Significance
Numerous noncoding RNA molecules serve important functional and regulatory roles. How new RNA structures with novel functions emerge through the forces of evolution remains poorly understood. Here, we show how distinct units of structure (modules) within an individual RNA molecule can facilitate this evolution. We show that stress-exposed mutational effects can produce altered RNA structures, providing the raw material for the evolution of new functions. The two modules of the studied RNA molecule serve different roles in this process. This previously unidentified intramolecular mechanism for the hiding and release of mutational effects provides insight into how such a process might have contributed to the evolution of enzyme functions since life’s origins.
Keywords: modularity, epistasis, phenotypic capacitance, ribozyme, evolution
Abstract
Phenotypic capacitance refers to the ability of a genome to accumulate mutations that are conditionally hidden and only reveal phenotype-altering effects after certain environmental or genetic changes. Capacitance has important implications for the evolution of novel forms and functions, but experimentally studied mechanisms behind capacitance are mostly limited to complex, multicomponent systems often involving several interacting protein molecules. Here we demonstrate phenotypic capacitance within a much simpler system, an individual RNA molecule with catalytic activity (ribozyme). This naturally occurring RNA molecule has a modular structure, where a scaffold module acts as an intramolecular chaperone that facilitates folding of a second catalytic module. Previous studies have shown that the scaffold module is not absolutely required for activity, but dramatically decreases the concentration of magnesium ions required for the formation of an active site. Here, we use an experimental perturbation of magnesium ion concentration that disrupts the folding of certain genetic variants of this ribozyme and use in vitro selection followed by deep sequencing to identify genotypes with altered phenotypes (catalytic activity). We identify multiple conditional mutations that alter the wild-type ribozyme phenotype under a stressful environmental condition of low magnesium ion concentration, but preserve the phenotype under more relaxed conditions. This conditional buffering is confined to the scaffold module, but controls the catalytic phenotype, demonstrating how modularity can enable phenotypic capacitance within a single macromolecule. RNA’s ancient role in life suggests that phenotypic capacitance may have influenced evolution since life’s origins.
Phenotypic capacitance requires mutations that reversibly alternate between hidden and revealed states in response to environmental or genetic perturbations (1, 2). In the hidden or “cryptic” state the mutations can survive selection because they do not change the phenotype, and multiple such mutations can accumulate within individual genomes in a population. Perturbation after such accumulation can reveal the combined effects of multiple mutations. By exposing the phenotypic effects of mutations that may not have been beneficial individually, capacitance provides a mechanism of generating new phenotypes—from macromolecules to morphological traits—with novel functions (1–4). The term “phenotypic capacitance” is appropriate for situations where altered phenotypes with a genetic basis can be hidden and revealed, even when no adaptive potential of the revealed phenotypes is demonstrated (2). “Evolutionary capacitance,” on the other hand, is a term reserved for instances when an adaptive role is demonstrated. Phenotypic capacitance has been known in fruit flies since the 1950s (5), but demonstrations of adaptive potential (4, 6), and the various molecular mechanisms behind it (7–11) have been reported only relatively recently.
A module in a biological system is a group of system parts that interact more with each other than with parts outside of the module (12). In RNA molecules, the parts are nucleotides, and modules are units of tertiary structure that fold independently, and often perform different functions, such as binding to different proteins, RNAs, or small molecules. Modularity has been described in the telomerase RNA component (13), ribosomal RNA (14), long noncoding RNA (e.g., Xist, Hotair) (15), riboswitches (16), and self-splicing introns (17, 18). A link between modularity and phenotypic capacitance could provide a mechanism for the evolution of novel functions involving modules of RNA structure, and especially for functional innovations that require multiple simultaneous mutations. It is not known how modularity might change the potential to hide and reveal the effects of mutations in RNA structures, which is a prerequisite for phenotypic capacitance.
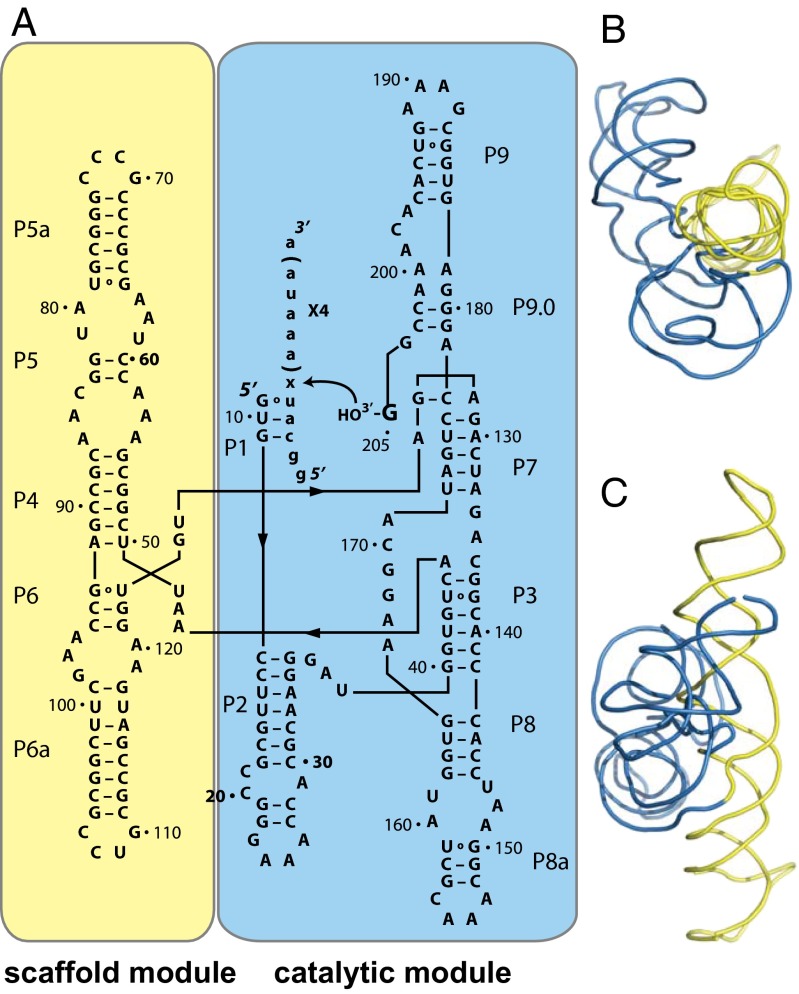
To investigate a possible link between modularity and phenotypic capacitance in RNA, we chose to study the Azoarcus group I RNA enzyme (ribozyme). Group I ribozymes such as this have two structural modules that are functionally distinct (Fig. 1). The first of them is a scaffold module (Fig. 1, yellow) that folds rapidly, and forms a nearly identical structure even when removed from the context of the rest of the ribozyme (19, 20). The scaffolding it provides facilitates the folding of the less thermodynamically stable catalytic module (Fig. 1, blue) (21). Biochemical studies have shown that the catalytic phenotype (protein-free splicing) resides in the catalytic module, which can maintain activity if the scaffold module is deleted, but only under conditions of very high magnesium (80 mM) and extended incubation times (16 h) (22). This instability caused by deleting the scaffold supports the idea that this module acts as an intramolecular chaperone.
Fig. 1.
The modular structure of the Azoarcus group I ribozyme. (A) The scaffold module (yellow) and the catalytic module (blue) are shown in the context of the ribozyme secondary structure. The regions that show base pairing (P) are numbered sequentially from the 5′ to the 3′ end, according to group I intron standards. The substrate for the ribozyme is written in lowercase letters. The modules are also shown in the context of the 3D crystal structure (PDB ID: 1ZZN), from a “top view” (B) and “side view” (C), with respect to the scaffold module.
The diversification of RNA functions has played an important role in the evolution of extant organisms, and may have been even more important at life’s beginnings when RNA enzymes (ribozymes) played a central role as catalysts in the RNA world scenario (23). In a previous publication using the Azoarcus ribozyme, we demonstrated an adaptive role for accumulated cryptic variation that was revealed when altered enzymatic activity was required (24). Here, we focus on how the functionally distinct modules of this structure might facilitate the occurrence of environmentally conditional mutations that enable phenotypic capacitance.
We aimed to identify mutations that maintain the catalytic phenotype under normal conditions but alter this phenotype under stressful conditions. The ideal stressful condition for our RNA system is a low concentration of magnesium ions. The concentration of magnesium ions inside of cells is maintained at a higher concentration than any other divalent ion due to its role in many cellular functions. Importantly for our current experiments, it is known that magnesium ions are critical for stabilizing the native structure of RNA molecules (25). Low concentrations of magnesium only allow folding of the most stable RNA structures and can lead to misfolding of less stable structures. In addition, many ribozymes, including group I introns, use highly coordinated magnesium ions in their active site (26). Based on previous reports on the magnesium dependence of the Azoarcus ribozyme (19), we here studied a stressful environment with low (2 mM) MgCl2, a relaxed environment with high (25 mM) MgCl2, and an intermediate environment (10 mM MgCl2). We note that magnesium availability is often limited in natural environments, a stressful condition that organisms have evolved adaptive responses to. For example, magnesium homeostasis in bacteria is maintained by the expression of ion transporters that are sometimes regulated by cis-acting RNA regulatory elements. These “magnesium riboswitch” elements control downstream mRNA expression by conformational changes induced by altered magnesium concentrations (27). This example and others (28) highlight the widespread importance of our chosen experimental stressor.
Results and Discussion
To determine the effects of mutations on our ribozyme phenotype in each environment, we carried out in vitro selection experiments that separate active from inactive molecules followed by deep sequencing (Materials and Methods). Previous publications have shown that the effect of a mutation on ribozyme activity in our biochemical selection assay correlates with the change in the relative frequency of the mutation in populations of molecules sequenced before and after one round of selection (24, 29). The selection assay enriches mutants that maintain ribozyme activity at the expense of mutations that decrease activity, which are purged and become less frequent. We can thus define the ribozyme fitness effect (w) of each individual point mutation as w = [frequency after selection]/[frequency before selection]. Our population before selection had an average of approximately six random mutations per molecule relative to the wild type (see circled data points in Fig. 3A). For any one point mutation, the ratio w reflects an average fitness effect over all of the genetic backgrounds in which the mutation occurs. Our population was generated from four consecutive rounds of mutagenic PCR (30) without any selection for ribozyme activity between rounds (Materials and Methods).
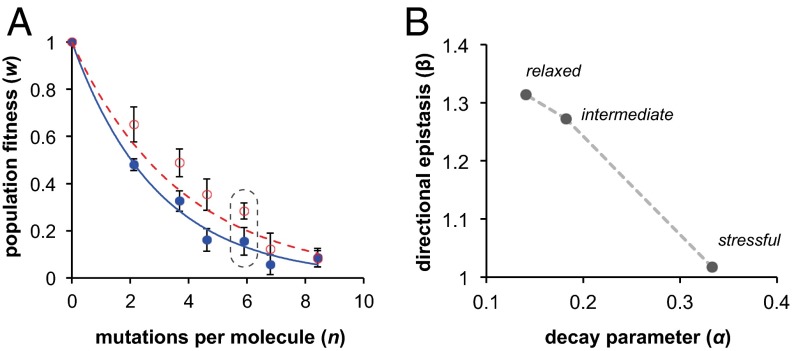
Fig. 3.
Decline in population fitness during mutation accumulation shows synergistic epistasis. (A) Mutations were accumulated through consecutive rounds of mutagenesis without any selection for activity (Materials and Methods). The average number of mutations per individual (n) was determined through deep sequencing. The population fitness (w) at each mutational distance was measured as the activity relative to the wild-type clone (n = 0) under relaxed conditions (25 mM MgCl2, red) and stressful conditions (2 mM MgCl2, blue) with both at 37 °C in 30 mM EPPS pH 7.5. Curves are best fits of an exponential equation w(n) = exp(−αn), based on a nonepistatic model. Error bars represent the SEM of three biological replicates. The population used for selections is encircled by a dashed line. Data for the intermediate condition (10 mM MgCl2) are also included in SI Appendix, Fig. S1. (B) The decline in population fitness w caused by mutation accumulation was fit to a directional epistasis model w(n) = exp(−αnβ) by a nonlinear least squares method (Matlab). The parameters describing the average deleterious mutational effect α (x axis) and directional epistasis β (y axis) were extracted from data obtained under relaxed, intermediate, and stressful conditions (SI Appendix, Fig. S2). A predominance of synergistic interactions between mutational effects is indicated by β > 1, antagonistic by β < 1, and no predominant interaction by β = 1.
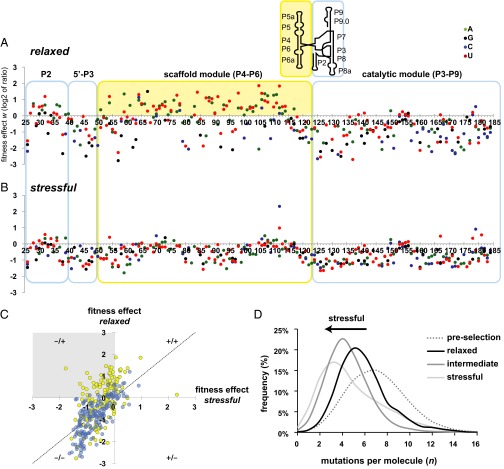
Mutations that make capacitance possible can survive selection under relaxed conditions, but alter the phenotype under environmental stress. In our data, we find that the mutations that fit this description are confined to the scaffold module (Fig. 2). To map mutations visually onto the modular structure, we show their fitness effects in relaxed (Fig. 2A) and stressful (Fig. 2B) conditions, so that the same position in the ribozyme sequence are vertically aligned. Each given module is enclosed in a box that is color coded as in Fig. 1. In both plots, mutations that maintain the phenotype are relatively enriched and located above the horizontal axis. Mutations that disrupt the phenotype are purged by selection and located below the horizontal axis. The vast majority of enriched mutations under relaxed conditions are located in the scaffold (Fig. 2A, yellow area), but most of these mutations are no longer enriched under stressful conditions; they are located below the horizontal axis in Fig. 2B.
Fig. 2.
The individual and combined effects of mutations change with environmental perturbation. Each data point represents an individual mutation effect under relaxed (A) and stressful (B) conditions. The values on the horizontal axes show the nucleotide position in the ribozyme. The vertical axis shows the logarithmically (base 2) transformed ratio w of a mutation’s population frequency after selection, divided by the frequency before selection. Mutations that are enriched by selection lie above the vertical axis, and mutations that are purged lie below. The colored boxes delineate the modules of the structure. The solid yellow background highlights conditional mutations in the scaffold module that are frequently enriched in relaxed conditions (A) but are purged under stressful conditions (B). The Inset above shows a schematic representation of the secondary structure of the Azoarcus ribozyme, indicating the location of the modules in this context. (C) Scatterplot highlighting the modular location of conditional mutations. The gray quadrant highlights a region where the same mutation is enriched under relaxed conditions (25 mM MgCl2), but is purged or neutral under stressful conditions (2 mM MgCl2). For pairwise comparisons of all three conditions, see SI Appendix, Fig. S4. (D) Mutations per molecule were counted using sequence alignment to the wild-type ribozyme reference for populations surviving selection in the relaxed, intermediate, and stressful conditions, as well as for the preselection population. The histograms show the relative frequency (%) with which each number of mutations per molecule was observed in the samples from each population. The sample size for each condition are as follows: relaxed n = 3,369 reads, intermediate n = 26,143 reads, and stressful n = 12,204 reads.
To highlight the difference between the modules, we show a scatterplot of fitness effects in the relaxed vs. stressful conditions with the module location indicated by color (Fig. 2C and SI Appendix, Fig. S4). The Upper Left quadrant of Fig. 2C (gray box “−/+”) contains mutations that maintain the phenotype and are enriched under relaxed conditions (y axis value > 0) but alter the phenotype and are purged under stressful conditions (x axis value ≤ 0). Note that the conditional mutations in this quadrant are almost exclusively yellow, indicating that they occur in the scaffold module. This illustrates the role of the scaffold modules in hiding mutations until environmental stress is introduced.
The previous data show that the average effects of individual mutations change in the transition from a relaxed to a stressful environment. Next we extended our observations to combinations of individual mutations. To this end, we first counted the number of mutations per individual molecule that survived selection in each condition. The surviving populations showed a broad distribution (Fig. 2D), whose peak depends on the environment. Specifically, the most frequent (modal) number of mutations per molecule decreases from five (relaxed) to four (intermediate) to three (stressful, Fig. 2D). This decrease indicates that the cumulative fitness effect of multiple mutations is deleterious and becomes more so as conditions become more stressful. Ribozymes with a greater numbers of mutations can fold into the native structure and survive selection, but only in more relaxed environments.
We biochemically tested the ribozyme activity effects of several individual mutations that show magnesium-dependent fitness effects in our sequence data (i.e., those in the Upper Left quadrant of Fig. 2C). Strikingly, in the wild-type background, in the absence of other mutations, these individual mutations are all nearly neutral and show no magnesium dependence (SI Appendix, Fig. S11). We further note that selected RNA sequences that contain these nearly neutral mutations each harbor only one extra mutation per molecule, on average (SI Appendix, Fig. S10). It thus appears that the neutrality of these extra mutations provides an advantage by effectively decreasing the mutational load of these individuals. In other words, their very neutrality may provide an advantage in the highly mutagenized population. These nearly neutral mutations in the selected ribozymes do not appear to be the kinds of “global suppressors” that occur in proteins following similar selections because these mutations do not suppress the effects of other, additional mutations (31). Ribozymes with these nearly neutral mutations do not harbor further, additional mutations beyond the population average, except for the single nearly neutral mutation itself.
Next, we constructed a series of mutations (single, double, and triple mutants) from a clone that showed prominent magnesium dependence (SI Appendix, Fig. S12). This ribozyme contained one scaffold mutation and two catalytic mutations. The analysis reveals two important points. First, each mutation individually is nearly neutral at 25 mM MgCl2. Second, the magnesium dependence increases only when all three mutations are present, demonstrating epistasis between mutations in the different modules. These results further support the conclusions from our sequence data that the scaffold module enables capacitance by buffering mutations under relaxed conditions, but result in a cumulative deleterious effect under stressful conditions (SI Appendix, SI Results and Discussion).
We further asked whether this cumulative mutational effect results from additive effects of individually deleterious mutations, or whether mutations interact in their effects when combined in the same molecule (epistasis). For this analysis, we first produced populations with incrementally increasing numbers of mutations n using consecutive rounds of a mutagenic PCR protocol, with no selection for ribozyme activity between rounds. Then, we biochemically assayed the population fitness w at each value of n that resulted following each round of mutagenesis (Fig. 3A). The resulting data from the relaxed environment showed a poor fit to a pure exponential (Fig. 3A, red and SI Appendix, Fig. S1), and we therefore fit the data from each environment to the more general equation w(n) = exp(−αnβ) (32, 33). In this equation, the parameter α reflects the average effects of individual mutations. The parameter β accounts for antagonistic or synergistic interactions between mutations (Fig. 3, legend). Contrary to several previous reports of this type pertaining to RNA structures (33–35), we do not find antagonistic epistatic interactions. In contrast, we find increasing amounts of synergistic interactions (β > 1) as the environment becomes less stressful (Fig. 3B and SI Appendix, Fig. S2).
The predominance of synergistic epistasis under relaxed conditions indicates that after a certain number of mutations n, the decline in ribozyme activity increases to a value that is greater than expected from the contribution of individual mutations (β > 1). However, the shape of this epistatic curve requires that for low n, before this accelerated decline, the effects of individual mutations α are smaller and fitness at low n is thus higher than expected from a pure exponential (Fig. 3A, red data and SI Appendix, Fig. S1C). The overall consequence is an inverse relationship between mutational effect α and the extent of synergistic epistasis β in response to our environmental stressor (Fig. 3B). It is the higher fitness of ribozymes with few mutations in relaxed conditions relative to stressful conditions that enables capacitance. This observation is consistent with the smaller number of surviving mutations per molecule under more stressful conditions (Fig. 2D) and with the environmentally conditional effects of individual mutations (Fig. 2 A and B).
Although the synergistic epistasis we observe is different from previous reports on RNA structures, it is surprisingly similar to a previous study of a protein enzyme (TEM-1 beta-lactamase) (36), which found synergistic β values (1.1 ≤ β ≥ 1.3) that are very similar to our findings (1.02 ≤ β ≥ 1.31) through an analogous stressful environmental modulation (SI Appendix, Fig. S3). This similarity is surprising because RNA and proteins differ in both the thermodynamics of folding and the size of their monomer alphabet (20 amino acids, 4 nucleotides), both of which are expected to shape the mutational neighborhood (37). However, somewhat similar to the Azoarcus ribozyme, the TEM-1 protein has a structure with flexible catalytic residues supported by a more rigid secondary structure “scaffold” (38). The finding of environmentally dependent synergistic mutational interactions in both a protein enzyme and a ribozyme supports the conclusion that structures with robust scaffolds can hide mutations in relaxed conditions and release them under stressful conditions. To the extent that robustness and epistasis are correlated (33), our results suggest that an intramolecular mechanism for phenotypic capacitance provided by robust structures may be general and warrant further investigation.
Our results demonstrate not only the potential for capacitance in catalytic RNA molecules, one of the evolutionarily oldest classes of enzymes, but they also highlight the importance of modularity for this capacitance. This is because the catalytic module is invariantly sensitive to mutations, whereas the scaffold module can tolerate mutations in a low-stress environment (Fig. 2 and SI Appendix, Fig. S11). More than that, the presence of these very mutations in the scaffold results in failure of the scaffolding function and decreased catalytic activity, which is evident by the purging of these mutations at low magnesium (Fig. 2B). In addition, we used a published experimental protocol (RNA SHAPE, selective 2′-hydroxyl acylation followed by primer extension) to analyze the structural variability of ribozymes in stressful low magnesium conditions (39). We performed this structural analysis on several ribozyme variants from our selection experiments (SI Appendix, Figs. S6 and S7). The results illustrate that the reduction in the native catalytic activity at low magnesium is coupled with greater variation in the novel structures formed upon environmental stress (SI Appendix, Figs. S8 and S9). Taken together, we conclude that the modular, scaffolded structure enables the phenotypic capacitance we observe.
Nature provides us with pertinent examples of how the kinds of altered RNA phenotypes we observe can produce new functions. The naturally occurring GIR1 ribozyme evolved from a group I self-splicing intron very similar to the Azoarcus ribozyme. This ribozyme lost self-splicing activity and gained a new 5′-capping activity after a period of relaxed evolution when it became embedded within a second self-splicing intron (40). Importantly, the scaffold module is still preserved in the GIR1 ribozyme (termed P4–P6), but significant rearrangements in the remainder of the molecule result in a new tertiary structure and new activity. Our results suggest that the chaperone function of the scaffold module was preserved despite mutation accumulation in this relaxed genetic environment and facilitated evolution of the new function. In another example, an internal ribosomal entry site (IRES) from a picornavirus requires a slightly unfolded structure that is only achieved at low magnesium concentrations (41). At higher magnesium concentrations, additional RNA base pairs are observed, and this different structure is not able to bind its protein ligand. This example provides further evidence of an adaptive role of structural changes induced by low magnesium, such as those produced in our experiments.
Our results demonstrate that phenotypic capacitance does not require the evolution of dedicated chaperone machinery or the complex protein–protein interactions in the regulatory and signaling networks that help shape extant organisms (1, 42). Instead, our results show that individual macromolecules, even the most ancient of biological catalysts, can show phenotypic capacitance and that their capacitance can be modulated through the availability of a critical and ecologically significant chemical element. Because of the ancient and ubiquitous role of RNA in life, capacitance was likely important throughout life’s evolution, even during the chemical events leading up to replicating entities, before the rise of protein enzymes (43, 44).
Materials and Methods
Ribozyme Mutagenesis.
A mutagenic replication cycle without selection was used to produce populations of RNA molecules with an incrementally increasing average number of mutations per molecule. To start, dsDNA containing the wild-type ribozyme sequence and a T7 promoter was constructed from six overlapping synthetic DNA oligonucleotides (Microsynth) (45). This template was transcribed to RNA, which was PAGE purified and then reverse transcribed. The resulting cDNA was amplified by a mutagenic PCR protocol that introduced approximately one mutation per template per PCR (30) and reintroduced the T7 promoter. This mutagenesis was free of selection because during these cycles, ribozyme activity was not selected for. From each cycle, gel-purified RNA was saved for later functional characterization by a reverse-splicing reaction with a synthetic RNA substrate, and cDNA was saved for deep sequencing (Roche 454). Gel purification was required to prevent the accumulation of short artifact sequences produced during the replication cycles.
In Vitro Selection.
The RNA sample from the fourth round of mutagenesis, and with an average of six mutations per molecule (Fig. 3A), was used for in vitro selection reactions with a 3′-biotinylated substrate, under the three different MgCl2 concentrations. Reactions contained ∼1013 molecules (20 pmol) from the ribozyme population, 30 mM EPPS (pH 7.5), 100 pmol RNA oligonucleotide substrate with the sequence 5′-GGCAU(AAAU)4A-3′-biotin, and either 2 mM, 10 mM, or 25 mM MgCl2. After 60 min at 37 °C, the reactions were stopped with equimolar EDTA and incubation on ice. This reverse splicing assay results in the 3′ portion of the substrate—and biotin—becoming attached to the 3′ end of the reacted ribozymes. Half of this reaction was incubated with 50 μg streptavidin-coated magnetic beads (NEB) in a buffer containing 0.5 M NaCl, 20 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, and 0.1% Tween 20 (Sigma). Beads were washed three times with this buffer to prevent nonspecific binding and resuspended in 20 μL nuclease-free water. RNA was converted to cDNA with AMV Reverse Transcriptase (Fermentas) and a primer complementary to the substrate sequence.
Population Fitness Determination.
The fitness of populations produced during each mutagenesis cycle was determined as the activity of the sample relative to a clonal Azoarcus ribozyme sample. Reaction conditions were identical to the in vitro selection protocol (above) except that the substrate was not 5′ biotinylated. Reacted and unreacted molecules were separated on a 6% polyacrylamide gel containing 8 M urea. Following electrophoresis, gels were stained with GelRed (Biotium) and the activity was determined as the fraction of fluorescence in the upshifted band corresponding to reacted riboyzmes. The background fluorescence of the gel was subtracted from unreacted and reacted bands.
Sequencing.
In order to genotype the populations used for fitness assays and those surviving selection at each MgCl2 concentration (above), cDNA samples were PCR amplified with VENT polymerase (NEB) using primers that added adaptor sequences for Roche 454 sequencing (GS FLX Titanium). One of the primers also added a unique “barcode” sequence (multiplex identifier) to one end of the dsDNA product. A different primer with a different unique sequence was used for each sample, which was mixed and sequenced on a single picotiter plate. Reads were mapped to the wild-type sequence and mismatches inside the 159-nt mutable region were counted using LASTZ (46). These counts were converted to relative frequencies to determine the fitness effects (w) reported in Results and Discussion. Reads were discarded if they did not cover the mutable positions or if they contained low quality bases. When computing the average number of mutations per molecule (n) used in curve fitting, we accounted for sequencing errors and errors introduced during reverse transcription and PCR (RT-PCR). To account for these mutational differences not present in the RNA samples, we also sequenced cDNA produced from “clonal” wild-type Azoarcus RNA, following RT-PCR. The average number of mutations per molecule in this clonal sample was subtracted from the averages in other samples, giving corrected values of n used for nonlinear curve fitting to the epistasis equation. Each value of n used in curve fitting was decreased by the mean number of mutations per molecule detected in the clonal wild-type sample. Descriptive statistics (mean, mode, stdev) of mismatch counts for each sample were computed in a spreadsheet program (Gnumeric).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.F. is a guest editor invited by the Editorial Board.
Data deposition: The sequence data reported in this paper are available in Dataset S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420902112/-/DCSupplemental.
References
- 1.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396(6709):336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 2.Masel J, Siegal ML. Robustness: Mechanisms and consequences. Trends Genet. 2009;25(9):395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner A. Robustness and Evolvability in Living Systems. Princton University Press; Princeton: 2005. [Google Scholar]
- 4.Rohner N, et al. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342(6164):1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7(2):118–126. [Google Scholar]
- 6.Halfmann R, et al. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482(7385):363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407(6803):477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi KH. Multiple capacitors for natural genetic variation in Drosophila melanogaster. Mol Ecol. 2013;22(5):1356–1365. doi: 10.1111/mec.12091. [DOI] [PubMed] [Google Scholar]
- 9.Tirosh I, Reikhav S, Sigal N, Assia Y, Barkai N. Chromatin regulators as capacitors of interspecies variations in gene expression. Mol Syst Biol. 2010;6:435. doi: 10.1038/msb.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freddolino PL, Goodarzi H, Tavazoie S. Fitness landscape transformation through a single amino acid change in the rho terminator. PLoS Genet. 2012;8(5):e1002744. doi: 10.1371/journal.pgen.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marciano DC, et al. Negative feedback in genetic circuits confers evolutionary resilience and capacitance. Cell Reports. 2014;7(6):1789–1795. doi: 10.1016/j.celrep.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007;8(12):921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- 13.Zappulla DC, Cech TR. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci USA. 2004;101(27):10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korostelev A, Noller HF. The ribosome in focus: New structures bring new insights. Trends Biochem Sci. 2007;32(9):434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441(7097):1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 17.Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994;1(5):273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- 18.Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J. 1998;17(23):7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci USA. 2003;100(4):1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cate JH, et al. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273(5282):1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 21.Doherty EA, Doudna JA. The P4-P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry. 1997;36(11):3159–3169. doi: 10.1021/bi962428+. [DOI] [PubMed] [Google Scholar]
- 22.Ikawa Y, Shiraishi H, Inoue T. Minimal catalytic domain of a group I self-splicing intron RNA. Nat Struct Biol. 2000;7(11):1032–1035. doi: 10.1038/80947. [DOI] [PubMed] [Google Scholar]
- 23.Ancel LW, Fontana W. Plasticity, evolvability, and modularity in RNA. J Exp Zool. 2000;288(3):242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 24.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474(7349):92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- 25.Draper DE. A guide to ions and RNA structure. RNA. 2004;10(3):335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309(5740):1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 27.Ramesh A, Winkler WC. Magnesium-sensing riboswitches in bacteria. RNA Biol. 2010;7(1):77–83. doi: 10.4161/rna.7.1.10490. [DOI] [PubMed] [Google Scholar]
- 28.Harder W, Dijkhuizen L. Physiological responses to nutrient limitation. Annu Rev Microbiol. 1983;37:1–23. doi: 10.1146/annurev.mi.37.100183.000245. [DOI] [PubMed] [Google Scholar]
- 29.Pitt JN, Ferré-D’Amaré AR. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330(6002):376–379. doi: 10.1126/science.1192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2(1):28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 31.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLOS Comput Biol. 2008;4(2):e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenski RE, Ofria C, Collier TC, Adami C. Genome complexity, robustness and genetic interactions in digital organisms. Nature. 1999;400(6745):661–664. doi: 10.1038/23245. [DOI] [PubMed] [Google Scholar]
- 33.Wilke CO, Adami C. Interaction between directional epistasis and average mutational effects. Proc Biol Sci. 2001;268(1475):1469–1474. doi: 10.1098/rspb.2001.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjuán R. Quantifying antagonistic epistasis in a multifunctional RNA secondary structure of the Rous sarcoma virus. J Gen Virol. 2006;87(Pt 6):1595–1602. doi: 10.1099/vir.0.81585-0. [DOI] [PubMed] [Google Scholar]
- 35.Wilke CO, Lenski RE, Adami C. Compensatory mutations cause excess of antagonistic epistasis in RNA secondary structure folding. BMC Evol Biol. 2003;3(1):3. doi: 10.1186/1471-2148-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444(7121):929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 37.Ferrada E, Wagner A. A comparison of genotype-phenotype maps for RNA and proteins. Biophys J. 2012;102(8):1916–1925. doi: 10.1016/j.bpj.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425(14):2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 39.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013;19(1):63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer M, et al. Speciation of a group I intron into a lariat capping ribozyme. Proc Natl Acad Sci USA. 2014;111(21):7659–7664. doi: 10.1073/pnas.1322248111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozano G, Fernandez N, Martinez-Salas E. Magnesium-dependent folding of a picornavirus IRES element modulates RNA conformation and eIF4G interaction. FEBS J. 2014;281(16):3685–3700. doi: 10.1111/febs.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424(6948):549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 43.Vaidya N, et al. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491(7422):72–77. doi: 10.1038/nature11549. [DOI] [PubMed] [Google Scholar]
- 44.Levy M, Ellington AD. The descent of polymerization. Nat Struct Biol. 2001;8(7):580–582. doi: 10.1038/89601. [DOI] [PubMed] [Google Scholar]
- 45.Rydzanicz R, Zhao XS, Johnson PE. Assembly PCR oligo maker: A tool for designing oligodeoxynucleotides for constructing long DNA molecules for RNA production. Nucleic Acids Res. 2005;33(Web Server):W521–W525. doi: 10.1093/nar/gki380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris RS. Pennsylvania State University; University Park, PA: 2007. Improved pairwise alignment of genomic DNA. PhD dissertation. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.