frame shifting null mutation in lnx2a did not mimic this phenotype, but a mutation that removed the exon 2 splice donor site did. We found that Lnx2b functions in a redundant manner with its paralog Lnx2a. Inhibition of lnx2a exon 2/3 splicing causes exon 2 skipping and leads to the production of an N-truncated protein that acts as an interfering molecule. Thus, the phenotype characterized by inhibition of exocrine cell differentiation requires inactivation of both Lnx2a and Lnx2b. Human LNX1 is known to destabilize Numb, and we show that inhibition of Numb expression rescues the Lnx2a/b-deficient phenotype. Further, Lnx2a/b inhibition leads to a reduction in the number of Notch active cells in the pancreas. We suggest that Lnx2a/b function to fine tune the regulation of Notch through Numb in the differentiation of cell types in the early zebrafish pancreas. Further, the complex relationships among genotype, phenotype, and morpholino effect in this case may be instructive in the ongoing consideration of morpholino use.
The pancreas is a vertebrate-specific bifunctional organ that is composed of exocrine tissue for secretion of digestive enzymes and endocrine tissue for production of hormones involved in regulating glucose homeostasis. Morphogenesis of the developing pancreas has been well characterized in amniotes and other vertebrates (1). The zebrafish has emerged as a useful model organism for studying pancreas formation. As in mammals, the zebrafish pancreas develops from two distinct pancreatic anlagen of the endoderm, the dorsal-posterior bud and the ventral-anterior bud, which subsequently fuse to form the definitive pancreas. In zebrafish, the dorsal bud gives rise to the primary islet, whereas the ventral bud gives rise to exocrine cells, the pancreatic duct, and secondary islets (2, 3).
Pancreas development is regulated by a network of transcription factors and signal transduction pathways. The Pdx1 homeobox factor is of critical importance to pancreas formation in the mouse (4, 5) and is the earliest marker for cells specified as pancreatic precursors in all animals studied including the zebrafish (6). The basic helix-loop-helix factor Ptf1a is essential for the development of exocrine precursor cells in the mouse (7) and zebrafish (8, 9) and, furthermore, represents a valuable early marker for exocrine precursors. Among multiple signaling pathways that have a role in the specification and differentiation of the pancreas, the Notch pathway has received particular attention. Studies in the mouse showed that artificial activation of the Notch pathway prevents precursors from differentiating into functional pancreatic cell types (10). Further, manipulation of Notch signaling affects the balance between endocrine and exocrine differentiation in the pancreas (11, 12). Several studies in zebrafish have focused on the secondary transition in which a specific population of ventral bud-derived cells differentiates into secondary islets that eventually account for the majority of endocrine cells in the mature organ (13–15). These studies conclude that cell differentiation in the pancreas depends on a reduction or cessation of Notch signaling, whereas, in turn, precursor maintenance requires a Notch signal. Further, specific levels of Notch signaling can direct precursor cells to distinct fates, and the Notch pathway affects cell proliferation in this system.
In previous work, we have studied the role of the E3 ubiquitin ligase Ligand of Numb protein-X (Lnx)2b (16, 17) in embryonic development (18–20). We proceeded to explore possible functions of the other lnx2 paralog in zebrafish, lnx2a. The lnx2a gene is expressed in the pancreas anlage in addition to the nervous system, and lnx2a knockdown mediated by a splice morpholino (MO) led to differential inhibition of exocrine cell differentiation. Because the role of ubiquitylation and protein stability in pancreas development has received little attention, we pursued these observations further. A null mutation in lnx2a did not mimic the MO-induced phenotype because of redundancy of the lnx2a and lnx2b genes. We could show that the splice MO led to exon 2 skipping and the production of an N-truncated Lnx2a protein that acts as an interfering factor. This effect could be demonstrated most clearly by studying a mutation that deletes the exon 2 splice donor site targeted by the MO. The mutant contained the N-truncated protein also seen in the morphant and fully reproduced the exocrine deficiency phenotype. Further we provide evidence that Lnx2a and Lnx2b act in pancreas development by destabilizing Numb, thereby affecting Notch signaling. We conclude that regulation of protein stability is an important mechanism in early pancreas development in zebrafish. Further, this example shows that nonreplication of an MO phenotype by a null mutation need not indicate off-target effects but, in this case, helped reveal a more complex underlying mechanism.
Results
Early Differential Expression of lnx2a in the Ventral Pancreatic Bud.
In the context of our interest in the function of Lnx family E3 ubiquitin ligases in embryonic development (18–20), we examined the expression of lnx2a in the zebrafish embryo. In addition to expression in the CNS, lnx2a is transiently expressed in the ventral, but not in the dorsal pancreatic bud from 19 h after fertilization (hpf; 20–25 somites) to 55 hpf. In addition to an embryo overview, Fig. 1 illustrates the relative expression domains of lnx2a and the marker genes pdx1 (pan-pancreas), insulin (β-cells), foxa3 (endoderm), and ptf1a (exocrine cells). SI Appendix, Fig. S1 shows a time course of lnx2a expression and a drawing of zebrafish pancreas development illustrating the lnx2a expression pattern. Ptf1a, the earliest ventral pancreas-specific transcription factor, is expressed from 33 hpf on (refs. 8 and 9). Lnx2a seems to be the earliest gene differentially expressed in the ventral pancreatic bud and, therefore, we tested whether lnx2a has a role in the specification of different pancreatic domains in the zebrafish embryo.
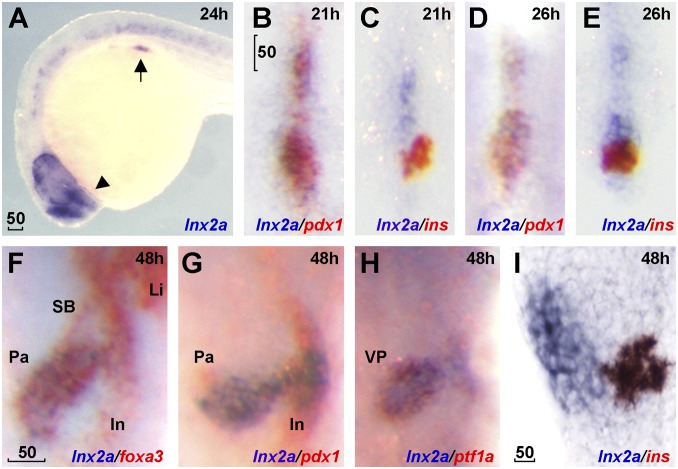
Fig. 1.
lnx2a is expressed in the ventral pancreas during early pancreas specification. Lateral views (A and I), dorsal views (B–E), and ventral views (F–H). (A) lnx2a transcripts (blue) are detected in regions of the endoderm (arrow), forebrain (arrowhead), and the spinal cord at 24 hpf. Expression of pdx1 (B, D, and G), ins (C, E, and I) and foxa3 (F) are in red. lnx2a expression is observed in the antero-ventral part of pdx1-positive pancreas precursors at 21 (B) and 26 (D) hpf, but excluded from β-cells (ins+) in the dorsal pancreas at 21 (C), 26 (E), and 48 (I) hpf. At 48 hpf, lnx2a expression is detected in the ventral pancreas but not in the intestine, swim bladder, or liver (F–H). SB, swim bladder; In, intestine; Li, liver; Pa, pancreas; VP, ventral pancreas. (Scale bars: 50 μm.)
lnx2a Knockdown Using a Splice MO Inhibits Exocrine Pancreas Differentiation.
To explore the pancreatic role of Lnx2a, we used a splice MO (hereafter named lnx2a-MO) that effectively blocks splicing between the first coding exon, exon 2, and exon 3 (Fig. 2A and SI Appendix, Figs. S2 and S3). Whole-mount in situ hybridization (WISH) using markers for ventral/exocrine (trypsin, ptf1a) and dorsal/endocrine cells (insulin, glucagon) revealed that injection of lnx2a-MO causes a severe deficit in ventral but not dorsal markers (Fig. 2 B–E). Reduction in ventral pancreas formation was confirmed by using WISH with additional markers at different stages of development (SI Appendix, Fig. S2 C and D). Using the transgenic strain Tg(ptf1a:EGFP), cell type-specific antibodies, and confocal laser scanning microscopy, we illustrate the reduction in ventral pancreatic markers at the protein level as well (SI Appendix, Fig. S2E). In contrast to the strong reduction of ventral pancreatic cells, other organ precursors in the endoderm formed normally, suggesting that Lnx2a is involved in the development of the ventral pancreas but not the initial specification of the endoderm (SI Appendix, Fig. S2 C and D).
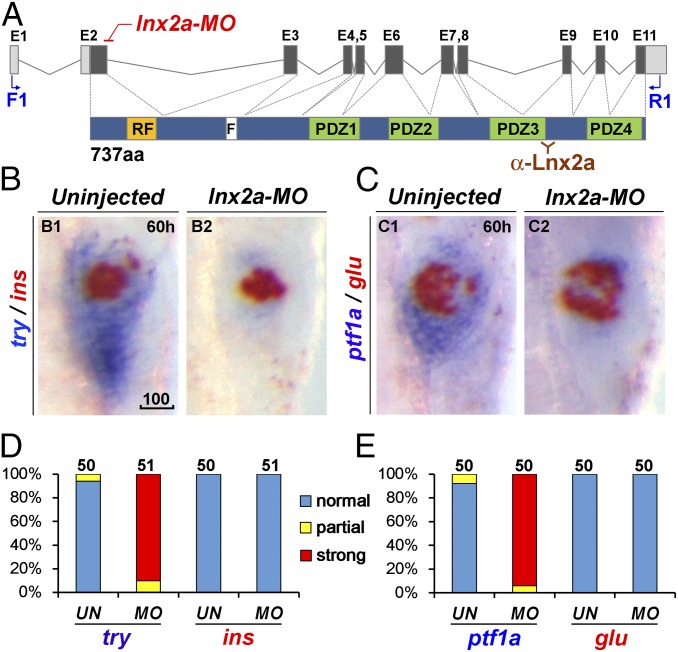
Fig. 2.
lnx2a knockdown causes defects in the exocrine pancreas. (A) Schematic drawing of the lnx2a locus containing 11 exons, and the Lnx2a protein (737 aa) containing a RING-finger domain (RF), the Numb binding NPAF motif (F), and four PDZ domains. The translation initiation site is located in exon 2, which encodes the RING-finger domain. The lnx2a-MO targets the splice donor site of exon 2. Primers in exon 1 (F1) and exon 11 (R1) are shown, as is the epitope for the Lnx2a antibody in the third PDZ domain. (B and C) The lnx2a-MO (5 ng) leads to inhibition of exocrine markers (try, ptf1a), but not endocrine markers (ins, glu), as seen by two-color WISH at 60 hpf. (D and E) Quantification of marker expression; effects were classified as strong, partial, and unaffected. MO, lnx2a-MO; UN, uninjected. (Scale bar: 100 μm.)
A lnx2a Null Mutant Does Not Recapitulate the Morphant Phenotype.
The validity of MO-induced phenotypes has been questioned recently (21), yet the high regional precision of the lnx2a-MO–induced defects seems to argue against a nonspecific effect. To investigate this issue, we generated lnx2a null mutants by using TALEN-mediated gene targeting (22). The lnx2aΔ70 mutation has a frame shift early in the protein coding region, leading to early termination of translation (Fig. 3 A and B), and contains no detectable Lnx2a protein (Fig. 3C and SI Appendix, Fig. S3A). Thus, lnx2aΔ70 is almost certainly a null mutation, yet the mutant embryos displayed no pancreatic phenotype (Fig. 3 D1, D3, and E). Further, homozygous mutant fish are morphologically normal, and are viable and fertile. Thus, we investigated the possible functional redundancy of lnx2a with lnx2b, a paralog encoding a similar protein (SI Appendix, Fig. S3B); lnx2b is expressed in the endoderm including the pancreas in zebrafish embryos (SI Appendix, Fig. S3C). To test for possible redundancy, we injected a validated (18) MO against lnx2b (SI Appendix, Fig. S3 D and E) into WT and lnx2aΔ70 mutant embryos, and found that the MO generated a ventral pancreas deficiency phenotype in mutant but not in WT embryos (Fig. 3 D2, D4, and E). Thus, redundant lnx2b function may account for the fact that lnx2a is dispensable for pancreatic development.
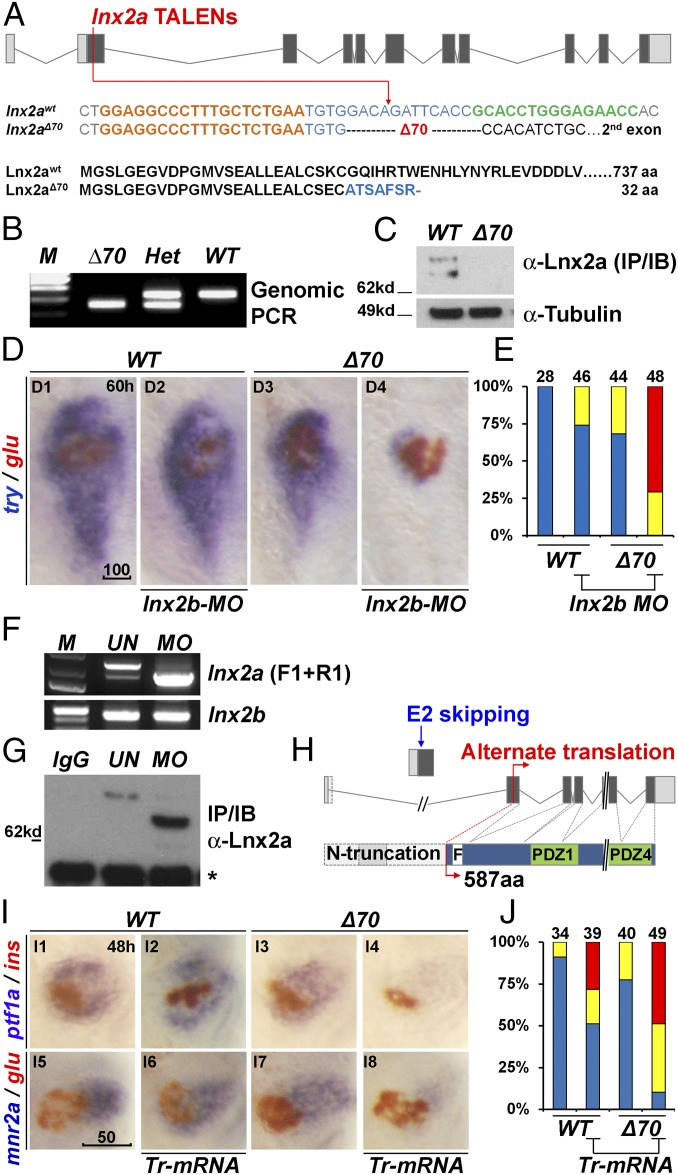
Fig. 3.
Generation of lnx2aΔ70 mutant using TALENs, and the functional redundancy of lnx2 genes. (A) Schematic representation of the lnx2a locus and the TALEN target site in the first protein coding exon (exon 2). TALEN targets (left, orange and right, green), and the lnx2aΔ70 mutation are shown below the drawing. Next, the protein sequences of WT and the lnx2aΔ70 frameshift allele are shown. Genotype of lnx2aΔ70 mutant was analyzed by the genomic PCR (B), and immunoblotting of endogenous Lnx2a protein (C). For validation of the anti-Lnx2a antibody, see SI Appendix, Fig. S2F. (D) Marker gene (try and glu) expression shows little effect in lnx2aΔ70 null mutants, and in lnx2b-MO–injected embryos at 60 hpf. However, lnx2b-MO injection into lnx2aΔ70 mutant embryos shows suppression of exocrine markers. (E) Quantification of pancreatic defects by analysis of trypsin expression, classified as in Fig. 2D. See SI Appendix, Fig. S3 D and E for effectiveness of lnx2b-MO. (F–H) lnx2a-MO leads to production of N-truncated Lnx2a protein. (F) RT-PCR using primers F1 and R1 (defined in Fig. 2A) and 48 hpf embryo RNA, followed by sequencing, shows that lnx2a-MO injection results in exon 2 skipping and stabilization of the resulting transcript. lnx2b expression was not changed by lnx2a-MO injection. (G) Endogenous Lnx2a showed the expected size in controls, but a smaller protein was seen in lnx2a-MO–injected embryos, which was identified by mass spectrometry (SI Appendix, Fig. S4) as an N-truncated protein (587 aa) arising from an alternate start site in exon 3. (H) Schematic drawing of exon 2 skipping, alternate translation start site, and N-truncated protein in lnx2a-MO–injected embryos. (I) N-truncated Lnx2a (exon 2-skipped, as shown in F–H) has interfering effect. Tr-mRNA was injected into the WT and lnx2aΔ70 mutant embryos and analyzed at 48 hpf. (J) Quantification of pancreatic defects in I by analysis of ptf1a expression. Injection of Tr-mRNA into WT embryos has little effect, but leads to the exocrine pancreas phenotype in lnx2aΔ70 mutants. (Scale bars: D, 100 μm; I, 50 μm.)
Because a block to exon 2/3 splicing disconnects the standard initiation site in exon 2 from downstream coding, the lnx2a morphant cannot produce full-length Lnx2a protein (Fig. 2A and SI Appendix, Fig. S2 A and B). Nevertheless, RT-PCR and antibody blotting showed that morphant embryos produce exon 2 skipped RNA (Fig. 3 F and H) and, importantly, abundant levels of Lnx2a immunoreactive protein with a molecular size smaller than the wild type (Fig. 3G). Using mass spectrometry, we showed that the short form of Lnx2a represents an N-terminally truncated form that is initiated from a cryptic start site in exon 3 (Fig. 3H and SI Appendix, Fig. S4). Truncated Lnx2a has lost the RING domain that has been shown previously to be required for E3 ubiquitin ligase activity of Lnx proteins (16–18), as confirmed in SI Appendix, Fig. S5 A and B. The N-truncated protein retains the NPAF motif and the PDZ domains that mediate protein–protein interactions (Figs. 2A and 3H), suggesting that it could act as a negative interfering form. This suggestion is borne out by the observation that injection of the truncated mRNA into lnx2aΔ70 mutant embryos re-created the loss of exocrine marker phenotype (Fig. 3 I and J). A weak phenotype was seen after truncated mRNA injection into WT embryos, most likely because the truncated protein has only a moderately strong interfering effect. We conclude that lnx2a and lnx2b have redundant function in pancreas development so that loss of expression of one paralog has no phenotype but sensitizes the organism to further interference with Lnx2 protein function.
The lnx2aΔ329 Mutant Recapitulates the Morphant Phenotype.
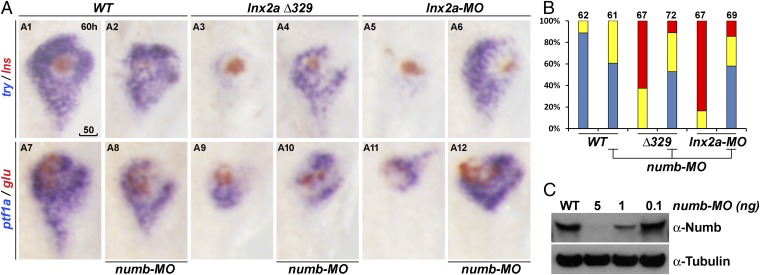
Using the same TALEN pair described above, we isolated a larger deletion, lnx2aΔ329 (Fig. 4 A and B). In this mutation, the exon 2 splice donor site has been lost, providing a definitive genetic model for the putative effect of the lnx2a-MO. As predicted, lnx2aΔ329 mutant embryos express the same truncated form of Lnx2a protein as the morphant (Fig. 4C and SI Appendix, Fig. S4). Most critically, lnx2aΔ329 mutants shows the same pancreatic phenotype as the morphant, with specific inhibition of exocrine marker expression, whereas endocrine markers are essentially unaffected (Fig. 4 D and E). This fact is true for both zygotic and maternal/zygotic mutants (Fig. 4E). It should be noted that homozygous lnx2aΔ329 zebrafish show high embryonic lethality, but some mutant fish survive, allowing the cross illustrated in bars 2 and 3 of Fig. 4E. The behavior of the lnx2aΔ329 mutant provides support for the conclusion that the phenotype is due to loss of functional Lnx2a combined with interference with the function of Lnx2b, and that the lnx2a morphant phenotype is based on the same mechanism. These results legitimize the use and interpretation of the lnx2a-MO.
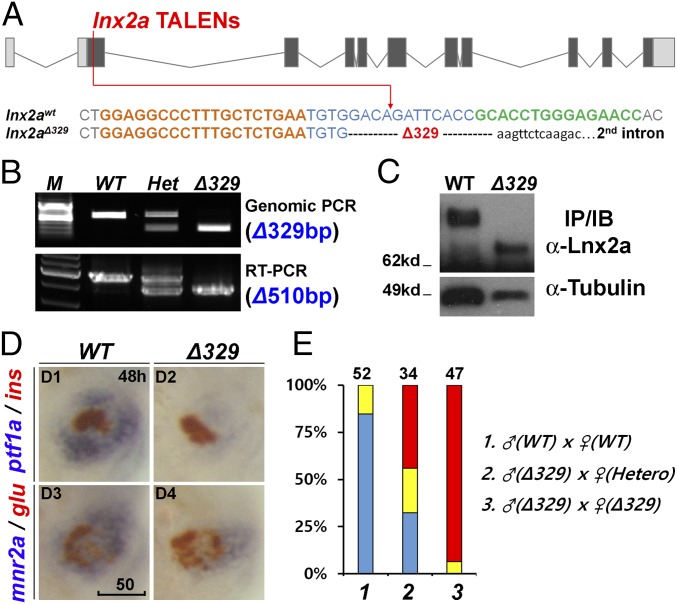
Fig. 4.
The lnx2aΔ329 mutant recapitulates the morphant phenotype. (A) Sequence of the lnx2aΔ329 mutation (Fig. 3A). The lowercase sequence in the mutant indicates that it is in intron 2, with lnx2aΔ329 having lost the exon 2 splice donor site. (B and C) Genotyping of lnx2aΔ329 by genomic PCR (Δ329), RT-PCR (Δ510 equaling exon 2) (B), and by Western blotting of embryo extracts showing the presence of N-truncated protein (C). (D) Phenotypic analysis of lnx2aΔ329 mutants at 48 hpf. (Scale bar: 50 μm.) (E) The quantification of pancreatic defects by analysis of ptf1a expression, classified as in Fig. 2D. Development of the exocrine pancreas is specifically suppressed in lnx2aΔ329.
The Ventral Pancreas Defects in lnx2aΔ329 Mutants and lnx2a Morphants Are Rescued by Knockdown of Numb.
Lnx is known to target Numb for ubiquitylation and degradation (16), and Numb is an inhibitor of Notch signaling (23–26). Because Notch signaling has a major role in the specification and differentiation of various pancreatic cell types (10, 13–15) we asked whether the Lnx2a morphant and mutant phenotype is due to a change in the activity of Numb. Numb is expressed in the embryonic endoderm including the pancreas, but whereas numb RNA could be detected in the entire pancreas, the protein was detected only in the dorsal bud-derived primary islet (SI Appendix, Fig. S5 C and D). Because the known ubiquitylation and degradation of Numb by Lnx proteins (16, 18) was confirmed for zebrafish Lnx2a (SI Appendix, Fig. S5 A and B), it seemed possible that Lnx2a reduces the level of Numb protein in the ventral pancreas where Lnx2a is expressed (Fig. 1). The lnx2a mutant/morphant phenotype might then be due to abnormal stabilization of Numb; we illustrate in the next section that there is, in fact, an increase of Numb-positive cells in lnx2a morphants. Therefore, we tested whether reduction of Numb expression could rescue the lnx2a-deficient phenotype. This prediction proved correct: Both lnx2aΔ329 mutant and lnx2a morphant phenotypes were substantially rescued by injection of numb-MO (Fig. 5 and SI Appendix, Fig. S5E). The absence of a gross morphological effect of the numb-MO itself is consistent with the work of Bresciani et al. (27). This agreement, and the fact that we observe a specific rescue phenotype rather than developmental malformation, should help alleviate concerns regarding the validity of the numb-MO in this context. These results support the view that Lnx2a and Lnx2b function in early pancreas development to limit the extent and area of Numb protein accumulation and activity.
Fig. 5.
Ventral pancreas defects in lnx2aΔ329 mutant and lnx2a-MO–injected embryos can be rescued by knockdown of Numb. (A) The translation blocking MO for Numb (5 ng) was injected into WT, lnx2aΔ329-, and lnx2a-MO–injected embryos, and pancreatic phenotype was examined at 60 hpf. (Scale bar: 50 μm.) (B) Quantification of pancreatic defects by analysis of ptf1a expression. Exocrine pancreas defects were substantially rescued by knockdown of Numb; numb-MO had little effect in WT embryos. (C) Immunoblotting of embryo extracts with Numb antibody shows the efficiency of the numb-MO.
Loss of Lnx2 Activity Causes a Reduction of Notch-Responsive Cells in the Pancreas.
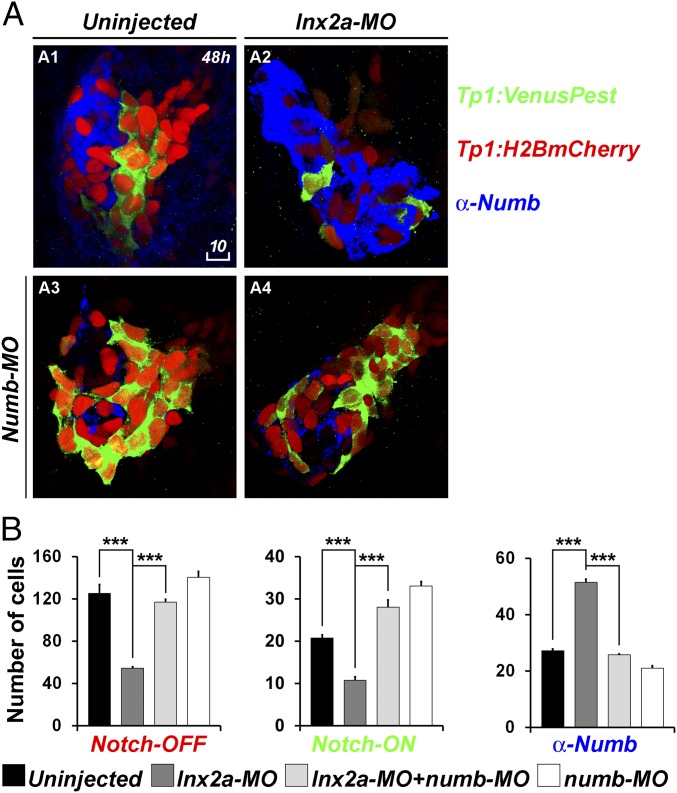
Notch signaling is a key regulator of pancreas formation, affecting specification of different cell types and controlling their quiescence or proliferation (10, 13–15, 28). We asked whether Lnx2a, through its modulation of Numb protein stability, can affect Notch activity in the embryonic pancreas. For this purpose, we used the transgenic strain Tg(Tp1:H2BmCherry); Tg(Tp1:VenusPest) in which a Notch-responsive element drives expression of stable nuclear mCherry and unstable Venus (13). In this strain, red nuclei signify cells in which Notch signaling was previously active but has now been turned off, whereas green and double-positive cells are active for Notch signaling when examined. We find that lnx2a-MO increased Numb-positive cells and reduced Notch-active cells in the pancreas (Fig. 6 A, 2 and B, 1 and 2). Furthermore, coinjection of numb-MO rescues the effect of lnx2a-MO on the abundance of Tg(Tp1:VenusPest) expressing Notch-ON cells (Fig. 6 A, 4 and B). We propose that numb is transcribed in the entire pancreatic anlage, but the protein is destabilized by Lnx2 in the ventral bud. Loss of Lnx2 allows Numb accumulation in the ventral bud, interfering with Notch regulation and cell differentiation.
Fig. 6.
lnx2a-MO injection causes a reduction of Notch-responsive cells in the pancreas. (A) Tg(Tp1:H2BmCherry); Tg(Tp1:VenusPest) embryos were injected with lnx2a-MO (A2), numb-MO (A3), or both (A4). At 48 hpf, embryos were stained with anti-Numb antibody and analyzed by confocal microscopy; Z projections are shown. (Scale bar: 10 μm.) (B) Total number of Notch-OFF (Tp1:H2BmCherry+) and Notch-ON (Tp1:H2BmCherry+ and Tp1:VenusPest+) cells, as well as Numb-positive cells, were counted in all Z sections of four embryos for each category. **P < 0.005; ***P < 0.002.
To test how Lnx2 affects ventral bud precursor populations, we tested Notch activity and Numb accumulation at earlier stages. In Lnx2a morphants at 36 hpf, the number of Tg(Tp1:VenusPest)-positive cells is reduced but not as strongly as at 48 hpf, and the number of Numb+ cells is increased (Fig. 6 and SI Appendix, Fig. S6). At 24 hpf, before the ventral bud marker ptf1a is expressed, no difference between wild type and morphant embryos was seen (SI Appendix, Fig. S6).
Reduced Cell Proliferation in Lnx2a-Deficient Pancreas.
Notch signaling affects the cell cycle in the early pancreas (13, 14). We used Tg(ptf1a:EGFP) embryos to visualize ventral pancreatic cells and BrdU to mark proliferating cells. Injection of lnx2a-MO resulted in a substantial reduction in EGFP+ cells and an increase in Numb+ cells, in agreement with the phenotype described earlier (Fig. 7 A, 1 and 3 and B). Further, the number of BrdU+/EGFP+ cells decreased in concert with the reduction of total EGFP+ cells, consistent with the view that proliferation of Ptf1a-expresssing cells is reduced in the Lnx2-deficient pancreas (Fig. 7).
Fig. 7.
lnx2a-MO injection results in impaired cell proliferation in the ventral pancreas. (A) Tg(ptf1a:EGFP) embryos were incubated in 10 mM BrdU from 46 to 47 hpf, fixed, stained with anti-BrdU and anti-Numb antibodies, and analyzed by confocal microscopy. For clarity, the same plane of the BrdU channel is shown separately in addition to the merged images. (Scale bar: 10 μm.) (B) The average number of ptf1:EGFP+, Numb+ and ptf1:EGFP+/BrdU+ double-positive cells in the pancreas of 10 uninjected and lnx2a-MO–injected embryos are shown (***P < 0.001).
Are fewer ventral bud precursors specified in mutants/morphants, or does the initial population fail to proliferate normally? Early ventral bud markers, ptf1a and mnr2a, are already reduced in the morphants at the earliest time when they can be detected, but the degree of reduction appears to increase as development proceeds (SI Appendix, Fig. S7). This fact suggests that Lnx2a regulates both the entry of cells into the ventral pancreas precursor pool and the expansion of this population during development.
Discussion
Functional Redundancy Between Lnx2a and Lnx2b.
The mammalian LNX family is composed of five members, but only LNX1 and LNX2 share an N-terminal RING-finger domain, a NPAY/F motif for NUMB binding, and four PDZ domains (29, 30). The in vivo functions of LNX1/2 remain incompletely known. Mammalian LNX1 was shown to act as an E3 ubiquitin ligase that mediates ubiquitylation and degradation of Numb in cultured cells (16, 17). Lnx1 and Lnx2 are expressed in the nervous system and may have a role in neuronal cell proliferation and differentiation (31, 32). Further, interactions of Lnx1/2 have been analyzed with individual proteins or at a global proteomic level (33–35). However, to our knowledge, no genetic test of Lnx1/2 function has been reported.
In zebrafish, three LNX1/2 homologs (lnx1, lnx2a, and lnx2b) have been identified (18), and Lnx2b affects dorso-ventral patterning by modulation of Dharma (Bozozok, Nieuwkoid) stability (18, 19). In eutherian mammals, LNX-2b has been lost by pseudogenization, contributing several exons to the noncoding Xist RNA (36, 37). We show here that zebrafish Lnx2a, like human LNX1, can mediate Numb ubiquitylation and degradation. In studying the zebrafish pancreas, we show that Lnx2a is expressed in the ventral bud at an early stage of embryogenesis. Further, Lnx2a and Lnx2b function redundantly in zebrafish pancreas development through the destabilization of Numb. It remains to be tested whether the single Lnx2 gene in mammals carries out an analogous function.
Control of Protein Stability as a Regulatory Mechanism in Pancreas Formation.
Our results support the view that Lnx2a and Lnx2b mediate the specification of exocrine pancreatic cells by limiting Numb stability. Numb has an inhibitory effect on Notch signaling (23–26, 38), and the Notch pathway has a major role in pancreas development. In the mouse, interfering with Notch activity in the early pancreas led to accelerated differentiation of endocrine cell types while inhibiting formation of exocrine cells (11), whereas overexpression of the constitutively active Notch intercellular domain led to inhibition of differentiation of all pancreatic cell types (10, 28). These observations led to the view that Notch signaling maintains precursor populations, whereas down-regulation of the pathway allows differentiation to proceed. In the formation of secondary islets in zebrafish, Notch activation inhibits exocrine differentiation (39), and specification of particular cell types depends on different Notch ligands (12). Secondary islet formation requires cells that are initially Notch active but undergo Notch down-regulation before differentiation (13, 14). Further, specific levels of Notch activity control quiescence or proliferation of precursor cells and their eventual entry into the endocrine differentiation pathway (13).
In our context, the above conclusions may be summarized as (i) Notch signaling is required in pancreatic cell differentiation; (ii) high Notch activity maintains precursor pools, lower activity allows differentiation, in certain cases preceded by proliferation; and (iii) cells that down-regulate Notch cannot maintain their precursor status. Our observations suggest that Lnx2a, together with Lnx2b, destabilizes Numb in ventral bud-derived cells, allowing Notch activity, which may affect specification and expansion of precursor cells. Loss of Lnx2 activity in the lnx2aΔ329 mutants or lnx2a morphants stabilizes Numb to inhibit Notch in cells where it is normally active, ultimately interfering with the normal developmental progression of these cells. We suggest that in normal development, the regulation of Numb protein stability by Lnx2a/b is an important component of the system that controls the levels of Notch activity during pancreas formation and the assignment of cells to different pancreatic lineages.
Morphant Versus Mutant Phenotypes.
Recent publications (21, 40) and much informal discussions have raised doubts about the validity of morphant phenotypes. Although this suspicion is undoubtedly well founded in certain instances, we show here that failure of a null mutant to replicate a morphant phenotype may have complex and ultimately insightful reasons. It should be noted that Kok et al. (21) considered the possibility of functional redundancy of paralogs and possible effects of exon skipping in the interpretations. Our example emphasizes the need to consider these and possibly other effects in evaluating mutant versus morphant phenotypes.
Materials and Methods
Animals.
Zebrafish AB* were maintained as described (41). Embryos were treated with 0.003% Phenylthiourea (Sigma) at 22–24 hpf to prevent pigmentation. Transgenic lines were as follows: Tg(ptf1a:EGFP) (42), Tg(Tp1bglob:H2BmCherry)S939 abbreviated Tg(Tp1:H2BmCherry), and Tg(Tp1bglob:venusPest)S940 abbreviated Tg(Tp1:venusPest) (13).
TALEN Construction.
TALENs were designed by using the TAL Effector-Nucleotide Targeter (TALE-NT) 2.0 (43) and assembled with Golden gate kits (22). Modified destination vectors, pCS2TAL3 KKR/ELD, were constructed by replacing RR and DD FokI domains in pCS2TAL3 RR/DD vectors (44) with KKR and ELD FokI domains by using site-directed mutagenesis of pMLM 290/292 (KK/EL) vectors (45) (Addgene nos. 21872/21873).
MO Injection.
MOs were purchased from Gene Tools: lnx2a splice MO, TTGAGAACTTTACCTGTGTTTGAGA, 5 ng per embryo; lnx2b splice MO, TGCAGCATGCACTAACCTGTTGTGC, as described (20); numb translation MO, AACTCTGCCGTAGCTTATTCATCGC, 5 ng per embryo. Random sequence MO from Gene Tools was used for control injections, and we also used uninjected embryos for comparison.
For additional materials and methods, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Greg Palardy and Damian Dalle Nogare for advice on microscopy; John Gonzales and Allisan Aquilina-Beck for help with fish husbandry; T.-Y. Choi and Jin-Gu Lee for helpful discussion; and T.-Y. Choi, N. Ninov, D. Y. Stainier, C.V. Wright, and L. Godinho for reagents and zebrafish lines. This work has been supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health, and by the Basic Science Research Program of the National Research Foundation of Korea, Ministry of Education (NRF-2015R1D1A4A01016532).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517033112/-/DCSupplemental.
References
- 1.Slack JM. Developmental biology of the pancreas. Development. 1995;121(6):1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 2.Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261(1):197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- 3.Kinkel MD, Prince VE. On the diabetic menu: Zebrafish as a model for pancreas development and function. BioEssays. 2009;31(2):139–152. doi: 10.1002/bies.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 5.Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122(3):983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 6.Argenton F, Zecchin E, Bortolussi M. Early appearance of pancreatic hormone-expressing cells in the zebrafish embryo. Mech Dev. 1999;87(1-2):217–221. doi: 10.1016/s0925-4773(99)00151-3. [DOI] [PubMed] [Google Scholar]
- 7.Krapp A, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12(23):3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JW, et al. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;274(2):491–503. doi: 10.1016/j.ydbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Zecchin E, et al. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268(1):174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apelqvist A, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400(6747):877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 12.Zecchin E, et al. Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish. Dev Biol. 2007;301(1):192–204. doi: 10.1016/j.ydbio.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 13.Ninov N, Borius M, Stainier DY. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139(9):1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126(10):898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih HP, et al. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development. 2012;139(14):2488–2499. doi: 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie J, Li SS, McGlade CJ. A novel PTB-PDZ domain interaction mediates isoform-specific ubiquitylation of mammalian Numb. J Biol Chem. 2004;279(20):20807–20815. doi: 10.1074/jbc.M311396200. [DOI] [PubMed] [Google Scholar]
- 17.Nie J, et al. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002;21(1-2):93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ro H, Dawid IB. Organizer restriction through modulation of Bozozok stability by the E3 ubiquitin ligase Lnx-like. Nat Cell Biol. 2009;11(9):1121–1127. doi: 10.1038/ncb1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ro H, Dawid IB. Lnx-2b restricts gsc expression to the dorsal mesoderm by limiting Nodal and Bozozok activity. Biochem Biophys Res Commun. 2010;402(4):626–630. doi: 10.1016/j.bbrc.2010.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ro H, Dawid IB. Modulation of Tcf3 repressor complex composition regulates cdx4 expression in zebrafish. EMBO J. 2011;30(14):2894–2907. doi: 10.1038/emboj.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kok FO, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32(1):97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284(39):26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278(25):23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 25.Beres BJ, et al. Numb regulates Notch1, but not Notch3, during myogenesis. Mech Dev. 2011;128(5-6):247–257. doi: 10.1016/j.mod.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhao C, et al. Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development. 2014;141(2):281–295. doi: 10.1242/dev.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bresciani E, et al. Zebrafish numb and numblike are involved in primitive erythrocyte differentiation. PLoS One. 2010;5(12):e14296. doi: 10.1371/journal.pone.0014296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hald J, et al. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260(2):426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 29.Dho SE, et al. The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J Biol Chem. 1998;273(15):9179–9187. doi: 10.1074/jbc.273.15.9179. [DOI] [PubMed] [Google Scholar]
- 30.Rice DS, Northcutt GM, Kurschner C. The Lnx family proteins function as molecular scaffolds for Numb family proteins. Mol Cell Neurosci. 2001;18(5):525–540. doi: 10.1006/mcne.2001.1024. [DOI] [PubMed] [Google Scholar]
- 31.Lenihan JA, Saha O, Mansfield LM, Young PW. Tight, cell type-specific control of LNX expression in the nervous system, at the level of transcription, translation and protein stability. Gene. 2014;552(1):39–50. doi: 10.1016/j.gene.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Yin FT, et al. Caspr4 interaction with LNX2 modulates the proliferation and neuronal differentiation of mouse neural progenitor cells. Stem Cells Dev. 2014;24(5):640–652. doi: 10.1089/scd.2014.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Agostino M, et al. Ligand of Numb proteins LNX1p80 and LNX2 interact with the human glycoprotein CD8α and promote its ubiquitylation and endocytosis. J Cell Sci. 2011;124(Pt 21):3545–3556. doi: 10.1242/jcs.081224. [DOI] [PubMed] [Google Scholar]
- 34.Wolting CD, et al. Biochemical and computational analysis of LNX1 interacting proteins. PLoS One. 2011;6(11):e26248. doi: 10.1371/journal.pone.0026248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, et al. Proteomics strategy to identify substrates of LNX, a PDZ domain-containing E3 ubiquitin ligase. J Proteome Res. 2012;11(10):4847–4862. doi: 10.1021/pr300674c. [DOI] [PubMed] [Google Scholar]
- 36.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312(5780):1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 37.Flynn M, Saha O, Young P. Molecular evolution of the LNX gene family. BMC Evol Biol. 2011;11:235. doi: 10.1186/1471-2148-11-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: Interaction of Numb and Notch. Neuron. 1996;17(1):27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 39.Esni F, et al. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131(17):4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- 40.Schulte-Merker S, Stainier DY. Out with the old, in with the new: Reassessing morpholino knockdowns in light of genome editing technology. Development. 2014;141(16):3103–3104. doi: 10.1242/dev.112003. [DOI] [PubMed] [Google Scholar]
- 41.Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th Ed Univ Oregon Press; Eugene, OR: 2000. The zebrafish book. [Google Scholar]
- 42.Godinho L, et al. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132(22):5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- 43.Doyle EL, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: Tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40(Web Server issue):W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahlem TJ, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8(8):e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.